Abstract
The Clp protease ATPase subunit and chaperone ClpX is dispensable in some bacteria, but it is thought to be essential in others, including streptococci and lactococci. We confirm that clpX is essential in the Rx strain of Streptococcus pneumoniae but show that the requirement for clpX can be relieved by point mutations, frame shifts, or deletion of the gene spr1630, which is found in many isolates of S. pneumoniae. Homologs occur frequently in Staphylococcus aureus as well as in a few strains of Listeria monocytogenes, Lactobacillus johnsonii, and Lactobacillus rhamnosus. In each case, the spr1630 homolog is accompanied by a putative transcriptional regulator with an HTH DNA binding motif. In S. pneumoniae, the spr1630-spr1629 gene pair, accompanied by a RUP element, occurs as an island inserted between the trpA and cclA genes in 15 of 22 sequenced genomes.
The ATP-dependent proteases play important roles in bacterial physiology. In addition to monitoring protein quality and clearing damaged or misassembled proteins, they also participate in numerous regulatory pathways via specific proteolysis of regulatory proteins (13, 26, 32, 40). One family of these proteases is composed of a peptidase core, ClpP, complexed with one of a few ATPase specificity factors that provide for transport of protein substrates into the otherwise inaccessible core for proteolysis. While it thus might be expected that Clp protease mutations would usually be highly pleiotropic, the most prominent phenotype associated with loss of ClpP varies from one species to another. Thus, for example, clpP mutants of a number of low-GC gram-positive species have reduced thermo-tolerance (7, 11, 20, 28, 29, 35). A loss of virulence is the most prominent phenotypic effect of mutation of ClpP in Staphylococcus aureus (8), while in Salmonella, altered expression of flagellar operons is a major effect (43). In contrast, inactivation of clpP in Caulobacter crescentus is lethal (15). Some of these phenotypes have been traced to roles of ATP-dependent proteases in controlling the half-life of bacterial regulatory proteins, including the stationary phase sigma factor σS and the heat shock sigma factor σ32 in Escherichia coli (41, 49) and the sporulation sigma factor σH (22), the competence transcription factor ComK (44), the RsiW antisigma factor (48), the regulator Spx (30), and the Clp protease regulator CtsR in Bacillus subtilis (18).
Among the Clp ATPase subunits, ClpX is widely conserved among bacteria, but its role in bacterial physiology varies from one species to another. In some, it carries out essential functions, while in others its roles are more subtle. In Caulobacter, critical cell cycle regulators depend on proteolysis by ClpXP for correct timing of cell division (15). In Synechococcus, Streptococcus, and Lactococcus, ClpX also appears to be essential (5, 9, 37), but it is dispensable in other bacteria. In Streptomyces, clpX mutants are viable but have a developmental defect revealed only at low pH (45). While clpX mutants of B. subtilis are heat sensitive (12), clpX mutants of Staphylococcus aureus are more tolerant of elevated temperatures (8, 10). The ClpX protein can also act in nonproteolytic roles, as when it retards assembly of the FtsZ ring (47), remodels the Mu phage replication complex (16, 21, 27, 46), or acts independently of ClpP in positive regulation of transcription of the protein A gene in S. aureus (8, 10).
The creation of ClpX mutants of Streptococcus pneumoniae (pneumococcus) was reported independently by Robertson et al. (35), using strain R6, and by Luo (23), using the related strain, Rx. However, Robertson et al. (36) subsequently reported that all apparent clpX mutants in their hands resulted from rare rearrangements in strain R6 that result in acquisition of a disrupted clpX gene but also in retention of an intact copy of the gene, creating a merozygote. Specifically, an R6 derivative was created that carries both a disrupted clpX gene and an ectopic copy of the intact gene expressed under the control of the fucose promoter. As the viability of this strain depends on the provision of fucose to ensure expression of the ectopic clpX, it is clear that clpX is an essential gene in strain R6, presumably reflecting some nonproteolytic role, as clpP is dispensable in this strain. We were curious as to whether CP1361, the clpX mutant obtained by replacing most of clpX with an erm marker in the Rx background (23), might have a similar complex explanation or might be truly deficient in clpX. Here we resolve this discrepancy by showing that the clpX deletion mutant CP1361 carries an unlinked suppressor mutation that permits normal growth in the absence of clpX and by mapping the suppressor mutation to a conserved genetic island found in most pneumococcal isolates.
MATERIALS AND METHODS
Strains used in this study.
Tables 1 and 2 list the strains and primers used in this study. CAT broth contained (per liter) 5 g of tryptone (Difco Laboratories), 10 g of enzymatic casein hydrolysate (ICN Nutritional Biochemicals), 1 g of yeast extract (Difco), and 5 g of NaCl. It was sterilized for 40 min at 121°C and then brought to 0.2% glucose and 1/60 (0.0167) M K2HPO4 before use. Antibiotics were used at the following concentrations: 2.5 μg/ml for novobiocin (Nov), 0.25 μg/ml for erythromycin (Em), 200 μg/ml for kanamycin (Kan), and 40 μg/ml for spectinomycin (Spc).
TABLE 1.
Strains used in this study
| Strain | Description | Reference, source, or constructiona |
|---|---|---|
| CP1250 | Rx derivative, low β-galactosidase activity; hex malM511 str-1 bgl-1; Hex− Mal− Smr Bga− | 33 |
| CP1361 | CP1250, but ΔclpX::erm slx-1; Emr | 23 |
| CP1500 | Rx derivative; hex novo-r brya-r str-r1 ery-r2 ery-r6; Novr Emr Strr | 4 |
| CP1973 | CP1361, but clpX+; Ems | This study |
| CP2008 | CP1250, but Δspr1630::kan; Kanr | This study |
| CP2086 | R6, but novo-r; Novr | R6 × CP1500 |
| CP2087 | R6, but Δspr1630::kan; Kanr | R6 × CP2008 |
| CP2088 | R6, but Δspr1630 ΔclpX::erm; Kanr Emr | CP2087 × CP1361 |
| CP2089 | TIGR4, but Δspr1630::kan; Kanr | TIGR4 × CP2008 |
| R6 | Rough derivative of D39 | 14 |
| TIGR4 | Serotype 4 isolate | 42 |
Strain construction by transformation is indicated as recipient × DNA donor.
TABLE 2.
Primers used in this study
| Primer name | Gene | Primer sequence (5′-3′)a |
|---|---|---|
| AP22 | clpX | GATAGAATGACAGTATAGAATGGAAGGAAATC |
| AP23 | clpX | ATTTGTAATGAATGCGTGGAGTTAGCTCAG |
| AP24 | clpX | ATGCGTGGAGTTAGCTCAGGAAATCATTCG |
| AP25 | clpX | CTAGGATCGGTTTATCCGTTCCATCGACAG |
| AP26 | clpX | TAGCAATCTCTTGAAGGGCTTCGTCGTCAAATTCC |
| AP27 | clpX | ACGCGCCCCTGTCTTCCGTTCGATTGCTTTATTAG |
| AP30 | spr1422 | CTTGCCTGACTCAGGGTCACGAATG |
| AP33 | lytC | AACGCAAACGAGCGCAGAAGTACAG |
| DAM301 | kan | CGCGCAAGCTGGGGATCCG |
| DAM302 | kan | acgtgggcccTAGGTACTAAAACAATTCATCCAGTAA |
| DAM850 | spr1620 | TGGAAGCTCTGATTCCGCAATTCCG |
| DAM851 | spr1627 | AGGCTTAGGTAGCCAGGCTTTAAGG |
| DAM852 | spr1623 | AGCTGGAGACAGCACTCCGTAGTTG |
| DAM853 | trpA | GGTGACCACGAGAAAGGTTTGGATG |
| DAM854 | spr1630 | GTAACTGACCAGTTCGTGCATCAGG |
| DAM855 | trpE | GAAAGCAACGCTTCCTGCTGGAACC |
| DAM856 | trpD | TCAAGCCAGCTTCATCCAACCCTTC |
| DAM857 | mgtC | TCTGACAACTGCAGCAGGCATTTGG |
| DAM858 | ccpA | AGAACGCACACCTCTTTCACGATCC |
| DAM859 | galEb | TGACAGCGCTTGACAAAGGAAACGG |
| DAM860 | spr1645 | TTGCTAGAATTGCCCGTCCTGTACC |
| DAM861 | spr1650 | ATCAACCGAGTGGGTACTGGATAGG |
| DAM862 | trpA | CACCAAGGAGAGCCGATTCAGGAC |
| DAM863 | spr1629 | CCATGCGCTATAAGCATCAGCTTGG |
| DAM864 | spr1630 | gcaggatccCCTGATGCACGAACTGGTCAGTTAC |
| DAM865 | spr1630 | acagggcccAGGTCGCGTCCATGCATAACCATTC |
| PL46 | erm | GGTCTAGA GGATCCGGGTACCGGGCCCAAAATTTGTTTGATTTGT |
| PL47 | erm | GGGGTACCGAAGCTTGGGATCCGGAATTCAGTCGGCAGCGACTCATAGAATTA |
| PL71 | clpX | gctcTAGACATGATTTCCTTCCATTCTATACTG |
| PL72 | dpr | CGTGTTCTTGTTATCTACCGTTACT |
| PL73 | clpX | cccaagctttcATGTTTGAGGTGCCGAGTCAGGA |
| PL74 | aldR | TTACATCACGAGGAAGACGAGCTAC |
| SA130 | spc | GATCTGTCAATGGTTCAGATAC |
| SA133 | spc | GAAGAGCCATTATGGATTCG |
Lowercase segments indicate 5′ extensions; underlined segments indicate restriction sites.
Primer binds upstream of galE.
Creation of strain CP1361 (ΔclpX::erm) was as described previously (23, 34). Briefly, a fragment upstream of clpX was amplified from strain CP1500 using primers PL72 and PL71, a downstream fragment was amplified from CP1500 using PL73 and PL74, and a third fragment containing the erm marker was amplified from amplicon aMSL1 (19) using PL46 and PL47. After digestion by XbaI and/or HindIII, the three fragments were purified, ligated, and used directly as donors for transforming strain CP1250, with selection for Emr. The mutation in a single clone, named CP1361, was confirmed by sequencing a fragment that was amplified using the outermost primers for creating the construct.
Strain CP2008 was created similarly by disruption of spr1630, except the upstream fragment was amplified using DAM862 and DAM865, the downstream fragment was amplified using DAM863 and DAM864, the kan insert was amplified from pR410 (38) using DAM301 and DAM302, and the fragments were digested by BamHI and/or ApaI. To create strain CP1973, a clpX+ revertant of strain CP1361, we took advantage of the high transformation efficiency of large donor DNA amplicons to accomplish allele replacement without selection. A 7,736-bp clpX+ donor fragment was amplified from CP1250 using AP30 and AP33. CP1361 was transformed using this fragment as a donor, as described below, but with an extended (3-h) period of incubation to allow for segregation of the new allele. Thirty resulting colonies were used individually to inoculate CAT and, in parallel, CAT with Em. Seven clones that grew in CAT, but not in the presence of Em, were analyzed by PCR, using primers AP22/25 and AP23/27 to determine which were clpX+. Of the seven clones, six appeared to be clpX+ and one was retained as CP1973.
PCR amplification.
PCR amplification was done by using commercially available PCR supermixes. PCR SuperMix high fidelity (Invitrogen) was used for fragments intended for sequencing or for transformation, while PCR SuperMix (Invitrogen) was used in other cases. Reaction mixtures (50 μl) contained 45 μl SuperMix, 0.5 μl of each primer (final concentration 500 nM), 1 μl of DNA (∼100 ng), or 1 μl cell culture at an optical density (OD) of 0.1, and 3 μl sterile water. The PCR conditions were 95°C for 30 s; 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C (68°C for fragments >5 kbp in length) for 1 min/kbp target length; 72°C (or 68°C) for 10 min.
slx marinerT7 library DNA.
To create an slx marinerT7 transposon (Ωspc) library, CP1973 was transformed with DNA from the marinerT7 transposon library (2), as described below. The cells were plated on the surface of CAT agar containing Spc. After overnight incubation, ∼150,000 Spcr colonies were washed off the surface of the agar and suspended in 300 ml complete CAT plus Spc. After growth for ∼five generations, the 0.4-OD550 culture was harvested by pelleting the cells (5,000 × g, 20 min, 4°C), washing them in 20 ml buffer A (0.1 M Tris-HCl [pH 7.6], 10 mM EDTA, 0.1 M NaCl), and resuspending them in 10 ml buffer A with Triton X-100 to 0.2%. After incubation at 37°C for 100 min, DNA was purified by chloroform extraction and ethanol precipitation, as described previously (24). DNA in the final 5-ml volume was estimated by quantitation of gel images after electrophoresis in 0.8% agarose gels.
Inverse PCR.
Mini-DNA extracts were prepared by a similar procedure using 10 ml of each culture. To determine the position of marinerT7 insertions, 1 μg of genomic DNA was digested with 10 units of HpyCHIV (New England BioLabs) in 25 μl NEBuffer 1. After heat inactivation of the enzyme at 65°C for 20 min and addition of 2.8 μl of 10× Ligase buffer (Fermentas), the DNA was treated with five units of T4 DNA ligase at room temperature overnight. A 1-μl sample from the ligation mix was amplified with insert-specific primers SA130 and SA133. The PCR product was sequenced using the same primers, and both marinerT7-pneumococcus junctions were determined by comparison with the R6 genome to locate the position of the insert. Each insert was mapped to a unique location, where it was accompanied by a TA target duplication.
Transformation.
Transformation of Rx derivatives was done by growing a 10-ml culture of CP1250 in CAT (19) plus 10 mM HCl to an OD550 of 0.06 and inducing development of competence with CaCl2 (to 0.5 mM), BSA (to 0.002%), and CSP-1 (Chiron Mimotopes, Raleigh, NC) (to 250 ng/ml). After 5 min, 1 ml of the induced cells was incubated with 100 ng of donor DNA for 70 min, diluted, and plated in CAT agar for challenge with selective antibiotic, as described previously (39). Transformation of R6, TIGR4, and their derivatives was done as described in reference 25, with the minor modifications described in reference 3.
RESULTS
A viable Rx mutant lacks clpX.
To investigate the genomic structure of strain CP1361 (ΔclpX::erm), primers were designed to amplify internal fragments of clpX (primer pairs AP23/AP27 and AP24/AP26) or nearly the entire clpX gene (primer pair AP22/AP25) (Fig. 1). These primers were specifically selected so that the internal primers would amplify part of clpX from CP1250 (the wild-type parent strain to CP1361) but would not be expected to match sites in CP1361. In contrast, the third primer pair would create a product from CP1250 encompassing 99% of clpX, as well as a smaller one encompassing the erm insert from CP1361. Both internal clpX fragments and the entire clpX gene were readily amplified from CP1250, but no internal clpX fragments were obtained from CP1361 (Fig. 1). Furthermore, the size of the fragment obtained from CP1361 with AP22/AP25 was smaller than that from CP1250, in accord with the gene replacement design. We conclude that CP1361 contains an altered clpX gene region, as reported previously (23), but appears to lack a residual intact clpX gene.
FIG. 1.
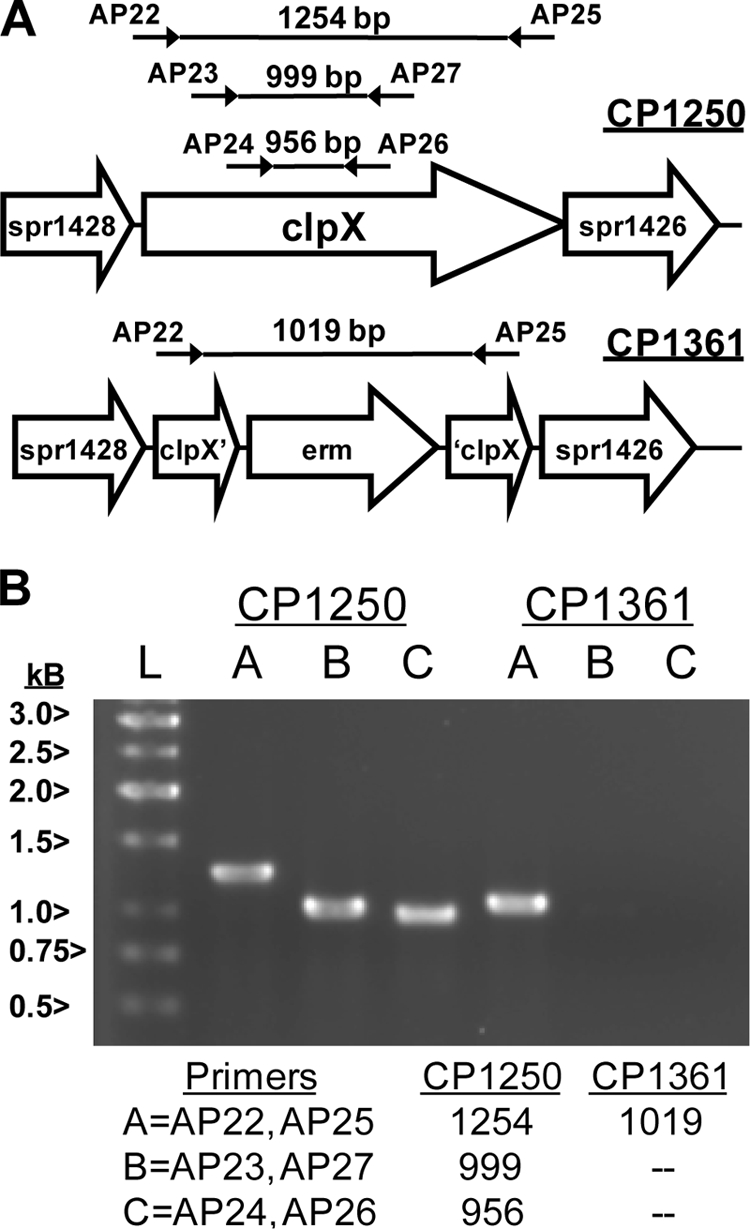
Structure of an erm deletion/replacement mutation of clpX and verification of the absence of clpX from the mutant strain CP1361. (A) Map of ORFs in the genome of R6 and design of the erm insertion in CP1361. Locations of primer pairs used to amplify internal fragments of clpX or to reveal the size of the entire clpX region are indicated, as well as the sizes of predicted PCR products. ORFs in the R6 genome (accession no. AE007317) are indicated by open arrows. ‘ and ’ indicate a truncated ORF. (B) Analysis of PCR products obtained using genomic DNA from the wild type and from CP1361 after electrophoresis on a 0.8% agarose gel. Sizes of fragments (bp) expected for the primer pairs indicated as A, B, or C, using template DNA from each strain are listed at the bottom. Sizes of reference DNA fragments (lane L) are indicated at the left.
The viable Rx clpX deletion mutant appears to carry an unlinked suppressor of ΔclpX lethality.
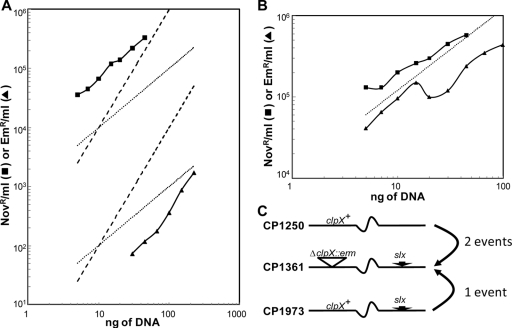
To seek an explanation for the discrepancy between the viability of strain CP1361 without a functional clpX gene and the lethality of clpX loss in R6, we performed a backcross that used DNA from CP1361 to transform CP1250. Emr transformants were obtained after the usual 18-h incubation of plates at 37°C, but in low yields. However, each of the five transformants analyzed carried the same molecular structure as CP1361 in the clpX region, as verified by PCR, and PCR failed to recover internal fragments of the clpX gene (data not shown). To determine whether CP1361 might carry an unlinked extragenic suppressor of ΔclpX lethality that could explain this pattern, the dose-response pattern for the CP1361 × CP1250 backcross was determined. As described by Kent and Hotchkiss (17), cotransformation by two unlinked markers in pneumococcus displays a quadratic dependence on the dose of donor DNA, while transfer of a single marker is more efficient and follows linear dependence on donor DNA dose. Production of Emr transformants followed quadratic kinetics in the backcross, while a point marker (novo-r) transformed the same culture of competent cells at a higher rate and with linear kinetics (Fig. 2). Thus, formation of viable Emr transformants required uptake and integration of two different fragments of donor chromosomal DNA, suggesting that CP1361 does carry both the (potentially lethal) ΔclpX::erm mutation and an unlinked suppressor of the lethal effect of loss of ClpX (Fig. 2).
FIG. 2.
Role of an unlinked suppressor in the low efficiency and two-hit kinetics of transformation of the wild type to ΔclpX::erm. (A) Transformation of wild-type CP1250 with the indicated amounts of CP1500 genomic DNA carrying a Novr marker (—▪—) or of genomic DNA from Emr strain CP1361 (—▴—). Theoretical slopes are shown for linear (dotted; y = ax, for a = 10 or 1,000) and quadratic (dashed; y = bx2, for b = 1 or 100) kinetics. (B) Transformation of the ClpX+Ems strain CP1973, using the same donor DNAs as those represented in panel A. Theoretical slope is shown for linear kinetics (dotted; y = 12,000×). (C) Interpretation of transformation kinetics. The hypothetical suppressor of lethality of ΔclpX (slx) is sufficiently distant from clpX that it is carried on a distinct DNA fragment and transferred independently from clpX. CP1973, retaining the slx suppressor, is transformed by CP1361 DNA via a single recombination event.
To verify the presence of a hypothetical unlinked suppressor, the ΔclpX::erm mutation in CP1361 was restored to wild type (clpX+) by high-efficiency transformation with a 7,736-bp PCR fragment carrying the wild-type clpX gene from CP1250. Restoration of an intact clpX gene in one clone (CP1973) was confirmed by PCR. To determine whether the clpX+ strain retained a suppressor of clpX lethality, it was transformed using the ΔclpX::erm marker in genomic DNA from CP1361. Emr transformants were obtained with single-hit kinetics and with a large increase in efficiency (Fig. 2). We conclude that CP1361 does indeed carry an unlinked suppressor of the lethal clpX phenotype and that it is retained in CP1973. We designate this mutation slx for suppressor of lethality of clpX and designate the allele in CP1361 slx-1.
The suppressor maps to spr1630.
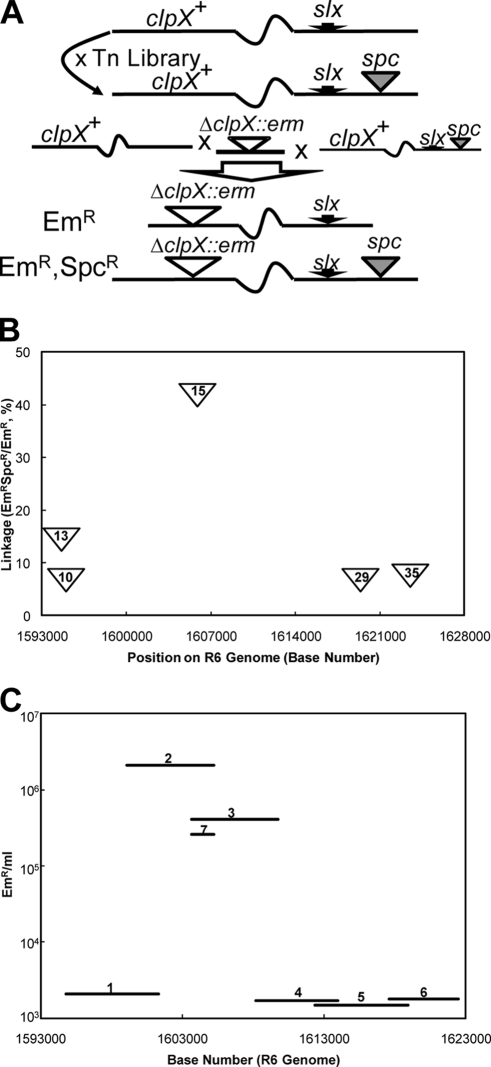
To identify the hypothetical suppressor, we employed a random insertion library as a mapping tool. Transformation of CP1973 with DNA from the marinerT7 (Ωspc) library described previously (2, 3), with selection for spectinomycin resistance, created a new slx Ωspc mutant library, containing 150,000 independent clones (Fig. 3). To identify Ωspc insertions in this library that are linked to the suppressor, wild-type CP1250 was cotransformed with a mixture of the 7,501-bp ΔclpX::erm amplicon and genomic DNA from the slx Ωspc library. By virtue of the lethal effect of a clpX deletion in the CP1250 background, only triple transformants which had acquired ΔclpX::erm, slx-1, and Ωspc would be expected to survive selection for Em and Spc. Since the transformation of three markers would be most efficient for DNA from library members in which Ωspc and slx-1 are carried by the same DNA fragment, we expected many of the Emr Spcr transformants to have Ωspc inserts located near slx-1. In accord with this expectation, among 40 of the resulting Emr Spcr transformants subsequently tested for linkage between Ωspc and slx, 10 exhibited elevated cotransformation of Emr with Spcr, in the range of 4 to 40% (data not shown).
FIG. 3.
Mapping of slx by linkage to marinerT7 inserts. (A) Mapping strategy. Top, preparation of a marinerT7 (Ωspc) library in the slx-1 background by transformation. Bottom, cotransfer of a ΔclpX::erm amplicon, slx-1, and Ωspc from the library. (B) Linkage of Ωspc inserts to slx. Frequency of cotransformation of Emr and Spcr from genomic DNA of five Emr Spcr clones that had been obtained by transformation of CP1250 with clpX::erm and the Ωspc slx-1 library donor DNA. Inserts in clones 10,13, 15, 29, and 35 were located at bp 1594927, 1594560, 1605766, 1619303, and 1623486, respectively. (C) Mapping the slx mutation by assay of the slx content of PCR products amplified from CP1361 genomic DNA. Fragments 1, 2, 3, 4, 5, 6, and 7 were amplified with primer pairs DAM850/851, DAM852/853, DAM854/855, DAM856/857, DAM858/859, DAM860/861, and DAM854/853, respectively. Bars indicate the genetic extent of each fragment and the yield of clpX::erm after cotransformation of CP1250 with the indicated fragment and a pure clpX::erm amplicon.
The five Ωspc insertions giving the highest yield of Emr Spcr triple transformants, i.e., those most tightly linked to slx-1, were mapped by inverse PCR and sequencing (Fig. 3). All five mapped to a 30-kbp region (bp 1593000 to 1623000) of the R6 genome (14).
To map the suppressor more precisely within this region, six amplicons were prepared from CP1361. Each such amplicon was cotransformed with the 7,501-bp ΔclpX::erm amplicon, as described above, to detect its slx content by a single-marker selection (for Emr) for the double transformation event (slx and ΔclpX). Two overlapping 6-kbp fragments exhibited the highest slx activity, while slx activity of the four others was indistinguishable from the background (Fig. 3). To test the inference that slx-1 is within the region of overlap, a smaller fragment (1,579 bp) representing the region of overlap was prepared and assayed for slx activity by cotransformation in the same way. As this new fragment also had high slx activity (Fig. 3), we conclude that mutation slx-1 lies in the 1,579-bp region between bp1603661 and bp1605240.
Loss-of-function mutations in spr1630 are suppressors of ΔclpX lethality.
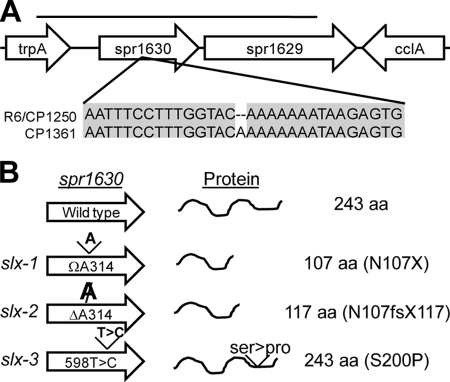
To identify the specific nature of slx-1, the 6-kbp amplicon exhibiting the highest slx activity was amplified from both the wild type and CP1361, and a portion of it was sequenced. A single-base difference between the strains was found: an insertion of A, at position 1603927 on the R6 genome, which is base 314 of the predicted open reading frame (ORF) spr1630 (Fig. 4). The mutant gene is predicted to encode a truncated protein of only 107 amino acids, suggesting that the loss of activity of spr1630 is the cause of suppression of clpX lethality. We infer that the slx-1 mutation arose spontaneously in an unstable A repeat in the course of the creation of strain CP1361, being selected because of the lethality of a clpX deletion in a Slx+ background.
FIG. 4.
Sequence changes in spr1630 in slx mutants. (A) Comparison of sequences of spr1630 in the wild type and the slx mutant CP1361. The sole difference found after sequencing both strands in the region indicated by the bar is shown, with identical bases shaded. (B) Sequence changes in three slx alleles and predicted protein products for each allele. Base changes in each allele are indicated at the left; predicted protein products are indicated at the right. fs, frameshift; X, stop codon.
Indeed, the selection was so strong that it allowed us to obtain additional suppressor alleles created during PCR amplification of wild-type DNA by cotransforming with the 7,501-bp ΔclpX::erm amplicon, as described above. Emr transformants were obtained by using this donor DNA, at a frequency 10-fold lower than that when using the same region amplified from CP1361, both with the 6,166-bp fragment (DAM852/853) and the 6,073-bp overlapping fragment (DAM854/855) (data not shown). Sequencing of the gene spr1630 in three transformants recovered from transformation by the 6,166-bp fragment revealed two new mutations in spr1630 and an independent creation of the original insertion of A314 found in CP1361. Two mutations (an insertion and a deletion) encode truncated proteins, while the third causes a replacement of a serine residue by proline (Fig. 4).
The ready recovery of slx mutations in spr1630, compared to the failure to obtain slx mutations from fragments elsewhere in the 30-kbp neighborhood around spr1630, suggested that any sufficient disruption of spr1630 might have suppressor activity. To determine if complete loss of spr1630 would also suppress the lethality of ΔclpX, a deletion/replacement of spr1630 was constructed by directed mutagenesis (see Materials and Methods). As this deletion also raised the transforming efficiency of the ΔclpX::erm marker 100-fold (Table 3), we conclude that in the Rx background the presence of a functional spr1630 protein is lethal in the absence of functional ClpX.
TABLE 3.
Transformation efficiency of ΔclpX::erm in spr1630 mutants in three genetic backgroundsa
| Recipient | DNA donor genotypeb
|
|
|---|---|---|
| novo-r (Novr/ml) | slxΔclpX::erm (Emr/ml) | |
| CP1250 (wild type) | 350,000 | 6,000 |
| CP1973 (slx-1) | 430,000 | 640,000 |
| CP2008 (Δspr1630) | 480,000 | 620,000 |
| R6 | 4,889 | 289 |
| CP2087 (R6 Δspr1630) | 3,511 | 14,000 |
| TIGR4 | 40 | 0.3 |
| CP 2089 (TIGR4 Δspr1630) | 148 | 557 |
Exposure (70 min) of cells at OD of 0.06 to 0.1 μg DNA/ml.
Novr donor DNA was from CP1500, but from CP2086 for R6 and TIGR4 recipients. Emr donor DNA was from CP1361, but from CP2088 for R6 and TIGR4 recipients.
To determine whether the relationship between spr1630 and loss of ClpX function described above is strain specific, we prepared Δspr1630 and clpX mutant transformants of strain R6, another widely used descendant of strain D39, the progenitor of strain Rx, as well as of the serotype 4 strain TIGR4, which is a virulent isolate. As shown in Table 3, in both of these genetic backgrounds, the clpX-deficient allele was transferred at very low efficiency, but this was alleviated by deletion of spr1630. Thus, both the lethality of ΔclpX and its suppression by loss of spr1630 are shared by Rx, R6, and TIGR4.
DISCUSSION
clpX mutations exhibit a variety of phenotypes in different bacterial species. When the phenotype is lethality, it may present an opportunity to identify a new critical cell function, but because of the many roles proteases may play simultaneously, unraveling such a phenotype can be difficult. For example, lethality of a clpX mutation has been traced to cell cycle regulation in Caulobacter but remains unexplained for Synechococcus, Streptococcus, and Lactococcus. In the present case, we have traced the lethality of clpX mutations to a dispensable gene and thus opened the way to the study of specific roles of ClpX in pneumococcus, as illustrated by the recent identification of ClpXP as the major player in degradation of pneumococcal SsrA-tagged proteins (1).
Inspection of the >1,000 publicly available bacterial genome sequences reveals that homologs of spr1630 occur as part of a bi-cistronic island among certain strains of several gram-positive species. spr1630 itself is found in 15 of 22 available pneumococcal genomes, where it is located adjacent to trpA. The gene is strongly conserved within this species, as the 15 cases include only six distinct alleles, differing by substitutions of 1 or 2 bp. A homolog (66% similarity) occurs in two of the three sequenced strains of Streptococcus suis and a few more distantly related homologs occur in Listeria monocytogenes (59% similarity), Lactobacillus rhamnosus (52% similarity), and Lactobacillus johnsonii (52% similarity). In Staphylococcus aureus, spr1630 homologs are found as three alleles that are divergent from one another in 30 to 40 bp, in 9 of 17 sequenced genomes. In R6, spr1630 is adjacent to spr1629, encoding a putative transcriptional regulator, which contains the HTH-3 and DUF955 Pfam domains (6). Interestingly, in the other species mentioned, homologs of spr1630 are also adjacent to homologs of spr1629. In pneumococcus, the pair of genes is always found between trpA and cclA, while in S. aureus, it is always found adjacent to adhE. Thus, spr1630 appears to have been subject to horizontal gene transfer as part of a bi-cistronic operon or gene cluster, which may be self-regulating. In no case, however, has a specific function been assigned to either gene.
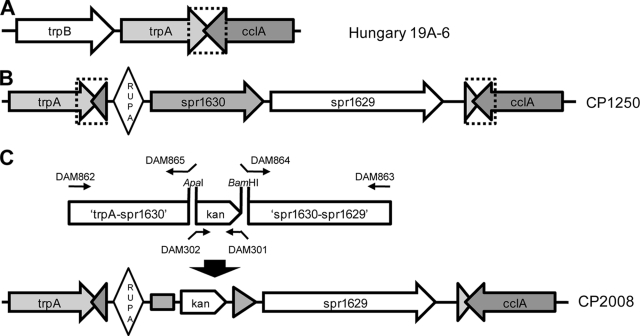
In pneumococcus, comparison of the genome sequence of strain R6 to those of strains that lack spr1630 alleles (including CCRI1974, CDC1873, CGSP14, Hungary 19A-6, SP14-BS69, and SP23-BS72) reveals a structure that may reflect the rearrangement(s) that accompanied the original acquisition of the element (Fig. 5). spr1630 and spr1629 are immediately adjacent to a copy of a 105-bp RUPA element (31) that is otherwise found near the L11 ribosomal protein gene. The three elements are further flanked by a 107-bp direct repeat that encompasses, in spr1630-free strains, the 3′ end of the trpA gene and the overlapping 3′ end of the convergently transcribed type IV prepilin peptidase gene known as cclA or cilC. The consequence of the apparent duplication event is that the spr1630 island is isolated between two convergently transcribed operons. Since RUPA is proposed to be a vestigial insertion element occasionally activated by the transposase of IS630-Spn1, it is worth considering whether insertion of RUPA, spr1630, and spr1629 may have been accomplished in a singe concerted event involving creation of the flanking duplication. This would be consistent with the proposal that RUP elements may specifically target entering foreign DNA and foster genetic rearrangements (31).
FIG. 5.
Organization of the Slx/spr1630 locus in two natural isolates and a synthetic deletion derivative. (A) Structure of the trpA-cclA region in Hungary19A-6 and other pneumococcal strains lacking spr1630. The segment duplicated in strains carrying spr1630 is indicated by a dotted bracket. (B) Elements adjacent to trpA and cclA in the R6 genome. Diamond, RUPA; triangles, gene remnants in direct repeat. (C) Strategy for deletion of spr1630. PCR products ligated to kan to target insertion by flanking homology are depicted. Small arrows represent primers; small arrows with tails represent primers with 5′ extension and restriction site.
As spr1630 is quite common in pneumococcus and Staphylococcus aureus, it may be advantageous to both of these human pathogens and should be considered a candidate virulence factor. The report that a ΔclpX mutant of S. aureus is viable (8) may suggest that the role of the spr1630 homolog in S. aureus or its interaction with ClpX differs significantly from that in pneumococcus; however, it is not known whether the strain in which the clpX mutation was evaluated carries a homolog of spr1630. While the function of spr1630 is at present entirely unknown, the special relation to ClpX implies that it acts in some critical aspect of physiology. In light of the relation to ClpX activity, it should be noted that transcription of spr1630 and spr1629 is elevated 10-fold in a clpP mutant (35). Because clpP mutants of R6, Rx, and TIGR4 are viable (but heat sensitive) the essential activity of ClpX in Slx+ strains is not likely to be proteolysis. A simple hypothesis to explain its relationship to ClpX would be that the spr1630 protein constitutes a toxin, unless processed to a harmless form by a ClpX chaperone activity. Of course, the participation of ClpX may be indirect and might involve some interaction with the product of spr1629, if indeed it acts to regulate expression of spr1630.
Acknowledgments
This material is based upon work supported in part by the National Science Foundation under grant no. MCB-0543187.
Footnotes
Published ahead of print on 22 May 2009.
REFERENCES
- 1.Ahlawat, S., and D. A. Morrison. 2009. ClpXP degrades SsrA-tagged proteins in Streptococcus pneumoniae. J. Bacteriol. 1912894-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bijlsma, J. J., P. Burghout, T. G. Kloosterman, H. J. Bootsma, A. de Jong, P. W. Hermans, and O. P. Kuipers. 2007. Development of genomic array footprinting for identification of conditionally essential genes in Streptococcus pneumoniae. Appl. Environ. Microbiol. 731514-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burghout, P., H. J. Bootsma, T. G. Kloosterman, J. J. Bijlsma, C. E. de Jongh, O. P. Kuipers, and P. W. Hermans. 2007. Search for genes essential for pneumococcal transformation: the RADA DNA repair protein plays a role in genomic recombination of donor DNA. J. Bacteriol. 1896540-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cato, A., and W. R. Guild. 1968. Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double cross-over model. J. Mol. Biol. 37157-178. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, A. K., J. Schelin, and J. Porankiewicz. 1998. Inactivation of the clpP1 gene for the proteolytic subunit of the ATP-dependent Clp protease in the cyanobacterium Synechococcus limits growth and light acclimation. Plant Mol. Biol. 37791-801. [DOI] [PubMed] [Google Scholar]
- 6.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, J. S. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of mis-folded protein in Lactococcus lactis. Mol. Microbiol. 3179-87. [DOI] [PubMed] [Google Scholar]
- 8.Frees, D., S. N. Qazi, P. J. Hill, and H. Ingmer. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 481565-1578. [DOI] [PubMed] [Google Scholar]
- 9.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, gram-positive bacteria. Mol. Microbiol. 631285-1295. [DOI] [PubMed] [Google Scholar]
- 10.Frees, D., K. Sørensen, and H. Ingmer. 2005. Global virulence regulation in Staphylococcus aureus: pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect. Immun. 738100-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 351286-1294. [DOI] [PubMed] [Google Scholar]
- 12.Gerth, U., E. Kruger, I. Derré, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28787-802. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S. 1999. Regulation by proteolysis: developmental switches. Curr. Opin. Microbiol. 2142-147. [DOI] [PubMed] [Google Scholar]
- 14.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 175658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, J. M., D. J. Welty, and H. Nakai. 1998. Versatile action of Escherichia coli ClpXP as protease or molecular chaperone for bacteriophage Mu transposition. J. Biol. Chem. 273459-465. [DOI] [PubMed] [Google Scholar]
- 17.Kent, J. L., and R. D. Hotchkiss. 1964. Kinetic analysis of multiple, linked recombinations in pneumococcal transformation. J. Mol. Biol. 9308-322. [DOI] [PubMed] [Google Scholar]
- 18.Kirstein, J., D. Zühlke, U. Gerth, K. Turgay, and M. Hecker. 2005. A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J. 243435-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 1815004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 1846357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levchenko, I., L. Luo, and T. A. Baker. 1995. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 92399-2408. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J., W. M. Cosby, and P. Zuber. 1999. Role of Lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol. Microbiol. 33415-428. [DOI] [PubMed] [Google Scholar]
- 23.Luo, P. 2003. Genetic transformation in Streptococcus pneumoniae: regulation by ComX an alternative sigma factor. Ph.D. thesis. University of Illinois—Chicago, Chicago, IL.
- 24.Marmur, J. 1961. A procedure for the isolation of DNA from microorganisms. J. Mol. Biol. 3208-218. [Google Scholar]
- 25.Martin, B., P. Garcia, M. P. Castanie, and J. P. Claverys. 1995. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol. Microbiol. 15367-379. [DOI] [PubMed] [Google Scholar]
- 26.Maurizi, M. R. 1992. Proteases and protein degradation in Escherichia coli. Experientia 48178-201. [DOI] [PubMed] [Google Scholar]
- 27.Mhammedi-Alaoui, A., M. Pato, M. J. Gama, and A. Toussaint. 1994. A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol. Microbiol. 111109-1116. [DOI] [PubMed] [Google Scholar]
- 28.Msadek, T., V. Dartois, F. Kunst, M. L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27899-914. [DOI] [PubMed] [Google Scholar]
- 29.Nair, S., C. Poyart, J. L. Beretti, H. Veiga-Fernandes, P. Berche, and P. Trieu-Cuot. 2003. Role of the Streptococcus agalactiae ClpP serine protease in heat-induced stress defence and growth arrest. Microbiology 149407-417. [DOI] [PubMed] [Google Scholar]
- 30.Nakano, S., M. M. Nakano, Y., Zhang, M. Leelakriangsak, and P. Zuber. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. USA 1004233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oggioni, M. R., and J.-P. Claverys. 1999. Repeated extra-genic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 1452647-2653. [DOI] [PubMed] [Google Scholar]
- 32.Oruga, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6575-597. [DOI] [PubMed] [Google Scholar]
- 33.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21853-862. [DOI] [PubMed] [Google Scholar]
- 34.Piotrowski, A., P. Luo, and D. A. Morrison. 2009. Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of the proteolysis of ComX and ComW. J. Bacteriol. 1913359-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 1843508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, G. T., W. L. Ng, R. Gilmour, and M. E. Winkler. 2003. Essentiality of clpX, but not clpP, clpL, clpC, or clpE, in Streptococcus pneumoniae R6. J. Bacteriol. 1852961-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schelin, J., F. Lindmark, and A. K. Clarke. 2002. The clpP multigene family for the ATP-dependent Clp protease in the cyanobacterium Synechococcus. Microbiology 1482255-2265. [DOI] [PubMed] [Google Scholar]
- 38.Sung, C. K., H. Li, J.-P. Claverys, and D. A. Morrison. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 675190-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sung, C. K., and D. A. Morrison. 2005. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J. Bacteriol. 1873052-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki, C. K., M. Rep., J. M. van Dijl, K. Suda, L. A. Grivell, and G. Schatz. 1997. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem. Sci. 22118-123. [DOI] [PubMed] [Google Scholar]
- 41.Tatsuta, T., T. Tomoyasu, B. Bukau, M. Kitagawa, H. Mori, K. Karata, and T. Ogura. 1998. Heat shock regulation in the ftsH null mutant of Escherichia coli: dissection of stability and activity control mechanisms of sigma32 in vivo. Mol. Microbiol. 30583-593. [DOI] [PubMed] [Google Scholar]
- 42.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 43.Tomoyasu, T., T. Ohkishi, Y. Ukyo, A. Tokumitsu, A. Takaya, M. Suzuki, K. Sekiya, H. Matsui, K. Kutsukake, and T. Yamamoto. 2002. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 176730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viala, J., and P. Mazodier. 2003. The ATPase ClpX is conditionally involved in the morphological differentiation of Streptomyces lividans. Mol. Genet. Genomics 268563-569. [DOI] [PubMed] [Google Scholar]
- 46.Wawrzynow, A., D. Wojtkowiak, J. Marszalek, B. Banecki, M. Jonsen, B. Graves, C. Georgopolous, and M. Zylicz. 1995. The ClpX heat shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 141867-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weart, R. B., S. Nakano, B. E. Lane, P. Zuber, and P. A. Levin. 2005. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol. Microbiol. 57238-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zellmeier, S., W. Schumann, and T. Wiegert. 2006. Involvement of Clp protease activity in modulating the Bacillus subtilis sigma stress response. Mol. Microbiol. 611569-1582. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, Y., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 1801154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]