Abstract
Shiga-toxigenic Escherichia coli (STEC) strains cause serious gastrointestinal disease, which can lead to potentially life-threatening systemic complications such as hemolytic-uremic syndrome. Although the production of Shiga toxin has been considered to be the main virulence trait of STEC for many years, the capacity to colonize the host intestinal epithelium is a crucial step in pathogenesis. In this study, we have characterized a novel megaplasmid-encoded outer membrane protein in locus of enterocyte effacement (LEE)-negative O113:H21 STEC strain 98NK2, termed Sab (for STEC autotransporter [AT] contributing to biofilm formation). The 4,296-bp sab gene encodes a 1,431-amino-acid protein with the features of members of the AT protein family. When expressed in E. coli JM109, Sab contributed to the diffuse adherence to human epithelial (HEp-2) cells and promoted biofilm formation on polystyrene surfaces. A 98NK2 sab deletion mutant was also defective in biofilm formation relative to its otherwise isogenic wild-type parent, and this was complemented by transformation with a sab-carrying plasmid. Interestingly, an unrelated O113:H21 STEC isolate that had a naturally occurring deletion in sab was similarly defective in biofilm formation. PCR analysis indicated that sab is present in LEE-negative STEC strains belonging to serotypes/groups O113:H21, O23, and O82:H8. These findings raise the possibility that Sab may contribute to colonization in a subset of LEE-negative STEC strains.
Shiga-toxigenic Escherichia coli (STEC) strains are prominent food-borne pathogens that cause watery or bloody diarrhea and hemorrhagic colitis, which can progress to the life-threatening hemolytic-uremic syndrome (HUS) (15, 21, 29). In order to establish and maintain an infection, STEC strains are equipped with a diverse array of virulence factors. Among these factors, Shiga toxin has been considered to be a sine qua non of virulence, as reviewed previously (21, 29). However, attachment of STEC to the human intestinal mucosa is a critical first step in pathogenesis. Many STEC strains, including those of the highly prevalent O157:H7 serotype, carry the locus of enterocyte effacement (LEE) pathogenicity island, which encodes the capacity to produce attaching and effacing (A/E) lesions on the intestinal epithelium, similarly to those produced by enteropathogenic E. coli strains (11, 35). These STEC strains are often referred to as enterohemorrhagic E. coli (EHEC), although this classification is ill defined. A/E lesions are characterized by ultrastructural changes including the remodeling of the host cell cytoskeleton and intimate attachment of the bacteria to the cell surface (11, 35). The process of the generation of A/E lesions involves the expression of the eae gene, which encodes intimin, an outer membrane surface adhesin, and the delivery of the intimin receptor Tir and several other effector proteins into host cells via the LEE-encoded type III secretion apparatus (reviewed in references 5 and 11).
However, many STEC isolates from cases of severe disease, including HUS, lack the LEE locus yet are clearly capable of efficient colonization of the human gut (28, 29). Several candidate adhesins have been identified in these strains, including the megaplasmid-encoded autoagglutinating adhesin Saa (26), the long polar fimbriae encoded by the lpf operon (10) (two distinct homologues of which are also present in STEC O157:H7 strains [39, 40]), and the immunoglobulin-binding protein EibG, which contributes to a chain-like adherence phenotype on HEp-2 cells (18). Tarr et al. (38) also previously identified Iha, a homologue of Vibrio cholerae IrgA, which promotes the adherence of STEC O157:H7 to HeLa cells and is widely distributed in LEE-positive and LEE-negative strains. STEC O157:H7 strains also produce a type IV pilus, HCP (47), and an E. coli common pilus, ECP (30), both of which contribute to in vitro adherence to intestinal epithelial cells. Additional putative adhesins from LEE-positive STEC strains include Efa1, which mediates the attachment of O111:NM STEC strains to Chinese hamster ovary cells (23). In addition, Torres et al. (41) previously identified a calcium-binding and heat-extractable AT protein of EHEC, termed Cah, which mediates aggregation and participates in biofilm formation. Recently, Wells et al. (45) also characterized the EHEC-encoded AT protein EhaA, which contributes to adherence to primary bovine epithelial cells (but not HeLa cells) and promotes biofilm formation as well.
The AT proteins referred to above belong to a rapidly growing family of gram-negative surface proteins that are exported across the periplasmic space and either attached to the external face of the outer membrane or released by proteolysis into the environment (13). These large proteins share a characteristic structure comprising three distinct domains, namely, an N-terminal signal peptide, a divergent functional passenger domain (α-domain), and a conserved C-terminal domain which forms a β-barrel pore in the outer membrane (13, 46). This unique protein structure provides all the information required for transport to the cell surface, with the N-terminal sequence directing the protein to the periplasm via the sec pathway and the C-terminal domain mediating the translocation of the passenger domain to the external surface (14). The various α-domains confer a broad range of functions and/or phenotypes including aggregation, biofilm formation, adherence, invasion, serum resistance, and protease or esterase activity (7, 12).
Research in our laboratory has focused on the identification of novel virulence factors of LEE-negative STEC strains associated with human disease. In this study, we describe the identification and characterization of a member of the AT family produced by hypervirulent LEE-negative O113:H21 STEC strain 98NK2, which confers adherence to human epithelial cells and mediates biofilm formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and routine molecular biological techniques.
The E. coli strains and plasmids used in this study are listed in Table 1. Cells were routinely grown in Luria-Bertani (LB) medium with or without 1.5% Bacto agar. All bacteria were grown at 37°C except the temperature-sensitive strain 98NK2(pkD46). When required, antibiotics were used at 50 μg/ml for kanamycin and 100 μg/ml for ampicillin. SOC medium (20 g/liter l-tryptone, 5 g/liter yeast extract, 20 mM glucose, 8.6 mM NaCl, 2.5 mM KCl, 20 mM MgSO4) was used as the recovery medium for transformants.
TABLE 1.
Bacteria and plasmids used in this study
| E. coli strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| 98NK2 | STEC O113:H21 (HUS isolate) | 28 |
| MW10 | STEC O113:H21 (food isolate) | 27 |
| JM109 | E. coli K-12 derivate | 48 |
| JM109(pB) | JM109 carrying pB; Apr | This study |
| JM109(pBsab) | JM109 carrying pBsab; Apr | This study |
| 98NK2sab::kan | 98NK2 sab deletion mutant; Knr | This study |
| 98NK2sab::kan(pB) | 98NK2sab::kan carrying pB; Knr Apr | This study |
| 98NK2sab::kan(pBsab) | 98NK2sab::kan with pBsab; Knr Apr | This study |
| M15(pREP4) | E. coli K-12 derivate | Qiagen |
| M15(pREP4)(pQESab) | M15(pREP4) carrying pQESab; Apr | This study |
| Plasmids | ||
| pB | pBluescript II SK(−) | Fermentas |
| pBsab | sab cloned into pBluescript II SK(−) | This study |
| pQE30 | His6 expression vector | Qiagen |
| pQESab | pQE30 with nucleotides 94 to 2676 of sab | This study |
| pkD4 | kan template for mutagenesis | 9 |
| pkD46 | Red recombinase expression vector | 9 |
Knr, kanamycin resistance; Apr, ampicillin resistance.
Routine DNA methods such as restriction, ligation, and transformation of DNA were carried out as described previously by Maniatis et al. (19). Nucleotide sequences were derived from E. coli K-12 or E. coli O113:H21 strain 98NK2 and strain E41 sequences deposited in the National Center for Biotechnology Information (NCBI) database. Purification of PCR products and isolation of plasmid DNA were carried out using a MinElute PCR purification kit (Qiagen) and a Qiaprep Spin minikit (Qiagen), respectively. All oligonucleotide primers were obtained from Sigma Genosys. Restriction enzymes (New England Biolabs) were used as recommended by the manufacturer.
Mutagenesis of 98NK2 and plasmid constructs.
The deletion of the LH0147 (sab) gene and insertion of a kanamycin resistance cassette were performed using the Lambda Red recombinase system as described previously (9). In order to do so, a kanamycin resistance gene was amplified from plasmid pKD4 using the Expand Long Template PCR system (Roche) and primer pairs d-LH0147-F (5′-ATAGCTCCGGTTGCTATGACCTTTGCAGAAAAGAATACGTCTCG TGTAGGCTGGAGCTGCTTC-3′) and d-LH0147-R (5′-GGGTGTTCTGACCTGCATTCAGTTTCATTATCACCTTACCGTTCATATGAATATCCTCCT TAG-3′). The resultant purified PCR product was electroporated into E. coli strain 98NK2(pkD46). Kanamycin-resistant, ampicillin-sensitive recombinants were checked by PCR using the respective flanking region primers and kanamycin-specific primers K1, K2, and Kt, which were described previously (9). Successful mutagenesis was confirmed by PCR and sequence analysis. The absence or reduced size of PCR products obtained using primer pairs LH0147-f (5′-GGTGGATACAGCAGGTAATG-3′) and LH0147-r (5′-TATCTCACCACCTGCTATCG-3′), or LH0147-fla-f (5′-CCTTTGTGTTTCTATTGCGC-3′) and LH0147-fla-r (5′-CACCGACGGAGAAATTACCC-3′), respectively, confirmed that all megaplasmid copies lacked the entire sab gene. Confirmed mutants were designated 98NK2sab::kan.
The complete sab gene was also amplified from 98NK2 genomic DNA using primer pair LH0147-BamHI (5′-CCCGGATCCGGAAACTCCAAGAGTATTGC-3′) and LH0147-EcoRI (5′-CCCGAATTCCCTTGCTTTTCCCTGTTACC-3′). The resultant PCR product was purified, digested with BamHI and EcoRI, and ligated into BamHI and EcoRI restriction sites of pBluescript SK(−) to generate pBsab. The plasmid was then transformed into E. coli JM109 and confirmed by sequence analysis. For complementation of the 98NK2sab::kan mutant, plasmid pBsab was introduced by transformation to generate 98NK2sab::kan(pBsab).
Purification of Sab and preparation of anti-Sab serum.
Purification of Sab was carried out using a Qiaexpressionist kit (Qiagen). In order to construct an N-terminal His6-Sab fusion protein lacking the signal sequence and the translocator domain, the region from nucleotides 94 to 2676 was amplified using the Expand Long Template PCR system (Roche) and primer pair R1-LH0147-F (5′-GGGGGATCCGAGGTTGATGAAGCTCCGGTG-3′) and R1-LH0147-R (5′-GGGGCATGCCAGGTCACCGATATTTGCCGC-3′) with restriction sites BamHI and SphI, respectively. The purified PCR product was digested and ligated into pQE30 (Qiagen). Correct in-frame insertion was confirmed by restriction and sequence analysis. The recombinant plasmid (pQESab) was transformed into the expression host E. coli M15(pREP4), and large-scale purification of Sab was performed as follows. A log-phase LB broth culture of E. coli M15(pREP4)(pQESab) containing 100 μg/ml of ampicillin and 25 μg/ml kanamycin was induced by the addition of 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Inalco Pharmaceutical, CA) and incubated for 3 h with shaking at 37°C. The cells were harvested by centrifugation (6,000 × g for 15 min at 4°C) and resuspended in 10 ml sonication buffer (50 mM Na2HPO4, 1 M NaCl, 40 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml pepstatin, 10 μg/ml leupeptin). Cells were lysed using an SLM Aminco French pressure cell at 12,000 lb/in2. The cell lysate was then centrifuged at 100,000 × g for 1 h at 4°C, and the supernatant was loaded onto a Ni-nitrilotriacetic acid (HisLink; Promega) column preequilibrated with 10 ml sonication buffer at a rate of 15 ml/h. After washing the column with 15 ml of wash buffer (50 mM Na2HPO4, 300 mM NaCl, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, 1 μg/ml leupeptin), the protein was eluted using 30 ml of a 0 to 500 mM imidazole gradient. Ten 3-ml fractions were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Peak fractions were pooled, and purified protein was stored in phosphate-buffered saline (PBS) containing 50% glycerol at −20°C.
To generate antiserum, five male BALB/c mice were immunized by intraperitoneal injection of three 10-μg doses of purified Sab in alum adjuvant at 1-week intervals, as described previously (26). Mice were exsanguinated by cardiac puncture 1 week after the third immunization, and serum was stored at 4°C.
E. coli membrane isolation.
E. coli strains were grown in LB broth, inner and outer membranes were isolated, and proteins in the latter were fractionated on the basis of solubility in 1% Zwittergen 3-14 (Calbiochem), as described previously by Veiga et al. (43).
Western blot analysis.
Bacterial lysates and protein fractions were separated by SDS-PAGE using a 4 to 20% Bis-Tris gradient gel (Invitrogen) according to the manufacturer's instructions. Loadings were standardized on the basis of initial CFU for whole bacterial lysates (108 CFU per lane) or total protein. Immunodetection was performed following transfer onto polyvinylidene difluoride membranes (Hybond; Amersham). Membranes were blocked for 30 min with 1× Tris-buffered saline-0.05% Tween 20 containing 5% skim milk and probed with mouse anti-Sab serum, followed by incubation with goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Bio-Rad). Bands were detected using a chromogenic nitroblue tetrazolium-5-bromo-4-chloro-3-indolyl phosphate substrate.
HEp-2 and Henle 407 cell adherence assay.
HEp-2 or Henle 407 cells in Dulbecco's modified Eagle's medium (DMEM; Gibco) (with 10% fetal calf serum [FCS]) were seeded onto 24-well tissue culture plates and used when approximately 90% confluent. E. coli cells were grown overnight in LB medium at 37°C and diluted to a density of approximately 104 CFU/ml in DMEM (without FCS or antibiotics). Washed monolayers were infected with 1 ml bacterial suspension and incubated for 3 h at 37°C (in 5% CO2). Following this step, culture medium was removed, and monolayers were washed four times with PBS to remove nonadherent bacteria. To detach and lyse cells, monolayers were treated with 100 μl of 0.025% trypsin-0.02% EDTA and 400 μl of 0.025% Triton X-100. Aliquots and serial dilutions thereof were plated onto LB agar to determine the total number of adherent bacteria (CFU/well). In parallel, strains were incubated likewise but without epithelial cells, and growth of bacteria was monitored by measurements of CFU/well. Results were expressed as a percentage of adherent bacteria relative to total bacteria for each strain. Assays were carried out in quadruplicate on two different days.
For Giemsa staining, 50% confluent HEp-2 cells grown on coverslips in 24-well tissue trays were washed and infected with approximately 106 CFU of the appropriate strain suspended in 1 ml DMEM. After incubation at 37°C for 3 h, monolayers were washed four times with PBS, fixed with 70% methanol for 10 min, and stained with Giemsa stain. Finally, washed coverslips were mounted onto glass slides and examined by light microscopy.
ELISA.
Surface expressions of Sab in 98NK2 and various recombinant strains were compared using a whole unfixed bacterial cell enzyme-linked immunosorbent assay (ELISA), as described elsewhere previously (1), with the following modifications. Strains were grown overnight in LB medium with the appropriate antibiotics at 37°C and subcultured in the same medium for 2 h. The equivalent of 1 × 108 bacteria was then pelleted by centrifugation (4,000 × g for 10 min at 4°C), washed once with PBS, and blocked in 200 μl PBS-10% FCS for 30 min. After removing the blocking reagent, bacteria were incubated with mouse anti-Sab diluted 1:200 in 10% FCS in PBS for 1 h at room temperature, washed three times with PBS, and resuspended in 200 μl 10% FCS in PBS containing goat anti-mouse immunoglobulin G-alkaline phosphatase conjugate (1:5,000, enzyme immunoassay grade; Bio-Rad). After incubation at 37°C for 1 h, bacteria were washed three times with PBS, resuspended in 300 μl substrate buffer (1 M diethanolamine, 3 mM NaN3, and 1 mM MgCl2 containing p-nitrophenyl phosphate) (one tablet dissolved in 5 ml substrate buffer; Sigma), and transferred onto a 96-well plate (Sarstedt). Following 1 h of incubation at 37°C, the A405 was measured. Each strain was analyzed in triplicate.
Protein binding assay.
Purified Sab was concentrated using a Nanosep instrument (Pall Corporation) as recommended by the manufacturer and labeled with Oregon Green 488 using a FluoReporterOregon Green 488 protein labeling kit (Molecular Probes) according to the manufacturer's instructions. For protein-binding assays, HEp-2 cells were grown on coverslips in 24-well plates until 50% confluent. Medium was then removed, HEp-2 cells were washed twice with PBS, and fresh DMEM was added. Cells were exposed to 0 or 20 μg/ml purified labeled Sab for 2.5 h at 37°C. Unbound protein was removed by washing with PBS three times. Cells were fixed with formalin (10% in PBS) for 10 min and washed with PBS and water. Samples were mounted onto glass slides using Prolong Gold antifade and cured for 2 h. All samples were viewed using a 60× 1.4-numerical-aperture oil objective in an Olympus IMT-2 inverted microscope coupled to a Bio-Rad MRC-600 dual-laser confocal microscope, as described previously (6).
Immunofluorescence microscopy.
The equivalent of 1 × 108 CFU of log-phase bacteria grown in LB medium or DMEM was pelleted by centrifugation (4,000 × g for 10 min at 4°C), washed once with PBS, and fixed in 500 μl 3.7% paraformaldehyde in 0.85% saline for 15 min at room temperature. Sterile coverslips were placed into 24-well plates and coated with 200 μl 10% poly-l-lysine for 1 min. Fifty microliters of fixed bacteria was applied onto the coverslip by centrifugation (2,000 × g for 5 min), and unbound bacteria were removed by washing once with PBS. Bacteria were incubated with polyclonal mouse anti-Sab (1:100 in PBS-10% FCS) for 15 min, washed three times with PBS, and incubated with Alexa 594-conjugated donkey anti-mouse secondary antibody (Molecular Probes) for another 15 min. Finally, coverslips were washed three times with PBS, mounted with Prolong Gold antifade reagent, and visualized by fluorescence microscopy.
Biofilm assay on polystyrene surfaces.
Biofilm formation assays on polystyrene surfaces using 96-well Maxisorb plates (Nunc) were carried out as described previously (42, 44), with the following modifications. Briefly, 200 μl of 1:40-diluted cultures of each strain grown overnight was inoculated into 96-well plates (24 wells for each strain), incubated for 18 h at 37°C, washed twice with 300 μl PBS to remove unbound cells, and stained with 200 μl 0.1% crystal violet for 5 min. Unbound stain was removed by washing twice with PBS, and quantification of biofilm formation was carried out by the addition of 200 μl of 96% ethanol and measurement of the A570.
RESULTS
Identification of an ORF of E. coli O113:H21 98NK2 encoding a predicted AT.
In a previous study comparing adherence phenotypes of different LEE-negative STEC isolates, we observed that an O113:H21 food isolate, strain MW10, exhibited significantly lower adherence to Henle 407 cells than other O113:H21 isolates from HUS cases, such as strain 98NK2 (27). Human-virulent O113:H21 STEC strains are known to carry putative adherence-associated genes such as saa and iha on their megaplasmid (pO113) (26, 38). We therefore used the known DNA sequence of pO113 from another O113:H21 STEC strain (EH41) (22) to design PCR primers to detect the presence of these putative adhesin genes as well as several other pO113 genes of unknown function in adherent versus nonadherent strains (98NK2 and MW10, respectively). Interestingly, the only difference noted was the absence in MW10 of a PCR product generated from an open reading frame (ORF) designated LH0147 in the published sequence (data not shown). Further PCR and sequence analyses confirmed that the regions of pO113 DNAs from 98NK2 and EH41 containing this ORF and flanking regions are essentially identical, but the LH0147 ORF in MW10 contains a deletion (result not shown). This raised the possibility that a lack of a functional LH0147 product might account for the adherence defect in MW10.
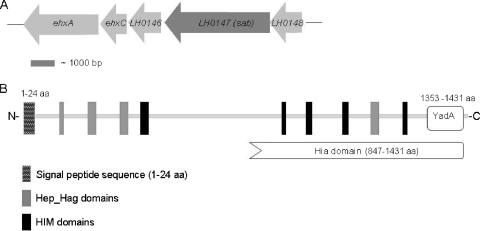
The 4,296-bp ORF LH0147 from E. coli 98NK2 is located upstream of the pO113 ehx locus, which encodes the EHEC hemolysin (Fig. 1A). LH0147 encodes a 1,431-amino-acid (aa) (146-kDa) protein with features characteristic of members of the AT protein family. This, as well as the functional properties of ORF LH0147 presented below, led us to designate the gene product of LH0147 as Sab (for STEC AT mediating biofilm formation). In silico analysis using SignalP 3.0 (3) and Pfam (2) indicated the presence of a signal sequence with a predicted cleavage site after aa 24 and a large N-terminal passenger domain containing four Hep-Hag repeats and five Him motifs, as depicted in Fig. 1B. These motifs are often found in bacterial invasins and hemagglutinins, respectively (42). After cleavage of the putative signal sequence, the theoretical molecular mass of mature Sab is 142 kDa. The C-terminal region (aa 1353 to 1431) comprises a putative translocator domain, which exhibits the features of the pfam03895 superfamily represented by the trimeric AT YadA of Yersinia enterocolitica. Moreover, the region from aa 847 to 1431 has multidomain similarity with the C termini of Haemophilus influenzae Hia-like proteins (Fig. 1B). The trimeric ATs are a novel AT family characterized by a very short translocator domain that forms stable trimers in the outer membrane. All members of the trimeric AT family examined to date appear to have adhesin-like activity, mediating adherence to eukaryotic cells and extracellular matrix proteins, such as collagen and fibronectin, or binding to circulating host factors, such as immunoglobulins or complement-inhibitory proteins (7, 17). BlastP analysis indicated that Sab is most similar to an AT adhesin of Actinobacillus pleuropneumoniae (GenBank accession number YP_001968314), which exhibits 33% identity and 48% similarity (15% gaps) to aa 601 to 1431 of Sab. Furthermore, aa 601 to 1010 of Sab exhibit 30% identity and 48% similarity (with 12% gaps) to the collagen adhesin EmaA of Aggregatibacter actinomycetemcomitans (accession number AAQ22366), another member of the trimeric AT family. However, at the nucleotide sequence level, the entire sab gene showed negligible homology to any other entries in the NCBI database.
FIG. 1.
Genetic organization of the sab gene and domain structure of the Sab protein. (A) The sab locus is flanked on one side by LH0146 (encoding a putative OmpA family lipoprotein) and the ehx locus (encoding STEC hemolysin) and on the other side by LH0148 (unknown function). The region shown corresponds to nucleotides 114000 to 124000 of the pO113 sequence from STEC EH41 (GenBank accession number NC_007365). (B) The domain structure of the Sab protein (aa 1 to 1431) was predicted using SignalP (3), Pfam (2), and NCBI Protein BLAST (20).
Presence of sab in other STEC strains.
The prevalence of sab in a selection of STEC strains from our collection was then determined by PCR analysis by using a primer pair that amplifies a 163-bp product from within the region of the sab ORF encoding the passenger domain. Both LEE-positive and LEE-negative strains were tested, and the distribution of sab was compared with those of eae (a marker for LEE) and the known STEC megaplasmid-borne genes ehxA, saa, and subA (Table 2). The sab gene was not present in any of the LEE-positive strains tested, nor was sab present in four LEE-negative STEC strains which also lacked any of the other known plasmid-borne genes, consistent with absence of a megaplasmid altogether. The remaining 14 LEE-negative strains were all positive for ehxA, indicating the likely carriage of a megaplasmid. Of these strains, six were positive for sab by PCR: four of these strains were of serotype O113:H21, one was of serotype O82:H8, and one was of serotype O23. These strains were all positive for the three other plasmid-carried putative virulence genes tested (ehxA, saa, and subA) (Table 2). Several of the sab-negative strains carried two or three of the other genes, implying that sab is associated with a specific subset of LEE-negative STEC strains.
TABLE 2.
Presence of sab and other STEC virulence genes in various E. coli strains
| E. coli strain | Serotypee | Sourcea | Presence of gene:
|
||||
|---|---|---|---|---|---|---|---|
| eaeb | ehxAb | saab | subAb,d | sabc | |||
| 98NK2 | O113:H21 | HUS | − | + | + | + | + |
| 96GR1 | O157:H− | HUS | + | + | − | − | − |
| 95ZG1 | O26 | BD | + | + | − | − | − |
| 96RO1 | O111:H− | HUS | + | + | − | − | − |
| 95NR1 | O111:H− | HUS | + | + | − | − | − |
| 99BG1 | O111:H− | BD | + | + | − | − | − |
| 97MW1 | O113:H21 | BD/M/T | − | + | + | + | + |
| 3848 | O113:H21 | HUS | − | + | + | + | + |
| 1183 | O113:H21 | HUS | − | + | + | + | + |
| MW10 | O113:H21 | Food | − | + | + | + | − |
| 94CR | O48:H21 | HUS | − | + | + | + | − |
| 99AM1 | O23 | BD | − | + | + | + | + |
| 00MC2 | O6 | BD | − | + | + | ND | − |
| B2F1 | O91:H21 | HUS | − | + | + | − | − |
| MW13 | O98 | Food | − | + | + | − | − |
| MW9 | OR | Food | − | + | + | + | − |
| MW3 | O82:H8 | Food | − | + | + | + | + |
| MW7 | Ont:H11 | Food | − | + | + | + | − |
| 95HE4 | O91:H7 | D | − | + | + | + | − |
| MW15 | O141 | Food | − | − | − | ND | − |
| MW4 | Ont | Food | − | − | − | − | − |
| 95AS1 | O128 | D | − | − | − | − | − |
| MW12 | O159 | Food | − | − | − | − | − |
STEC strains were isolated either from foods or from the feces of patients with uncomplicated diarrhea (D), bloody diarrhea (BD), microangiopathic hemolytic anemia and thrombocytopenia (M/T), or HUS.
Determined by PCR using primers LH0147-f (5′-GGTGGATACAGCAGGTAATG-3′) and LH0147-r (5′-TATCTCACCACCTGCTATCG-3′) (163-bp PCR product).
ND, not determined.
Ont, O nontypeable; OR, O rough.
Cloning of the sab gene and construction of a 98NK2 sab deletion mutant.
PCR was used to amplify a 4,368-bp fragment containing the complete sab ORF, and this was cloned as a BamHI/EcoRI fragment into pBluescript SK(−) to obtain pBsab. The construct was then transformed into E. coli JM109 [JM109(pBsab)].
A 98NK2 sab-negative derivate was constructed by the deletion of the sab gene and insertion of a kanamycin cartridge using the Lambda Red recombinase system, as described in Materials and Methods. Successful mutagenesis was confirmed by PCR and sequence analysis. The confirmed mutant was designated 98NK2sab::kan. To complement the 98NK2 sab deletion mutant, pBsab and the empty vector control were transformed into 98NK2sab::kan to generate 98NK2sab:: kan(pBsab) and 98NK2sab::kan(pB), respectively.
Expression and location of Sab.
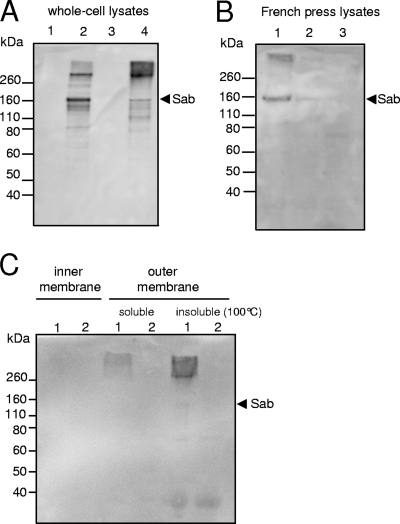
BlastP analysis of the mature Sab protein predicted that Sab belongs to the AT family and, hence, should be exposed on the bacterial surface. To confirm this, and to examine Sab expression, Sab from 98NK2 was purified as an N-terminal His6-Sab fusion protein lacking the signal sequence and the translocator domain, as described in Materials and Methods. The purified protein was used to raise a polyclonal mouse antiserum, enabling Western immunoblot analysis of cell lysates of wild-type 98NK2, 98NK2sab::kan, 98NK2sab::kan(pB), 98NK2sab::kan(pBsab), JM109(pB), and JM109(pBsab). Immunoreactive bands were not seen in unconcentrated lysates of 98NK2, suggesting low baseline levels of expression of Sab, at least in vitro (result not shown). However, strong immunoreactive bands with sizes of approximately 160 kDa and >260 kDa, as well as several other minor species, were seen in lysates of 98NK2sab::kan(pBsab) but not 98NK2sab::kan(pB) (Fig. 2A). The same two immunoreactive species were seen in lysates of JM109(pBsab) except that the larger species predominated and there was an additional smear of immunoreactive material with even greater molecular size. However, there were no immunoreactive bands in the JM109(pB) lysate (Fig. 2A). To overcome the inability to detect Sab in wild-type 98NK2, we analyzed a French press lysate of concentrated cells from a 500-ml culture grown in DMEM. A clear immunoreactive band of approximately 160 kDa, as well as higher-molecular-mass material, was detected in the lysate of 98NK2 but not in that from the otherwise isogenic mutant 98NK2sab::kan (Fig. 2B). To further probe the cellular location of anti-Sab-reactive material, inner and outer membranes from 98NK2 and 98NK2sab::kan were prepared, and proteins were solubilized, as described in Materials and Methods. These extracts, along with the insoluble pellet remaining after detergent extraction of the outer membrane fraction, were subjected to SDS-PAGE and Western blotting. A smear of high-molecular-mass immunoreactive material was detected in both the soluble and insoluble outer membrane protein fractions of 98NK2, and in the case of the boiled insoluble fraction, a faint band with a mass of approximately 160 kDa was also seen. No immunoreactive bands were detected in the inner membrane protein fraction of 98NK2 or in any of the fractions from 98NK2sab::kan (Fig. 2C). In order to verify appropriate fractionation, replicate filters were also probed with an antibody specific for Iha, which is known to be located exclusively in the outer membrane (38). Immunoreactive bands of the expected sizes were seen in the outer membrane fractions of both 98NK2 and 98NK2sab::kan but not in any of the other fractions (data not shown). Collectively, these data show that the expression of the sab gene in either the 98NK2 or JM109 background results in the production of a 160-kDa protein species, which is slightly larger than that predicted by sequence analysis (142 kDa). Given the high specificity of the antiserum, the higher-molecular-mass immunoreactive material is likely to represent Sab multimers, as previously observed for other AT proteins (1, 32, 42). Bands with reduced masses detected in 98NK2sab::kan(pBsab) and JM109(pBsab) are presumably a consequence of proteolytic degradation during the isolation process.
FIG. 2.
Western blot analysis using anti-Sab. Strains were grown in LB medium (A and C) or DMEM (B) at 37°C, and lysates or protein extracts were prepared and separated by SDS-PAGE, blotted, and probed with anti-Sab, as described in Materials and Methods. (A) Whole-cell lysates. Lanes: 1, 98NK2sab::kan(pB); 2, 98NK2sab::kan(pBsab); 3, JM109(pB); 4, JM109(pBsab). (B) French press lysates. Lanes: 1, 98NK2; 2, 1:10 dilution of 98NK2; 3, 98NK2sab::kan. (C) Inner and outer membrane fractions. Lanes: 1, 98NK2; 2, 98NK2sab::kan. The arrow indicates the mobility of the 160-kDa Sab species.
Surface exposure of Sab.
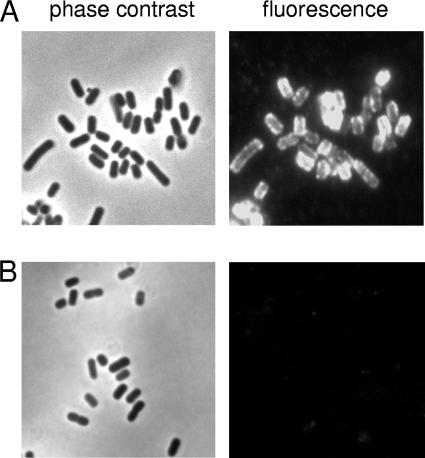
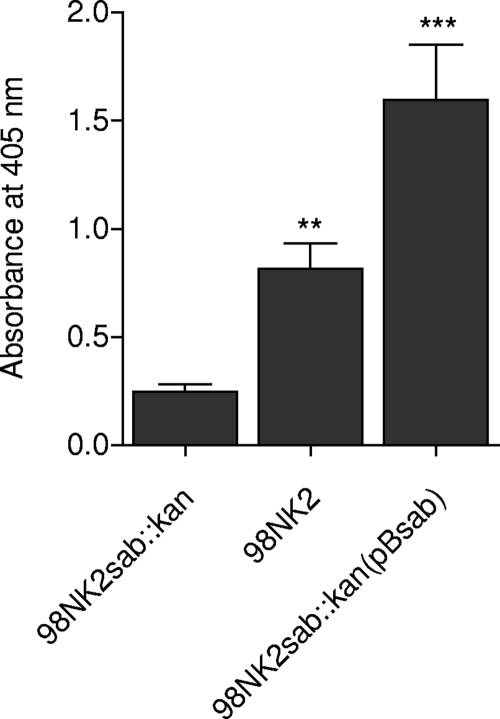
Surface exposure of Sab was then examined by indirect immunofluorescence microscopy with anti-Sab, in the first instance using cultures of wild-type strain 98NK2 and its isogenic mutant. The total expression level in 98NK2 was too low in either LB medium or DMEM to enable detection by immunofluorescence (data not shown). However, strong immunofluorescence labeling was observed in cultures of 98NK2sab::kan(pBsab) but not in cultures of 98NK2sab::kan(pB) (Fig. 3). The accessibility of Sab on whole unfixed cells to exogenous antibody was also examined by ELISA (Fig. 4). This showed significantly greater labeling in 98NK2 cells than in 98NK2sab::kan cells (P < 0.01), while labeling of 98NK2sab::kan(pBsab) cells was significantly greater again (P < 0.005). In contrast, there were no significant differences in labeling among the three strains when whole-cell ELISAs were performed using the same dilution of either nonimmune mouse serum or anti-Iha as the primary antibody (result not shown). Collectively, these findings confirm that Sab is exposed on the surface of intact 98NK2 cells and is accessible to exogenous antibody. However, the baseline level of expression in wild-type cells is low compared to that in 98NK2 derivatives carrying sab on a multicopy plasmid.
FIG. 3.
Immunofluorescence microscopy. Log-phase cultures of 98NK2sab::kan(pBsab) (A) and 98NK2sab::kan(pB) (B) were formalin fixed and labeled with anti-Sab followed by Alexa 594-conjugated donkey anti-mouse secondary antibody (see Materials and Methods). The fluorescent image for each strain is accompanied by the phase-contrast image for the corresponding field.
FIG. 4.
Surface expression of Sab on whole unfixed cells judged by use of ELISA. Cells from log-phase cultures of 98NK2, 98NK2sab::kan, and 98NK2sab::kan(pBsab) were probed with anti-Sab and secondary goat anti-mouse immunoglobulin G-alkaline phosphatase antibody, as described in Materials and Methods. Surface expression was quantitated by measuring the A405. Data are means ± standard deviations of data from three independent experiments (**, P < 0.01; ***, P < 0.005 by Student's unpaired, two-tailed t test [relative to 98NK2sab::kan]).
Sab contributes to epithelial cell adherence.
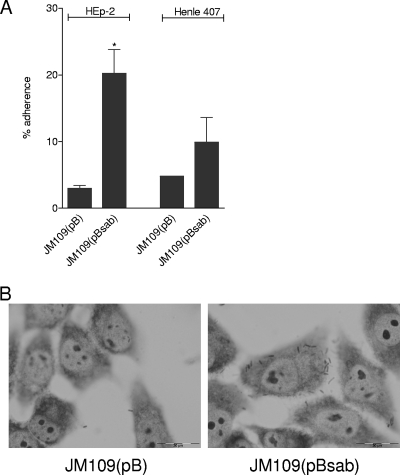
Many ATs, especially members of the trimeric AT family, mediate adherence to epithelial cells. Accordingly, we examined the adherence of JM109(pBsab) or JM109(pB) to human epithelial cell lines (HEp-2 and Henle 407 cells) using a quantitative assay (see Materials and Methods). A significant increase in the level of adherence to HEp-2 cells was observed for JM109(pBsab) relative to that of JM109(pB) (20.2% versus 3.0% of the total number of bacteria, respectively; P < 0.05) (Fig. 5A). A similar trend was observed for Henle 407 cells [9.9% versus 4.8% of the total numbers of bacteria were adherent for JM109(pBsab) and JM109(pB), respectively], although this difference did not reach statistical significance (Fig. 5A). Interestingly, no difference in adherence was observed when similar experiments were conducted with Hct-8 (human colonic) cells (data not shown).
FIG. 5.
Adherence assays. (A) Quantitative adherence of JM109(pBsab) and JM109(pB) to HEp-2 and Henle 407 cells (see Materials and Methods). Data correspond to means ± standard deviations of data from quadruplicate assays from two independent experiments. (*, P < 0.05 by Student's unpaired, two-tailed t test). (B) Giemsa staining of adherent bacteria on HEp-2 cell monolayers visualized by light microscopy.
Examination of Giemsa-stained monolayers after 3 h of incubation revealed a diffuse pattern of adherence for JM109(pBsab) but not for JM109(pB), as shown in Fig. 5B.
Interaction of purified Sab with HEp-2 cells.
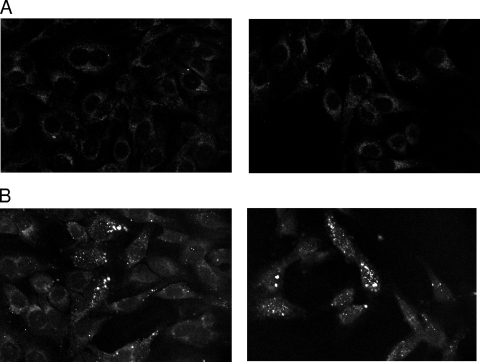
To determine whether Sab binds directly to human cell lines, HEp-2 cell monolayers were incubated with Oregon Green-labeled Sab in DMEM at 37°C. After 2.5 h of incubation, unbound proteins were washed, and samples were examined with a dual-laser confocal microscope. Substantial binding of Sab was observed (Fig. 6).
FIG. 6.
Protein-binding assay. HEp-2 monolayers were exposed to 0 μg/ml (A) or 20 μg/ml (B) Oregon Green-labeled Sab protein for 2.5 h at 37°C. Unbound protein was removed by washing, and fixed cells were viewed by confocal microscopy.
Sab mediates biofilm formation.
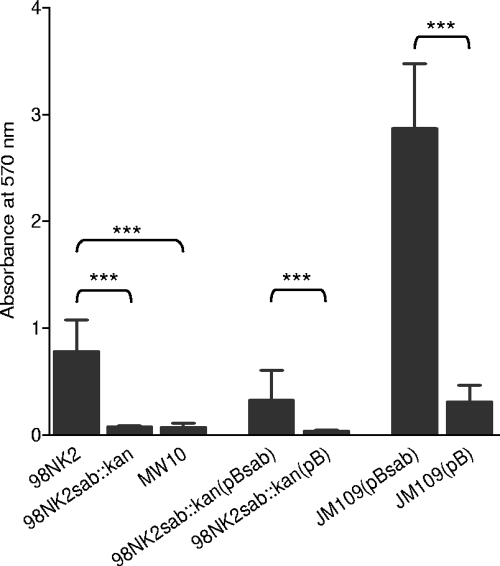
To assess whether Sab promotes biofilm formation, wild-type MW10, 98NK2, 98NK2sab::kan, 98NK2sab::kan(pBsab), 98NK2sab::kan(pB), JM109(pB), and JM109(pBsab) were tested for their capacities to form biofilms on polystyrene surfaces. As shown in Fig. 7, significant biofilm formation was observed for 98NK2, whereas biofilm formation was negligible for the otherwise isogenic mutant 98NK2sab::kan (P < 0.005) as well as sab-negative STEC strain MW10 (P < 0.005). 98NK2sab::kan(pBsab) also exhibited significantly increased levels of biofilm formation relative to that of 98NK2sab::kan(pB) (P < 0.005), as did JM109(pBsab) relative to that of JM109(pB) (P < 0.005).
FIG. 7.
Biofilm formation on polystyrene plates. The indicated strains were grown in 96-well polystyrene plates for 18 h. Plates were then washed and stained with crystal violet (see Materials and Methods). Biofilm formation was quantitated by measuring the A570 after solubilization of the stained bacteria. Data are means ± standard deviations (n = 24) (***, P < 0.005 by Student's unpaired, two-tailed t test [relative to the respective negative control]).
DISCUSSION
Although Shiga toxin is the definitive virulence factor of STEC, the capacity to compete with commensal flora and colonize the human intestinal epithelium is critical for pathogenesis. The mechanism by which LEE-positive STEC strains adhere intimately to the enterocyte surface has been studied extensively, whereas the mechanism(s) by which LEE-negative STEC strains adhere to the intestinal mucosa is less well understood. Nevertheless, the importance of adhesins is reflected in the broad range of such proteins discovered in LEE-positive as well as in LEE-negative STEC strains, including Saa, (26), Lpf (10, 39, 40), EibG (18), Iha (38), Efa1 (23), Cah (41), ECP (30), HCP (47), and EhaA (45). Nearly all LEE-positive as well as many LEE-negative STEC strains carry megaplasmids, which encode putative accessory virulence factors, including some of the above-mentioned proteins. Significantly, the megaplasmids of highly virulent LEE-negative STEC strains such as O113:H21 strains EH41 and 98NK2 are much larger than those of the classical LEE-positive STEC O157:H7 strains and encode a broad range of additional virulence factors, many of which appear to be unique to LEE-negative strains (22, 25).
In this study, we have characterized one such putative virulence factor, the AT Sab, which is present in some but not all LEE-negative STEC serotypes/groups including multiple isolates of O113:H21 and single isolates of O23 and O82:H8. In both 98NK2 and EH41, the sab gene is located on megaplasmid pO113, approximately 1.3 kb downstream of the hemolysin locus ehx. All sab-positive STEC strains tested were also positive for ehxA as well as the autoagglutinating adhesin gene saa and the subtilase cytotoxin gene subA; the latter genes are located 20 and 28 kb, respectively, downstream of sab on pO113. However, the absence of sab in several other LEE-negative STEC strains that carry one or more of the other megaplasmid-borne genes tested (ehxA, saa, or subA) underscores the heterogeneity of STEC strains with respect to their complement of megaplasmid-encoded accessory virulence factors.
Western blot analysis indicated that the level of expression of the Sab protein in wild-type 98NK2 is low under the in vitro conditions employed in this study (growth in LB medium or DMEM). Indeed, it was detectable only when concentrated cell lysates were examined. It is possible that Sab is expressed at high levels in vivo, although we found no difference in sab gene expression levels by real-time reverse transcription-PCR when cells were grown in LB medium or DMEM in the presence or absence of HEp-2 cells (data not shown). Nevertheless, the introduction of sab on a high-copy-number plasmid in either the JM109 or 98NK2sab::kan background resulted in much higher levels of expression. SDS-PAGE and Western blotting results suggested that the protein is capable of forming multimers. Moreover, Sab was present exclusively in the outer membrane protein fraction and was exposed on the surface of intact E. coli cells, as judged by the accessibility to exogenous antibody using immunofluorescence or ELISA.
The surface localization of Sab is consistent with our observation that it confers the capacity to adhere diffusely to HEp-2 cells when expressed in JM109 cells. Moreover, the purified fluorescently labeled passenger domain of Sab bound to the surface of HEp-2 cells, suggesting that Sab may act as a direct adhesin rather than as an indirect facilitator of adherence. Sab also appears to be largely responsible for the capacity of wild-type 98NK2 cells to form a biofilm on polystyrene surfaces. In contrast, biofilm formation was negligible in 98NK2sab::kan as well as in the poorly adherent STEC food isolate MW10, which lacks an intact sab gene. Furthermore, the biofilm formation defect in 98NK2sab::kan was complemented by transformation with pBsab but not with empty vector. Likewise, biofilm formation by JM109(pBsab) was significantly greater than that by JM109(pB).
The C terminus of Sab shares homology with the prototypic trimeric ATs Hia and YadA, and given its mobility on SDS-PAGE gels, which is indicative of multimerization, its location in the outer membrane, its exposure on the E. coli surface, and its unequivocal role in adherence to epithelial cells and biofilm formation, we hypothesize that Sab is also a member of the trimeric AT family. Several other ATs were previously reported to promote biofilm formation, for example, Cah (41), EhaA (45), UpaG (42), Ag43 (8), and AIDA (34). The capacity to form biofilms may enhance the survival of pathogens in different environmental niches, such as in food products or in the gastrointestinal tracts of humans or animal reservoirs of infection. It is particularly noteworthy that even the low baseline level of sab expression in wild-type 98NK2 was sufficient to confer a substantial level of biofilm formation, and this was completely abolished in the otherwise isogenic mutant 98NK2sab::kan. Other studies have shown that the trimeric AT proteins YadA, NhhA, BadA, and UpaG promote binding to extracellular matrix proteins, such as fibronectin, laminin, or collagen (31, 33, 37, 42). However, we found no evidence that either purified Sab protein or E. coli JM109 carrying sab was capable of binding to any of these extracellular matrix proteins (result not shown). Some ATs have also been shown to mediate autoagglutination, but this was not observed for Sab (result not shown).
In summary, we have shown that the STEC megaplasmid-encoded putative trimeric AT family protein Sab promotes adherence to human epithelial cells and mediates biofilm formation. The distribution of sab, although based on an analysis of a relatively small number of clinical and environmental STEC isolates, indicates that Sab, like the autoaggregative adhesin Saa (26), is associated with LEE-negative STEC strains. Thus, both adhesins may contribute to intestinal colonization in STEC strains that lack the capacity to form A/E lesions on enterocytes. It is also noteworthy that apart from encoding Sab and Saa, as well as additional putative accessory virulence factors including subtilase cytotoxin (SubAB) (25) and two additional SPATE family secreted serine protease ATs, EspP (4) and EpeA (16), the megaplasmids of highly virulent STEC O113:H21 strains 98NK2 and EH41 are self-transmissible (36). Thus, the assembly of such a diverse payload of virulence factors onto a single mobile DNA element is likely to have contributed to the evolution of hypervirulent LEE-negative STEC strains.
Acknowledgments
This work was supported by program grant 284214 and project grant 565359 from the National Health and Medical Research Council of Australia (NHMRC) (to J.C.P. and A.W.P.). S.H. is a recipient of a German Research Foundation (Deutsche Forschungsgemeinschaft) research fellowship; J.C.P. is an NHMRC Australia Fellow.
We are also grateful to Kerrie May and Marcin Grabowicz for helpful discussions and to Ursula Talbot for technical assistance.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 1 June 2009.
REFERENCES
- 1.Ackermann, N., M. Tiller, G. Anding, A. Roggenkamp, and J. Heesemann. 2008. Contribution of trimeric autotransporter C-terminal domains of oligomeric coiled-coil adhesin (Oca) family members YadA, UspA1, EibA, and Hia to translocation of the YadA passenger domain and virulence of Yersinia enterocolitica. J. Bacteriol. 1905031-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340783-795. [DOI] [PubMed] [Google Scholar]
- 4.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24767-778. [DOI] [PubMed] [Google Scholar]
- 5.Caron, E., V. F. Crepin, N. Simpson, S. Knutton, J. Garmendia, and G. Frankel. 2006. Subversion of actin dynamics by EPEC and EHEC. Curr. Opin. Microbiol. 940-45. [DOI] [PubMed] [Google Scholar]
- 6.Chong, D. C., J. C. Paton, C. M. Thorpe, and A. W. Paton. 2008. Clathrin-dependent trafficking of subtilase cytotoxin, a novel AB5 toxin that targets the endoplasmic reticulum chaperone BiP. Cell. Microbiol. 10795-806. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13199-205. [DOI] [PubMed] [Google Scholar]
- 8.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37424-432. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doughty, S., J. Sloan, V. Bennett-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 706761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30911-921. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 691231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6370-378. [DOI] [PubMed] [Google Scholar]
- 15.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 215-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leyton, D. L., J. Sloan, R. E. Hill, S. Doughty, and E. L. Hartland. 2003. Transfer region of pO113 from enterohemorrhagic Escherichia coli: similarity with R64 and identification of a novel plasmid-encoded autotransporter, EpeA. Infect. Immun. 716307-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linke, D., T. Riess, I. B. Autenrieth, A. Lupas, and V. A. Kempf. 2006. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 14264-270. [DOI] [PubMed] [Google Scholar]
- 18.Lu, Y., S. Iyoda, H. Satou, H. Satou, K. Itoh, T. Saitoh, and H. Watanabe. 2006. A new immunoglobulin-binding protein, EibG, is responsible for the chain-like adhesion phenotype of locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 745747-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 20.Marchler-Bauer, A., J. B. Anderson, M. K. Derbyshire, C. DeWeese-Scott, N. R. Gonzales, M. Gwadz, L. Hao, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, D. Krylov, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, N. Thanki, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2007. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35D237-D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton, H. J., J. Sloan, D. M. Bulach, T. Seemann, C. Allison, M. Tauschek, R. M. Robins-Browne, J. C. Paton, T. S. Whittam, A. W. Paton, and E. L. Hartland. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg. Infect. Dis. 15372-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35275-288. [DOI] [PubMed] [Google Scholar]
- 24.Paton, A. W., and J. C. Paton. 2005. Multiplex PCR for direct detection of Shiga toxigenic Escherichia coli producing the novel subtilase cytotoxin. J. Clin. Microbiol. 432944-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paton, A. W., P. Srimanote, U. M. Talbot, H. Wang, and J. C. Paton. 2004. A new family of potent AB5 cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 20035-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paton, A. W., P. Srimanote, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 696999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paton, A. W., E. Voss, P. A. Manning, and J. C. Paton. 1997. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect. Immun. 653799-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 373357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rendón, M. A., Z. Saldaña, A. L. Erdem, V. Monteiro-Neto, A. Vázquez, J. B. Kaper, J. L. Puente, and J. A. Girón. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. USA 10410637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riess, T., S. G. Andersson, A. Lupas, M. Schaller, A. Schafer, P. Kyme, J. Martin, J. H. Walzlein, U. Ehehalt, H. Lindroos, M. Schirle, A. Nordheim, I. B. Autenrieth, and V. A. Kempf. 2004. Bartonella adhesin A mediates a proangiogenic host cell response. J. Exp. Med. 2001267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 1853735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scarselli, M., D. Serruto, P. Montanari, B. Capecchi, J. Adu-Bobie, D. Veggi, R. Rappuoli, M. Pizza, and B. Arico. 2006. Neisseria meningitidis NhhA is a multifunctional trimeric autotransporter adhesin. Mol. Microbiol. 61631-644. [DOI] [PubMed] [Google Scholar]
- 34.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 1868058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spears, K. J., A. J. Roe, and D. L. Gally. 2006. A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol. Lett. 255187-202. [DOI] [PubMed] [Google Scholar]
- 36.Srimanote, P., A. W. Paton, and J. C. Paton. 2002. Characterization of a novel type IV pilus locus encoded on the large plasmid of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 703094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamm, A., A. M. Tarkkanen, T. K. Korhonen, P. Kuusela, P. Toivanen, and M. Skurnik. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10995-1011. [DOI] [PubMed] [Google Scholar]
- 38.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 681400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torres, A. G., J. A. Giron, N. T. Perna, V. Burland, F. R. Blattner, F. Avelino-Flores, and J. B. Kaper. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 705416-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres, A. G., K. J. Kanack, C. B. Tutt, V. Popov, and J. B. Kaper. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol. Lett. 238333-344. [DOI] [PubMed] [Google Scholar]
- 41.Torres, A. G., N. T. Perna, V. Burland, A. Ruknudin, F. R. Blattner, and J. B. Kaper. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol. Microbiol. 45951-966. [DOI] [PubMed] [Google Scholar]
- 42.Valle, J., A. N. Mabbett, G. C. Ulett, A. Toledo-Arana, K. Wecker, M. Totsika, M. A. Schembri, J. M. Ghigo, and C. Beloin. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J. Bacteriol. 1904147-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veiga, E., S. Etsuko, H. Nikaido, V. de Lorenzo, and L. A. Fernández. 2002. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 212122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakimoto, N., J. Nishi, J. Sheikh, J. P. Nataro, J. Sarantuya, M. Iwashita, K. Manago, K. Tokuda, M. Yoshinaga, and Y. Kawano. 2004. Quantitative biofilm assay using a microtiter plate to screen for enteroaggregative Escherichia coli. Am. J. Trop. Med. Hyg. 71687-690. [PubMed] [Google Scholar]
- 45.Wells, T. J., O. Sherlock, L. Rivas, A. Mahajan, S. A. Beatson, M. Torpdahl, R. I. Webb, L. P. Allsopp, K. S. Gobius, D. L. Gally, and M. A. Schembri. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 10589-604. [DOI] [PubMed] [Google Scholar]
- 46.Wells, T. J., J. J. Tree, G. C. Ulett, and M. A. Schembri. 2007. Autotransporter proteins: novel targets at the bacterial cell surface. FEMS Microbiol. Lett. 274163-172. [DOI] [PubMed] [Google Scholar]
- 47.Xicohtencatl-Cortes, J., V. Monteiro-Neto, M. A. Ledesma, D. M. Jordan, O. Francetic, J. B. Kaper, J. L. Puente, and J. A. Girón. 2007. Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J. Clin. Investig. 1173519-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]