Abstract
In Streptococcus pneumoniae, stkP and phpP, encoding the eukaryotic-type serine-threonine kinase and PP2C phosphatase, respectively, form an operon. PhpP has the features of a so-called “soluble” protein, whereas StkP protein is membrane associated. Here we provide the first genetic and physiological evidence that PhpP and StkP, with antagonist enzymatic activities, constitute a signaling couple. The StkP-PhpP couple signals competence upstream of the competence-specific histidine kinase ComD, receptor for the oligopeptide pheromone “competence stimulating peptide.” We show that PhpP activity is essential in a stkP+ genetic background, suggesting tight control of StkP activity by PhpP. Proteins PhpP and StkP colocalized to the cell membrane subcellular fraction and likely belong to the same complex, as revealed by coimmunoprecipitation in cellular extracts. Specific coimmunoprecipitation of the N-kinase domain of StkP and PhpP recombinant proteins by PhpP-specific antibodies demonstrates direct interaction between these proteins. Consistently, flow cytometry analysis allowed the determination of the cytoplasmic localization of PhpP and of the N-terminal kinase domain of StkP, in contrast to the periplasmic localization of the StkP C-terminal PASTA (penicillin-binding protein and serine-threonine kinase associated) domain. A signaling route involving interplay between serine, threonine, and histidine phosphorylation is thus described for the first time in this human pathogen.
Signal transduction via the transfer of phosphoryl groups and transient protein phosphorylation involves the concerted activities of kinases and phosphatases and controls various cellular functions (for a review, see reference 18). In eubacteria, the histidine kinases of two-component systems (TCSs) and the phosphatases with which they interact are involved in cellular adaptation to environmental conditions (for a review, see reference 44). Serine-threonine kinases and PP2C phosphatases also contribute to regulatory and developmental processes, as described in Bacillus subtilis (1, 2, 13, 23, 29, 33, 43). In mycobacteria, a membrane-associated PP2C phosphatase controls the activity of several serine-threonine kinases (4, 6, 52), and the transcriptional activator EmbR has been identified as a phosphorylation target for PknH kinase (3, 4, 42). In Streptococcus agalactiae, the signaling pair Stk1-Stp1 plays a role in virulence, by modifying cytotoxin production and purine metabolism (34, 35, 36). In Streptococcus pyogenes, a histone-like protein is phosphorylated by the serine-threonine kinase, generating a substrate for the kinase-related phosphatase (21). The human pathogen Streptococcus pneumoniae has only one gene encoding a PP2C-type phosphatase, PhpP, located upstream from the stkP gene, which encodes the only membrane-associated serine-threonine kinase, StkP (17). The phpP and stkP genes overlap by 4 bp and form an operon (31). In a mouse model of infection, null mutations affecting StkP greatly attenuate tissue and bloodstream invasion (17). In cultures, these mutations are highly pleiotropic, presenting a notably important impact on competence development for genetic transformation (17, 40). In vitro studies have shown that autophosphorylated recombinant StkP (StkP-P) is a substrate for recombinant PhpP, suggesting that StkP and PhpP may function in a coordinated manner (31). However, the regulators controlling the cellular level of StkP-P and the network involving StkP in growing cultures remain ill defined. Recent studies of strain TIGR4 revealed that the transcriptional regulator RitR (45) is phosphorylated by StkP and dephosphorylated by PhpP (50). In order to get better insight into StkP signaling, we investigated the specific role of PhpP phosphatase on bacterial growth and on the development of competence for genetic transformation in cultures, by using genetic analysis. Specific null mutation in PhpP was not obtained, indicating the essentiality of the phosphatase in the control of StkP activity. It is thus likely that PhpP and StkP constitute a functional couple. Data on cellular organization and coimmunoprecipitation suggest that PhpP and StkP proteins can interact within a submembrane complex facing the cytoplasm. Furthermore, results from physiological and genetic studies demonstrate that the StkP-PhpP couple belongs to the competence-signaling network, upstream of the ComDE TCS target for the competence stimulating peptide pheromone, CSP.
(Part of this work was presented at the Europneumo meeting, Oreiras, Portugal, April 2007; at the Gram+ meeting, Pisa, Italy, June 2007; and at the Experimental Workshop on Signal Transduction in Host-Bacterial Interactions, Jerusalem, Israel, October 2007.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Streptococcus pneumoniae strains described in Table 1 were grown in casein tryptone medium (CAT), and the cultures were stored at −80°C in 12% (vol/vol) glycerol. For competence studies, bacteria from frozen cultures were grown in CAT transformation medium (CTM), made from CAT medium supplemented with 0.2% (wt/vol) bovine serum albumin (BSA) and 0.1 mM CaCl2. The pH was then adjusted to 6.5 or 8 (5, 9, 49). S. pneumoniae transformants were selected on appropriate antibiotics: kanamycin, 50 mg liter−1; rifampin (rifampicin), 2 mg liter−1; and chloramphenicol, 10 mg liter−1. The Escherichia coli strains described in Table 1 were propagated in Luria broth in the presence of appropriate antibiotics: ampicillin, 100 mg liter−1; kanamycin, 50 mg liter−1; and rifampin, 400 mg liter−1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype | Phenotype | Source or reference |
|---|---|---|---|
| Strain | |||
| E. coli | |||
| XL1-Blue | F′::Tn10 proA+B+ lacIqΔ(lacZ)M15/recA1 endA1 gyrA96 (Nalr) thi hsdR17(rK− mK+) supE44 relA1 lac | Stratagene | |
| BL21(DE3) | F−ompT gal [dcm] [lon] hsdSB(rB− mB−) (DE3) | Novagen | |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen | |
| S. pneumoniae | |||
| Cp1015 | RX derivative; str1 hexA | Smr | 30 |
| Cp1016 | Cp1015, but rif23 and stgF | Rifr, Stgr | 5 |
| R119 | R6 hex, but rif23 stgF str1 fusA nov1 opt12 gln tdr | Smr, Fusr, Rifr, Stgr, Novr, Optr | 46 |
| Cp1008 | Cp1015, but comA::spc | Spr | 30 |
| Cp1095 | Cp1015, but lytA::ery | Eryr | 47, 49 |
| Cp7000 | Cp1015, but ΔstkP::cat | Camr | 31 |
| Cp9000 | Cp1015, but stkP::aphA-3 | Kmr | 17 |
| Cp9090 | Cp1095, but stkP::aphA-3 | Kmr, Eryr | 17 |
| Cp7004 | Cp7000, but phpPD1 | Camr | This work |
| Cp9091 | Cp9090, but stkP+ | Kans | This work |
| Cp6600 | Cp1015, but comD(D299N) rif23 | Rifr | 14 |
| Cp6607 | Cp6600, but ΔstkP::cat | Rifr, Camr | This work |
| Plasmid | |||
| pCR2.1TOPO | Cloning vector | Ampr, Kmr | Invitrogen |
| pWSK29 | Cloning vector derived from pBluescript | Ampr | 51 |
| pET28b | His tag expression vector; Kanr, f1 ori | Kmr | Novagen |
| pBluescript ISK+/KS+ | Ampr, lacZ, f1± ori, ColE1 ori | Ampr | Stratagene |
| pPJ1 | pUC derivative containing a HincII fragment carrying the kanamycin resistance gene | Ampr, Kmr | 32 |
| pPHK29 | 2.96-kb EcoRI/SalI amplicon (primers PHL and PCR) from S. pneumoniae Cp1015 DNA containing phpP and stkP genes cloned into pWSK29 | Ampr | 17 |
| pPK29 | 2.3-kb EcoRI/SalI amplicon (primers PCL and PCR) from S. pneumoniae Cp1015 DNA containing stkP gene in pWSK29 | Ampr | This work |
| pEXstkP-N | 0.825-kb NdeI/EcoRI amplicon (primers STKP-F and STKP-RT) from pPHK29 containing fragment (kinase domain) of stkP gene inserted into pET28b | Kmr | This work |
| pEX-PhpP | 0.74-kb amplicon (phpP) inserted into pET28b | Kmr | 31 |
| pEX-PASTA | 0.9-kb NcoI/EcoRI amplicon (primers STKP-PF and STKP-R2) from pPHK29 containing stkP gene inserted into pET28b | Kmr | This work |
| pMUT1 | 2.6-kb amplicon of phpPD1 stkP+ | Kmr | This work |
| pMUT2 | 2.6-kb amplicon of phpPD2 stkP+ | Kmr | This work |
| pMUT3 | 2.6-kb amplicon of phpPD1 ΔstkP::cat | Kmr | This work |
| pMUT4 | 2.6-kb amplicon of phpPD2 ΔstkP::cat | Kmr | This work |
Construction of mutagenic plasmids and mutant strains.
Escherichia coli strains XL1-Blue and Top10 were used as hosts for recombinant vectors (Table 1). Isogenic mutant strains of S. pneumoniae were generated by allelic exchange. Briefly, the S. pneumoniae recipient strain was transformed with nonreplicative chimeric plasmids carrying the relevant pneumococcal allele(s). In the absence of direct selection with antibiotics, 40 clones were selected and the chromosomal DNA of each clone was extracted with Instagene Matrix (Bio-Rad) for diagnostic PCR and restriction analysis of the corresponding amplicon (5). One clone carrying the relevant mutation was verified by DNA sequencing with the primers listed in Table 2 and was maintained under standard growth conditions. The insertion/deletion mutation of stkP has been described elsewhere (31) and was used to construct strain Cp7000, carrying the ΔstkP::cat allele. Plasmids pMUT1 to -4 were used as DNA donors in transformation of the wild-type strain Cp1015, in order to obtain the single mutants phpPD1 and phpPD2 and the double mutants phpPD1 stkP::cat and phpPD2 stkP::cat. The phpPD1 allele containing the point mutation D241A and a StuI restriction site was associated with stkP+ or ΔstkP::cat DNA fragments, respectively, and the phpPD2 allele carrying only the StuI restriction site was associated with either stkP+ or ΔstkP::cat DNA fragments to create plasmids pMUT1 to -4 (Table 1). Plasmids pMUT1 to -4 were constructed using standard protocols (39). The 5′ region of phpP was amplified from the genomic DNA of Cp1015 with the UFKFP and MUT4 primers, and the 3′ region of phpP was amplified with the MUT2 and DSKR primers. The amplified fragments were mixed and fused with UFKFP and DSKR by a second PCR. The fused fragment was inserted into pCR2.1TOPO to yield pMUT1. Similarly, a DNA fragment carrying only the StuI restriction site was amplified by PCR from the genomic DNA of Cp1015, using the UFKFP-plus-MUT3 and MUT5-plus-DSKR primer combinations, to yield pMUT2. Similarly, using Cp7000 genomic DNA as a template and primers MUT2 to -5, UFKFP, and DFKRP, we constructed pMUT3 and pMUT4. The sequences of all mutated DNA fragments were verified using universal primers and specific primers of the generated sequences (Table 2).
TABLE 2.
Primers
| Name | Sequence | Unique restriction site |
|---|---|---|
| DSKR | 5′-CGGATAAATTTTTCCACATAGAGG | |
| UFKFP | 5′-CGCAAGATATCGGATTAGGAA | |
| MUT2 | 5′-GCAGGAGGCCTAGCCAACATTACG | StuI |
| MUT4 | 5′-AATGTTGGCTAGGCCTCCTGC | StuI |
| MUT3 | 5′-GCAGGAGGCCTAGACAACATTACG | StuI |
| MUT5 | 5′-AATGTTGTCTAGGCCTCCTGC | StuI |
| DFKRP | 5′-CGCGGATCCTCATAATATCACGGACCGCAT | |
| stkP-PF | 5′-AGGATGCCATATGAGATCTCCTGCAACCATTGCCATT | NdeI |
| stkP-R2 | 5′-TTGATTATGAATTCGCTTTTAAGGAGTAGCTGAAGTTG | EcoRI |
| STKP-RT | 5′-GTAGGACAGAATTCAAGACAAGTCTACATAA | EcoRI |
| STKP-F | 5′-AGGATGCCATATGATCCAAATCGGCAA | NdeI |
| PCL | 5′-CCAGCTGAATCCAGGCAGTGAGATTCGTG | EcoRI |
| PCR | 5′-CCATACGTCGACCGGCATTTAATTCTTTTG | SalI |
Production of specific rabbit antibodies.
Specific antibodies against PhpP phosphatase, the kinase domain (StkP-N), and the PASTA (penicillin-binding protein and serine-threonine kinase associated) domain of StkP were produced by Eurogenentec (France), using the corresponding recombinant proteins produced in E. coli. Recombinant proteins were obtained as oligohistidine fusions in the E. coli BL21(DE3) strain (Table 1) and purified on TALON metal affinity resin (Clontech) according to the manufacturer's protocol. Antibodies that rose against PhpP or the kinase or PASTA domain and the corresponding preimmune sera were purified on Tgel (Pierce) according to the manufacturer's instructions.
Cellular fractionation and localization of StkP and PhpP.
Cultures of the wild-type strain and the StkP null mutant were collected by centrifugation at 4°C, washed with phosphate-buffered saline, resuspended in phosphate-buffered saline, and sonicated. Subcellular fractions were obtained by centrifugation (100,000 × g for 1 h), resulting in a pellet fraction containing cellular envelopes and in a supernatant fraction containing the cytoplasm. Proteins (10 μg) from each fraction were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto nitrocellulose, probed with specific antibodies, and detected with an ECL system (Amersham). Specific membrane localization of StkP has already been demonstrated (31) and has been considered the fractionation control. Nox immunoblots have been used as the loading control, because this protein is found in both the membrane and the cytoplasmic fractions (5; also our unpublished data).
Immunoprecipitation.
For cellular extract analysis, clarified lysates (500 μg of proteins in lysis buffer: 50 mM Tris-HCl, pH 8, 1% Igepal, and antiprotease cocktail from Roche) were incubated in the presence or absence of PhpP-specific antibodies (2 μg) for 18 h at 4°C. Recombinant proteins (2 μg in 150 μl of lysis buffer containing 0.5% [wt/vol] BSA) were incubated during 90 min at 4°C before the addition of 20 μg of PhpP antibodies or the corresponding preimmune serum and incubation at 4°C under gentle agitation was allowed during 18 h. Protein A/G-coated Bio-Adembeads were subsequently added and used as recommended by the supplier (Ademtech), by performing washing steps with the lysis or alternatively the RipA buffer from Ademtech. Elution was obtained in loading buffer in denaturizing and reducing conditions. Rabbit preimmune serum was used as a specificity control of immunoprecipitation, and nonspecific binding to the Bio-Adembeads was evaluated in antibody-free assays when indicated. Total samples and the fraction eluted from the Bio-Adembeads were loaded onto a 4 to 20% gradient gel and analyzed by Western blotting with antibodies raised against the recombinant PASTA and N-kinase domain proteins.
Flow cytometry analysis.
Bacteria and protoplasts were prepared from fresh cultures in CAT medium at an optical density at 400 nm (OD400) of 0.2. Protoplasts were obtained by treating exponentially growing cultures for 30 min at room temperature with 500 mg liter−1 of lysozyme, 20% (wt/vol) sucrose, 0.1 M Tris, and 10 mM MgSO4 at pH 7, as described elsewhere (48). Where required, permeabilization of the bacteria was established by treatment of a suspension of heat-killed bacteria (60°C for 30 min) with a Cytofix/Cytoperm kit from BD Biosciences, according to the manufacturer's instructions. Cell permeability was evaluated by assessing the chromosome labeling induced by incubation with fluorescent propidium iodide. For flow cytometry analysis, suspensions of 1 × 107 bacteria and protoplasts in 100 μl of 0.1 M Tris, 20% (wt/vol) sucrose, and 10 mM MgSO4 at pH 7 were incubated for 30 min at 4°C with antibodies (5 μg/ml) raised against the kinase domain, the PASTA domain, and PhpP phosphatase. Cells and protoplast suspensions were washed three times with 1 ml of Tris-sucrose-MgSO4 buffer and subsequently incubated for 30 min in the dark at 4°C with goat anti-rabbit-specific immunoglobulin G labeled with phycoerythrin. Two washes were performed with a 1-ml solution of Tris, sucrose, and MgSO4, and fluorescence was assessed in a Becton Dickinson FACSCalibur spectrofluorimeter (excitation at 488 nm and analysis at 585 nm).
Competence measurements.
The level of competence development in cultures was evaluated using the “transformation test,” as described elsewhere (14, 30). In brief, cultures in CTM rendered either acidic or alkaline (9) were incubated at 37°C. At an OD of 0.1 to 0.2, 100-μl aliquots were withdrawn and mixed with the DNA from strain Cp1016 or R119 (1 μg ml−1) used for transformation (Table 1). Cells were incubated for 20 min at 20°C, mixed with blood agar medium, and incubated for 2 h at 37°C. A layer of agar with or without selective antibiotics was then poured over the surface of the plate. Colonies were counted after overnight incubation at 37°C, and transformation efficiencies were calculated as the ratio of resistant transformed colonies to total viable counts. For CSP-induced competence, cultures in the early exponential growth phase (OD400 of 0.05) in CTM were activated by incubation with 0.1 to 100 ng/ml of CSP for 10 min at 37°C. Competence was assessed with the “transformation test,” as described above.
Mutation rate determination.
The mutation rates of strains Cp1015 and Cp7000 were evaluated with the P0 test of Luria and Delbruck (27). For each strain, 50 independent 1-ml cultures in CAT medium at pH 7 with a starting inoculum of 1 × 102 CFU (N0) and containing zero mutants resistant to 2 μg/ml of rifampin were cultured for 8 h at 37°C. At the end of the growth period, the total CFU content of the culture (N) was determined by plating serial dilutions from one culture of each strain and by plating the other cultures on blood agar supplemented with rifampin. The frequency of cultures containing 0 CFU on rifampin plates was determined (P0) and the mutation frequency (μ) was calculated using the following equation: μ = −ln [P0/(N − N0)].
Genes stkP/phpP were given NCBI registration number AF285441 on 7 July 2000. They are named pkn2/spr1577 (and phpP is named pppL/spr1578) in the R6 genome databases.
RESULTS
The essential nature of PhpP.
Biochemical studies of recombinant proteins have shown that phosphorylated StkP is a substrate for PhpP (31), although the cellular function of PhpP has not been elucidated. We carried out a mutational analysis to specifically assess the physiological role of PhpP phosphatase in S. pneumoniae. Attempts to construct an in-frame deletion of the phpP gene were unsuccessful, a result reminiscent of previous observations in S. pyogenes (21). In contrast, phpP mutants carrying cassette insertion and stop codon mutations in phpP leading to a downstream polar effect on stkP could be obtained (data not shown). These results suggested that functional PhpP was probably essential in the wild-type but not in the StkP− genetic background. In order to test this hypothesis, the wild-type strain Cp1015 was transformed with pMUT1 to -4 plasmids encoding either D231A PhpP (phpPD1), which has been shown to have no phosphatase activity in vitro (31), or a functional D231D PhpP (phpPD2) protein, associated with both the wild-type stkP and ΔstkP::cat allele, in the vicinity of a newly created StuI restriction site (see Materials and Methods). Transformed clones resulting in allelic exchange (“transformants”) harboring the StuI restriction site were detected and quantified. Transformants carrying the phpPD2 allele in addition to StuI were obtained with high frequency, regardless of the wild-type or mutant nature of the associated stkP allele on plasmids pMUT2 and pMUT4 (Table 3). In contrast, phpPD1 transformants were exclusively recovered from the transformations involving the pMUT3 plasmid carrying the ΔstkP::cat allele. Moreover, all these transformants were also resistant to chloramphenicol, demonstrating cotransformation of StuI and ΔstkP::cat mutations (Table 3). These results provide the genetic demonstration that phpPD1 encoding the D231A PhpP protein is lethal, specifically in the stkP+ genetic background, suggesting that the phosphatase activity of PhpP is essential in wild-type but not in StkP mutant bacteria. Our data imply that uncontrolled cellular levels of StkP-P are toxic in S. pneumoniae RX derivatives (Table 1). During bacterial growth, PhpP phosphatase may therefore play a key role in controlling phosphorylation levels of StkP and of its targets. This hypothesis incited us to assess the cellular organization of the membrane-associated serine-threonine kinase and the “so-called” soluble PhpP phosphatase in order to get further insight into the cellular role of the PhpP phosphatase with regard to StkP kinase.
TABLE 3.
Allele phpPD1 encoding inactive D231A PhpP phosphatase is lethal in the RX stkP+ genetic backgrounda
| Donor DNA | Transformed colonies for the relevant allele (% CFU)
|
||
|---|---|---|---|
| phpP | cat | rif23 | |
| pMUT1 phpPD1 stkP+ | 0 (40) | ||
| pMUT2 phpPD2 stkP+ | 75 (40) | ||
| pMUT3 phpPD1 ΔstkP::cat | 91 (35) | 100 (32) | |
| pMUT4 phpPD2 ΔstkP::cat | 97 (40) | ||
| R119 chromosomal DNA rif23 | 3 | ||
The recipient bacteria was Cp1015 in all cases. Parentheses indicate the number of colonies submitted to diagnostic PCR and StuI restriction (CFU). Cam, chloramphenicol; Rif, rifampin (both antibiotics were used at 2 μg/ml).
The kinase domains of StkP and PhpP proteins interact within a submembrane complex.
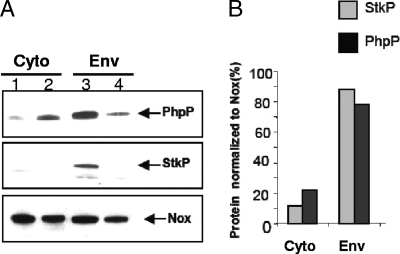
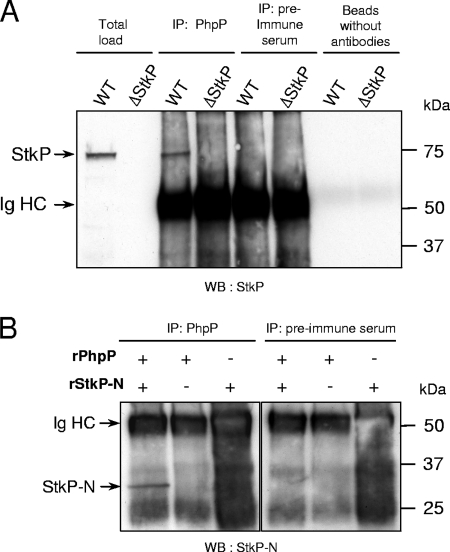
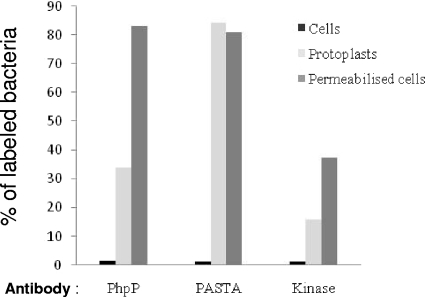
The membrane localization of StkP protein has been demonstrated elsewhere. In contrast, PhpP protein showed the features of a soluble protein (31). Western blot analysis of cellular envelopes and cytoplasmic subcellular fractions from the wild-type strain Cp1015, using specific antibodies directed against StkP or PhpP proteins, revealed that PhpP was mainly present in the envelopes in a manner similar to StkP. In contrast, in extracts from the stkP deletion strain Cp7000, PhpP was mainly present in the cytoplasmic fraction (Fig. 1). Cellular distribution of the Nox protein present both in the envelopes and in the cytoplasm (5; also our unpublished data) was not altered due to StkP mutation (Fig. 1), indicating that StkP protein affects the distribution of PhpP protein selectively. These results raised the hypothesis that StkP and PhpP may form a protein complex in living bacteria. This hypothesis was tested by coimmunoprecipitation experiments. Cleared bacterial lysates from exponential growing cultures of the wild-type and stkP mutant strains were incubated with antibodies directed against PhpP or the corresponding preimmune serum as indicated in Materials and Methods. Analysis of the immunocomplexes by Western blotting using StkP-specific antibodies clearly revealed a band at 72 kDa, corresponding to the StkP protein (calculated molecular mass of 72.4 kDa), specifically in the PhpP antibody assay. This result indicates coimmunoprecipitation of StkP protein by PhpP antibodies in cellular extracts (Fig. 2A). In a parallel experiment, we observed coimmunoprecipitation of the recombinant PhpP and N-kinase domain but not the C-terminal PASTA domain proteins by PhpP-specific antibodies, suggesting direct and specific interaction between the kinase module of StkP and PhpP proteins (Fig. 2B; also data not shown). Cellular localization of the proteins by flow cytometry analysis using suspensions of whole bacteria, protoplasts, and permeabilized cells (see Materials and Methods) provides credence to this hypothesis. The results presented in Fig. 3 show, on the one hand, that in the wild-type strain, the C-terminal PASTA domain of StkP can be efficiently labeled in protoplasts as well as in permeabilized cells, whereas the N-terminal kinase domain of StkP and PhpP proteins were labeled specifically in permeabilized cells. On the other hand, labeling efficiencies are very low in nonpermeabilized cells, suggesting that antibodies were hampered by the cell wall. These results suggest that the C-terminal PASTA domain of StkP sticks out in the periplasmic space, beneath the peptidoglycan cell wall, and the kinase domain is located in the cytoplasm, like PhpP is. Such a colocalization of the StkP kinase domain and PhpP proteins is consistent with the proposal of PhpP and its kinase module target being organized close together and interacting within a membrane-associated protein complex facing the cytoplasm. However, localization of PhpP protein in the cytoplasmic fraction of StkP mutant strains (Fig. 1) raises the question of the StkP-independent activity of PhpP accounting for their phenotype (17, 40).
FIG. 1.
Colocalization of StkP and PhpP in wild-type S. pneumoniae. (A) Cytoplasm (Cyto) and envelopes (Env) from sonicated bacteria were obtained as described in Materials and Methods. Proteins (10 μg) of each fraction from the wild-type strain Cp1015 (lanes 1 and 3) and the ΔstkP::cat mutant strain Cp7000 (lanes 2 and 4) were analyzed by Western blotting with kinase- and phosphatase-specific antibodies. Nox immunoblots were taken as controls. (B) Quantification of the immunoreactive material corresponding to StkP and PhpP in extracts from the wild-type strain Cp1015 has been obtained by reference to Nox-immunoreactive material. The data are representative of two independent experiments.
FIG. 2.
Coimmunoprecipitation of StkP and PhpP proteins. (A) Cellular lysates from the wild-type strain Cp1015 and the ΔstkP::cat mutant strain Cp7000 (Table 1), containing 500-μg proteins, were incubated with PhpP-specific antibodies (2 μg) or their corresponding preimmune serum in 150 μl of lysis buffer. The immunocomplexes were analyzed by Western blotting with N-kinase-specific antibodies as described in Materials and Methods. The “Beads” control was obtained in the absence of PhpP antibodies. The total load represents 0.4% of the assay. The data are representative of five independent experiments. (B) Recombinant PhpP and N-kinase domain proteins (2 μg) were mixed in 150 μl of lysis buffer supplemented with BSA [0.5% (wt/vol)] and incubated with PhpP antibodies (20 μg) or their corresponding preimmune serum. Controls were obtained with single protein assays containing either recombinant N-kinase protein or recombinant PhpP protein. Twenty percent of each sample was used for Western blot analysis with N-kinase domain-specific antibodies. The data are representative of two independent experiments. Ig, immunoglobulin; WT, wild type; IP, immunoprecipitation; r, recombinant; WB, Western blot.
FIG. 3.
Cellular organization of StkP kinase and PhpP phosphatase. Flow cytometry was performed with bacterial suspensions from exponentially growing cultures of the wild-type strain Cp1015. Nonpermeabilized and permeabilized bacteria as well as protoplast suspensions were analyzed as described in Materials and Methods, using specific antibodies directed against PhpP and the N-kinase (“Kinase”) and C-PASTA (“PASTA”) domains of StkP protein. Bacteria labeling was obtained as described in Materials and Methods. Nonspecific labeling was evaluated by measurements on the ΔstkP::cat mutant strain Cp7000 and represents 3.5% ± 5% of total binding on wild-type bacteria. Specific labeling of the wild-type bacteria was the difference between total labeling and nonspecific labeling. In each run, 10,000 cells were analyzed. Rates of permeability to propidium iodide were 5% for bacterial suspensions, 40% for protoplast suspensions, and 95% for permeabilized bacteria. The data are representative of two independent experiments.
Physiological characterization of the StkP-PhpP signaling system.
The loss-of-function mutation in StkP kinase is pleiotropic, and we have demonstrated a strong reduction of competence expression in cultures from stkP mutant strains (17). In brief, competence regulation involves signalization in response to the peptide pheromone CSP targeting the histidine kinase receptor of the TCS ComDE (19). The specific role of the phosphatase in this regulation has been evaluated by genetic dissection, using the isogenic strains Cp7000, Cp9090, Cp1095, and Cp7004 carrying the mutated alleles ΔstkP::cat, stkP::Apha3, stkP+, and ΔstkP::cat plus phpPD1, respectively (Table 1). Strain Cp9091 carrying a genetic reversion of stkP::Apha3 into stkP+ (Table 1) was added in the study because loss-of-function mutation in StkP has been related to a fivefold-increased mutation rate of the mutated—compared to the wild-type—strain and might culminate in genetic drift. Indeed, the calculated rates of mutation (27) conferring resistance to rifampin, in two independent experiments (see Materials and Methods), were 9.9 × 10−7 and 8.1 × 10−7 in the stkP mutant strain and 2.1 × 10−7 and 1.4 × 10−7 in the wild-type strain Cp1015.
The results presented in Table 4 show that a competence defect resulting from a loss-of-function mutation in StkP is not suppressed by the additional mutation PhpP(D231A) in strain Cp7004 but is reversed due to allelic exchange at the stkP::Apha3 locus in the stkP+ strain Cp9091 (Table 1). Taken together, our results for strains Cp7004 and Cp9091 demonstrate that the transformation-negative phenotype of strain Cp9090 (17) is a direct result of the StkP loss-of-function mutation rather than a consequence of the activity of PhpP in the cytoplasm of stkP mutant cells. Therefore in this model, the phenotype of StkP mutant strains is not related to StkP-independent PhpP activity but to StkP loss-of-function mutation. It is thus likely that the dedicated PhpP target, StkP-P, is required for the trigger of competence.
TABLE 4.
Impact of stkP and phpP mutations on culture transformabilitya
| Strain | Relevant genotype | n | Transformed colonies (% Rifr/total CFU) |
|---|---|---|---|
| Cp1015 | WT | 6 | 0.86 |
| Cp9090 | stkP::Apha-3 | 5 | <0.01 |
| Cp7000 | ΔstkP::cat | 6 | <0.01 |
| Cp9091 | stkP+ | 2 | 1.05 |
| Cp7004 | ΔstkP::cat phpD1 | 2 | <0.01 |
Donor DNA (1 μg/ml) was from strain Cp1016 carrying the rif23 allele, conferring resistance to rifampin (Rifr). Values are representative of “n” independent determinations. WT, wild type.
It has already been established that the expression of competence in growing cultures represents a global response to several signals transduced by TCSs ComDE, CiaRH, and MicAB (10, 11, 14, 15, 16, 17, 25, 41). We further investigated the role of the StkP-PhpP couple in the competence-signaling network by introducing ΔstkP::cat mutations into strain Cp6600 expressing the comD(D299N) allele to obtain strain Cp6607 (Table 1). Mutation D299N in ComD is a gain-of-function mutation conferring CSP-independent competence development in various conditions that are nonpermissive for the wild-type strain Cp1015, including growth in CTM at pH 6.5 (9, 25). Transformation tests on strain Cp6607 indicated pH-independent competence development similar to that of strain Cp6600 (Table 5), and similar results were obtained for the Cp6600 derivative carrying the stkP::Apha-3 allele (our unpublished data). Thus, the ComD(D299N) mutation has an epistatic effect on StkP loss-of-function mutations, suggesting that StkP intervenes upstream from the CSP receptor ComD. This strongly suggests that ComD activation is sufficient to bypass the effects of the StkP mutation. We tested this hypothesis by subjecting StkP mutant strains Cp7000 (ΔstkP::cat) and Cp7004 [ΔstkP::cat phpP(D241A)] to “transformation tests” in CSP-supplemented CTM (see Materials and Methods). Strain Cp1008, carrying the comA::spc mutation abolishing ComAB-dependent CSP export and maturation (7, 9, 11, 20, 30), was used as a control, in addition to the wild-type strain Cp1015. Dose-response tests clearly show that the threshold CSP concentration activating strains Cp7000 and Cp7004 is similar to that for strain Cp1008 and 20- to 100-fold higher than that for the wild type (see the supplemental material). Taken together, the results in Table 5 and the data presented in the supplemental material indicate that activation of ComD is sufficient to trigger competence development in StkP mutant strains. Complementation of StkP deficiency by synthetic CSP targeting the ComD receptor suggests that during bacterial growth, StkP, under the control of PhpP, transduces signals culminating directly or indirectly in ComD activation.
TABLE 5.
Impact of gain-of-function mutation in the CSP receptor ComD on the transformability of stkP mutant strainsa
| Strain | Relevant genotype | Transformed colonies (% Novr/total CFU)
|
|
|---|---|---|---|
| pH 6.5 | pH 8 | ||
| Cp1015 | WT | <0.01 | 0.31 |
| Cp6600 | comD(D299N) | 0.30 | 0.52 |
| Cp6607 | comD(D299N) ΔstkP::cat | 0.25 | 0.62 |
| Cp7000 | ΔstkP::cat | <0.01 | <0.01 |
Donor DNA (1 μg/ml) was from strain R119 carrying a nov1 mutation, conferring resistance to 2 μg/ml of novobiocin. Results are representative of three independent experiments. WT, wild type.
DISCUSSION
Adaptation to environmental conditions represents a major challenge for living organisms. Transient histidine phosphorylation plays a key role in TCS-mediated signaling in eubacteria. We provide biological and genetic evidences in favor of a role for transient serine-threonine phosphorylation in the regulation of growth and competence in Streptococcus pneumoniae. The proteins encoded by the stkP-phpP operon (31) constitute a signaling couple acting upstream of the competence-specific histidine kinase receptor ComD. ComD is a target for the oligopeptide pheromone CSP and belongs to the TCS ComDE. It is thus likely that transient serine-threonine phosphorylation is involved in the activation of a TCS signaling pathway in Streptococcus pneumoniae.
PhpP contributes to the pattern of competence development through its essential control of StkP activity.
We previously established, using recombinant proteins, that StkP-P is a substrate for PhpP (31). The essentiality of PhpP phosphatase in the RX stkP+ genetic background (Table 3) suggests strong control on the level of StkP-P during bacterial growth. In growing bacteria, cellular localization of PhpP protein has been found to be dependent on the presence or absence of the membrane-associated protein kinase StkP (Fig. 1). Consistently, in StkP+ cells, proteins PhpP and StkP coimmunoprecipitate, suggesting that they belong to the same complex. Furthermore, utilization of recombinant proteins allows us to propose direct interaction between recombinant StkP kinase domain and recombinant PhpP proteins (Fig. 2). The transmembrane organization of StkP and intracellular localization of PhpP (Fig. 3) suggest a submembrane complex facing the cytoplasm, involving the N-terminal kinase domain of StkP and PhpP phosphatase. Mutations in StkP are highly pleiotropic. Cytoplasmic relocalization of PhpP as a soluble protein in StkP-deficient bacteria (Fig. 1) has not been related to the competence-negative phenotype of the strain and did not reveal StkP-independent activity of PhpP. Rather, physiological and genetic analyses in the RX strain Cp1015 and its isogenic derivatives highlight the role of StkP and PhpP as a functional couple. This couple is involved in competence signaling. CSP supplementation experiments on stkP mutant strains (see the supplemental material) and the epistasis of the ComD(D299N) gain-of-function mutation on the StkP loss-of-function mutation in strain Cp6607 (Table 5) demonstrate that StkP kinase activates competence upstream of the histidine kinase receptor, ComD. This finding allows the exclusion of the hypothesis of competence inhibition, due to PhpP phosphatase activity in the absence of its specific substrate(s) StkP-P- and StkP-dependent phosphorylated targets. In the TIGR4 strain of S. pneumoniae, in vitro experiments on recombinant proteins suggest that PhpP and StkP interact competitively with RitR, modulating RitR-dependent Piu heme transporter expression (50). It is possible that some phenotypes of the StkP mutant strains involve alteration of RitR regulation; however, so far the role of RitR in competence remains uncertain (50). We present the hypothesis that in exponentially growing cultures, permissive for the development of a competence peak(s), the time window of cellular StkP-P, resulting in the antagonistic activities of StkP kinase and PhpP phosphatase, might determine the competence burst. During the course of this work, a report was published showing that StkP mutation leads to CSP-independent competence development, under conditions nonpermissive for the wild-type strain (40). Considering the mutator phenotype of StkP mutant strains, genetic drift might account for the results in reference 40. Indeed, alleles ciaR::spc (17) and comD(D299N) (this work) were shown to mask the phenotype of stkP mutations; other unidentified mutations might interfere with the phenotypic expression of StkP loss-of-function mutations and account for the results presented in reference 40.
StkP-PhpP signaling.
The StkP-PhpP signaling couple transduces signals culminating in the activation of ComD, as shown by the epistatic relationship of comD(D299N) mutation on StkP0 mutations (Table 5) and by the CSP complementation test. Flow cytometry analysis using antibodies specific to the C-terminal PASTA domain or the N-terminal kinase domain of the StkP protein indicated a transmembrane organization of the protein with its PASTA domain located beneath the cell wall, e.g., in the periplasmic space (Fig. 3). This cellular compartment contains the penicillin-binding proteins involved in the last steps of peptidoglycan biosynthesis in both gram-negative and gram-positive bacteria (28). Furthermore, PASTA domains are signatures for penicillin-binding proteins and serine-threonine kinases in gram-positive species, leading to speculations that the PASTA-containing kinases could constitute receptors for unlinked peptidoglycan stem peptides (12, 53). Consistently, the StkP mutations confer hypersensitivity to inhibitors of cell wall metabolism in S. pneumoniae (17, 40), potentially reflecting an involvement of StkP in cell wall homeostasis, as proposed elsewhere (22). This observation is reminiscent of results obtained in Enterococcus faecalis, suggesting a role for the serine-threonine kinase in the response to cell wall stress (24). In S. pneumoniae, downregulation of several genes, including competence genes, was observed in cultures containing sublethal concentrations of penicillin, indicating that periplasmic signals may control competence (37). We postulate that StkP activity might allow the entry of “periplasmic signals” into the competence regulatory network, culminating in a competence trigger. In addition to the ComDE TCS, ComAB, the CSP-autoregulated signaling network, TCSs CiaRH and MicAB (11, 15, 26, 41), the global regulator RegR (8), and the putative AI2 autoinducer system (38) have been shown to modulate competence expression in S. pneumoniae, implying that integration of different signals determines the pattern of competence development in growing cultures. Taking into account that PhpP is essential in the wild-type stkP genetic background, indicating strong control on StkP activity (Table 3), the StkP-PhpP signaling couple also represents an important actor determining the kinetics of competence development during bacterial growth in Streptococcus pneumoniae.
In conclusion, the results presented in this work provide physiological and molecular evidence for a signaling system based on transient serine-threonine phosphorylation in the regulation of competence for genetic transformation in S. pneumoniae. This system acts upstream from the competence-specific receptor ComD of the TCS ComDE. The biochemical nature of the gating signals, determining the time window of cellular StkP-P in growing cultures and culminating in the trigger of the TCS competence-signaling pathway, remains to be elucidated. At any rate, it is proposed as a working hypothesis that during bacterial growth the cellular level of StkP-P is finely tuned to the specific cellular physiology by PhpP phosphatase. Given the pleiotropic aspect of loss-of-function mutations in StkP, with effects on in vitro growth (17, 40) and virulence in mice (17), the transient serine-threonine phosphorylation, depending on both StkP and PhpP activities, plays a key role in the homeostasis of this pathogen in cultures and in animals.
Supplementary Material
Acknowledgments
We thank Jean-Claude Lepert (Flow Cytometry Platform, IFR31) and Marc Trombe for helpful suggestions concerning the flow cytometry experiments. We also thank Lila Delbos and Camille Rouviere (master's students at Paul Sabatier University, Toulouse, France), and Nicolas Gay from INSA Toulouse for their contributions. Alexandre Dubrac was helpful in Western blot analysis, and Ursula Liebl kindly helped in editing the manuscript.
This work was funded by INSERM Unité 858, Equipe 15 in France and by NARO Japan to M.O.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Absalon, C., M. Obuchowski, E. Madec, D. Delattre, B. Holland, and S. J. Seror. 2009. CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology 155932-963. [DOI] [PubMed] [Google Scholar]
- 2.Adler, E., A. Donella-Deana, F. Arigoni, L. A. Pinna, and P. Stragier. 1997. Structural relationship between a bacterial developmental protein and eukaryotic PP2C protein phosphatases. Mol. Microbiol. 2357-62. [DOI] [PubMed] [Google Scholar]
- 3.Alderwick, L. J., V. Molle, L. Kremer, A. J. Cozzone, T. R. Dafforn, G. S. Besra, and K. Fütterer. 2006. Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 1032558-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Av-Gay, Y., and M. Everett. 2000. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8238-244. [DOI] [PubMed] [Google Scholar]
- 5.Auzat, I., S. Chapuy-Regaud, G. Le Bras, D. Dos Santos, A. D. Ogunniyi, I. Le Thomas, J.-R. Garel, J. C. Paton, and M.-C. Trombe. 1999. The NADH oxidase of Streptococcus pneumoniae, its role in competence and virulence. Mol. Microbiol. 341018-1028. [DOI] [PubMed] [Google Scholar]
- 6.Boitel, B., M. Ortiz-Lombardía, R. Durán, F. Pompeo, S. T. Cole, C. Cerveñansky, and P. M. Alzari. 2003. PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol. Microbiol. 491493-1508. [DOI] [PubMed] [Google Scholar]
- 7.Chandler, M. S., and D. A. Morrison. 1988. Identification of two proteins encoded by com, a competence control locus of Streptococcus pneumoniae. J. Bacteriol. 1703136-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapuy-Regaud, S., A. D. Ogunniyi, N. Diallo, Y. Huet, J.-F. Desnottes, J. C. Paton, S. Escaich, and M.-C. Trombe. 2003. RegR, a global LacI/GalR, modulates virulence and competence in Streptococcus pneumoniae. Infect. Immun. 712615-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J. D., and D. A. Morrison. 1987. Modulation of competence for genetic transformation in Streptococcus pneumoniae. J. Gen. Microbiol. 1331959-1967. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, Q., E. A. Campbell, A. M. Naughton, S. Johnson, and H. R. Masure. 1997. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23683-692. [DOI] [PubMed] [Google Scholar]
- 11.Dagkessamanskaia, A., M. Moscoso, V. Hénard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 511071-1086. [DOI] [PubMed] [Google Scholar]
- 12.Dessen, A., N. Mouz, E. Gordon, J. Hopkins, and O. Dideberg. 2001. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate. J. Biol. Chem. 27645106-45112. [DOI] [PubMed] [Google Scholar]
- 13.Deutscher, J., R. Herro, A. Bourand, I. Mijakovic, and S. Poncet. 2005. P-Ser-HPr—a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim. Biophys. Acta 1754118-125. [DOI] [PubMed] [Google Scholar]
- 14.Echenique, J. R., S. Chapuy-Regaud, and M.-C. Trombe. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36688-696. [DOI] [PubMed] [Google Scholar]
- 15.Echenique, J. R., and M.-C. Trombe. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 1834599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Echenique, J. R., and M.-C. Trombe. 2001. Competence modulation by the NADH oxidase of Streptococcus pneumoniae involves signal transduction. J. Bacteriol. 183768-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echenique, J., A. Kadioglu, S. Romao, P. W. Andrew, and M.-C. Trombe. 2004. Protein serine/threonine kinase StkP positively controls virulence and competence in Streptococcus pneumoniae. Infect. Immun. 722434-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter, T. 1995. Protein-kinases and phosphatases: The yin and yang of protein p phosphorylation and signaling. Cell 80225-236. [DOI] [PubMed] [Google Scholar]
- 19.Iannelli, F., M. R. Oggioni, and G. Pozzi. 2005. Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptococcus pneumoniae. FEMS Microbiol. Lett. 252321-326. [DOI] [PubMed] [Google Scholar]
- 20.Ishii, S., T. Yano, and H. Hayashi. 2006. Expression and characterization of the peptidase domain of Streptococcus pneumoniae ComA, a bifunctional ATP-binding cassette transporter involved in quorum sensing pathway. J. Biol. Chem. 284726-4731. [DOI] [PubMed] [Google Scholar]
- 21.Jin, H., and V. Pancholi. 2006. Identification and biochemical characterization of a eukaryotic-type serine/threonine kinase and its cognate phosphatase in Streptococcus pyogenes: their biological functions and substrate identification. J. Mol. Biol. 3571351-1372. [DOI] [PubMed] [Google Scholar]
- 22.Jones, G., and P. Dyson. 2006. Evolution of transmembrane protein kinases implicated in coordinating remodeling of gram-positive peptidoglycan: inside versus outside. J. Bacteriol. 1887470-7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, T. J., T. A. Gaidenko, and C. W. Price. 2004. In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J. Bacteriol. 1866124-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristich, C. J., C. L. Wells, and G. M. Dunny. 2007. A eukaryotic-type Ser/Thr kinase in Enterococcus faecalis mediates antimicrobial resistance and intestinal persistence. Proc. Natl. Acad. Sci. USA 1043508-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacks, S. A., and P. Greenberg. 2001. Constitutive competence for genetic transformation in Streptococcus pneumoniae caused by mutation of a transmembrane histidine kinase. Mol. Microbiol. 421035-1045. [DOI] [PubMed] [Google Scholar]
- 26.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237223-234. [DOI] [PubMed] [Google Scholar]
- 27.Luria, S. E., and M. Delbruck. 1943. Mutation of bacteria from virus sensitivity to virus resistance. Genetics 28491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macheboeuf, P., C. Contreras-Martel, V. Job, O. Dideberg, and A. Dessen. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30673-691. [DOI] [PubMed] [Google Scholar]
- 29.Madec, E., A. Laszkiewicz, A. Iwanicki, M. Obuchowski, and S. Serror. 2002. Characterisation of a membrane-linked Ser/Thr protein kinase in Bacillus subtilis implicated in developmental processes. Mol. Microbiol. 46571-586. [DOI] [PubMed] [Google Scholar]
- 30.Morrison, D. A., M.-C. Trombe, M. K. Hayden, G. A. Waszak, and J. D. Chen. 1984. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAMβ1. J. Bacteriol. 159870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nováková, L., L. Sasková, P. Pallová, J. Janecek, J. Novotná, A. Ulrych, J. Echenique, M.-C. Trombe, and P. Branny. 2005. Characterization of a eukaryotic type serine/threonine protein kinase and protein phosphatase of Streptococcus pneumoniae and identification of kinase substrates. FEBS J. 2721243-1254. [DOI] [PubMed] [Google Scholar]
- 32.Peeters, B. P., J. H. de Boer, S. Bron, and G. Venema. 1988. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol. Gen. Genet. 212450-458. [DOI] [PubMed] [Google Scholar]
- 33.Perego, M. 1998. Kinase and phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 6366-370. [DOI] [PubMed] [Google Scholar]
- 34.Rajagopal, L., A. Clancy, and C. E. Rubens. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 27814429-14441. [DOI] [PubMed] [Google Scholar]
- 35.Rajagopal, L., A. Vo, A. Silvestroni, and C. E. Rubens. 2005. Regulation of purine biosynthesis by a eukaryotic-type kinase in Streptococcus agalactiae. Mol. Microbiol. 561329-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajagopal, L., A. Vo, A. Silvestroni, and C. E. Rubens. 2006. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol. Microbiol. 62941-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogers, P. D., T. T. Liu, K. S. Barker, G. M. Hilliard, B. K. English, J. Thornton, E. Swiatlo, and L. S. McDaniel. 2007. Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob. Chemother. 59616-626. [DOI] [PubMed] [Google Scholar]
- 38.Romao, S., G. Memmi, M. R. Oggioni, and M.-C. Trombe. 2006. LuxS impacts on LytA-dependent autolysis and on competence in Streptococcus pneumoniae. Microbiology 152333-341. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Sasková, L., L. Nováková, M. Basler, and P. Branny. 2007. Eukaryotic-type serine/threonine protein kinase StkP is a global regulator of gene expression in Streptococcus pneumoniae. J. Bacteriol. 1894168-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sebert, M. E., K. P. Patel, M. Plotnick, and J. N. Weiser. 2005. Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J. Bacteriol. 1873969-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma, K., M. Gupta, A. Krupa, N. Srinivasan, and Y. Singh. 2006. EmbR, a regulatory protein with ATPase activity, is a substrate of multiple serine/threonine kinases and phosphatase in Mycobacterium tuberculosis. FEBS J. 2732711-2721. [DOI] [PubMed] [Google Scholar]
- 43.Singh, K. D., S. Halbedel, B. Görke, and J. Stülke. 2007. Control of the phosphorylation state of the HPr protein of the phosphotransferase system in Bacillus subtilis: implication of the protein phosphatase PrpC. J. Mol. Microbiol. Biotechnol. 13165-171. [DOI] [PubMed] [Google Scholar]
- 44.Stock, A. M., V. L Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 45.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35566-576. [DOI] [PubMed] [Google Scholar]
- 46.Tiraby, G., M. S. Fox, and H. Bernheimer. 1975. Marker discrimination in deoxyribonucleic acid-mediated transformation of various Pneumococcus strains. J. Bacteriol. 121608-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomasz, A., P. Moreillon, and G. Pozzi. 1988. Insertional inactivation of the major autolysin gene of Streptococcus pneumoniae. J. Bacteriol. 1705931-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trombe, M.-C. 1981. Characterisation de mutants de résistance à l'améthopterine chez Streptococcus pneumoniae, alteration du potential transmembranaire; mise en évidence d'une cible membranaire pour l'amethoptérine, Ph.D. thesis. Université Paul Sabatier, Toulouse, France.
- 49.Trombe, M.-C., C. Clavé, and J. M. Manias. 1992. Calcium regulation of growth and differentiation in Streptococcus pneumoniae. J. Gen. Microbiol. 13877-84. [DOI] [PubMed] [Google Scholar]
- 50.Ulijasz, A. T., S. P. Falk, and B. Weisblum. 2009. Phosphorylation of the RitR DNA-binding domain by a Ser-Thr phosphokinase: implications for global gene regulation in the streptococci. Mol. Microbiol. 71382-390. [DOI] [PubMed] [Google Scholar]
- 51.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
- 52.Wehenkel, A., M. Bellinzoni, M. Graña, R. Duran, A. Villarino, P. Fernandez, G. Andre-Leroux, P. England, H. Takiff, C. Cerveñansky, S. T. Cole, and P. M. Alzari. 2008. Mycobacterial Ser/Thr protein kinases and phosphatases: physiological roles and therapeutic potential. Biochim. Biophys. Acta 1784193-202. [DOI] [PubMed] [Google Scholar]
- 53.Yeats, C., R. D. Finn, and A. Bateman. 2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci. 27438-440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.