Abstract
The observation that Borrelia burgdorferi-induced arthritis is severe in C3H mice and milder in C57BL/6 (B6) mice has allowed a forward genetics approach for the identification of genetic elements that regulate the arthritis response. Quantitative trait loci (QTL) on five chromosomes (Chr) were identified previously in segregating crosses between C3H and B6 mice and collectively designated B. burgdorferi arthritis-associated (Bbaa) QTL. Reciprocal interval-specific congenic lines (ISCL) that encompass Bbaa1, Bbaa2-Bbaa3, Bbaa4, Bbaa6, and Bbaa12 on Chr 4, 5, 11, 12, and 1, respectively, have now been generated. Bidirectional transfer of the arthritis severity phenotype in association with Bbaa2-Bbaa3 and Bbaa4 was observed, and unidirectional transfer with the B6 allele of Bbaa6 was noted. These findings confirm the existence of polymorphic loci within Bbaa2-Bbaa3, Bbaa4, and Bbaa6 that regulate the severity of B. burgdorferi-induced arthritis. ISCL were used to assess the regulation of a previously identified interferon transcriptional profile associated with severe disease in C3H mice. The regulation of this transcriptional signature was found to be independent of penetrant Bbaa QTL, both in joint tissues and in isolated macrophages. These results clearly demonstrate the utility of forward genetics for the discovery of novel genes and pathways involved in the regulation of the severity of Lyme arthritis and predict the involvement of regulatory elements not evident from other experimental approaches.
Infection by the tick-borne spirochete Borrelia burgdorferi is responsible for Lyme disease, which is associated with an array of symptoms of differing severities in infected individuals (7). Although a self-limiting, acute form of arthritis occurs in 60% of individuals not treated at the time of the tick bite, serological evidence indicates that other individuals become infected without the development of clinical arthritis (24, 30). Additionally, in a small percentage of patients with acute disease, the symptoms are prolonged and found to be refractory to antibiotic therapy (8, 16, 27). Together, these findings suggest that the degree of pathology developing in infected hosts is genetically regulated. The genetic elements responsible for the differing severities of acute arthritis in patients have not been identified, although it is known that acute Lyme arthritis is not linked to the major histocompatibility complex (MHC) (29). Interestingly, data in several reports have indicated an association of prolonged Lyme arthritis with MHC class II alleles also associated with rheumatoid arthritis (16, 29). Because these prolonged symptoms arise in individuals with the most severe acute disease, the identification of genetic elements that regulate the severity of acute Lyme arthritis may also provide insight into genetic elements that set the stage for chronic disease in susceptible individuals (30).
Barthold and colleagues initially reported that inbred strains of mice differed in the severity of arthritis and carditis following infection with B. burgdorferi, providing a model to study the differences in responses associated with a spectrum of pathological outcomes (1). The results of additional studies have supported the conclusion that the genetic background of the mouse regulates the severity of disease, and several strains of mice (C3H/HeN, C3H/HeJ, and AKR/J) that reproducibly develop severe disease when infected with a variety of strains of B. burgdorferi via different inoculum regimens have been identified, while other mouse strains (C57BL/6 [B6], C57BL/6J, DBA/2J, DBA/1J, and 129SV) consistently develop milder disease (1, 3, 4, 25). BALB/cAN mice appear to be unique in that the severity of disease and the level of bacteria in tissues are directly correlated with the initial inoculum dose (20).
The characterization of mouse strains with differing phenotypes related to Lyme arthritis led to the identification and mapping of quantitative trait loci (QTL) within the mouse genome controlling a variety of disease-related phenotypes, and these loci were collectively designated B. burgdorferi arthritis-associated (Bbaa) QTL. Using microsatellite-based QTL linkage analysis and segregating crosses between B6 or BALB/c and C3H mice, we identified 23 Bbaa QTL distributed across 13 chromosomes (Chr) (26, 34). Six of the Bbaa QTL on five different Chr were identified in different segregating populations, with Bbaa on Chr 5 identified in four separate analyses (26). Further analysis using composite interval mapping predicted that multiple linked Bbaa QTL on Chr 5 might contribute to the greater severity of arthritis seen in C3H mice (26). Importantly, although H-2, the murine MHC, was linked to the magnitude of the antibody response in infected mice (Bbaa5), there was no linkage to arthritis severity; this finding is consistent with the lack of HLA association with the more common acute arthritis seen in humans (29).
To evaluate the six Bbaa QTL with the broadest arrays of arthritis-associated phenotypes exhibiting linkage in multiple analyses, and as the first step toward positionally cloning the respective genes, we generated reciprocal interval-specific congenic lines (ISCL) encompassing Bbaa1 (Chr 4), Bbaa2-Bbaa3 (Chr 5), Bbaa4 (Chr 11), Bbaa6 (Chr 12), and Bbaa12 (Chr 1). Of the five Bbaa QTL studied, Bbaa2-Bbaa3, Bbaa4, and Bbaa6 were associated with significant differences in disease severity between ISCL and the parental background strains. A previous microarray analysis had identified the selective upregulation of a number of interferon (IFN)-inducible genes, most of which do not reside within the Bbaa2-Bbaa3 or Bbaa4 intervals, in the joint tissues of disease-susceptible C3H mice (11). The contribution of Bbaa QTL with penetrant phenotypes to the regulation of IFN profile transcripts was assessed by using infected joint tissues and isolated macrophages.
MATERIALS AND METHODS
Generation of mouse ISCL.
Six distinct QTL on five Chr regulating the severity of murine Lyme arthritis were identified previously using segregating crosses between B6 and C3H mice (34). Standard nomenclature for the mouse lines is used herein, as follows: the background strain (e.g., C3H, abbreviated C3 in the ISCL designations) is listed first, followed by the donor strain (e.g., B6) and the introgressed Bbaa QTL (QTL positions are listed in parentheses below). C3.B6-Bbaa2-Bbaa3 and B6.C3-Bbaa2-Bbaa3 congenic mouse lines were generated by the introgression of the region of Chr 5 spanning both Bbaa2 and Bbaa3 from C3H/HeNCr (Mbp 72.29 to 141.16) or C57BL/6NCr (Mbp 72.29 to 137.53) mice (National Cancer Institute) into the reciprocal parental strain, C57BL/6NCr or C3H/HeNCr, respectively. Similarly, C3.B6-Bbaa1 (Mbp 3.58 to 150.08), B6.C3-Bbaa1 (Mbp 9.32 to 94.07), C3.B6-Bbaa4 (Mbp 25.86 to 103.24), B6.C3-Bbaa4 (Mbp 17.18 to 101.75), C3.B6-Bbaa6 (Mbp 73.39 to 116.11), B6.C3-Bbaa6 (Mbp 82.01 to 111.54), C3.B6-Bbaa12 (Mbp 116.02 to 196.25), and B6.C3-Bbaa12 (Mbp 171.14 to 196.25) ISCL were generated by marker-assisted selection. Heterozygous ISCL were screened after seven backcross generations with 280 informative microsatellites equally dispersed throughout the genome. Homozygous progeny derived from matings of heterozygous male and female ISCL mice that were free of background donor strain contamination were used to fix the lines.
Culture of B. burgdorferi and infection of mice.
Mice between 5 and 6 weeks of age were infected by intradermal injection with 2 × 103 bacteria of the B. burgdorferi N40 isolate (provided by S. Barthold at the University of California). B. burgdorferi cells were cultured in Barbour-Stoenner-Kelly II medium containing 6% rabbit serum (Sigma-Aldrich, St. Louis, MO).
Arthritis analysis.
Rear ankle joints were measured at the time of infection and at 4 weeks after infection by using a metric caliper. Measurements of the thickest anteroposterior portion of the ankle with the joint extended were taken and are reported as the change in ankle swelling over time (20). A histological assessment of arthritis severity was performed with the most swollen ankle joint. Tissues were fixed in 10% neutral buffered formalin, decalcified, embedded in paraffin, and cut into 5-μm-thick sections, and the sections were mounted onto glass slides and stained with hematoxylin and eosin. The joint sections were evaluated blindly and scored for the severity of injury according to a subjective scale ranging from 0 to 5. A score of 0 indicated no lesions, and scores of 1, 2, 3, 4, and 5 indicated minimal, mild, moderate, marked, and severe lesions, respectively (5). The overall lesion score represented a combined assessment of neutrophil infiltration, mononuclear cell infiltration, tendon sheath thickness, and reactive-reparative responses.
Isolation of RNA.
Total RNA from rear ankle tissues was isolated by acid guanidine extraction as described previously (9). Flash-frozen tissue was homogenized in cold acid guanidine by using an Ultra-Turrax disperser (IKA Works), and RNA was separated by cesium chloride pellet centrifugation. Recovered RNA was applied to an RNeasy kit (Qiagen). RNA was extracted from cultured cells with Trizol reagent according to the instructions of the manufacturer (Invitrogen).
Gene expression analysis.
Gene expression analyses of B. burgdorferi-infected C3H and B6 mice were performed as described previously, using ankle tissues from either uninfected mice or mice at 1 week postinfection (11). Equal amounts of total RNA from the ankle tissues of five to eight individual mice of each genotype were pooled into a single sample that was prepared for hybridization to an array according to the instructions of the manufacturer (Affymetrix). All samples were hybridized to the GeneChip Mouse Genome 430 2.0 array (Affymetrix) and were preprocessed using the affy and gcRMA (robust multiarray average) packages in R (10). Transcripts found to have a change in levels in infected tissues of twofold or greater relative to the levels in uninfected tissues were considered to be differentially expressed, whereas other transcripts were considered to be unchanged.
Design of the microarray experiment.
Gene expression profiling with joint tissues from mice infected with B. burgdorferi compared to those from uninfected mice was performed previously using Affymetrix GeneChip microarrays (11). The most striking distinctions in expression profiles between severely arthritic C3H and mildly arthritic B6 mice were found at 1 week of infection, a time point prior to the development of the inflammatory cell infiltrate characteristic of Lyme arthritis in mice. To determine if the genes within the Bbaa QTL regulated the gene expression profiles associated with the two contrasting arthritis phenotypes, we performed additional microarray analyses, focusing on the 1-week time point. Previously, we found no technical variation among triplicate GeneChips; therefore, in these experiments a single GeneChip was probed with pooled RNAs from at least five mice for each congenic mouse genotype (11). Expression profiling was also performed with C3H mice using the GeneChip Mouse Genome 430 2.0 array, not available for the previous study. Changes in expression in infected mice were confirmed by quantitative reverse transcriptase PCR (RT-PCR) analyses of selected transcripts in the individual RNA samples used to generate the pools.
Cultures of BMMs.
Bone marrow-derived macrophages (BMMs) were isolated from the femurs and tibias of mice, as described previously (21), by culture in RPMI 1640 medium (Invitrogen) supplemented with 30% L929 conditioned medium and 20% horse serum (HyClone). Complete RPMI 1640 medium was demonstrated to be free of Mycoplasma species with the MycoProbe Mycoplasma detection kit (R&D Systems). Harvested macrophages were replated into six-well dishes at a density of 6 × 105/ml in medium lacking serum and containing 1% Nutridoma. After overnight incubation, the medium was removed and replaced with medium alone or medium containing 5 μg/ml sonicated B. burgdorferi cloned N40 bacteria. Macrophages were stimulated at 37°C and 5% CO2 for 24 h, after which the cells were harvested for RNA extraction.
Real-time PCR analysis.
RT-PCR analysis of 5 μg of total RNA was performed using random primers and Moloney murine leukemia virus RT (Invitrogen Life Technologies). Quantitative PCR was performed using LightCycler Sybr Plus master mix and the LightCycler PCR system (Roche Applied Science). The oligonucleotide primers used to detect each transcript were as follows: β-actin transcript, bactinF (forward; 5′-GTAACAATGCCATGTTCAAT-3′) and bactinR (reverse; 5′-CTCCATCGTGGGCCGCTCTAG-3′); Iigp1 transcript, iigp1F (5′-GTAGTGTGCTCAATGTTGCTGTCAC-3′) and iigp1R (5′-TACCTCCACCACCCCAGTTTTAGC-3′); Tgtp transcript, tgtpF (5′-AGGGCTGTGTGTGGACAAGTAAGG-3′) and tgtpR (5′-TGGGTGTGGGTATGGAGTTCTC-3′); Cxcl9 transcript, cxcl9F (5′-TTGGGCATCATCTTCCTGGAGCAG-3′) and Ccxcl9R (5′-GAGGTCTTTGAGGGATTTGTAGTGG-3′); Ifit1 transcript, Ifit1F (5′-GTCAACTGTGAGTGCTTCCATCC-3′) and ift1R (5′-TCAGGGCAGAAAAGTCAAGGC-3′); Mpa2l transcript, mpa2lF (5′-CAGGAAGCCATAGAGATTCTGGAC-3′) and mpa2lR (5′-TGCCCTTGGTTTGAGACTGC-3′); Igtp transcript, igtpF (5′-TAGAGCAGACCCACAGAGTTCAGG-3′) and igtpR (5′-CAGCAGTCATAGATTTAGACCACGG-3′); and Cxcl10 transcript, cxcl10F (5′ GAAATCATCCCTGCGAGCCTATCC 3′) and cxcl10R (5′ GCAATTAGGACTAGCCATCCACTGGG 3′).
Statistical analysis.
Student's t test or the Mann-Whitney U test (as noted) for two-group comparisons and one-way analysis of variance, followed by either Tukey's honestly significant difference post hoc test (equal variances assumed) or Tamhane's T2 post hoc test (equal variances not assumed), for three-group comparisons were performed using GraphPad InStat3 version 3.0b for Macintosh (GraphPad Software, San Diego, CA; http://www.graphpad.com). Values of P of <0.05 were considered significant. The Mann-Whitney U test was performed for comparisons of categorical scores for histopathological lesions, and Student's t test was used for parameters with continuous values.
Microarray data accession number.
Microarray data from this study have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE16195.
RESULTS
Arthritis phenotypes in congenic mouse lines infected with B. burgdorferi.
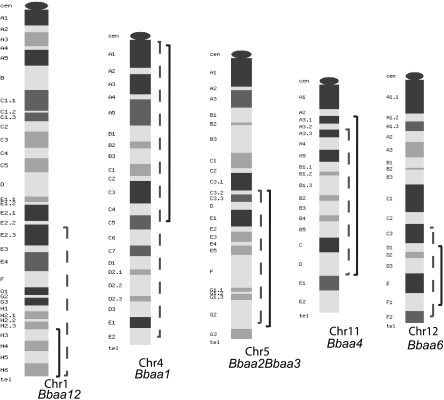
We reported previously that penetrant Lyme arthritis phenotypes were seen in reciprocal Bbaa2-Bbaa3 ISCL (12). Compared to B6 mice, B6.C3-Bbaa2-Bbaa3 mice displayed increased arthritis severity at 4 weeks following B. burgdorferi infection, whereas C3.B6-Bbaa2-Bbaa3 mice had less severe arthritis than infected C3H mice. Both Bbaa2-Bbaa3 ISCL had intermediate degrees of arthritis severity relative to those seen in the parental strains. In this study, arthritis severity in reciprocal Bbaa1, Bbaa4, Bbaa6, and Bbaa12 ISCL mice was assessed. The locations of the introgressed chromosomal regions for each Bbaa QTL are shown in Fig. 1. An overview of the phenotypes of all 10 ISCL is presented in Table 1.
FIG. 1.
Approximate boundaries of introgressed regions for each Bbaa QTL. Bbaa QTL were identified on Chr 1, 4, 5, 11, and 12. QTL were introduced into reciprocal backgrounds by backcrossing, with the introgressed regions from B6 mice indicated by solid lines and those from C3H mice indicated by dashed lines. Introgressed-region boundaries were determined by microsatellite marker analysis as described in Materials and Methods. The cytomaps used to generate this figure were retrieved from the Mouse Genome Database (Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, ME; http://www.informatics.jax.org) (6) in February 2009. cen, centromere; tel, telomere.
TABLE 1.
Arthritis phenotypes of B. burgdorferi-infected ISCL
| ISCLa | Relevant Chr (interval in Mbp) | Phenotypeb relative to that of background strain (C3H or B6) |
|---|---|---|
| B6.C3-Bbaa1 | 4 (9.32-94.07) | Unchanged |
| C3.B6-Bbaa1 | 4 (3.58-150.08) | Ankle swelling suppressed (males only); no change in histopathology |
| B6.C3-Bbaa2-Bbaa3 | 5 (72.29-141.16) | Ankle swelling and histopathology increased relative to those in B6 mice |
| C3.B6-Bbaa2-Bbaa3 | 5 (72.29-137.53) | Ankle swelling and histopathology decreased relative to those in C3H mice |
| B6.C3-Bbaa4 | 11 (17.18-101.75) | Ankle swelling and histopathology increased relative to those in B6 mice |
| C3.B6-Bbaa4 | 11 (25.86-103.24) | Ankle swelling and histopathology decreased relative to those in C3H mice |
| B6.C3-Bbaa6 | 12 (82.01-111.54) | Unchanged |
| C3.B6-Bbaa6 | 12 (73.39-116.11) | Ankle swelling and histopathology decreased relative to those in C3H mice |
| B6.C3-Bbaa12 | 1 (171.14-196.25) | Unchanged |
| C3.B6-Bbaa12 | 1 (116.02-196.25) | Unchanged |
Congenic lines are described in Materials and Methods. Boldface indicates lines exhibiting significant transfer of several arthritis traits.
Arthritis was assessed at 4 weeks postinfection.
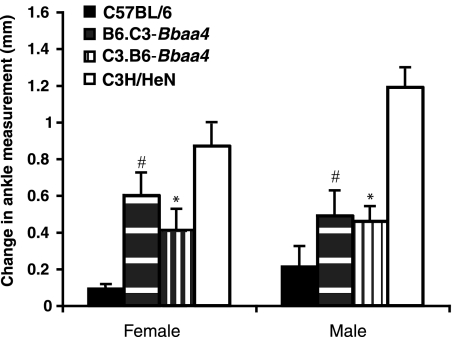
As shown in Fig. 2 and Table 2, B6.C3-Bbaa4 mice infected with B. burgdorferi developed significantly more severe arthritis than did B6 mice. Ankle swelling data are shown in Fig. 2, with statistical significance achieved following the infection of both male and female B6.C3-Bbaa4 mice. Ankle tissue samples mounted onto slides were scored blindly and also revealed increased arthritis severity for the categories of tendon sheath thickness, polymorphonuclear leukocyte (PMN) infiltration, reactive-reparative responses, and overall lesion severity (Table 2). In the reciprocal ISCL, C3.B6-Bbaa4, the degree of arthritis severity at 4 weeks postinfection was significantly lower than that seen in C3H mice. Similarly, multiple measures of arthritis severity, including ankle swelling, sheath thickness, PMN infiltration, reactive-reparative responses, and the overall lesion score, were significantly reduced. Like Bbaa2-Bbaa3 ISCL, reciprocal Bbaa4 ISCL exhibited intermediate arthritis severity relative to C3H and B6 mice. In most cases, Bbaa2-Bbaa3 and Bbaa4 ISCL mice had one rear ankle joint with greater swelling and higher histopathological scores than the other joint, a pattern characteristic of C3H mice. Initial experiments established concordance in ankle swelling measurements and histopathological scores for 85% of Bbaa2-Bbaa3 and Bbaa4 ISCL mice (data not shown).
FIG. 2.
Lyme arthritis phenotypes in C3H, B6, B6.C3-Bbaa4, and C3.B6-Bbaa4 mice infected with B. burgdorferi. Ankle measurements were taken as described in the text and used for an assessment of arthritis severity at 4 weeks after B. burgdorferi infection. Changes in measurements between the start of infection and 4 weeks postinfection are expressed as means ± standard errors. #, significantly increased (P < 0.05; Student's t test) for female (n = 18) and male (n = 16) B6.C3-Bbaa4 mice compared to those for B6 (10 female and 10 male) mice; *, significantly decreased (P < 0.05; Student's t test) for female (n = 10) and male (n = 7) C3.B6-Baab4 mice compared to those for C3H/HeN (10 female and 9 male) mice. Results are representative of three independent experiments.
TABLE 2.
Histological assessment of arthritis severity in B. burgdorferi-infected B6.C3-Bbaa4 and C3.B6-Bbaa4 ISCLa
| Mouse strain (no. of mice) | Overall lesion score | Score for:
|
Total arthritis score | ||
|---|---|---|---|---|---|
| Neutrophil infiltration | Tendon sheath thickness | Reactive-reparative response | |||
| B6 (20) | 1.05 ± 0.23 | 0.50 ± 0.22 | 0.8 ± 0.19 | 0.75 ± 0.18 | 3.25 ± 0.75 |
| B6.C3-Bbaa4 (34) | 2.07 ± 0.23 | 1.14 ± 0.20 | 1.90 ± 0.24 | 1.83 ± 0.24 | 6.83 ± 0.85 |
| C3.B6-Bbaa4 (17) | 1.93 ± 0.26 | 1.64 ± 0.29 | 1.64 ± 0.23 | 1.14 ± 0.21 | 6.14 ± 0.91 |
| C3H/HeN (19) | 3.05 ± 0.19 | 2.21 ± 0.21 | 2.79 ± 0.14 | 2.79 ± 0.21 | 10.68 ± 0.67 |
Histopathology was assessed as described in Materials and Methods. Values shown are means ±standard errors. Boldface indicates significant difference (P < 0.05) from the value for the background strain, as determined by a Mann-Whitney U test.
C3.B6-Bbaa6 mice also displayed reduced arthritis severity compared with that in C3H mice (Table 3). Arthritis severity in infected B6.C3-Bbaa6 mice was not increased relative to that in wild-type B6 mice, as determined in a separate experiment and summarized in Table 1. Male C3.B6-Bbaa1 ISCL mice exhibited significant suppression of ankle swelling; however, differences in other traits did not reach statistical significance (Table 1). Three additional ISCL, B6.C3-Bbaa1, B6.C3-Bbaa12, and C3.B6-Bbaa12, failed to display a consistent, significant, penetrant phenotype, although in some cases a trend toward altered arthritis traits was noted (Table 1).
TABLE 3.
Ankle swelling and histological assessment of arthritis severity in B. burgdorferi-infected C3.B6-Bbaa6 congenic micea
| Mouse strain (no. of mice) | Change (mm) in joint measurement | Overall lesion score | Score for:
|
Total arthritis score | ||
|---|---|---|---|---|---|---|
| Neutrophil infiltration | Tendon sheath thickness | Reactive-reparative response | ||||
| B6 (20) | 0.30 ± 0.10 | 2.20 ± 0.58 | 1.20 ± 0.58 | 2.00 ± 0.32 | 2.20 ± 0.58 | 7.60 ± 1.91 |
| C3.B6-Bbaa6 (27) | 0.62 ± 0.05 | 2.19 ± 0.21 | 1.73 ± 0.24 | 2.00 ± 0.20 | 1.58 ± 0.23 | 7.35 ± 0.75 |
| C3H/HeN (20) | 1.06 ± 0.14 | 3.20 ± 0.20 | 2.40 ± 0.20 | 3.00 ± 0.0 | 3.60 ± 0.24 | 11.80 ± 0.20 |
Histopathology was assessed as described in Materials and Methods. Values shown are means ± standard errors. Boldface indicates significant difference (P < 0.05) from the value for the C3H/HeN parent strain, as determined by a Mann-Whitney U test.
Gene expression profiling of infected Bbaa2-Bbaa3 and Bbaa4 ISCL mice.
A previous microarray analysis of C3H and B6 mice developing Lyme arthritis identified a unique set of transcripts linked to IFN-mediated responses in joint tissues of C3H mice that were not present in joint tissues of B6 mice 1 week following infection (11). This finding has now been repeated for B. burgdorferi-infected C3H mice by using the more complete Affymetrix GeneChip mouse 2.0 array, and the results of the two independent experiments, that in the previous study and the analysis in the present study, are shown side by side in Table 4. Strong concordance between results for 1-week samples from the two experiments was achieved: 91 genes identified previously (using the GeneChip 2.0A array) as being induced more than twofold compared to those in samples from uninfected mice were also identified in the present study (using the GeneChip 2.0 array) as being upregulated more than twofold. In fact, 14 of the 20 transcripts most highly upregulated in the previous study were ranked in the top 20 in this study, and 3 others were in the top 30. As found previously, transcripts of genes annotated as IFN inducible dominated the profile at 1 week postinfection (Table 4) (11). Thus, robust induction of a set of IFN-inducible genes is a consistent feature of the arthritis-associated C3H response to infection with B. burgdorferi.
TABLE 4.
Comparison of gene induction following B. burgdorferi infection of C3H and ISCL mice
| Probe set identification no. | Gene product description | Gene product designation | Chromosomal location of gene | Rank for gene in C3H mice (by 2.0 chip results)a | Upregulation (n-fold) at 1 wk postinfectionb in:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C3H micec
|
C3.B6 mice (2.0 chip result)
|
B6.C3 mice (2.0 chip result)
|
B6 mice (2.0 chip result) | ||||||||
| 2.0 chip result | 2.0A chip result | CB-Bb2-Bb3 | CB-Bb4 | BC-Bb2-Bb3 | BC-Bb4 | ||||||
| 1424923_at | Serine (or cysteine) peptidase inhibitor, clade A | Serpina3g | Chr 12, E | 1 | 45.92 | 6.64 | 8.24 | 55.17 | NCd | NC | NC |
| 1419043_a_at | IFN-inducible GTPase 1 | Iigp1 | Chr 18, E1 | 2 | 45.53 | 165.31 | 24.03 | 46.97 | NC | 2.15 | NC |
| 1449009_at | T-cell-specific GTPase | Tgtp | Chr 11, B1.2 | 3 | 36.81 | 42.31 | 21.01 | 34.30 | NC | 2.44 | NC |
| 1420549_at | Guanylate nucleotide binding protein 1 | Gbp1 | Chr 3, H1 3, 67.4 cM | 4 | 32.70 | 30.74 | 39.02 | 181.00 | NC | NC | NC |
| 1417141_at | IFN-γ-induced GTPase | Igtp | Chr 11, B1.3 11, 32.0 cM | 5 | 32.64 | 128.48 | 12.52 | 57.84 | NC | NC | NC |
| 1418652_ate | Chemokine (C-X-C motif) ligand 9 | Cxcl9 | Chr 5, E2 5, 53.0 cM | 6 | 31.53 | 17.14 | 33.08 | 67.86 | NC | 2.26 | NC |
| 1450783_at | IFN-induced protein with tetratricopeptide repeats 1 | Ifit1 | Chr 19, C1 | 7 | 28.87 | 8.82 | 10.90 | 85.66 | 3.61 | 2.52 | NC |
| 1418930_at | Chemokine (C-X-C motif) ligand 10 | Cxcl10 | Chr 5, E2 5, 53.0 cM | 8 | 26.31 | 2.58 | 16.96 | 147.58 | NC | 3.73 | NC |
| 1423555_a_at | IFN-induced protein 44 | Ifi44 | Chr 3, H3 | 9 | 22.10 | 5.72 | 7.75 | 55.32 | NC | NC | NC |
| 1438676_at | Macrophage activation protein 2-like protein | Mpa2l | Chr 5, E5 | 10 | 19.36 | 35.91 | 6.63 | 29.38 | NC | NC | NC |
| 1417292_at | IFN-γ-inducible protein 47 | Ifi47 | Chr 11, B1.2 | 11 | 18.37 | 6.55 | 8.31 | 20.72 | NC | NC | NC |
| 1417793_at | IFN-inducible GTPase 2 | Iigp2 | Chr 11, B1.3 | 12 | 16.37 | 122.78 | 15.95 | 29.97 | NC | NC | NC |
| 1418392_a_at | Guanylate nucleotide binding protein 4 | Gbp4 | Chr 3, H1 | 13 | 15.50 | 14.42 | 9.12 | 29.86 | 2.58 | 2.56 | NC |
| 1425225_at | Fc fragment of immunoglobulin G, low affinity IIIa, receptor | Fcgr3a | Chr 1, H3 1, 92.29 cM | 14 | 15.34 | 36.40 | 3.92 | 33.72 | NC | 2.76 | NC |
| 1419714_at | CD274 antigen | Cd274 | Chr 19, C2 | 15 | 14.94 | 32.80 | 7.32 | 56.27 | NC | NC | NC |
| 1428007_at | Keratin-associated protein 13-1 | Krtap13-1 | Chr 16, C3.3 | 16 | 14.89 | NC | NC | NC | NC | NC | NC |
| 1421596_s_at | Histocompatibility protein 28 | H28 | Chr 3, H3 3, 82.5 cM | 17 | 13.81 | 4.80 | 9.41 | 26.86 | NC | NC | NC |
| 1450033_a_at | Signal transducer and activator of transcription 1 | Stat1 | Chr 1, C1.1 1, 25.9 cM | 18 | 12.65 | 22.73 | 12.76 | 31.89 | 3.26 | NC | NC |
| 1418240_at | Guanylate nucleotide binding protein 2 | Gbp2 | Chr 3, H1 3, 67.4 cM | 19 | 11.41 | 12.15 | 6.95 | 28.35 | 2.25 | 2.14 | NC |
| 1418825_at | Immunity-related GTPase family, M | Irgm | Chr 11, B1.2 | 20 | 11.38 | 38.84 | 8.70 | 18.06 | NC | NC | NC |
| 1416016_at | Transporter 1, ATP binding cassette, subfamily B | Tap1 | Chr 17, B1 17, 18.6 cM | 21 | 9.92 | 15.24 | 5.40 | 19.91 | 2.18 | NC | NC |
The 21 transcripts most highly upregulated in C3H mice, according to the results of Affymetrix GeneChip 2.0 analysis, are ranked in order from that with the highest level of induction to that with the lowest. Values for the 20 transcripts also upregulated >2-fold in congenic mice with the C3H background are indicated in italics.
Increase in transcript levels in samples from infected mice at 1 week postinfection compared to those in samples from uninfected mice of each genotype. The gene transcripts most highly induced in the joints of C3H mice following infection with B. burgdorferi were highly upregulated in C3.B6-Bbaa2-Bbaa3 (CB-Bb2-Bb3) and C3.B6-Bbaa4 (CB-Bb4) ISCL but not in B6.C3-Bbaa2-Bbaa3 (BC-Bb2-Bb3) and B6.C3-Bbaa4 (BC-Bb4) ISCL.
The levels of induction (n-fold) of the top 21 transcripts upregulated in the joint tissues of C3H mice following infection with B. burgdorferi, as determined in the present study using Affymetrix GeneChip 2.0, are compared with those determined in the previous study (11) using Affymetrix GeneChip 2.0A.
NC, no change (indicates that transcript level was altered <2-fold compared with that in samples from uninfected mice).
Data in boldface correspond to genes mapping to Bbaa QTL.
To assess the association of IFN-inducible transcripts with arthritis in ISCL and to determine whether altered expression was consistent with the transfer of Bbaa QTL associated with disease, gene expression profiling was performed with tissues from infected ISCL. Joint tissues were collected at 1 week postinfection of ISCL to identify genes induced early in the response and to identify similarities between the ISCL and the parental strains at this critical time point. Our goal was to determine if the IFN response to B. burgdorferi infection observed in C3H mice was regulated by either Bbaa2-Bbaa3 or Bbaa4. RNAs from the joint tissues of at least five infected and five uninfected mice of each genotype were pooled to provide sufficient samples for GeneChip analysis. A cutoff of twofold upregulation was used to identify transcripts that were induced by infection, and commonly induced genes were identified by comparison with findings for C3H and B6 mice. The number of upregulated transcripts in common between each of the ISCL and the two parental strains is shown in Table 5. All four infected Bbaa2-Bbaa3 and Bbaa4 ISCL displayed a gene profile more similar to that of C3H mice than that of B6 mice at 1 week postinfection. However, more infection-induced transcripts were shared between C3H mice and ISCL with the C3H background (C3.B6-Bbaa2-Bbaa3 and C3.B6-Bbaa4) than between C3H mice and ISCL with the B6 background (B6.C3-Bbaa2-Bbaa3 and B6.C3-Bbaa4), somewhat surprising as the degrees of arthritis severity in the four ISCL at 4 weeks of infection were similar and intermediate relative to those in the parental strains. These results suggest that the upregulation of a portion of the transcripts from the C3H IFN profile is associated with arthritis development in ISCL but that the complete profile is not required for arthritis development.
TABLE 5.
Number of transcripts upregulated in joint tissues of ISCL and C3H or B6 parental mice at 1 week following infectiona
| Mouse strain | Arthritis phenotypeb | No. of transcripts upregulated in both ISCL mice and:
|
|
|---|---|---|---|
| C3H mice (characterized by severe arthritis phenotype) | B6 mice (characterized by mild arthritis phenotype) | ||
| C3.B6-Bbaa2-Bbaa3 | Intermediate | 114 | 6 |
| C3.B6-Bbaa4 | Intermediate | 208 | 8 |
| B6.C3-Bbaa2-Bbaa3 | Intermediate | 25 | 5 |
| B6.C3-Bbaa4 | Intermediate | 33 | 6 |
The number of transcripts upregulated >2-fold at 1 week postinfection, relative to transcripts in samples from uninfected controls, was determined for each ISCL and for wild-type C3H and B6 mice. The transcripts upregulated by infection in each ISCL were then compared with those induced in either C3H or B6 mice, and the number upregulated in both groups in each comparison is indicated. Numbers were determined using a single probe set for each gene or expressed sequence tag with NetAfx intersection analysis.
Phenotype relative to those of wild-type C3H and B6 mice.
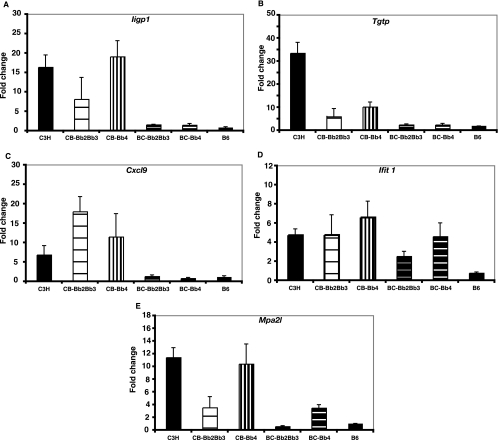
The transcripts most highly induced in C3H mice were also highly upregulated following the infection of C3.B6-Bbaa2-Bbaa3 and C3.B6-Bbaa4 congenic mice (Table 4). In contrast, very few of these transcripts were upregulated following the infection of ISCL mice with the B6 background, the B6.C3-Bbaa2-Bbaa3 and B6.C3-Bbaa4 mice, and none were upregulated in infected B6 mice. This trend in expression for many of the transcripts listed in Table 4 was confirmed by RT-PCR analyses of individual RNA samples used to construct the pools, and results for Iigp, Tgtp, Cxcl9, Ifit1, and Mpa2l are shown in Fig. 3. Thus, neither Bbaa2-Bbaa3 nor Bbaa4 regulates the induction of the robust IFN profile seen in C3H mice at 1 week of infection. Furthermore, the robust induction of the complete profile of IFN-inducible genes is not required for the increased arthritis severity seen in B6.C3-Bbaa2-Bbaa3 and B6.C3-Bbaa4 mice. Only two of the robustly upregulated transcripts identified in Table 4, those for Cxcl9 and Cxcl10, both found within Bbaa2-Bbaa3, were encoded by penetrant QTL. The robust induction of both genes was dependent on the C3H background and was not determined by the C3H Bbaa2-Bbaa3 allele.
FIG. 3.
Quantification of selected gene transcripts in the joints of individual mice. Expression levels of the robustly induced IFN-related transcripts listed in Table 4 were determined using quantitative RT-PCR. The amount of change (n-fold) was determined based on the difference between the value for a pooled sample from five individual mice and the average for the uninfected controls and is expressed as the mean ± the standard error. ISCL designations are abbreviated as follows: CB-Bb2Bb3, C3.B6-Bbaa2-Bbaa3; BC-Bb2Bb3, B6.C3-Bbaa2-Bbaa3; CB-Bb4, C3.B6-Bbaa4; and BC-Bb4, B6.C3-Bbaa4.
The microarray intersection analysis summarized in Table 5 suggested that there might be a core group of transcripts identified in C3H joints that were modestly upregulated in all ISCL with intermediate arthritis. Indeed, transcripts for guanylate nucleotide binding proteins 2 and 4 (encoded by Gbp2 and Gbp4), members of the p65 family of IFN-inducible GTPases, and the IFN-induced protein with tetratricopeptide repeats 1 (encoded by Ifit1) were robustly upregulated in C3H mice and in the C3H background ISCL and were also modestly increased (>2-fold) in B6.C3-Bbaa2-Bbaa3 and B6.C3-Bbaa4 mice following infection, but not in B6 mice. The results of PCR confirmation of Ifit1 expression in joint tissues from individual mice are shown in Fig. 3. The induction of Ifit1 transcripts in all four Bbaa2-Bbaa3 and Bbaa4 ISCL developing Lyme arthritis is interesting, as it has recently been associated with the earliest macrophage response to Listeria infection, occurring simultaneously with type I IFN expression, and therefore may be a marker of the early inflammatory response in the joint tissue (18). Thus, microarray analysis findings suggest that the modest expression of a small number of genes is a consistent feature preceding the development of Lyme arthritis that is absent in B6 mice developing milder disease.
Genetic regulation of IFN profile transcripts in BMMs.
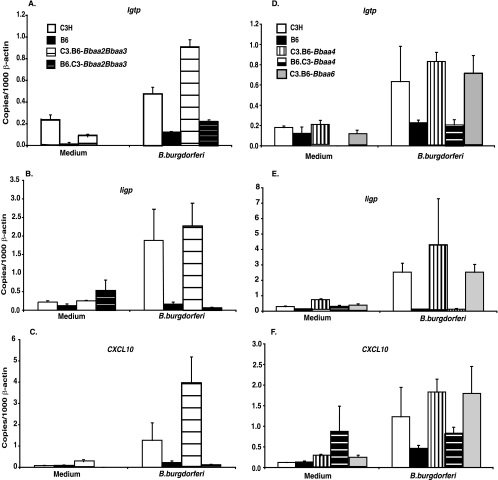
B. burgdorferi is a strong stimulant of chemokines and other inflammatory mediators, including transcripts for IFN-inducible genes, in both human and murine macrophages (13, 15, 17, 19, 23, 28, 31, 32). Additionally, the induction of the IFN profile transcripts by the treatment of BMMs with B. burgdorferi was found to be dependent on the type I IFN receptor, IFNAR1, thereby implicating type I IFN feedback in this response (22). Because the complexity of ankle tissue may mask the contribution of a subpopulation of cells such as macrophages to the inflammatory response, we assessed the responses of pure cultures of BMMs generated from the five ISCL with penetrant phenotypes and from the parental strains. BMMs were incubated with B. burgdorferi for 24 h, and the levels of three of the robustly induced IFN profile transcripts, those for Igtp, Iigp, and Cxcl10, were assessed by RT-PCR. The level of transcript production was strongly influenced by the background of the ISCL, with the transcript levels found in macrophages from C3.B6-Bbaa2-Bbaa3, C3.B6-Bbaa4, and C3.B6-Bbaa6 mice resembling the high levels produced by macrophages from C3H mice while the levels found in B6.C3-Bbaa2-Bbaa3 and B5.C3-Bbaa4 macrophages were similar to the low levels produced by B6 macrophages (Fig. 4). This result further confirms the appropriateness of the macrophage for analysis of the regulation of the B. burgdorferi-induced IFN profile and indicates that selective induction of IFN-inducible transcripts was not missed in the analysis of whole joint tissue. These findings provide further support for the conclusion that differential regulation of the IFN profile observed in joint tissues of infected C3H and B6 mice is not due to alleles of genes residing within the penetrant QTL Bbaa2-Bbaa3, Bbaa4, and Bbaa6.
FIG. 4.
Quantification of IFN profile transcripts of BMMs from ISCL and parental mice treated with B. burgdorferi. The expression of transcripts was evaluated by RT-PCR 24 h following the addition of B. burgdorferi to confluent cultures of BMMs. Transcript levels in duplicate wells treated with medium or B. burgdorferi were normalized to β-actin transcript levels. Values shown in this figure are means ± standard deviations and are representative of results from three experiments. One-way analysis of variance revealed that values for macrophages from congenic mice were not different from values for macrophages from the predominant wild-type parent, as confirmed with the Bonferroni multiple-comparisons test.
DISCUSSION
An enigmatic feature of infection with B. burgdorferi in humans is the spectra of symptoms and disease severities observed. The assessment of arthritis in infected patients provides a clear example of this range of disease severity, as some patients develop arthritis with profound swelling in joint tissues while other individuals with demonstrable serologic evidence of infection do not display arthritic symptoms (30). Seminal studies by Barthold and colleagues using inbred strains of mice demonstrated that the genetic background of the murine host dramatically influences the severities of infection-induced arthritis and carditis (2). Our studies have capitalized on these observations by applying forward genetics with the goal of characterizing the genetic regulation of Lyme arthritis severity. These studies would be difficult to perform with humans due to the small number of patients who fail to receive early treatment and in whom, therefore, infection is allowed to progress to the late-stage symptom of arthritis. Additionally, it has proven very difficult to ascribe common variants of genes to complex traits such as arthritis development in humans (14).
This paper reports a significant step toward the goal of identifying candidate genes that regulate the severity of Lyme arthritis. The development of 10 ISCL, involving the reciprocal transfer of five Bbaa QTL into C3H and B6 parent strains, has allowed confirmation of regulatory elements for arthritis severity. The transfer of arthritis severity phenotypes in four of the ISCL, those with Bbaa2-Bbaa3 on Chr 5 and Bbaa4 on Chr 11, was bidirectional and resulted in arthritis of intermediate severity (Fig. 1 and 2; Tables 1 and 2). A fifth ISCL, C3.B6-Bbaa6, also displayed intermediate arthritis severity, but in this case the transfer of the phenotype was unidirectional (Table 3). The infection of these five ISCL with penetrant phenotypes revealed the alteration of multiple arthritis traits, including ankle swelling, PMN infiltration, tendon sheath thickness, reactive-reparative events, and the overall lesion score, relative to those in the predominant parent. Interestingly, earlier QTL analyses of intercross populations identified significant linkages of rear ankle swelling and histopathological scoring to distinct QTL on different Chr (34). This difference may reflect increased statistical significance for arthritis traits in the ISCL populations, in which individual mice are identical in both the parental background and the introgressed Bbaa regions, compared to that for arthritis traits in the intercross populations in which genomic regions outside the identified QTL are randomly assorted. An important interpretation based on this finding is that all measurable traits associated with B. burgdorferi-induced arthritis result from a common mechanistic response to infection.
ISCL with introgressed Bbaa1 or Bbaa12 failed to display altered Lyme arthritis severity compared with that in the background parent strain. This result was surprising, as the strength of the initial linkage of these QTL was as great as that found for Bbaa4, which did transfer the arthritis severity phenotype (26, 34). One explanation for the lack of an altered phenotype is that other genomic regions in C3H or B6 mice were required for the full effects of Bbaa1 and Bbaa12 and that these regions were lost in the development of ISCL. Our results clearly illustrate the importance of ISCL development for confirmation that isolated alleles of genes can regulate arthritis severity. The transfer of phenotypes in five ISCL provides critical documentation of the utility of ISCL in the ultimate goal of identifying the responsible genes.
The generation of congenic mouse lines should allow genetic linkage of physiological responses associated with the development of Lyme arthritis to particular QTL. We had previously observed robust induction of a group of IFN-inducible signature transcripts in the joint tissues of C3H mice which were absent from the joint tissues of B6 mice at 1 week of infection (11). B6 mice lacking either interleukin-10 or Toll-like receptor 2 develop more severe Lyme arthritis than wild-type mice and display exaggerated expression of IFN-induced transcripts in joint tissues, further implicating IFN in the arthritic response to infection (11, 33). The findings of recent studies with blocking antibodies indicated that both IFN-γ and type I IFN contribute to the induction and magnitude of the IFN profile but that type I IFN is uniquely required for the development of arthritis in infected C3H mice (22). The ISCL described in this study allowed us to assess whether Bbaa2-Bbaa3 or Bbaa4 regulated the expression of the IFN profile during mouse infection. Transcripts related to the IFN profile were robustly upregulated following the infection of C3.B6-Bbaa2-Bbaa3 and C3.B6-Bbaa4 mice; however, most of these transcripts were not upregulated in the B6 background ISCL, B6.C3-Bbaa2-Bbaa3 and B6.C3-Bbaa4 (Tables 4 and 5). Taken in the context of the similar arthritis severities observed in these four ISCL, these findings indicate that the induction of the complete IFN profile is not required for the development of Lyme arthritis in mice. Additionally, the genes regulating the robust IFN-inducible response to B. burgdorferi do not reside within Bbaa2-Bbaa3 or Bbaa4.
It was possible that the production of IFN by a small number of cells in the joint was masked by the assessment of transcripts in the whole joint tissue. To circumvent the possible dilution effect of other cells in the joint, we assessed IFN-inducible transcripts in macrophages derived from ISCL and wild-type parental strains, as this cell type responds intensely to B. burgdorferi (13, 15, 17, 19, 23, 28, 31, 32). The induction of the IFN profile in BMMs treated with B. burgdoferi is independent of Toll-like receptor signaling but dependent on IFNAR1 (22). There was a demonstrably greater magnitude of response in BMMs derived from C3H background ISCL than in BMMs derived from B6 background ISCL, with C3H background ISCL resembling wild-type C3H mice and B6 background ISCL resembling wild-type B6 mice (Fig. 4). These findings were in concordance with results from assessments of whole joint tissue samples (Table 4; Fig. 3) and indicate that the reduced transcript induction in B6 background ISCL is not due to masking by irrelevant cells in the joint tissues but rather points to genetic regulation of the IFN profile by genes residing outside Bbaa2-Bbaa3, Bbaa4, and Bbaa6. This finding is extremely important, as it provides further justification for the forward genetics approach and predicts that additional, novel pathways that regulate the severity of Lyme arthritis will be identified.
The identification of three transcripts that were modestly induced in the joint tissues of all mouse strains destined to develop arthritis but not in infected B6 mice may be relevant to the characterization of other pathways contributing to arthritis development. However, further studies will be required to distinguish direct involvement in arthritis development from bystander activation of inflammatory responses in the joint tissue. The association of these transcripts, those for Gbp2, Gbp4, and Ifit1, with the early response of macrophages to the intracellular pathogen Listeria monocytogenes indicates the need for further investigation of their site of production and mechanism of activation (18).
The findings in this report demonstrate that a variety of approaches will be useful in the identification of candidate genes associated with Lyme arthritis. The development of ISCL has confirmed the transfer of the arthritis phenotype for three of five initially identified QTL (Bbaa4, Bbaa2-Bbaa3, and Bbaa6) and indicated that all arthritis-associated traits are coregulated, regardless of the genetic pathway implicated in localized regulation. Gene expression profiling of joint tissues from infected ISCL mice or their BMMs indicates that a previously identified IFN-induced profile is not regulated by Bbaa2-Bbaa3 or Baab4 (or Bbaa6) in joints or macrophages. This approach would be virtually impossible with human patients, given the rapid treatment that individuals infected with B. burgdorferi currently experience. Our findings strongly suggest that the forward genetics approach is likely to identify additional, IFN-independent pathways involved in the development of Lyme arthritis. Furthermore, this linkage is likely to provide insight into other inflammatory pathologies.
Acknowledgments
This work was supported by Public Health Services grants AR-43521 to J.J.W. and C.T. and AI-24158 to J.H.W., the Training Program in Microbial Pathogenesis grant 5T32-AI055434, and an Arthritis Foundation award to J.C.M.
We thank Jiantao Bian for technical assistance.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 1 June 2009.
REFERENCES
- 1.Barthold, S. W., D. S. Beck, G. M. Hansen, G. A. Terwilliger, and K. D. Moody. 1990. Lyme borreliosis in selected strains and ages of laboratory mice. J. Infect. Dis. 162133-138. [DOI] [PubMed] [Google Scholar]
- 2.Barthold, S. W., C. L. Sidman, and A. L. Smith. 1992. Lyme borreliosis in genetically resistant and susceptible mice with severe combined immunodeficiency. Am. J. Trop. Med. Hyg. 47605-613. [DOI] [PubMed] [Google Scholar]
- 3.Brown, C. R., and S. L. Reiner. 1998. Activation of natural killer cells in arthritis-susceptible but not arthritis-resistant mouse strains following Borrelia burgdorferi infection. Infect. Immun. 665208-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, C. R., and S. L. Reiner. 1998. Clearance of Borrelia burgdorferi may not be required for resistance to experimental Lyme arthritis. Infect. Immun. 662065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, J. P., J. F. Zachary, C. Teuscher, J. J. Weis, and R. M. Wooten. 1999. Dual role of interleukin-10 in murine Lyme disease: regulation of arthritis severity and host defense. Infect. Immun. 675142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bult, C. J., J. T. Eppig, J. A. Kadin, J. E. Richardson, J. A. Blake, and the Mouse Genome Database Group. 2008. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 36D724-D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science (New York, NY) 2161317-1319. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, D., J. Hernandez, B. J. Bloom, J. Coburn, J. M. Aversa, and A. C. Steere. 1999. Lack of Borrelia burgdorferi DNA in synovial samples from patients with antibiotic treatment-resistant Lyme arthritis. Arthritis Rheum. 422705-2709. [DOI] [PubMed] [Google Scholar]
- 9.Chirgwin, J. M., A. E. Przybyla, R. J. MacDonald, and W. J. Rutter. 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 185294-5299. [DOI] [PubMed] [Google Scholar]
- 10.Cope, L. M., R. A. Irizarry, H. A. Jaffee, Z. Wu, and T. P. Speed. 2004. A benchmark for Affymetrix GeneChip expression measures. Bioinformatics 20323-331. [DOI] [PubMed] [Google Scholar]
- 11.Crandall, H., D. M. Dunn, Y. Ma, R. M. Wooten, J. F. Zachary, J. H. Weis, R. B. Weiss, and J. J. Weis. 2006. Gene expression profiling reveals unique pathways associated with differential severity of Lyme arthritis. J. Immunol. 1777930-7942. [DOI] [PubMed] [Google Scholar]
- 12.Crandall, H., Y. Ma, D. M. Dunn, R. S. Sundsbak, J. F. Zachary, P. Olofsson, R. Holmdahl, J. H. Weis, R. B. Weiss, C. Teuscher, and J. J. Weis. 2005. Bb2Bb3 regulation of murine Lyme arthritis is distinct from Ncf1 and independent of the phagocyte nicotinamide adenine dinucleotide phosphate oxidase. Am. J. Pathol. 167775-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis, V. A., S. Dixit, S. M. O'Brien, X. Alvarez, B. Pahar, and M. T. Philipp. 2009. Live Borrelia burgdorferi spirochetes elicit inflammatory mediators from human monocytes via the Toll-like receptor signaling pathway. Infect. Immun. 771238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frazer, K. A., S. S. Murray, N. J. Schork, and E. J. Topol. 2009. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10241-251. [DOI] [PubMed] [Google Scholar]
- 15.Giambartolomei, G. H., V. A. Dennis, B. L. Lasater, and M. T. Philipp. 1999. Induction of pro- and anti-inflammatory cytokines by Borrelia burgdorferi lipoproteins in monocytes is mediated by CD14. Infect. Immun. 67140-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalish, R. A., J. M. Leong, and A. C. Steere. 1993. Association of treatment-resistant chronic Lyme arthritis with HLA-DR4 and antibody reactivity to OspA and OspB of Borrelia burgdorferi. Infect. Immun. 612774-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarus, J. J., M. A. Kay, A. L. McCarter, and R. M. Wooten. 2008. Viable Borrelia burgdorferi enhances interleukin-10 production and suppresses activation of murine macrophages. Infect. Immun. 761153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leber, J. H., G. T. Crimmins, S. Raghavan, N. P. Meyer-Morse, J. S. Cox, and D. A. Portnoy. 2008. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 4e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, N., R. R. Montgomery, S. W. Barthold, and L. K. Bockenstedt. 2004. Myeloid differentiation antigen 88 deficiency impairs pathogen clearance but does not alter inflammation in Borrelia burgdorferi-infected mice. Infect. Immun. 723195-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, Y., K. P. Seiler, E. J. Eichwald, J. H. Weis, C. Teuscher, and J. J. Weis. 1998. Distinct characteristics of resistance to Borrelia burgdorferi-induced arthritis in C57BL/6N mice. Infect. Immun. 66161-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meerpohl, H. G., M. L. Lohmann-Matthes, and H. Fischer. 1976. Studies on the activation of mouse bone marrow-derived macrophages by the macrophage cytotoxicity factor (MCF). Eur. J. Immunol. 6213-217. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. C., Y. Ma, J. Bian, K. C. Sheehan, J. F. Zachary, J. H. Weis, R. D. Schreiber, and J. J. Weis. 2008. A critical role for type I IFN in arthritis development following Borrelia burgdorferi infection of mice. J. Immunol. 1818492-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore, M. W., A. R. Cruz, C. J. LaVake, A. L. Marzo, C. H. Eggers, J. C. Salazar, and J. D. Radolf. 2007. Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect. Immun. 752046-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nocton, J. J., and A. C. Steere. 1995. Lyme disease. Adv. Intern. Med. 4069-117. [PubMed] [Google Scholar]
- 25.Potter, M. R., S. R. Rittling, D. T. Denhardt, R. J. Roper, J. H. Weis, C. Teuscher, and J. J. Weis. 2002. Role of osteopontin in murine Lyme arthritis and host defense against Borrelia burgdorferi. Infect. Immun. 701372-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roper, R. J., J. J. Weis, B. A. McCracken, C. B. Green, Y. Ma, K. S. Weber, D. Fairbairn, R. J. Butterfield, M. R. Potter, J. F. Zachary, R. W. Doerge, and C. Teuscher. 2001. Genetic control of susceptibility to experimental Lyme arthritis is polygenic and exhibits consistent linkage to multiple loci on chromosome 5 in four independent mouse crosses. Genes Immun. 2388-397. [DOI] [PubMed] [Google Scholar]
- 27.Shin, J. J., L. J. Glickstein, and A. C. Steere. 2007. High levels of inflammatory chemokines and cytokines in joint fluid and synovial tissue throughout the course of antibiotic-refractory Lyme arthritis. Arthritis Rheum. 561325-1335. [DOI] [PubMed] [Google Scholar]
- 28.Shin, O. S., R. R. Isberg, S. Akira, S. Uematsu, A. K. Behera, and L. T. Hu. 2008. Distinct roles for MyD88 and Toll-like receptors 2, 5, and 9 in phagocytosis of Borrelia burgdorferi and cytokine induction. Infect. Immun. 762341-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steere, A. C., E. Dwyer, and R. Winchester. 1990. Association of chronic Lyme arthritis with HLA-DR4 and HLA-DR2 alleles. N. Engl. J. Med. 323219-223. [DOI] [PubMed] [Google Scholar]
- 30.Steere, A. C., R. T. Schoen, and E. Taylor. 1987. The clinical evolution of Lyme arthritis. Ann. Intern. Med. 107725-731. [DOI] [PubMed] [Google Scholar]
- 31.Wang, G., Y. Ma, A. Buyuk, S. McClain, J. J. Weis, and I. Schwartz. 2004. Impaired host defense to infection and Toll-like receptor 2-independent killing of Borrelia burgdorferi clinical isolates in TLR2-deficient C3H/HeJ mice. FEMS Microbiol. Lett. 231219-225. [DOI] [PubMed] [Google Scholar]
- 32.Wang, G., M. M. Petzke, R. Iyer, H. Wu, and I. Schwartz. 2008. Pattern of proinflammatory cytokine induction in RAW264.7 mouse macrophages is identical for virulent and attenuated Borrelia burgdorferi. J. Immunol. 1808306-8315.18523297 [Google Scholar]
- 33.Wang, X., Y. Ma, A. Yoder, H. Crandall, J. F. Zachary, R. S. Fujinami, J. H. Weis, and J. J. Weis. 2008. T cell infiltration is associated with increased Lyme arthritis in TLR2−/− mice. FEMS Immunol. Med. Microbiol. 52124-133. [DOI] [PubMed] [Google Scholar]
- 34.Weis, J. J., B. A. McCracken, Y. Ma, D. Fairbairn, R. J. Roper, T. B. Morrison, J. H. Weis, J. F. Zachary, R. W. Doerge, and C. Teuscher. 1999. Identification of quantitative trait loci governing arthritis severity and humoral responses in the murine model of Lyme disease. J. Immunol. 162948-956. [PubMed] [Google Scholar]