Abstract
Symbiosis between Rhizobium and its leguminous host requires elaborate communication between the partners throughout the interaction process. A calmodulin-like protein, termed calsymin, was identified in Rhizobium etli; a calmodulin-related protein in a Gram-negative bacterium had not been described previously. Calsymin possesses three repeated homologous domains. Each domain contains two predicted EF-hand Ca2+-binding motifs. Ca2+-binding activity of calsymin was demonstrated on purified protein. R. etli efficiently secretes calsymin without N-terminal cleavage of the protein. The gene encoding calsymin, casA, is exclusively expressed during colonization and infection of R. etli with the host. Expression of casA is controlled by a repressor protein, termed CasR, belonging to the TetR family of regulatory proteins. Mutation of the casA gene affects the development of bacteroids during symbiosis and symbiotic nitrogen fixation.
The symbiotic interaction between leguminous plants and rhizobia leads to the formation of a new plant organ, the nodule. Within this structure, the bacteria differentiate into bacteroids that fix molecular nitrogen for the benefit of the plant. A sustained molecular dialogue between both partners during the infection process is a prerequisite for a successful symbiosis. During the initial step of the interaction, the presence of plant root-secreted flavonoids induces the expression of rhizobial nodulation (nod) genes. The nod genes encode enzymes involved in the synthesis of lipochitooligosaccharide molecules, also termed Nod factors (1–3). These rhizobial Nod factors are secreted and act as signal molecules that provoke several physiological and morphological alterations in the roots of the host plant (4, 5). During infection of the root, the bacteria are released from the unwalled tip of the infection thread in the plant cell cytoplasm by endocytosis of the membrane surrounding the infection thread. As a result, the bacteria, now termed bacteroids, are enclosed by a plant membrane. This new subcellular compartment is named a symbiosome. The generation and maintenance of this subcellular compartment is an essential facet of the coexistence between rhizobia and living plant cells.
Ca2+ is a common intracellular second-messenger molecule in eukaryotic cells and modulates a myriad of cellular processes (6). The earliest responses of root hairs to Nod factors include influx of Ca2+ and efflux of chloride and potassium ions (7) and intracellular calcium spiking (8). Changes in intracellular Ca2+ concentrations in eukaryotes are transduced through high-affinity Ca2+-binding proteins. Calmodulin, a primary Ca2+ receptor, is the most ubiquitous member of the largest family of Ca2+-binding proteins. It is a small acidic protein present in all eukaryotes, regulating the activity of many vital enzymes (9). Calmodulin has a dumbbell-shaped structure with two globular domains connected by a flexible central tether. Each of these lobes contains two Ca2+-binding motifs consisting of two nearly perpendicular α-helices separated by a 12-residue loop. This structure is known as the EF-hand helix–loop–helix motif (10). Ca2+ binding induces a large conformational change in calmodulin and causes the exposure of two hydrophobic patches involved in target recognition.
Although the presence of bacterial proteins with calmodulin-like properties has been reported repeatedly, calmodulin-like genes with authentic EF-hands appear to be very uncommon in prokaryotes (11). The only example so far described is a calmodulin-like protein from the Gram-positive bacterium Saccharopolyspora erythraea that contains four EF-hands (12). The biological role for this protein is presently unknown. Here we describe the isolation of a calmodulin-like gene from Rhizobium etli, casA, that is implicated in symbiosis and regulated by a TetR-type of repressor.
Materials and Methods
Growth Conditions.
R. etli strains were routinely grown in liquid tryptone/yeast extract (TY) or acid minimal salts (AMS) medium at 30°C and maintained on yeast-mannitol agar plates (13). Escherichia coli was grown in Luria–Bertani medium at 37°C.
Screening of a Mutant Expression Library.
The R. etli mutant library was constructed as described by Xi et al. (14). Approximately 4,000 mutants were screened for differential induction of the gusA gene when the cultures were grown in a microoxic environment with 0.3% oxygen and/or in the presence of nodule extracts. Details on the screening procedure are described by Xi et al. (15).
Plant Culture and Bacteroid Isolation.
Phaseolus vulgaris cv. Limburgse vroege plants were grown in the plant growth room essentially as described by Michiels et al. (16) and analyzed 3 weeks after inoculation. For symbiotic expressions, bacteroids were purified from plant material by differential centrifugation (13).
Cloning of the casA–casR Gene Region.
Standard methods were used for in vitro DNA manipulations (17). Total DNA from the selected mutant strain FAJ1806 was digested with XhoI and ligated into the SalI site of pUC18. Inserts containing part of the mTn5gusA-pgfp21 transposon were selected. From the cloned fragment, the partial sequence flanking the mTn5gusA-pgfp21 insertion was determined, using the gusA primer 5′-GATTTCACGGGTTGGTTCT-3′. To isolate the corresponding wild-type R. etli CNPAF512 gene, the sequenced fragment was amplified by PCR using the primers OJM142 (5′-GGCTGTCGTCAATGCTCTCCGATCTCGATACCG-3′) and OJM143 (5′-CATGAACTCTTCCAACGACACCAGCCCATCCC-3′). The resulting 360-bp PCR fragment was used as a probe to hybridize a λ phage EMBL3 gene library of CNPAF512. DNA from a positive plaque was hybridized with the 360-bp digoxigenin-labeled probe. Two positive SalI bands (1.8 kb and 0.7 kb) were cloned in pUC18 and sequenced.
Construction of Mutants.
To construct R. etli casR mutants, the 1.8-kb SalI fragment containing the complete casR gene was first cloned in the SalI site of pUC19, yielding plasmid pFAJ1822. The 2.1-kb spectinomycin resistance (spcR) cartridge from pHP45Ω (18) was removed with HindIII and ligated in the FseI and PshAI sites of pFAJ1822 to obtain plasmids pFAJ1824 (the orientation of the spcR gene is opposite to casR) and pFAJ1825 (the orientation of the spcR gene is the same as casR) after blunting of the fragments. The resulting SalI fragments from pFAJ1824 and pFAJ1825 were cloned in the SalI site of the suicide plasmid pJQ200SK (19), generating plasmids pFAJ1826 and pFAJ1827, respectively. These plasmids were introduced into strain CNPAF512 and double recombinants were selected as described (13). The casR mutants were FAJ1802 (spcR gene in FseI site in the same orientation as casR) and FAJ1803 (spcR gene in PshAI site, opposite orientation to casR).
For the construction of an R. etli casA mutant strain, a 2.5-kb fragment containing the casA and casR genes was amplified by PCR using primers OJM161 (5′-GATCCTCGAGCGCCGGTCAGGCGAGGCAACAGCC-3′) and OJM162 (5′-GATCCTCGAGGAAGTCGCCGGCCCGCGTCAGTACCG-3′) and, after digestion with XhoI, ligated into the SalI site of pUC19-ΔE (a pUC19 derivative from which the EcoRI site was removed), yielding plasmid pFAJ1828. Plasmid pFAJ1828 was digested with EcoRI, thereby removing the casA internal EcoRI fragment, and ligated to the 2.2-kb HindIII fragment from pHP45Ω-km, containing a kanamycin resistance (kmR) gene, by blunt-end ligation, generating plasmids pFAJ1829 (the orientation of the kmR gene is the same as casA) and plasmid pFAJ1830 (the kmR gene and casA have opposite orientations). The BamHI fragments containing the inserts of pFAJ1829 and pFAJ1830 were cloned into the BamHI site of pJQ200SK to obtain plasmids pFAJ1831 and pFAJ1832, respectively. Plasmids pFAJ1831 and pFAJ1832 were used to mutate the wild-type strain CNPAF512, yielding the casA mutant strains FAJ1804 and FAJ1805, respectively. R. etli casAR double mutants are described in the section on the construction of casA–gusA and casR–gusA fusions.
Construction of gusA Fusions.
The R. etli casA∷gusA strain FAJ1806 carries the insertion of mTn5gusA-pgfp21 between the nucleotides 838 and 839 from the ATG start codon of the casA gene.
To construct the R. etli casA∷mTn5gusA-pgfp21 casR.∷Ω-Spc strains FAJ1807 and FAJ1808, plasmids pFAJ1826 and pFAJ1827 were introduced into the R. etli mutant strain FAJ1806 and double recombinants were selected, generating the casAR double mutant strains FAJ1807 and FAJ1808, respectively.
To construct a genomic casR∷gusA fusion, the 4.5-kb BamHI fragment from pWM6 containing a promoterless gusA gene and a kmR cassette were ligated into the PshAI site of pFAJ1822, yielding plasmid pFAJ1836. The resulting 6.3-kb KpnI fragment from pFAJ1836 was cloned in the SmaI site of pJQ200-UC1 by blunt-end ligation, yielding plasmid pFAJ1837. This plasmid was introduced into the R. etli wild-type strain CNPAF512 and double recombinants were selected to obtain the casR∷gusA mutant strain FAJ1809.
Plasmid-borne PcasA–gusA (pFAJ1842) and PcasR–gusA (pFAJ1843) fusions were constructed. For this, the 450-bp casAR intergenic region, amplified by PCR using the primers OJM163 (5′-GATCTCTAGAGGATCCGGTGGAGACTTTCGGCCGGCCTCG-3′) and OJM164 (5′-GATCTCTAGAGACATCGGACGGCCGGGCCGCGACG-3′), was digested with XbaI and ligated into the XbaI site of pUC18NotI (20), yielding plasmid pFAJ1839. The 4.5-kb gusA-kmR SmaI fragment from pWM6 was inserted into the SmaI and SalI sites of pFAJ1839 to obtain plasmids pFAJ1840 and pFAJ1841, respectively. The NotI fragments containing the PcasA–gusA and PcasR–gusA fusions from the respective plasmids were inserted into the BamHI site of pLAFR3 (21) by blunt-end ligation, yielding plasmids pFAJ1842 and pFAJ1843.
Constructs for Complementation Analysis.
A 1.6-kb fragment containing the complete casA gene sequence but not the casR gene was obtained by PCR using primers OJM162 (5′-GATCCTCGAGGAAGTCGCCGGCCCGCGTCAGTACCG-3′) and OJM163 (5′-GATCTCTAGAGGATCCGGTGGAGACTTTCGGCGGCCTCG-3′). This fragment was digested with XbaI and XhoI and ligated with XbaI/SalI-digested pUC18NotI, yielding plasmid pFAJ1833. To facilitate subsequent cloning in pLAFR3, the 2.1-kb SmaI fragment containing the spcR gene from pHP45Ω was inserted into the SmaI site of pFAJ1833. From the resulting plasmid pFAJ1834, the 3.8-kb NotI fragment containing the casA gene was cloned into the BamHI site of pLAFR3 by using blunt-end ligation, yielding pFAJ1835. Plasmid pFAJ1838 was obtained by inserting the 4.2-kb BamHI fragment from pFAJ1829 containing the complete casR gene into the BamHI site of pLAFR3.
Light and Electron Microscopy.
For light microscopic analysis, 3-μm sections of 3-week-old nodules were prepared as described (16).
To obtain transmission electron micrographs, 3-week-old nodules were fixed overnight at 4°C in 2% cold glutaraldehyde solution in 10 mM sodium cacodylate buffer (pH 7.3) and postfixed in osmium tetroxide in the same buffer. Then the sample was block-stained in uranyl acetate (in 10% aqueous acetone) and dehydrated in a graded acetone series, followed by embedding in Araldite. Serial semithin sections were stained with methylene blue and thionin. Thin sections, made with a Reichert Ultracut E microtome, were stained with uranyl acetate and lead citrate in an LKB 2168 Ultrostainer and examined in a Zeiss EM 900 electron microscope.
Purification of Proteins.
Bacterial cells and culture supernatant were collected from the wild type or the CasA-overproducing casR strain FAJ1802 grown overnight at 30°C in TY medium. Proteins were precipitated from the growth medium supernatant by incubation with trichloroacetic acid (10%, wt/vol) for 2 h on ice and pelleted by centrifugation at 15,000 × g for 20 min. The cell pellets were washed twice with PBS at pH 6.8 and resuspended in the same buffer. The mixture was passed three times through a French press (SLM Instruments, Rochester, NY) at 10,000 psi (69 MPa). Cell debris was removed by centrifugation for 20 min at 10,000 rpm. The proteins were precipitated as described above. Proteins were separated by electrophoresis on SDS/polyacrylamide gels (5% stacking gel, 15% separating gel, 0.1% SDS). After electrophoresis, proteins were transferred to poly(vinylidene difluoride) (PVDF) membranes by electroblotting.
To overexpress calsymin in E. coli, a 900-bp fragment was amplified by PCR using primers OJM186 (5′-GTCACTCGAGATGACGACCATTTTCTGCTGCAACATC-3′) and OJM187 (5′-GTCACTGCAGCTCGCGGCGGACTGGACGG-3′) and cloned as a XhoI/PstI fragment in the corresponding sites of pBAD/HisA, yielding pFAJ1845. To express the recombinant gene, E. coli Top10 cells carrying pFAJ1845 were grown exponentially and subsequently induced for 6 h in the presence of 0.2% arabinose. The recombinant calsymin fusion protein carrying the N-terminal polyhistidine tag was purified under denaturing conditions on ProBond as recommended by the manufacturer (Invitrogen).
Calcium-Binding Assay.
After electroblotting of the proteins on PVDF membranes, the same membrane was first used in a 45Ca2+-binding assay, then stained with ruthenium red, and finally stained with Coomassie brilliant blue. The 45Ca2+-binding assay was performed essentially as described by Maruyama et al. (22). Ruthenium red staining was carried out as described by Charuk et al. (23). To confirm the identity of the ≈45-kDa 45Ca2+-binding band, it was excised (50 μg) from several lanes from PVDF blots and N-terminally sequenced by Edman degradation on a pulsed liquid-phase 477A/120A protein sequencer (Perkin–Elmer) using N-methylpiperidine as a coupling base.
The NAD kinase activity assay was performed according to Harmon et al. (24), using purified chicken NAD kinase (Sigma). Calsymin was prepared from the culture supernatant of FAJ1803 as described above. Aliquots were tested for induction of NAD kinase activity.
Results
Sequence of casA and casR.
R. etli is the nodulating symbiont of P. vulgaris, the common bean plant. To identify new symbiotic genes in this species, R. etli was mutagenized with the miniTn5 transposon derivative mTn5gusA-pgfp21 (14) carrying a promoterless gusA gene suitable for promoter trapping. Expression of the gusA gene in the mutants was assessed under conditions of aerobiosis or microaerobiosis in the presence or absence of nodule extracts. One of the strains displaying a differential induction pattern was selected for further characterization.
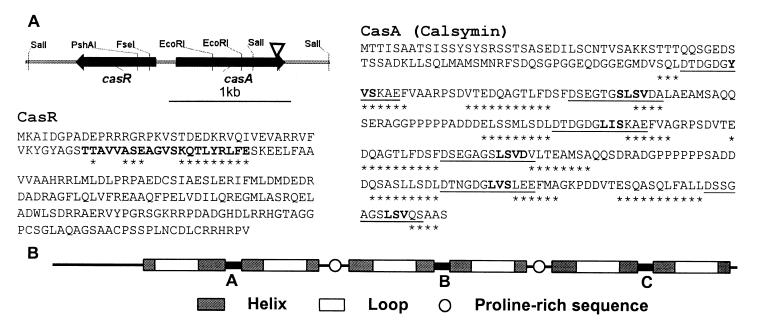
Sequence analysis of a 2442-bp segment containing the region flanking the mTn5gusA-pgfp21 insertion in mutant FAJ1806 indicated the presence of two divergently transcribed genes (Fig. 1A). The casA gene (calmodulin-like symbiosis gene A), which was inactivated in FAJ1806, codes for a protein of 293 amino acids (Mr 29,891) that is highly acidic (pI 3.6) (Fig. 1A). This protein, named calsymin, is composed of three domains (A, B, and C), containing 57 or 58 amino acids, structurally similar to the two domains of calmodulin, and a fourth nonhomologous N-terminal domain (Fig. 1B). Each of the three domains of calsymin contains two EF-hand helix–loop–helix structural motifs (Fig. 1). The 12-residue Ca2+-binding loops I, III, and V of calsymin contain the calmodulin hallmark residues at positions 1, 3, 5, 7, 9, and 12. These residues provide one (positions 1, 3, 5, 7, and 9) or two (position 12) oxygen ligands for the coordination of Ca2+ in calmodulin. Loops II, IV, and VI are atypical because of the absence of the conserved glutamic acid bidendate ligand at position 12. The conserved glycine residue at position 6, enabling side-chain and main-chain ligations at positions 5 and 7, respectively, is found in the six Ca2+-binding loops of calsymin. Structure predictions indicate the presence of short β-strands of 3 or 4 residues in calsymin at positions 7–11 in all loops (Fig. 1A). In calmodulin, two β-strands (residues 7–9 in both loops) from each domain interact and form an antiparallel sheet structure allowing cooperativity of Ca2+ binding. The amino acid sequence is more conserved between alternate loops (I, III, V and II, IV, VI) than between adjacent loops, similar to what is observed in calmodulin. In addition, highly similar linkers connect domains A to B (20 amino acids between loops II and III) and B to C (22 amino acids between IV and V). Calsymin may therefore have arisen by successive duplications and fusions of an ancestral gene coding for one Ca2+-binding domain giving rise to a protein composed of three homologous domains.
Figure 1.
Sequence and structure of the R. etli CasA (calsymin) and CasR proteins. (A) Schematic representation and physical map of the divergently transcribed casA and casR genes. The arrowhead indicates the position of the mTn5 insertion in the casA mutant FAJ1806. Predicted products of the casA and casR genes showing the six putative EF-hand calcium-binding sites of CasA and the helix–turn–helix structure of CasR. The secondary structure predictions are based on the algorithm of Geourjon and Deleage (28). Amino acids in boldface in the CasR sequence represent the conserved helix–turn–helix structure as found in TetR-related proteins (27). The amino acids in the CasR and CasA proteins marked with asterisks are part of predicted α-helices. Underlined amino acids in CasA are similar to EF-hand calcium-binding motifs (10). Amino acids forming putative short β-strands within the loops are in boldface. (B) Schematic representation of the three homologous calcium-binding domains of calsymin and localization of the EF-hand helix–loop–helix calcium-binding motifs and proline-rich domains.
Calsymin also possesses two proline-rich stretches, between amino acids 148–152 and 226–231, with five and six consecutive proline residues, respectively. These sequences contain the SH3 domain core recognition consensus sequence Pro-Xaa-Xaa-Pro (Xaa is any amino acid) (25). In the case of class I peptides, an arginine residue is found three amino acids N-terminal from the core recognition sequence. Also in calsymin, arginines are located N-terminal from the core sequences; however, they are separated by three amino acids instead of two. Differences in the number of arginine residues are known to affect the affinity for the SH3 domain (26).
A divergently transcribed gene, casR (R, repressor), is located in the region upstream from casA (Fig. 1A). The casR gene product contains 214 amino acids (Mr 24,240) and is homologous to transcriptional repressors belonging to the TetR family of bacterial regulatory proteins [21% identical and 28% conserved residues with TetR(E), accession no. X14035.1] (Fig. 1A). Sequence conservation is particularly strong in the helix–turn–helix structure localized in the N-terminal region of the protein and responsible for DNA binding in the TetR protein (27).
Symbiotic Phenotype.
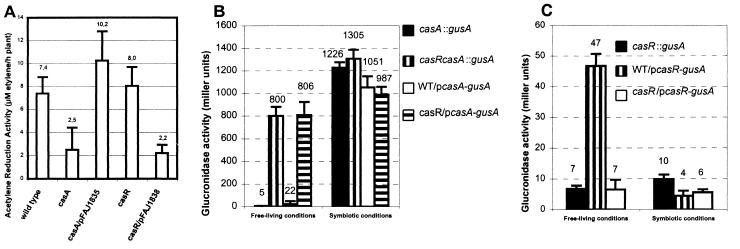
To study the role of calsymin during symbiosis with P. vulgaris, R. etli casA and casR insertional mutants were constructed and tested on plant. Symbiotic acetylene reduction activity (ARA), a measure for nitrogen fixation, was reduced by approximately 70% in nodules infected with the casA mutant strain versus wild type (Fig. 2A). A similar phenotype was observed when multiple copies of the casR gene, supplied on a multicopy plasmid, were present (Fig. 2A), which is in agreement with the postulated role of CasR as a repressor of CasA (see below). The ARA phenotype of a casA mutant could be restored to the wild-type level by complementation. No effect of a casR mutation on ARA was observed (Fig. 2A). Nodule numbers on plants inoculated with R. etli wild type or casA or casR mutants were not statistically different.
Figure 2.
Symbiotic phenotype of mutants and expression of R. etli casA and casR genes. (A) Nitrogen fixation of P. vulgaris plants inoculated with the R. etli wild-type strain, casA (FAJ1804) and casR (FAJ1802) mutant strains, and their complemented derivatives. Values are the mean ± SD (n = 10). The means are indicated above the bars. Plasmids pFAJ1835 and pFAJ1838 carry casA and casR genes, respectively. Similar results were obtained between strains FAJ1802 and FAJ1803 (casR mutants) and between strains FAJ1804 and FAJ1805 (casA mutants). (B and C) Expression of fusions between gusA and the casA (B) and casR (C) genes in free-living aerobic cultures (AMS) medium (13) supplemented with 10 mM sodium succinate, OD595 = 0.1–0.2, read with 100 μl culture in a Versamax microplate reader (Molecular Devices) and during symbiosis. Direct comparison of GusA activities under the two conditions may be difficult. Expressions were determined by using plasmid-borne PcasA–gusA (pFAJ1842) and PcasR–gusA (pFAJ1843) fusions and chromosomally located casA∷gusA (FAJ1806 and FAJ1807) and casR∷gusA (FAJ1809) fusions. β-Glucuronidase activities are expressed in Miller units. Values are the mean ± SD (n = 3).
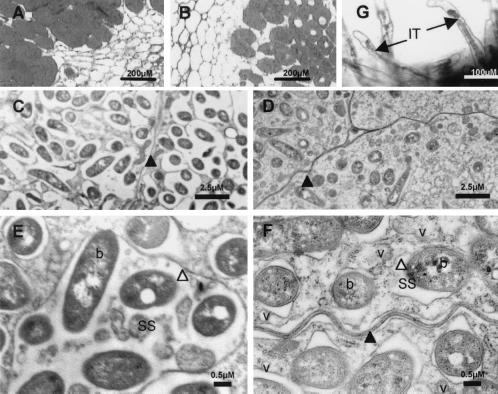
Light microscopic examination of stained nodule sections suggested a clearly reduced number of bacteroids in nodules infected with casA mutants compared with those infected with wild-type R. etli (Fig. 3 A and B). This was confirmed by a transmission electron microscopic analysis showing a reduction of the number of bacteroids by approximately 40% in plant cells colonized by the casA mutant (3.3 bacteroids per 10 μm2, sample standard deviation 1.1, 3 samples of 750 μm2 each analyzed) compared with the wild type (5.7 bacteroids per 10 μm2, standard deviation 0.7), 12 days after inoculation. Furthermore, these images indicated that casA mutant bacteroids are packed individually in the symbiosome membrane (SM), whereas wild-type symbiosomes often contain multiple (2–10) bacteroids (Fig. 3 C–F). The SM was always found closely associated with the bacteroid membrane of casA mutants. As a result, the casA mutant symbiosomes were virtually lacking any symbiosome space (Fig. 3F). We can, however, not exclude that the observed differences are reinforced by the experimental protocol used.
Figure 3.
Microscopic analysis of nodules and infection threads formed by R. etli wild-type (A, C, and E) and casA mutant FAJ1805 (B, D, F, and G) strains. (A and B) Toluidine blue stainings of 3-μm-thick sections of P. vulgaris nodules. (C–F) Transmission electron micrographs of 3-week-old nodules. (G) GusA staining of FAJ1806 bacteria expressing a casA–gusA fusion inside the infection threads. Black arrowhead, plant plasma membrane; white arrowhead, symbiosome membrane; black arrow, infection thread (IT); b, bacteroid; SS, symbiosome space; v, vesicle.
Expression Patterns.
The expression patterns of casA are consistent with a role during symbiosis. In the absence of the plant, expression of chromosomally integrated as well as plasmid-borne fusions between the casA promoter and gusA (PcasA–gusA) in a wild-type or a casA mutant background was repressed under aerobic or microaerobic (0.3% oxygen) conditions in the absence or presence of nodule extracts (Fig. 2B). Inhibition of casA expression under free-living conditions is relieved by the inactivation of casR, indicating that the expression of casA is negatively controlled by CasR (Fig. 2B). At the moment we cannot exclude that the casA gene could be induced under yet-unknown specific free-living growth conditions.
In contrast, plasmid-borne or chromosomal casA-gusA fusions in a wild-type or a casA mutant background are strongly expressed on the root surface of P. vulgaris plants approximately 1 day after inoculation (not shown). The expression is specific for P. vulgaris plants as no induction of the fusions was observed under similar conditions on Medicago sativa or wheat rootlets. GusA activity of PcasA–gusA was maintained in the infection threads (Fig. 3G) and inside the nodules of P. vulgaris (Fig. 2B).
Under free-living conditions, a plasmid-borne casR-gusA fusion is expressed in the wild type but GusA activity is strongly reduced in the casR mutants. Therefore, expression of casR is positively autoregulated permitting efficient repression of casA in the absence of the plant (Fig. 2C). The expression of casR-gusA was also reduced during symbiosis both in the wild type and in the casR mutant. Likely, under these conditions, the activator function of CasR on casR expression is inhibited. Expression of the casR gene occurred independently of the oxygen tension (results not shown).
Plant root expression of casA occurs independently of nod gene regulation as the activity of a PcasA–gusA (pFAJ1842) fusion is not reduced in a pSym-cured R. etli background whereas, under the same conditions, expression of a PnodA–gusA fusion is reduced to background levels (results not shown). Also, no effect of a mutation in fixL, nifA, and fnrN, regulators of symbiotic gene expression in R. etli, on casA expression was observed. In addition, expression of the casA–gusA fusion was not induced by a low oxygen tension as it is the case with nif/fix genes (results not shown).
Calsymin Is a Calcium-Binding and Secreted Protein.
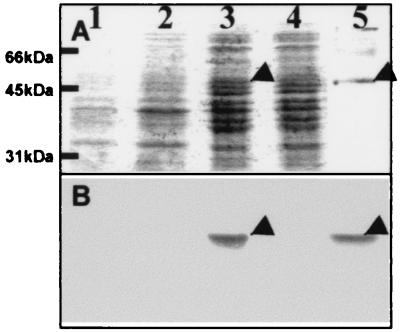
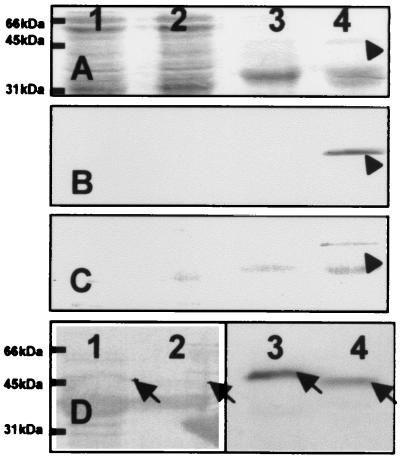
The Ca2+-binding activity of calsymin was demonstrated on overexpressed and purified protein from E. coli after polyacrylamide gel electrophoresis (PAGE) and transfer to a nylon membrane, using a 45Ca2+ overlay technique (22) (Fig. 4). Ca2+-binding activity of calsymin was not abolished/or could be restored after treatment by several protein denaturing treatments, including boiling, trichloroacetic acid precipitation, and denaturing PAGE. Unexpectedly, when overproduced in an R. etli casR mutant, 45Ca2+-binding activity was detected only in the growth-medium supernatant, indicating that the protein was secreted (Fig. 5B). The same result was obtained when a ruthenium red binding assay was used (Fig. 5C). The identity of the secreted 45Ca2+-binding protein as calsymin was confirmed by N-terminal sequence analysis. The amino acid sequence obtained, TTISAATSISSYSY, corresponds to the predicted N terminus of the protein based on the DNA sequence. Calsymin is therefore secreted from R. etli without cleavage of the casA gene product. Calsymin migrates on PAGE with a molecular mass of approximately 45 kDa, slightly higher than the calculated molecular mass. Aberrant migration may be caused by the acidity of the protein as noticed previously for other calmodulin-like proteins. Secretion of calsymin is efficient, because the protein could never be detected inside of cultured cells carrying a casR mutation. In agreement with the expression analysis of casA, no calsymin was secreted from R. etli wild-type cultures. In the mutant strain FAJ1808 (casR mutant derived from FAJ1806), the miniTn5 insertion is located in codon 280 in the loop of the sixth calcium-binding site of the casA gene and therefore lacks the 13 C-terminal amino acids found in wild-type calsymin. As a result, the truncated form has a slightly reduced molecular mass (Fig. 5D). However, this truncated protein is still able to bind calcium and is secreted in the growth medium (Fig. 5D). Overproduction of CasA in free-living R. etli cells does not produce any obvious phenotype. Calsymin protein, purified from the casR mutant FAJ1803 culture supernatant, was not able to activate chicken NAD kinase.
Figure 4.
Overexpression and calcium binding of recombinant calsymin in E. coli. Coomassie brilliant blue staining (A) and autoradiograph (B) after incubation with 45Ca2+, of protein profiles immobilized on a PVDF membrane. Crude protein extracts were obtained from the culture supernatant (lanes 1 and 2) or from the cell pellet (lanes 3 and 4). Exponential cell cultures were either uninduced (lanes 2 and 4) or induced with 0.2% arabinose for 6 h (lanes 1 and 3) before harvesting. The recombinant protein was extracted under denaturing conditions from a crude protein extract of E. coli Top10 cells (Invitrogen) induced with arabinose and purified by Ni-chelation chromatography (lane 5). The calsymin band is marked with an arrowhead.
Figure 5.
Calcium binding and secretion of calsymin. (A–C) Staining of whole-cell proteins (lanes 1 and 2) and proteins secreted into the culture supernatant (lanes 3 and 4) from the R. etli wild-type CNPAF512 (lanes 1 and 3) and the casR mutant strain FAJ1803 (lanes 2 and 4). Proteins were subjected to electrophoresis on an SDS/polyacrylamide gel and transferred to PVDF membranes by electroblotting. The membranes were stained with Coomassie brilliant blue (A) or incubated with 45Ca2+ and autoradiographed (B), or the calcium-binding proteins were stained with ruthenium red (C). The protein band indicated with an arrow was identified as calsymin by N-terminal sequence analysis of the isolated protein. (D) Calcium binding and secretion of a truncated calsymin protein from R. etli cultures. Proteins were isolated from the culture supernatant of R. etli FAJ1803 (lanes 1 and 3) and strain casA∷mTn5gusA-pgfp21 casR∷Ω-Spc FAJ1808 (lanes 2 and 4). The membrane was stained with Coomassie brilliant blue (lanes 1 and 2) or incubated with 45Ca2+ and autoradiographed (lanes 3 and 4). The wild-type (lanes 1 and 3) and truncated (lanes 2 and 4) calsymin proteins are marked with an arrow.
Discussion
Two divergently transcribed genes, casA and casR, were identified in R. etli. Transcription of the casA gene is negatively controlled by the gene product of casR. The CasR repressor belongs to the TetR family of regulators. In the case of TetR, a helix–turn–helix motif on the N terminus is responsible for protein binding to an operator sequence in the tetA promoter, which inhibits transcription of the latter gene (27). Recognition of the antibiotic metabolite tetracycline by TetR causes dissociation of the repressor–DNA complex and induces transcription of the tetA gene, conferring tetracycline resistance. Other members of this family, including BarA from Streptomyces virginiae and ArpA from Streptomyces griseus, also bind to small effector molecules such as the butyrolactone autoregulators IM-2 and the A-factor. The casA gene is expressed on the roots of P. vulgaris plants, in the infection threads, and inside the bacteroids but not under the free-living conditions tested, suggesting that the inducing compound is plant derived. The expression pattern of casA is not altered in a pSym-cured R. etli strain and was not affected by mutations in nif and fix regulatory genes. Therefore, casA regulation occurs independently of known regulatory mechanisms of nodulation and nitrogen fixation genes and constitutes a type of symbiotic regulation in R. etli that has not been described previously.
The casA mutant was affected in the production of a calmodulin-like protein named calsymin. Reports on this type of protein in prokaryotes are very uncommon, and, to our knowledge, calsymin constitutes the first example of a calmodulin-like protein with canonical EF-hand motifs in a Gram-negative bacterium. In other prokaryotes, proteins with calmodulin-like properties have been reported previously (11, 29). However, the amino acid sequences of the corresponding proteins are still unknown. In the Gram-positive bacterium Saccharopolyspora erythraea, a 20-kDa calcium-binding protein, named calerythrin, was previously shown to possess a structural organization similar to calmodulin with four potential helix–loop–helix EF-hand motifs (12). More specifically, the protein belongs to the subfamily of eukaryotic sarcoplasmic Ca2+-binding proteins and might therefore function as an intracellular calcium buffer (30).
Calsymin belongs to the calmodulin superfamily. Calsymin possesses a three-domain structure similar to the organization of calbindin D28k and calretinin. Pairs of EF hands in each of the domains of calsymin may interact through short β-strands present in the loops and form a compact structure. Proteins belonging to the calmodulin superfamily are often divided into sensor and buffer proteins. The latter class may act as calcium buffers in the cell and control the concentration of free cytosolic Ca2+. The sensor proteins change their conformation upon binding Ca2+ and interact directly with target proteins to modulate their activity. It is presently unclear to which class calsymin belongs.
Other calcium-binding rhizobial proteins have been implicated in symbiosis previously (31, 32). Secretion of the calcium-binding NodO protein requires a specialized secretion system homologous with ABC transporters involved in the secretion RTX proteins (32). N-terminal amino acid sequence analysis of calsymin isolated from the culture supernatant indicated that, similarly to NodO, calsymin is secreted without cleavage of an N-terminal transit peptide. The mechanism by which calsymin is secreted is presently unknown. However, the observation that calsymin possesses an N-terminal region of approximately 100 amino acids that is different from the three other homologous Ca2+-binding domains suggests that determinants for secretion might be located within the N-terminal part of the protein. Also, in mutant FAJ1808, a truncated calsymin protein lacking part of the C terminus is still efficiently secreted.
Calcium has been implicated in a number of symbiosome functions, arguing for an important role of this ion in this compartment (33, 34). Complexation of Ca2+ by calsymin could have a direct or indirect effect on Ca2+-dependent processes in the plant. The identified gene may therefore constitute a valuable tool to study the communication between plant and bacteria at the late stages of symbiosis.
Acknowledgments
We thank Dr. W. Broekaert, Dr. J. Cox, and Dr. F. Wuytack for the critical reading of the manuscript and their valuable comments, and Dr. P. Proost for the sequence analysis of the N terminus of calsymin. C.X. is a recipient of a doctoral scholarship from the Research Council of the Katholieke Universiteit Leuven. J.M. and E.S. are postdoctoral fellows of the Fund for Scientific Research–Flanders.
Abbreviations
- spcR
spectinomycin resistance
- kmR
kanamycin resistance
- PVDF
poly(vinylidene difluoride)
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF288533).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210181097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210181097
References
- 1.Dénarié J, Debellé F, Promé J C. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 2.Long S R. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Rhijn P, Vanderleyden J. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohn J, Day R B, Stacey G. Trends Plant Sci. 1998;3:105–110. [Google Scholar]
- 5.Schultze M, Kondorosi A. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- 6.Clapham D E. Cell. 1995;80:259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- 7.Felle H H, Kondorosi E, Kondorosi A, Schultze M. Plant J. 1998;13:455–463. [Google Scholar]
- 8.Ehrhardt D W, Wais R, Long S R. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 9.Crivici A, Ikura M. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 10.Strynadka N C, James M N. Annu Rev Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 11.Norris V, Grant S, Freestone P, Canvin J, Sheikh F N, Toth I, Trinei M, Modha K, Norman R I. J Bacteriol. 1996;178:3677–3682. doi: 10.1128/jb.178.13.3677-3682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swan D G, Hale R S, Dhillon N, Leadlay P F. Nature (London) 1987;329:84–85. doi: 10.1038/329084a0. [DOI] [PubMed] [Google Scholar]
- 13.Michiels J, Van Soom T, D'hooghe I, Dombrecht B, Benhassine T, de Wilde P, Vanderleyden J. J Bacteriol. 1998;180:1729–1740. doi: 10.1128/jb.180.7.1729-1740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi C, Lambrecht M, Vanderleyden J, Michiels J. J Microbiol Methods. 1999;35:85–92. doi: 10.1016/s0167-7012(98)00103-1. [DOI] [PubMed] [Google Scholar]
- 15.Xi, C., Dirix, G., Hofkens, J., De Schrijver, F. C., Vanderleyden, J. & Michiels, J. (2000) Microb. Ecol., in press. [DOI] [PubMed]
- 16.Michiels J, Dombrecht B, Vermerien N, Xi C, Luyten E, Vanderleyden J. FEMS Microbiol Ecol. 1998;26:193–205. [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Fellay R, Frey J, Krisch H. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 19.Quandt J, Hynes M F. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 20.Herrero M, de Lorenzo V, Timmis K N. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staskawicz B, Dahlbeck D, Keen N, Napoli C. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruyama K, Mikawa T, Ebashi S. J Biochem (Tokyo) 1984;95:511–519. doi: 10.1093/oxfordjournals.jbchem.a134633. [DOI] [PubMed] [Google Scholar]
- 23.Charuk J H, Pirraglia C A, Reithmeier R A. Anal Biochem. 1990;188:123–131. doi: 10.1016/0003-2697(90)90539-l. [DOI] [PubMed] [Google Scholar]
- 24.Harmon A C, Jarrett H W, Cormier M J. Anal Biochem. 1984;141:168–178. doi: 10.1016/0003-2697(84)90441-x. [DOI] [PubMed] [Google Scholar]
- 25.Ren R, Mayer B J, Cicchetti P, Baltimore D. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 26.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 27.Hillen W, Berens C. Annu Rev Microbiol. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 28.Geourjon C, Deleage G. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 29.Onek L A, Smith R J. J Gen Microbiol. 1992;138:1039–1049. doi: 10.1099/00221287-138-6-1039. [DOI] [PubMed] [Google Scholar]
- 30.Cox J A, Bairoch A. Nature (London) 1988;331:491. doi: 10.1038/331491a0. (lett.). [DOI] [PubMed] [Google Scholar]
- 31.Smit G, Logman T J, Boerrigter M E, Kijne J W, Lugtenberg B J. J Bacteriol. 1989;171:4054–4062. doi: 10.1128/jb.171.7.4054-4062.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finnie C, Hartley N M, Findlay K C, Downie J A. Mol Microbiol. 1997;25:135–146. doi: 10.1046/j.1365-2958.1997.4471803.x. [DOI] [PubMed] [Google Scholar]
- 33.Weaver C D, Roberts D M. Biochemistry. 1992;31:8954–8959. doi: 10.1021/bi00152a035. [DOI] [PubMed] [Google Scholar]
- 34.Tyerman S D, Whitehead L F, Day D A. Nature (London) 1995;378:629–632. [Google Scholar]