Abstract
The LspA1, LspA2, and LspB proteins of Haemophilus ducreyi comprise a two-partner secretion system that has been shown to be necessary for H. ducreyi to inhibit phagocytosis by immune cells in vitro. Inactivation of lspA1 resulted in increased levels of LspA2, suggesting that these two proteins are differentially controlled (C. J. Ward et al., Infect. Immun. 71:2478-2486, 2003). Expression of LspA2 but not LspA1 was shown to be both growth phase dependent and affected by the presence of fetal calf serum (FCS) in the growth medium. In addition, neither LspA1 nor LspA2 could be detected in culture supernatant fluid in the absence of FCS. DNA microarray analysis revealed that 324 H. ducreyi genes were differentially regulated after growth in the presence of FCS. Among these, the CpxRA two-component sensory transduction system was downregulated by the presence of FCS. Inactivation of cpxR resulted in increased expression of both LspB and LspA2. Electrophoretic mobility shift assays showed that a recombinant H. ducreyi CpxR protein bound the promoter region of the lspB-lspA2 operon. The cpxR and cpxA genes were shown to be part of an operon containing two additional genes in H. ducreyi 35000HP. This is the first description of a two-component sensory transduction system regulating a proven virulence factor of H. ducreyi.
Haemophilus ducreyi is a gram-negative coccobacillus and the causative agent of the sexually transmitted genital ulcer disease (GUD) chancroid (1, 8). Globally, chancroid is a significant sexually transmitted disease, with more than 6 million cases reported in 1997 (60). In the United States, several outbreaks were reported between 1980 and 2000 (24, 36, 51), but since then the number of cases has greatly diminished, and today the soft chancres characteristic of H. ducreyi infection occur only in isolated cases that are typically associated with the sex trade industry (57). Chancroid is endemic in some developing countries in Africa, Asia, and South America, where it accounts for almost half of all GUD cases, although these numbers could be higher as H. ducreyi cases remain poorly documented (55, 57). GUD is a recognized cofactor for human immunodeficiency virus acquisition and transmission (25, 37), and a better understanding of H. ducreyi pathogenesis is necessary to allow a rational approach to the identification of vaccine candidates that could be used to prevent chancroid.
H. ducreyi is a strict human pathogen, and it is likely that during the different stages of ulcer production (i.e., progression of a papule into a pustule followed by frank ulceration [1, 8, 50]), this pathogen controls gene expression to enhance its growth and to evade the host immune system. Information about regulatory networks that might control the expression of H. ducreyi virulence factors is very limited at present. Although nucleotide sequence analysis of the H. ducreyi 35000HP genome (GenBank accession no. NC002940) revealed the presence of several genes encoding predicted proteins with homology to known bacterial regulators, to date, only the fur gene (12), the groE operon (44), and the dnaK-dnaJ operon (43) have been studied in any detail.
The LspA1, LspA2, and LspB proteins constitute a two-partner secretion system in H. ducreyi (65). Expression of either LspA1 or LspA2 has been shown to be necessary for H. ducreyi to inhibit phagocytosis by immune cells in vitro (61). LspA1 and LspA2 have 86% identity (64), and their respective open reading frames (ORFs) are located very distant from each other in the H. ducreyi chromosome. These proteins are also required for full virulence of this pathogen in both the human (30) and the temperature-dependent rabbit (63) models of experimental H. ducreyi infection. Previous studies in our laboratory indicated that wild-type H. ducreyi apparently expressed more LspA1 than LspA2 and that a lspA1 mutant expressed increased amounts of LspA2 (63). Taken together, these results suggested that these two proteins might be under the control of different, unidentified regulatory factors. In the present study, we report that inactivation of the gene encoding the response regulator CpxR resulted in increased expression of both LspB and LspA2, suggesting that the H. ducreyi lspB-lspA2 operon is under the control of the CpxRA two-component regulatory system (18, 46, 47) homolog present in H. ducreyi.
MATERIALS AND METHODS
Bacteria and culture conditions.
Wild-type H. ducreyi strains were routinely recovered from frozen stock on chocolate agar plates that were incubated at 33°C in a humidified atmosphere containing 95% air and 5% CO2. Strains were grown at 33°C in a gyratory water bath at 100 rpm in a Columbia broth-based (CB) medium (35 g of Columbia broth [Difco, Detroit, MI] per liter, 0.1% [wt/vol] Trizma base [Sigma], equine hemin [25 μg/ml; Sigma], 1% [vol/vol] IsoVitaleX [Becton Dickinson, Franklin Lakes, NJ]) with or without 2.5% (vol/vol) heat-inactivated fetal calf serum (FCS). Escherichia coli DH5α was used as the host for general cloning manipulations. E. coli strains were grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (30 μg/ml) when appropriate for maintenance of the plasmids. For protein expression experiments, E. coli M15(pREP4) (Qiagen, Valencia, CA) was used. Plasmid constructs used for complementation studies were transformed into and isolated from E. coli HB101 before they were electroporated into H. ducreyi.
Development of a polyclonal CpxR antibody.
The peptide TPSNHSPEDSNKQLSFGGVE, corresponding to amino acids 129 to 148 of the H. ducreyi 35000HP CpxR protein, was synthesized by the Protein Technology Center at the University of Texas Southwestern Medical Center. This peptide was coupled to maleimide-activated keyhole limpet hemocyanin (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. Polyclonal rabbit antibody to this keyhole limpet hemocyanin-peptide conjugate was produced by Rockland Immunochemicals (Boyertown, PA). Protein G-beads (GE Healthcare, Piscataway, NJ) were used to purify immunoglobulin G (IgG) from the immune rabbit serum. This purified IgG was used at a concentration of 230 ng/ml as a primary antibody in Western blot analysis.
Antigen preparation and Western blot analysis.
Proteins present in H. ducreyi whole-cell lysates and concentrated culture supernatant fluids (CCS) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 3 to 8%, 7.5%, or 12.5% (wt/vol) polyacrylamide separating gels and blotted onto polyvinylidene difluoride membranes. Each whole-cell lysate was standardized by using optical density at 600 nm (OD600) values to obtain cell suspensions, which were then diluted such that 5 × 107 CFU equivalents were loaded per lane. H. ducreyi CCS was prepared and concentrated at least 40-fold as described previously (64). Membranes were then incubated in phosphate-buffered saline-Tween containing 10% (wt/vol) nonfat milk for 1 h at room temperature or overnight at 4°C. The membranes were then incubated for 3 to 4 h at room temperature or overnight at 4°C in primary antibody at the appropriate dilution, followed by a 1-h incubation at room temperature in a 1:5,000 dilution of either goat anti-mouse IgG-horseradish peroxidase or goat anti-rabbit IgG-horseradish peroxidase (Bio-Rad, Hercules, CA). The LspA1-specific monoclonal antibody (MAb) 40A4 (64), the LspA2-specific MAb 1H9 (64), and the PAL-specific MAb 3B9 (54) have been described. Western blots were developed using the Western Lightning Chemiluminescence Reagent Plus (New England Nuclear, Boston, MA).
RNA isolation.
Total RNA was extracted from broth-grown bacteria by using the RiboPure Kit (Ambion, Austin, TX) according to the manufacturer's instructions. At the required time points, stop buffer (200 mM Tris-HCl [pH 8], 20 mM EDTA, 20 mM sodium azide) was added (final concentration, 5%) to a 5- to 10-ml sample of bacterial culture prior to collection by centrifugation. After isolation, RNA was treated twice with DNase I (Ambion) for 1 h at 37°C and then purified by using the RNeasy system (Qiagen). Samples were checked for residual DNA by PCR and retreated with DNase I if necessary.
DNA microarray analysis.
The nucleotide sequence of the genome of H. ducreyi 35000HP (GenBank accession no. NC002940) was used to develop the DNA microarrays used in the present study. Probe design parameters were the same as described previously for development of M. catarrhalis DNA microarrays (62). The 70-mer oligonucleotides were synthesized (Qiagen) and spotted onto Corning UltraGAP II slides in triplicate (Microarrays, Inc., Nashville, TN). Using the FindGDPs program (7), 57 genome-directed primers were designed; these primers were synthesized by Integrated DNA Technologies and used for cDNA synthesis from total RNA. For each experiment, 20-μg quantities of total RNA extracted from cells grown for 8 h in CB+FCS or CB-FCS were used for first-strand cDNA synthesis, using Amino Allyl cDNA labeling kit reagents (Ambion). Template RNA in the reverse transcriptase (RT) reaction was removed, and the amino-allyl-labeled cDNA was further purified with QIAquick Clean-Up columns (Qiagen). Samples in each experimental replicate were labeled with either Cy3-dCTP or Cy5-dCTP, unincorporated dye was removed, and samples were concentrated by using YM-30 Microcon centrifugal filter devices (Millipore, Billerica, MA). One sample pair was used in a dye swap experiment. Cy-labeled cDNA was quantified by using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE), and equivalent amounts of the labeled cDNA from each growth condition were mixed thoroughly and vacuum dried in the dark. Prior to hybridization, the Cy-labeled mixture was resuspended in 14 μl of DNase/RNase-free water, denatured at 94°C for 3 min, allowed to cool to 25°C, and added to a hybridization mix (10 μl of 4× hybridization buffer [GE Healthcare] and 16 μl of formamide). This mixture was applied to the DNA microarray slide, which was then incubated in the dark at 50°C for 16 h. After this incubation, the slides were washed (in the dark with gently rocking) by sequential incubations in 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) containing 0.01% Tween 20 (SSPE-T) at 50°C, 0.8× SSPE-T at 50°C, and 0.6× SSPE at 25°C. After a low-speed centrifugation step (5 min at 900 × g), the slides were kept in the dark and immediately scanned by using a GenePix 4100A microarray reader (Molecular Devices, Sunnyvale, CA). GenePix Pro 5.0 and Acuity 4.0 software packages (Axon Instruments, Inc.) were used for image and data analyses, respectively.
The data from four biological replicates (dye-swap included) were individually subjected to two types of normalization, a ratio-based normalization so that the mean or median intensities are the same across the array, and a nonlinear locally weighted scatterplot smoothing (LOWESS) normalization, which corrects intensity-dependent variation in dye bias, before being combined into a single data set for further analysis. After normalization, the results indicated that 73.4% of the total H. ducreyi genome was represented in the analysis. Differential expression was defined as a minimum of a twofold change in expression in the cells grown in CB+FCS relative to cells grown in CB-FCS. The data were further scrutinized so as to only include expression profiles that were observed in at least three of the four experiments and had a P value of ≤0.05 after a one-sample t test analysis. The data from these DNA microarray experiments were deposited at the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE13851.
Real-time RT-PCR.
Selected oligonucleotide primers used in the present study are listed in Table 1 and were derived from the nucleotide sequence of the H. ducreyi 35000HP genome (GenBank accession no. NC002940). Oligonucleotide primer pairs were designed using Primer3 (5). Primers were run through BLAST using the nonredundant database, as well as the H. ducreyi genome, to rule out possible false positives. Primers were also tested for the presence of “primer dimers” by means of a dissociation curve analysis. Seventeen genes were selected for analysis of their relative transcription levels by two-step real-time RT-PCR. The RT reaction was performed with 2 μg of RNA in a 20-μl reaction volume, using TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions and using gene-specific primers at a final concentration of 400 nM. After the RT reaction, residual RNA was removed by treatment with RNase for 20 min at 37°C, and the cDNAs were cleaned by using QiaPrep columns (Qiagen). Each real-time RT-PCR was performed in a 20-μl volume containing 50% (vol/vol) SYBR master mix, the two oligonucleotide primers at final concentrations of 400 nM each, and 10 ng of cDNA. PCR amplification was accomplished with a model 7500 real-time PCR system (Applied Biosystems). Assays were performed in triplicate independent experiments, using HD1643 (gyrB) to normalize the amount of cDNA per sample, because its transcript levels did not change in any of the DNA microarray experiments performed. The fold change of each gene was calculated by using the 2−ΔΔCT method.
TABLE 1.
Oligonucleotides used in this study
| Method and oligonucleotide | Sequence (5′-3′)a |
|---|---|
| Standard methods | |
| P1 | ACGCGATATCAAACTATTATTAAGTAAAACTAG |
| P2 | ACGCGCTAGCCTTCAAGTTGATCAATAATTGAT |
| P3 | TGGTGTTAAACGCTTACCTAACCCGGGACCATTATGCACGCAATGAATATG |
| P4 | TTAGGTAAGCGTTTAACACCA |
| P5 | ATGCCTAGAATTTTACTC |
| P6 | GTTATTTCTCAGTCACTAA |
| P7 | ACGCAAGCTTAGATGCTGAAAACTGTTTAC |
| P8 | ACGCAAGCTTAGGGATTCGTTCAATCGACA |
| P9 | ACGCGAATTCATTTGGAATCAGGTGAAGATC |
| P10 | ACGCGAATTCCCCGAACTTGCGAGGGATTCG |
| catP(F) | ACGCCCCGGGGTTGATACCGGGAAGCCCTGG |
| catP(R) | GTATATCCAGTGATTTTTTTCTCCATTTTAGCTTCCTTAGCTCCTGAAAA |
| cat(F) | ATGGAGAAAAAAATCACTGGATATAC |
| catSL33(R) | ACGCCCCGGGCATTATTCCCTCCAAAAATTA |
| DeltaEcat(F) | CCGTTTTTATCAGGCTCTGG |
| DeltaEcat(R) | CTTTTGCCGTTACGCACCAC |
| pQE30cpxR-F | ACGCGGATCCATGCCTAGAATTTTACTC |
| pQE30cpxR-R | ACGCCTGCAGGTTATTTCTCAGTCACTAA |
| Real-time RT-PCR | |
| HD1505F | AAAGTTTCAGCAGGGACAGC |
| HD1505R | TGCCGACAGTTTGGATGTAT |
| HD1155F | ATATACCGTCCGCGGTTTTA |
| HD1155R | CCATGCCGACAAGTATATCG |
| HD1156F | TGAATGGGCGAAAAATGATA |
| HD1156R | AACGCCCGGTACAGTAATTT |
| HD2025F | GCCGTTTATTTAGCCTCGAA |
| HD2025R | AGAACGAAGGATGCTGGAAT |
| HD1643F | TCCGTTAAAGTACCCGATCC |
| HD1643R | GCACGAGCAGCATCAATAAT |
| HD1312F | TTCAGCGACTGAAAGTTTACG |
| HD1312R | GCCATTAGCGCTACTTATACCA |
| HD0198F | TGAACGTTTAACCGCACAAT |
| HD0198R | ATAACCATACGCATCGTGGA |
| HD0046F | GGTTGGGCAAGTTTCCATAG |
| HD0046R | TAATTCAGCCGCTAAACCTG |
| HD0045F | AGTTCACTACCTGGGGCATT |
| HD0045R | ATAGGGGGTCTTGGTCCT |
| HD1433F | CCACCGAATCCAAAGAAACA |
| HD1433R | TGGCAGTAATACCCAAACGA |
| HD0769F | TGCAAACTGGTTTAGCCAAC |
| HD0769R | ATGCCGCCATTGTAGGTATT |
| HD1435F | GCCTCAGCAGTTACGCTTTA |
| HD1435R | GGGCTGATAAACCAATACGG |
| HD1772F | GATGCTATGGCGGTTAATCA |
| HD1772R | GTTCATCCGCATGTCCTGTA |
| HD0902F | CTGATAGTGGTGCGGTTGTC |
| HD0902R | CTAATACAGGGGGAGCGAGA |
| HD1470F | CAAGAAAGCCGTCAAACAGA |
| HD1470R | GGTTCAAGTGGTTGTGC |
| HD1469F | TAAGTGCCCGAGATGATGAA |
| HD1469R | ATGGTTTGATGGCGTGACTA |
| EMSA | |
| HD1155P(F) | AGTAAATTTCTTCAAAAATGT |
| HD1155P(R) | TGAAAGCATAAATAAATAAGA |
| HD1505P(F) | AAAGATTTAATTAAACAGGCT |
| HD1505P(R) | AGTTTATAACGTTTGTTGTTC |
| HD1643P(F) | GCTTGAATATTGCAAAGATTC |
| HD1643P(R) | TTAATACTCGAAGAATCATAA |
| RT-PCR | |
| HD1468-cpxR F | ATCGCAGTCAAAAAGCAAGGA |
| HD1468-cpxR R | TGCATCTTGACCATTATGCAC |
| cpxR-cpxA F | CGTAATGATAACCTCCCATGG |
| cpxR-cpxA R | TAAGCCAAAATAGGCAAAGAG |
| cpxA-HD1471 F | GGTTTGGGCCTTGCGATTGTA |
| cpxA-HD1471 R | CGGCAATGAAGCAGTCACACC |
Boldface text indicates the complementary sequence used in overlapping extension PCR. Underlining indicates the restriction site as described in Materials and Methods.
Construction of an H. ducreyi cpxR mutant.
An ∼1.5-kb fragment corresponding to the 5′ upstream region of the H. ducreyi cpxR ORF was PCR amplified from chromosomal DNA with ExTaq DNA polymerase (Takara Bio, Inc., Shiga, Japan) and the primers P1 and P3 (Table 1). Another ∼2-kb fragment corresponding to the 3′ downstream region of the H. ducreyi cpxR ORF was PCR amplified with the primers P4 and P2. Primers P3 and P4 shared a 21-nucleotide (nt) complementary sequence (indicated in boldface in Table 1), and a SmaI site (underlined in Table 1) was included in primer P3. The two PCR fragments were gel purified, and equal amounts were mixed and used as the templates in overlapping extension PCR (28) with the primers P1 and P2. The resultant ∼3.5-kb PCR product had a ∼500-bp deletion within the cpxR ORF, with a SmaI site in the center. This fragment was cloned into pCR2.1 (Invitrogen) to obtain pML115. The ∼200-bp promoter region of the pACYC184 cat gene (13) was PCR amplified by using the primers catP(F) and catP(R). The nonpolar cat cartridge from pSL33 (32) was amplified by using the primers cat(F) and catSL33(R). Primer cat(F) was complementary to 26 nt at the 5′ end of catP(R) (indicated in boldface in Table 1). Both amplicons were gel purified, and equal amounts were mixed and used as the templates in overlapping extension PCR with the primers catP(F) and catSL33(R). This PCR generated a ∼800-bp fragment flanked by SmaI sites. This amplicon was digested with SmaI and ligated to SmaI-digested pML115 to obtain pML116. The primers P1 and P2 were used to amplify a ∼4.5-kb fragment from pML116 containing the cpxR deletion construct described above. The amplicon was DpnI digested to cut any residual plasmid DNA, followed by gel purification. A 90-μg amount of purified PCR fragment was used to electroporate H. ducreyi 35000HP as previously described (23), with some modifications. Briefly, bacterial growth from three chocolate agar plates was collected into 5 ml of cold 10% (vol/vol) glycerol. These H. ducreyi cells were washed four times and suspended in a final volume of 200 μl. A 50-μl portion of the final cell suspension was mixed with the purified PCR product and placed on ice for 20 min. The mixture of DNA and cells was transferred to a cold 0.1-cm electroporation cuvette, and a 2.5-kV pulse was applied. The cells were placed in 5 ml of CB and incubated at 33°C in a gyratory water bath at 100 rpm. After a 5-h recovery period, the cells were harvested by centrifugation and plated on chocolate agar containing chloramphenicol (1 μg/ml). Nucleotide sequence analysis of the relevant chromosomal region in the chloramphenicol-resistant transformant, designated 35000HPΔcpxR, indicated the presence of a 7-nt insertion within the remainder of the cpxR ORF and two nucleotide changes in the cpxA gene that did not affect the predicted amino acid sequence.
Southern blot analysis.
Purified chromosomal DNA preparations from H. ducreyi strains were digested to completion with EcoRV and used in Southern blot analysis. The probe for the cpxR gene was obtained by PCR, using the primers P5 and P6 (Table 1) to amplify an ∼700-bp fragment from H. ducreyi 35000HP chromosomal DNA. The probe for the cat cartridge was obtained by using the primers DeltaEcat(F) and DeltaEcat(R) (Table 1) to amplify an ∼1.3-kb fragment from pACYC184 containing the cat gene. Both probes were labeled with digoxigenin (DIG) by using a random-primed DNA labeling kit (Roche Applied Sciences, Indianapolis, IN).
Complementation of the H. ducreyi 35000HPΔcpxR mutant.
The wild-type cpxR gene from H. ducreyi, together with 100 nt upstream of the translational initiation codon, was amplified from 35000HP chromosomal DNA by using the primers P9 and P10 (Table 1). The amplicon was digested with EcoRI and ligated to EcoRI-digested pLS88 (66) to obtain pML125. A 100-ng quantity of pML125 DNA was used to transform 35000HPΔcpxR (as described above) to obtain the kanamycin- and chloramphenicol-resistant strain 35000HPΔcpxR(pML125). Plasmid pLS88 was also introduced into 35000HPΔcpxR for use as a negative control.
Construction and purification of a histidine-tagged CpxR fusion protein.
A DNA fragment corresponding to the cpxR ORF was amplified from 35000HP chromosomal DNA by PCR using the primers pQE30cpxR-F and pQE30cpxR-R (Table 1), which added BamHI and PstI sites, respectively, to the fragment. The amplicon was digested with these two restriction enzymes and ligated to BamHI- and PstI-digested pQE30 (Qiagen) to obtain pML123. E. coli M15 cells harboring pML123 were cultured to an OD600 of ∼0.5 and, after the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.1 mM, the cultures were shaken for 4 h. Cells were harvested by centrifugation, suspended in buffer A (50 mM sodium phosphate, 200 mM sodium chloride, 10 mM imidazole) containing protease inhibitors, and frozen at −20°C until further use. After disruption of these cells by sonication, a soluble crude extract was obtained by centrifugation (15,000 × g for 30 min), and the extract was filtered through a 0.22-μm-pore-size filter before using Ni2+-chelate chromatography to purify the His-tagged CpxR protein. This fusion protein was further purified by anion-exchange chromatography and stored at −70°C.
Electrophoretic mobility shift assay (EMSA).
The ability of CpxR to recognize and bind the lspB promoter was examined by using a DIG gel shift kit from Roche. The lspA1 and gyrB promoter regions were used as negative controls. The lspB DNA promoter region containing the putative CpxR consensus binding sequence in the center of the fragment (∼200 bp) was generated by PCR (the primer sequences are given in Table 1). The 3′ termini of 4 pmol of each probe were labeled with 1 nmol of digoxigenin (DIG)-11-ddUTP by using 1 U of terminal transferase at 37°C for 30 min in labeling buffer (200 mM potassium cacodylate, 25 mM Tris-HCl [pH 6.6], 0.25 mg of bovine serum albumin per ml, 5 mM CoCl2). The purified, His-tagged CpxR protein (50 pmol) was incubated in binding buffer [20 mM HEPES (pH 7.6) containing 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol, 0.2% (wt/vol) Tween 20, 30 mM KCl, 1 μg of poly(I-C), and 0.1 μg of poly-l-lysine] in a final volume of 22 μl at 37°C for 15 min. Then, a 10-fmol portion of the DIG-labeled probe was added, and the reaction mixture was incubated at 25°C for 15 min. After the addition of 5 μl of loading buffer (0.25× Tris-borate-EDTA, 40% [vol/vol] glycerol, 0.2% [wt/vol] bromophenol blue), samples were run on a 4.5% polyacrylamide gel (29:1 acrylamide-bis-acrylamide) in 0.5× Tris-borate-EDTA, followed by electroblotting (Transblot; Bio-Rad) onto a nylon membrane (Hybond-N+; Amersham, Piscataway, NJ) and fixation by UV cross-linking. Detection of DNA fragments was performed as directed by the manufacturer (Roche).
RESULTS
Expression of LspA2 but not of LspA1 is growth phase dependent.
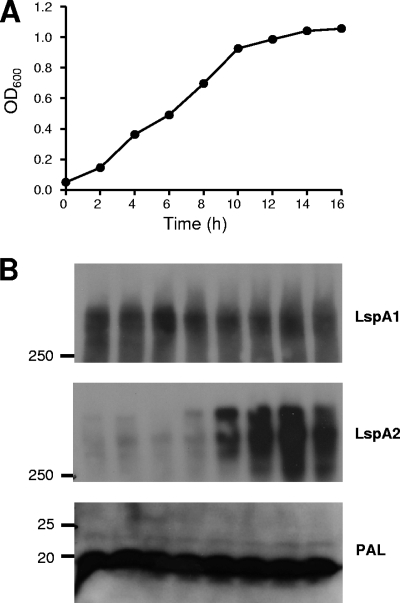
A previous study indicated that the level of expression of LspA2 by broth-grown H. ducreyi cells was lower than that of LspA1 (63), suggesting that the lspA1 and lspA2 genes could be regulated differently. In order to test this hypothesis, Western blot analysis was used to determine the expression profiles of LspA1 and LspA2 throughout the different stages of growth. Cultures were inoculated at low densities (OD600 ∼0.05), and growth was monitored for 16 h (Fig. 1A). At each time point, proteins present in whole-cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the expression of LspA1 and LspA2 was determined by using LspA1- and LspA2-specific MAbs (Fig. 1B). In addition, at 8, 12, and 16 h, total RNA was extracted, and the transcript levels of lspA1 and lspA2 were determined by using real-time RT-PCR. LspA1 appeared to be constitutively expressed (Fig. 1B, upper panel), although slightly increased expression was sometimes observed during stationary phase (data not shown). LspA2 has a delayed expression pattern, first appearing in exponential phase between the 8- and 10-h time points and peaking in the stationary phase (Fig. 1B, middle panel). Transcript levels correlated with protein levels (data not shown). These data suggested that lspA1 and lspA2 are under the control of different regulatory factors and are therefore differentially regulated.
FIG. 1.
Expression of LspA1 and LspA2 during growth of H. ducreyi in broth. (A) Growth of wild-type H. ducreyi 35000HP in CB. (B) Western blot-based detection of LspA1 and LspA2 proteins in whole-cell lysates using the LspA1-specific MAb 40A4 (top panel), the LspA2-specific MAb 1H9 (middle panel), and the PAL-specific MAb 3B9 (bottom panel). This latter antigen was used as a loading control. Cells were sampled every 2 h, beginning with the 2-h time point. Molecular mass position markers (in kilodaltons) are present on the left sides of these three panels.
Expression of LspA2 but not of LspA1 is affected by FCS.
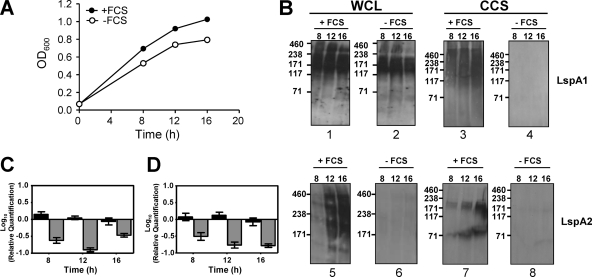
H. ducreyi is a fastidious bacterium that requires a rich and complex medium for growth in vitro (2, 15, 22, 33). One medium constituent that facilitates the optimal growth of this pathogen in vitro is FCS. Preliminary experiments in this laboratory indicated that the absence of FCS in the CB-based growth medium resulted in a lack of detectable LspA1 and LspA2 in CCS (data not shown). To further investigate this phenomenon, whole-cell lysates, CCS, and relevant transcript levels from H. ducreyi grown in the presence or absence of FCS were analyzed. The growth of H. ducreyi in the absence of FCS was slightly slower than growth in the presence of FCS (Fig. 2A). Analysis of whole-cell lysates by Western blotting indicated that, in the presence of FCS, both LspA1 and LspA2 were synthesized (Fig. 2B, panels 1 and 5, respectively). In contrast, in the absence of FCS, LspA2 expression (Fig. 2B, panel 6) was highly reduced, while LspA1 was present at all of the time points tested (Fig. 2B, panel 2). This pattern of expression was corroborated at the transcriptional level by using real-time RT-PCR. In the absence of FCS, expression from lspA1 was relatively unaffected, whereas expression from lspA2 was greatly reduced (Fig. 2C and D).
FIG. 2.
Effect of FCS on growth and LspA protein expression. (A) Wild-type H. ducreyi 35000HP cells were grown in CB with FCS (•) and without FCS (○). (B) Western blot-based detection of LspA1 and LspA2 proteins in whole-cell lysates (WCL) and CCS from H. ducreyi 35000HP cells grown in CB with FCS (panels 1, 3, 5, and 7) or without FCS (panels 2, 4, 6, and 8) using the LspA1-specific MAb 40A4 (panels 1 to 4) and the LspA2-specific MAb 1H9 (panels 5 to 8). Molecular mass position markers (in kilodaltons) are present on the left sides of these eight panels. It should be noted that both LspA1 and LspA2 frequently appeared smeared or as multiple bands in Western blot analysis (63). (C and D) Real-time RT-PCR measurement of relative expression levels of lspA1 (▪) and lspA2 (░⃞) in wild-type H. ducreyi 35000HP cells grown in CB−FCS compared to CB+FCS in two separate experiments. Expression of gyrB was used to normalize the amount of cDNA per sample. These two experiments were performed independent of those used to measure protein expression. The bracket bars denote the highest and lowest values.
In the absence of FCS, neither LspA1 nor LspA2 could be detected in CCS (Fig. 2B, panels 4 and 8, respectively), whereas both proteins could be detected in CCS (Fig. 2B, panels 3 and 7, respectively) in the presence of FCS. Taken together, these data indicate that LspA1 expression is not affected by the presence or absence of FCS, whereas the release of LspA1 and LspA2 from the H. ducreyi cell is mediated directly or indirectly by a component(s) present in the serum. In addition, the presence of FCS significantly affected the expression of LspA2, suggesting that a factor(s) in serum may regulate the expression of LspA2.
Analysis of the H. ducreyi transcriptome in the presence of FCS.
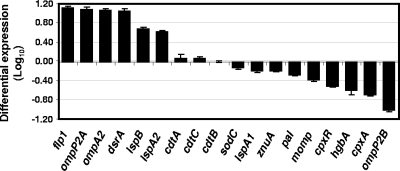
In an attempt to identify the regulatory mechanism(s) involved in the expression of LspA2, we performed DNA microarray analysis, using a microarray based on the nucleotide sequence of the H. ducreyi 35000HP genome. A total of 324 genes were differentially regulated (i.e., at least a twofold increase or decrease in expression) after growth in the presence of FCS. Of these, 163 genes were upregulated, and 161 were downregulated. The 70 most significantly upregulated or downregulated genes are listed in Table 2. Among the upregulated genes were several encoding known or putative H. ducreyi virulence factors including lspA2 (30), dsrA (9), ompP2A (29, 45), ompA2 (58), and flp1 (53), suggesting that the presence of serum might be a cue for the expression of certain bacterial products necessary for the infection process. When real-time RT-PCR was used to determine relative transcript levels for a subset of these genes under the same two sets of conditions, the majority of the expression patterns (Fig. 3) correlated well with the DNA microarray results.
TABLE 2.
Genes in H. ducreyi whose expression is most affected by growth in the presence of FCSa
| ORF | Gene | Description | log2(+FCS/-FCS)b | SD |
|---|---|---|---|---|
| HD0344 | nrfA | Nitrate reductase, cytochrome c552 | 3.93 | 0.17 |
| HD0233 | carB | Carbamoyl-phosphate synthase, large subunit | 3.75 | 0.07 |
| HD0235 | carA | Carbamoyl-phosphate synthase, small subunit | 3.72 | 0.14 |
| HD1433 | ompP2A | Outer membrane protein P2 homolog | 3.61 | 0.26 |
| HD1311 | flp2 | flp operon protein Flp2 | 3.56 | 0.21 |
| HD0347 | nrfB | Nitrate reductase, cytochrome c type protein | 3.46 | 0.17 |
| HD1312 | flp1 | flp operon protein Flp1 | 3.33 | 0.18 |
| HD0349 | nrfC | Nitrate reductase, Fe-S protein | 3.26 | 0.31 |
| HD0998 | uraA | Uracil permease | 3.07 | 0.18 |
| HD1310 | flp3 | flp operon protein Flp3 | 3.06 | 0.26 |
| HD0232 | arcB1 | Ornithine carbamoyltransferase | 3.05 | 0.17 |
| HD1985 | Possible DNA transformation protein | 2.84 | 0.10 | |
| HD1306 | rcpB | Rough colony protein B | 2.75 | 0.41 |
| HD0769 | dsrA | Serum resistance protein DsrA | 2.74 | 0.50 |
| HD0427 | comA | Possible competence protein A homolog | 2.52 | 0.45 |
| HD1309 | flp operon protein B | 2.47 | 0.46 | |
| HD1078 | ompP1 | Outer membrane protein P1 | 2.40 | 0.19 |
| HD1326 | hhdB | Hemolysin activation/secretion protein | 2.37 | 0.28 |
| HD1305 | flp operon protein D | 2.36 | 0.20 | |
| HD1304 | tadA | Tight adherence protein A | 2.35 | 0.30 |
| HD1307 | rcpA | Rough colony protein A | 2.34 | 0.36 |
| HD1156 | lspA2 | Large supernatant protein 2 | 2.33 | 1.99 |
| HD0046 | ompA2 | Major outer membrane protein homolog | 2.32 | 0.28 |
| HD0350 | nrfD | Nitrate reductase, transmembrane protein | 2.31 | 0.37 |
| HD1290 | msrB | Peptide methionine sulfoxide reductase MsrB | 2.27 | 0.22 |
| HD1278 | Possible serine protease | 2.27 | 0.24 | |
| HD0073 | napD | Possible NapD protein | 2.18 | 0.15 |
| HD1921 | recG | ATP-dependent DNA helicase RecG | 2.07 | 0.17 |
| HD0072 | napF | Ferredoxin-type protein NapF | 2.03 | 0.21 |
| HD1155 | lspB | Large supernatant protein exporter | 2.01 | 0.18 |
| HD1303 | tadB | Tight adherence protein B | 1.78 | 0.28 |
| HD1221 | ykgE | Conserved putative dehydrogenase subunit | 1.75 | 0.23 |
| HD1302 | tadC | Tight adherence protein C | 1.74 | 0.10 |
| HD1666 | Probable phosphatase | 1.64 | 0.10 | |
| HD1291 | gapA | Glyceraldehyde 3-phosphate dehydrogenase | 1.61 | 0.13 |
| HD1435 | ompP2B | Outer membrane protein P2 homolog | -3.74 | 0.50 |
| HD1512 | Acriflavine resistance protein | -2.81 | 0.05 | |
| HD1163 | ribAB | Riboflavin biosynthesis protein RibA | -2.80 | 0.24 |
| HD1513 | Putative RND efflux membrane fusion protein | -2.67 | 0.30 | |
| HD1161 | ribD | Riboflavin-specific deaminase | -2.28 | 0.21 |
| HD1162 | ribE | Riboflavin synthase, alpha chain | -2.24 | 0.19 |
| HD1109 | Putative oxalate/formate antiporter | -2.18 | 0.19 | |
| HD0282 | fimB | Possible fimbrial structural subunit | -2.16 | 0.17 |
| HD1816 | yfeA | Iron (chelated) ABC transporter | -2.04 | 0.11 |
| HD2025 | hgbA | Hemoglobin-binding protein HgbA | -2.02 | 0.19 |
| HD0648 | tnaB | Tryptophan-specific transport protein | -2.02 | 0.25 |
| HD1024 | yfeD | Iron (chelated) transport system | -2.01 | 0.46 |
| HD1470 | cpxA | Sensor kinase CpxA | -2.00 | 0.05 |
| HD1165 | ribH | 6,7-Dimethyl-8-ribityllumazine synthase | -1.96 | 0.06 |
| HD1179 | ksgA | Dimethyladenosine transferase | -1.91 | 0.11 |
| HD0723 | trmA | tRNA (uracil-5-)-methyltransferase | -1.88 | 0.30 |
| HD1817 | yfeB | Iron (chelated) transporter, ATP-binding protein | -1.81 | 0.11 |
| HD1511 | glmU | Bifunctional GlmU protein | -1.77 | 0.13 |
| HD1025 | yfeC | Iron (chelated) transport system | -1.74 | 0.25 |
| HD1010 | prc | Tail-specific protease | -1.73 | 0.28 |
| HD1469 | cpxR | Transcriptional regulatory protein CpxR | -1.69 | 0.20 |
| HD0454 | waaA | 3-Deoxy-d-manno-octulosonic acid transferase | -1.68 | 0.10 |
| HD1084 | HesB family protein | -1.68 | 0.14 | |
| HD1815 | Putative sulfite reductase | -1.68 | 0.25 | |
| HD1629 | lolB | Outer membrane lipoprotein LolB | -1.68 | 0.23 |
| HD2012 | lipA | Lipoic acid synthetase | -1.63 | 0.19 |
| HD0791 | ccmE | Cytochrome c-type biogenesis protein | -1.62 | 0.27 |
| HD0328 | exbD | Biopolymer transport protein | -1.60 | 0.08 |
| HD1342 | recJ | Single-stranded-DNA-specific exonuclease RecJ | -1.54 | 0.18 |
| HD1191 | Outer membrane protein D15 | -1.54 | 0.16 | |
| HD1628 | ispE | 4-Diphosphocytidyl-2-C-methyl-d-erythritol kinase | -1.54 | 0.08 |
| HD1806 | thiI | Thiamine biosynthesis protein ThiI | -1.54 | 0.20 |
| HD1094 | Possible outer membrane serine protease | -1.52 | 0.10 | |
| HD0329 | exbB | Biopolymer transport protein | -1.49 | 0.12 |
| HD1085 | hscB | Chaperone protein HscB | -1.49 | 0.14 |
The table lists the 35 most upregulated genes and the 35 most downregulated genes, not including ORFs described as hypothetical or conserved hypothetical proteins.
That is, the median log2 ratio of expression when grown in the presence of FCS relative to expression in the absence of FCS from four experiments (P < 0.05).
FIG. 3.
Real-time RT-PCR analysis of the relative levels of expression of selected H. ducreyi genes in the presence of FCS. The data indicate relative expression levels in H. ducreyi 35000HP cells grown in the presence of FCS compared to cells grown in the absence of FCS.
Deletion of cpxR increases expression of LspB and LspA2.
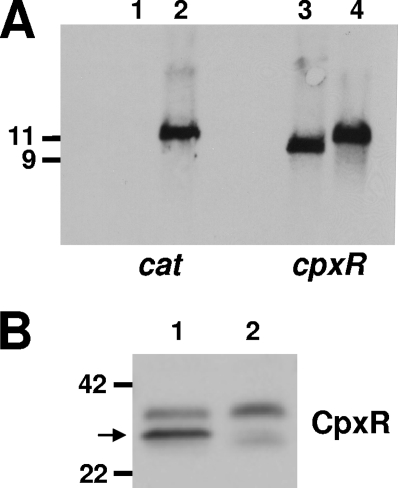
DNA microarray results were scrutinized to look for up- or downregulated ORFs annotated as encoding homologs of known or putative regulators. The homologs to the E. coli CpxAR two-component system (47) were found to be downregulated in the presence of serum (Table 2). The CpxAR system responds to cell envelope stress (46, 47, 49) and has been shown to positively or negatively regulate some virulence determinants in certain bacteria (4, 19, 34, 39, 41). To determine whether expression of LspA2 was under the control of the CpxRA system, a cpxR deletion mutation was generated in H. ducreyi 35000HP. The insertion of a single cat cartridge into the deletion site within cpxR in the 35000HPΔcpxR mutant was confirmed by Southern blotting (Fig. 4A) and nucleotide sequence analyses. Lack of expression of the CpxR protein by this mutant was confirmed by Western blot analysis (Fig. 4B). Real-time RT-PCR analysis indicated that the expression of cpxA (located immediately downstream of cpxR) was only minimally affected in this mutant (data not shown).
FIG. 4.
Characterization of the H. ducreyi 35000HP cpxR deletion mutant. (A) Southern blot analysis of EcoRV-digested chromosomal DNA from 35000HP (lanes 1 and 3) and the 35000HPΔcpxR mutant (lanes 2 and 4) probed with a cat gene fragment (lanes 1 and 2) and with a cpxR gene fragment (lanes 3 and 4). Size markers (in kb) are present on the left side of this panel. (B) Western blot analysis of whole-cell lysates from 35000HP (lane 1) and the 35000HPΔcpxR mutant (lane 2) probed with polyclonal antibody to the H. ducreyi CpxR protein. The arrow indicates the position of the CpxR protein. Molecular mass position markers (in kilodaltons) are present on the left side of this panel.
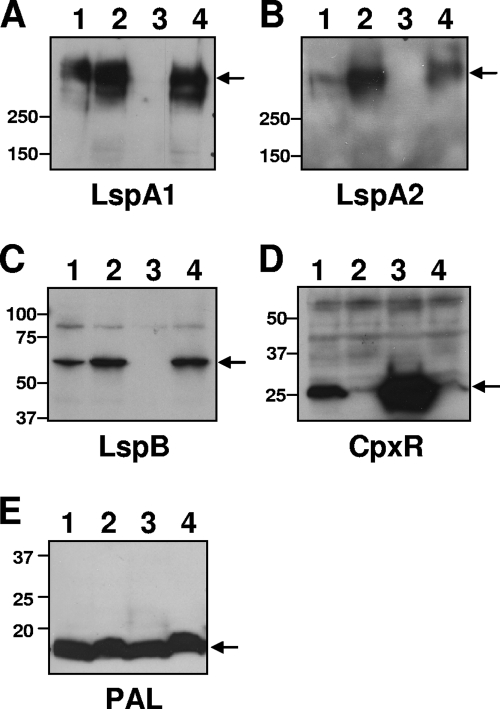
Western blot analysis of whole-cell lysates indicated that the 35000HPΔcpxR mutant expressed more LspA2 (Fig. 5B, lane 2) and more LspB (Fig. 5C, lane 2) than the parental wild-type strain (Fig. 5B and C, lanes 1). It should be noted here that it was previously shown that lspB and lspA2 comprise an operon in H. ducreyi (74). Detection of PAL (54) was used to ensure equivalent loading of samples of the four different strains (Fig. 5E). When the cpxR mutation was complemented with a plasmid carrying cpxR [35000HPΔcpxR(pML125)], the levels of LspB (Fig. 5C, lane 3) and LspA2 (Fig. 5B, lane 3) expression were greatly reduced compared to wild-type (Fig. 5B and C, lanes 1). Unexpectedly, the level of LspA1 expression was also greatly reduced in the complemented mutant 35000HPΔcpxR(pML125) (Fig. 5A, lane 3). Analysis of CpxR expression in all four strains showed that CpxR was more highly expressed in strain 35000HPΔcpxR(pML125) (Fig. 5D, lane 3) compared to wild-type levels (Fig. 5D, lane 1). Taken together, these data indicate that CpxR negatively regulates expression of LspB and LspA2. In addition, overexpression of CpxR can result in inhibition of expression of LspA1.
FIG. 5.
Protein expression by wild-type, mutant, and complemented mutant strains of H. ducreyi. Whole-cell lysates of 35000HP (lane 1), 35000HPΔcpxR (lane 2), 35000HPΔcpxR(pML125) (lane 3), and 35000HPΔcpxR(pLS88) (lane 4) were probed in Western blot analysis with the LspA1-specific MAb 40A4 (A), the LspA2-specific MAb 1H9 (B), polyclonal antiserum against LspB (C), polyclonal antiserum against CpxR (D), and the PAL-specific MAb 3B9 (E). The arrows indicate the relevant antigen in each panel. Molecular mass position markers (in kilodaltons) are present on the left sides of these five panels.
Interaction of CpxR with the promoter region of lspB.
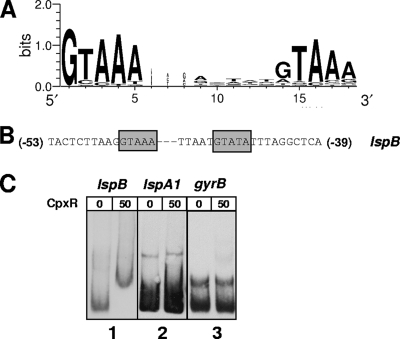
Inspection of the lspB promoter region revealed the presence of a putative CpxR-binding consensus sequence based on the E. coli CpxR binding motif (Fig. 6A) (17). This upstream region contained two conserved pentamers separated by a 5-nt linker (indicated in italics) [5′-(−53)GTAAATTAATGTATA(−39)-3′] (Fig. 6B). The presence of a T instead of an A in the second pentamer (underlined) should not interfere with the binding of CpxR, since this substitution has been observed in other CpxR recognition sites (67).
FIG. 6.
Interaction of CpxR with the promoter region of lspB. (A) Sequence logo for the CpxR recognition site based on 24 published E. coli CpxR binding sequences (17, 67). (B) Putative CpxR-binding consensus sequence upstream from the lspB ORF. (C) EMSA using 50 pmol purified His-tagged CpxR, together with the DIG-labeled lspB promoter region DNA (nt −157 to +43; panel 1), the DIG-labeled lspA1 promoter region DNA (nt −179 to +21; panel 2), and the DIG-labeled gyrB promoter region DNA (nt −154 to +47; panel 3).
A His-tagged H. ducreyi CpxR protein was purified and used in EMSAs. A ∼200-bp fragment encompassing the putative lspB promoter region (nt −157 to +43), with the predicted CpxR binding motif in the center of the fragment, was used as the probe. The predicted promoter regions of lspA1 (nt −178 to +23) and gyrB (HD1643) (nt −154 to +47) were used as negative controls. Neither of these latter two promoter regions contains a predicted CpxR binding motif (data not shown). Incubation of the lspB promoter region with 50 pmol His-tagged CpxR (Fig. 6C, panel 1) resulted in a shift in the migration of the DNA probe, whereas no shift was observed when the promoter regions of lspA1 (Fig. 6C, panel 2) and gyrB (Fig. 6C, panel 3) were used. Taken together, these EMSA results and the LspB/LspA2 expression profile of the cpxR mutant strongly suggest that there is a direct negative effect of CpxR on expression of the lspB-lspA2 operon.
cpxR is located in an operon with three other ORFs.
Genome sequence comparisons of several bacterial species including representatives from the Escherichia, Salmonella, Yersinia, and Shigella genera indicate that the CpxRA two-component regulatory system is arranged in a two-gene operon (35). In the H. ducreyi genome, cpxR and cpxA are situated within a cluster of four genes, HD1468, cpxR, cpxA, and HD1471, which are on the same strand, are putatively transcribed in the same orientation, and have short intergenic regions (62, 64, and 85 nt, respectively). RT-PCR indicated that these four genes are transcribed into a continuous RNA molecule (data not shown), suggesting that they form an operon in H. ducreyi 35000HP.
DISCUSSION
Previous work in our laboratory suggested that lspA1 and lspA2 were under the control of different regulatory mechanisms, based on the observation that LspA1 was apparently more abundant than LspA2 in whole-cell lysates and CCS (64), together with the fact that inactivation of lspA1 resulted in the detection of increased levels of LspA2 in whole-cell lysates and CCS (63). Analysis of LspA1 and LspA2 expression profiles during growth in vitro in the present study indicated that LspA2 had a temporal pattern of expression, with relatively high levels being attained only in the late exponential and stationary phases (Fig. 1). In contrast, LspA1 appeared to be expressed throughout the growth period (Fig. 1). Taken together with our previously published findings (63, 64), these data supported the hypothesis that LspA1 and LspA2 are differentially regulated. In the present study, we obtained evidence strongly suggesting that the Cpx stress response system (47) is involved in the negative regulation of the lspB-lspA2 operon. In addition, we discovered that FCS might serve as an environmental signal to upregulate a variety of virulence factors that could potentially aid in the infection process.
It has been well established that bacteria are able to sense their environment and respond to different stimuli by altering gene expression (27). During the infection process, bacteria have been shown to adjust gene expression depending on the host, tissue, or cellular localization (for a review, see reference 10). The fact that H. ducreyi is an obligate human pathogen does not eliminate its need to be able to regulate gene expression, and there is ample evidence that other obligate human pathogens, including both Bordetella pertussis (52) and Neisseria gonorrhoeae (42), use two-component sensory transduction systems to control gene expression. Although there is some indirect evidence that heme restriction can regulate gene expression in H. ducreyi (20, 56), there have been no reports to date describing regulatory gene mutations in H. ducreyi.
In the present study, we found two different mechanisms involved in controlling expression of the LspA proteins. The first involved FCS, which H. ducreyi requires for optimal growth in vitro. During infection, H. ducreyi is likely to be exposed to serum components at the site of ulceration as a consequence of vascular leakage. Several pathogens have been shown to alter gene expression in response to plasma or serum, including Streptococcus pyogenes (31), Enterococcus faecalis (40), and Yersinia pestis (14). We had previously found that neither LspA protein could be detected in CCS when wild-type H. ducreyi was grown in the absence of FCS (data not shown). In the present study, we found that LspA2 is not expressed in the absence of FCS (Fig. 2B, panel 6), and this result was reflected by real-time RT-PCR analysis (Fig. 2C and 2D). In contrast, expression of LspA1 is not affected by FCS (Fig. 2B, panels 1 and 2). However, even though expression of LspA1 is not affected by FCS, LspA1 was not detectable in CCS derived from cultures grown in the absence of FCS (Fig. 2B, panel 4), suggesting that a factor(s) present in FCS is necessary for the processing and/or release of this very large protein into the medium. Whether this factor acts directly or indirectly to effect this release is not known at this time.
Global transcriptional analysis of H. ducreyi 35000HP cells grown in the presence or absence of FCS indicated that, besides the upregulation of the lspB-lspA2 operon, other genes including both dsrA and the tad operon were also upregulated (Table 2). Expression of these latter two genes or operons has been shown to be important for normal virulence of H. ducreyi in the human challenge model (9, 53). In contrast, expression of the hemoglobin-binding protein HgbA (20, 56), which has been previously shown to be a virulence factor for H. ducreyi (3), was downregulated in vitro by the presence of FCS (Table 2).
Among the H. ducreyi genes that were differentially expressed in the presence or absence of serum, it is interesting to note the dichotomy involving the ompP2A and ompP2B ORFs. Expression of ompP2A was upregulated by the presence of FCS, whereas the expression of ompP2B was downregulated (Fig. 3). Both of these genes encode porins, and H. ducreyi 35000HP expresses both OmpP2A and OmpP2B in vitro (45). However, the majority of H. ducreyi strains tested to date appear to express only OmpP2A and do not express OmpP2B as the result of different mutations in and near the ompP2B ORF. Why H. ducreyi 35000HP would differentially regulate these two ORFs in the presence of serum is not immediately apparent, but this could suggest that the OmpP2A protein may function better as a porin than does OmpP2B.
Further analysis of the transcriptional data indicated that the CpxRA two-component regulatory system was downregulated in the presence of FCS. The CpxRA regulatory system is one of the four cell envelope stress systems in gram-negative bacteria (the other three being the alternative sigma factor σE, BaeSR, and the phage shock response), which are induced by a variety of signals (for reviews, see references 16 and 48). Although initially thought to only respond to stress in the cell envelope, the CpxRA regulatory system has also been found to affect expression of virulence factors in several pathogens, including Legionella pneumophila (21), Salmonella enterica (38), enteropathogenic E. coli (41), and Yersinia species (11). The target genes controlled by the CpxRA system in the aforementioned bacteria are different but are mostly related to adhesion to and/or invasion of mammalian cells. The effect that this two-component regulatory system exerts on these target genes can also vary, in that in some cases it behaves as a positive modulator (21, 41), while in other instances the effect is negative (11, 26).
As described above, FCS enhanced the growth of H. ducreyi in CB (Fig. 2). Under these conditions, it could be inferred that there is less stress on the H. ducreyi cell. Consequently, a reduction in transcription of the cpxRA genes might be expected, as was detected in the DNA microarray experiments in the present study. This in turn could affect the expression of genes regulated by CpxR. The fact that LspA2 was only expressed when H. ducreyi was grown in CB with FCS (Fig. 2) suggested that CpxR might be involved in controlling the expression of this protein.
Inactivation of the cpxR ORF in H. ducreyi 35000HP resulted in increased expression of both LspB and LspA2 (Fig. 5). EMSA findings indicated that a recombinant H. ducreyi CpxR protein bound to the lspB promoter region between nt −157 and +43. This is the region that contains the putative CpxR recognition site, thus providing additional evidence for the involvement of CpxR in the regulation of expression of LspB and LspA2. What element(s) controls of expression of LspA1 remains to be determined. It appears that LspA1 is constitutively expressed by the wild-type strain in vitro (Fig. 1) and that expression of this protein is not affected by the presence of FCS (Fig. 2). However, overexpression of CpxR (in the complemented cpxR mutant) resulted in a dramatic reduction in LspA1 synthesis (Fig. 5A, lane 3). The DNA upstream from lspA1 apparently lacks a CpxR binding site consensus sequence, and the EMSA experiment indicated that this region does not bind CpxR (Fig. 6). It is possible that overexpression of CpxR in the complemented cpxR deletion mutant affected another regulatory pathway(s), which in turn affected LspA1 expression.
Two previous studies focused on identifying bacterial genes transcribed during experimental H. ducreyi infection. The first of these used RT-PCR to detect H. ducreyi transcripts present in biopsies from pustules produced in human volunteers by H. ducreyi 35000HP (59). Both lspA1 and lspA2 transcripts were detected in these in vivo-derived samples. The second study used the selective capture of transcribed sequences to identify genes expressed in vivo and found a cDNA containing a nucleotide sequence common to both lspA1 and lspA2 (6). The fact that both lspA1 and lspA2 transcripts were detected in the former study indicates that both of these genes are transcribed in vivo, although these data do not address protein expression. At the very least, the presence of both lspA1 and lspA2 transcripts in vivo raises the possibility that H. ducreyi synthesizes both of the encoded proteins, perhaps to optimize its ability to avoid phagocytosis.
In conclusion, the studies described here provide the first evidence for the involvement of the CpxRA system in the differential regulation of the LspA proteins. Although CpxR has been shown to be a negative regulator of LspB and LspA2 expression, it remains to be determined what gene product(s) control expression of LspA1. Similarly, the extent of involvement of the CpxRA system in controlling expression of other H. ducreyi virulence factors is not known. In addition, the identity of the factor(s) in FCS necessary for expression of LspA2 in vitro and for release of both LspA1 and LspA2 from the H. ducreyi cell surface is unknown at this time. These results and unanswered questions warrant more detailed investigation of the CpxRA two-component system in H. ducreyi.
Acknowledgments
This study was supported by U.S. Public Health Service grant AI32011 to E.J.H. J.R.M. was supported by U.S. Public Health Service training grant 5-T32-AI007520.
We thank Stanley Spinola for kindly providing MAb 3B9.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 18 May 2009.
REFERENCES
- 1.Albritton, W. L. 1989. Biology of Haemophilus ducreyi. Microbiol. Rev. 53377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton, W. L., I. W. MacLean, P. D. Bertram, and A. R. Ronald. 1981. Haemin requirements in Haemophilus with special reference to H. ducreyi, p. 75-82. In M. Kilian, W. Frederiksen, and E. L. Biberstein (ed.), Haemophilus, Pasteurella, and Actinobacillus. Academic Press, London, England.
- 3.Al Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 1811049-1054. [DOI] [PubMed] [Google Scholar]
- 4.Altman, E., and G. Segal. 2008. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J. Bacteriol. 1901985-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anonymous. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 6.Bauer, M. E., K. R. Fortney, A. Harrison, D. M. Janowicz, R. S. Munson, Jr., and S. M. Spinola. 2008. Identification of Haemophilus ducreyi genes expressed during human infection. Microbiology 1541152-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blick, R. J., A. T. Revel, and E. J. Hansen. 2003. FindGDPs: identification of primers for labeling microbial transcriptomes for DNA microarray analysis. Bioinformatics 191718-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bong, C. T., M. E. Bauer, and S. M. Spinola. 2002. Haemophilus ducreyi: clinical features, epidemiology, and prospects for disease control. Microbes Infect. 41141-1148. [DOI] [PubMed] [Google Scholar]
- 9.Bong, C. T., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 691488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyce, J. D., P. A. Cullen, and B. Adler. 2004. Genomic-scale analysis of bacterial gene and protein expression in the host. Emerg. Infect. Dis. 101357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsson, K. E., J. Liu, P. J. Edqvist, and M. S. Francis. 2007. Influence of the Cpx extracytoplasmic-stress-responsive pathway on Yersinia sp.-eukaryotic cell contact. Infect. Immun. 754386-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carson, S. D. B., C. E. Thomas, and C. Elkins. 1996. Cloning and sequencing of a Haemophilus ducreyi fur homolog. Gene 176125-129. [DOI] [PubMed] [Google Scholar]
- 13.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1341141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chauvaux, S., M. L. Rosso, L. Frangeul, C. Lacroix, L. Labarre, A. Schiavo, M. Marceau, M. A. Dillies, J. Foulon, J. Y. Coppee, C. Medigue, M. Simonet, and E. Carniel. 2007. Transcriptome analysis of Yersinia pestis in human plasma: an approach for discovering bacterial genes involved in septicaemic plague. Microbiology 1533112-3124. [DOI] [PubMed] [Google Scholar]
- 15.Dangor, Y., S. D. Miller, H. J. Koornhof, and R. C. Ballard. 1992. A simple medium for the primary isolation of Haemophilus ducreyi. Eur. J. Clin. Microbiol. Infect. Dis. 11930-934. [DOI] [PubMed] [Google Scholar]
- 16.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57621-628. [DOI] [PubMed] [Google Scholar]
- 17.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 27726652-26661. [DOI] [PubMed] [Google Scholar]
- 18.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 1852432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of Escherichia coli in biofilm formation. FEMS Microbiol. Lett. 178169-175. [DOI] [PubMed] [Google Scholar]
- 20.Elkins, C. 1995. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect. Immun. 631241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gal-Mor, O., and G. Segal. 2003. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J. Bacteriol. 1854908-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond, G. W., C. J. Lian, J. C. Wilt, W. L. Albritton, and A. R. Ronald. 1978. Determination of the hemin requirement of Haemophilus ducreyi: evaluation of the porphyrin test and media used in the satellite growth test. J. Clin. Microbiol. 7243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, E. J., J. L. Latimer, S. E. Thomas, M. Helminen, W. L. Albritton, and J. D. Radolf. 1992. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J. Bacteriol. 1745442-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haydock, A. K., D. H. Martin, S. A. Morse, C. Cammarata, K. J. Mertz, and P. A. Totten. 1999. Molecular characterization of Haemophilus ducreyi strains from Jackson, Mississippi, and New Orleans, Louisiana. J. Infect. Dis. 1791423-1432. [DOI] [PubMed] [Google Scholar]
- 25.Hayes, R. J., K. F. Schulz, and F. A. Plummer. 1995. The cofactor effect of genital ulcers on the per-exposure risk of HIV transmission in sub-Saharan Africa. J. Trop. Med. Hyg. 981-8. [PubMed] [Google Scholar]
- 26.Hernday, A. D., B. A. Braaten, G. Broitman-Maduro, P. Engelberts, and D. A. Low. 2004. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16537-547. [DOI] [PubMed] [Google Scholar]
- 27.Hoch, J. A., and Silhavy (ed.). 1995. Two-component signal transduction. American Society for Microbiology, Washington, DC.
- 28.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8528-535. [PubMed] [Google Scholar]
- 29.Janowicz, D., N. R. Luke, K. R. Fortney, B. P. Katz, A. A. Campagnari, and S. M. Spinola. 2006. Expression of OmpP2A and OmpP2B is not required for pustule formation by Haemophilus ducreyi in human volunteers. Microb. Pathog. 40110-115. [DOI] [PubMed] [Google Scholar]
- 30.Janowicz, D. M., K. R. Fortney, B. P. Katz, J. L. Latimer, K. Deng, E. J. Hansen, and S. M. Spinola. 2004. Expression of the LspA1 and LspA2 proteins by Haemophilus ducreyi is required for virulence in human volunteers. Infect. Immun. 724528-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson, B. P., F. Levander, U. von Pawel-Rammingen, T. Berggard, L. Bjorck, and P. James. 2005. The protein expression of Streptococcus pyogenes is significantly influenced by human plasma. J. Proteome Res. 42302-2311. [DOI] [PubMed] [Google Scholar]
- 32.Lukomski, S., R. A. Hull, and S. I. Hull. 1996. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J. Bacteriol. 178240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macdonald, K., D. W. Cameron, G. Irungu, L. J. D'Costa, F. A. Plummer, L. A. Slaney, J. O. Ndinya-Achola, and A. R. Ronald. 1989. Comparison of Sheffield media with standard media for the isolation of Haemophilus ducreyi. Sex. Transm. Infect. 1688-90. [DOI] [PubMed] [Google Scholar]
- 34.Macritchie, D. M., J. D. Ward, A. Z. Nevesinjac, and T. L. Raivio. 2008. Activation of the Cpx envelope stress response downregulates expression of several locus of enterocyte effacement-encoded genes in enteropathogenic Escherichia coli. Infect. Immun. 761465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNeil, L. K., C. Reich, R. K. Aziz, D. Bartels, M. Cohoon, T. Disz, R. A. Edwards, S. Gerdes, K. Hwang, M. Kubal, G. R. Margaryan, F. Meyer, W. Mihalo, G. J. Olsen, R. Olson, A. Osterman, D. Paarmann, T. Paczian, B. Parrello, G. D. Pusch, D. A. Rodionov, X. Shi, O. Vassieva, V. Vonstein, O. Zagnitko, F. Xia, J. Zinner, R. Overbeek, and R. Stevens. 2007. The National Microbial Pathogen Database Resource (NMPDR): a genomics platform based on subsystem annotation. Nucleic Acids Res. 35D347-D353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mertz, K. J., D. L. Trees, W. C. Levine, J. S. Lewis, B. Litchfield, K. S. Pettus, S. A. Morse, M. E. St. Louis, J. B. Weiss, J. Schwebke, J. Dickes, R. Kee, J. Reynolds, D. Hutcheson, D. Green, I. Dyer, G. A. Richwald, J. Novotny, M. Goldberg, J. A. O'Donnell, and R. Knaup. 1998. Etiology of genital ulcers and prevalence of human immunodeficiency virus coinfection in 10 US cities. J. Infect. Dis. 1781795-1798. [DOI] [PubMed] [Google Scholar]
- 37.Mohammed, T. T., and Y. M. Olumide. 2008. Chancroid and human immunodeficiency virus infection: a review. Int. J. Dermatol. 471-8. [DOI] [PubMed] [Google Scholar]
- 38.Nakayama, S., A. Kushiro, T. Asahara, R. Tanaka, L. Hu, D. J. Kopecko, and H. Watanabe. 2003. Activation of hilA expression at low pH requires the signal sensor CpxA, but not the cognate response regulator CpxR, in Salmonella enterica serovar Typhimurium. Microbiology 1492809-2817. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama, S., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 1803522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nallapareddy, S. R., and B. E. Murray. 2008. Role played by serum, a biological cue, in the adherence of Enterococcus faecalis to extracellular matrix proteins, collagen, fibrinogen, and fibronectin. J. Infect. Dis. 1971728-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevesinjac, A. Z., and T. L. Raivio. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 187672-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overton, T. W., R. Whitehead, Y. Li, L. A. Snyder, N. J. Saunders, H. Smith, and J. A. Cole. 2006. Coordinated regulation of the Neisseria gonorrhoeae-truncated denitrification pathway by the nitric oxide-sensitive repressor, NsrR, and nitrite-insensitive NarQ-NarP. J. Biol. Chem. 28133115-33126. [DOI] [PubMed] [Google Scholar]
- 43.Parsons, L. M., A. L. Waring, R. J. Limberger, and M. Shayegani. 1999. The dnaK/dnaJ operon of Haemophilus ducreyi contains a unique combination of regulatory elements. Gene 233109-119. [DOI] [PubMed] [Google Scholar]
- 44.Parsons, L. M., A. L. Waring, and M. Shayegani. 1992. Molecular analysis of the Haemophilus ducreyi groE heat shock operon. Infect. Immun. 604111-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prather, D. T., M. Bains, R. E. Hancock, M. J. Filiatrault, and A. A. Campagnari. 2004. Differential expression of porins OmpP2A and OmpP2B of Haemophilus ducreyi. Infect. Immun. 726271-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raivio, T. L. 2005. Envelope stress responses and gram-negative bacterial pathogenesis. Mol. Microbiol. 561119-1128. [DOI] [PubMed] [Google Scholar]
- 47.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 1797724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF s factors. Annu. Rev. Microbiol. 55591-624. [DOI] [PubMed] [Google Scholar]
- 49.Raivio, T. L., and T. J. Silhavy. 1999. The sE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2159-165. [DOI] [PubMed] [Google Scholar]
- 50.Ronald, A. R., and W. Albritton. 1990. Chancroid and Haemophilus ducreyi, p. 263-271. In K. K. Holmes, P.-A. Mardh, P. F. Sparling, and P. J. Wiesner (ed.), Sexually transmitted diseases. McGraw-Hill Book Co., New York, NY.
- 51.Schulte, J. M., F. A. Martich, and G. P. Schmid. 1992. Chancroid in the United States, 1981-1990: evidence for underreporting of cases. MMWR Morbid. Mortal. Wkly. Rep. 4157-61. [PubMed] [Google Scholar]
- 52.Smith, A. M., C. A. Guzman, and M. J. Walker. 2001. The virulence factors of Bordetella pertussis: a matter of control. FEMS Microbiol. Rev. 25309-333. [DOI] [PubMed] [Google Scholar]
- 53.Spinola, S. M., K. R. Fortney, B. P. Katz, J. L. Latimer, J. R. Mock, M. Vakevainen, and E. J. Hansen. 2003. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect. Immun. 717178-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spinola, S. M., T. J. Hiltke, K. R. Fortney, and K. L. Shanks. 1996. The conserved 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi has homology to PAL. Infect. Immun. 641950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steen, R. 2001. Eradicating chancroid. Bull. W. H. O. 79818-826. [PMC free article] [PubMed] [Google Scholar]
- 56.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. R. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 641724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor, S. N., and D. H. Martin. 2007. Chancroid, p. 69-74. In J. D. Klausner and E. W. I. Hook (ed.), Current diagnosis and treatment of sexually transmitted diseases. McGraw-Hill Companies, Columbus, OH.
- 58.Throm, R. E., J. A. Al Tawfiq, K. R. Fortney, B. P. Katz, A. F. Hood, C. A. Slaughter, E. J. Hansen, and S. M. Spinola. 2000. Evaluation of an isogenic major outer membrane protein-deficient mutant in the human model of Haemophilus ducreyi infection. Infect. Immun. 682602-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Throm, R. E., and S. M. Spinola. 2001. Transcription of candidate virulence genes of Haemophilus ducreyi during infection of human volunteers. Infect. Immun. 691483-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.UNAIDS. 1997. Sexually transmitted diseases: policies and principles for prevention and care. World Health Organization, New York, NY.
- 61.Vakevainen, M., S. Greenberg, and E. J. Hansen. 2003. Inhibition of phagocytosis by Haemophilus ducreyi requires expression of the LspA1 and LspA2 proteins. Infect. Immun. 715994-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, W., L. Reitzer, D. A. Rasko, M. M. Pearson, R. J. Blick, C. Laurence, and E. J. Hansen. 2007. Metabolic analysis of Moraxella catarrhalis and the effect of selected in vitro growth conditions on global gene expression. Infect. Immun. 754959-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward, C. K., J. L. Latimer, J. R. Nika, M. Vakevainen, J. R. Mock, K. Deng, R. J. Blick, and E. H. Hansen. 2003. Mutations in the lspA1 and lspA2 genes of Haemophilus ducreyi affect the virulence of this pathogen in an animal model system. Infect. Immun. 712478-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ward, C. K., S. R. Lumbley, J. L. Latimer, L. D. Cope, and E. J. Hansen. 1998. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J. Bacteriol. 1806013-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ward, C. K., J. R. Mock, and E. J. Hansen. 2004. The LspB protein is involved in the secretion of the LspA1 and LspA2 proteins by Haemophilus ducreyi. Infect. Immun. 721874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Willson, P. J., W. L. Albritton, L. Slaney, and J. K. Setlow. 1989. Characterization of a multiple antibiotic resistance plasmid from Haemophilus ducreyi. Antimicrob. Agents Chemother. 331627-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto, K., and A. Ishihama. 2006. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 701688-1695. [DOI] [PubMed] [Google Scholar]