Structural biology has been extremely successful in providing functional insights for a large number of proteins and macromolecular assemblies, but in some cases, the structure has contributed a three-dimensional (3D) framework to interpret years of accumulated biochemical and genetic knowledge. In these particular systems, structural information has allowed us to learn things that would have been difficult to learn with other techniques.
The periplasmic Escherichia coli DegP protease/chaperone exemplifies this scenario very well. This protein was initially identified in the 1980s (23, 24, 37, 38), and over more than two decades, several groups characterized its activities (2, 12, 16, 27). However, a comprehensive 3D functional model did not become apparent until the first DegP X-ray structure was revealed in 2002 (20). Following the discovery of this remarkable structure, a number of groups concentrated on testing the essentials of the functional model proposed from the crystal structure. Interestingly, this functional model was rewritten recently, after the structures of DegP in two additional oligomeric forms were resolved (13, 22). Current research efforts are now concentrated on probing the new functional model and also on answering new, interesting questions posed by these structures. Therefore, DegP provides an excellent example for the structure-driven study of protein function. This minireview aims to summarize how the functional model for DegP protein has evolved as the structures of the different oligomeric forms of the protein have been elucidated.
E. coli DegP (also called HtrA or protease Do) is an important periplasmic protein with the unusual property of functioning both as a protease and as a chaperone (36). Unlike the cytoplasmic compartment, the periplasm lacks ATP and does not support the function of large protein machines powered by this molecule. However, in this cellular compartment, DegP can still degrade and refold misfolded proteins in an ATP-independent manner (2). Although DegP is not an essential protein, its activity is required for bacterial survival at high temperatures (34) and under harsh environmental conditions. Consequently, its expression is upregulated by both the Cpx and σE protein quality control pathways under conditions of protein-folding stress (3, 29).
DegP homologs have been isolated from a variety of species, including gram-negative and -positive bacteria, plants, and mammals. All these proteins constitute the HtrA family of proteases (2). In bacteria, members of this family are key players mainly in protein quality control in the periplasmic space. In eukaryotic cells, these proteins are involved in functions as diverse as the regulation of apoptosis (1, 4, 9) and the delay of the aggregation process of intracellular amyloid peptides (19). HtrA proteins usually contain a protease domain and at least one C-terminal PDZ domain. In some cases, members of this family of proteins also include additional domains, such as transmembrane regions, located usually at the N terminus. Specifically, in E. coli, DegP, DegQ, and DegS compose the HtrA family. There is a high degree of homology among these three proteins, particularly in the protease domain. However, the number of PDZ domains is variable. DegP and DegQ contain two PDZ domains, whereas DegS contains only one (16).
EARLY STRUCTURAL STUDIES OF DegP
The initial insights into the architecture of the DegP structure and oligomeric state were provided by an electron microscopy study reported in 1999 (18). Negatively stained electron micrographs of a proteolytically inactive S210A variant of DegP (DegPS210A) showed ring-shaped particles and two-layered structures that represent end-on and side-view orientations of DegP oligomers. Subsequent rotational analyses identified statistically significant sixfold rotational symmetry, and the study concluded that in solution DegP behaves as a dodecamer consisting of two stacked hexameric rings (18).
This early study, although limited in resolution, already established that DegP has a structure consistent with a self-compartmentalized protease. Interestingly, many important proteases involved in protein quality control in bacteria and eukaryotic cells also have self-compartmentalized architecture. The common feature for all these enzymes is that the proteolytic site performing the substrate cleavage is sequestered within a digestion chamber and only substrates in an unfolded state reach this chamber. Quite commonly for these enzymes, substrate entry occurs through an axial pore.
Later studies (20) showed that the initial assignment of six subunits per ring in the DegP structure was incorrect. However, the finding that DegP is a self-compartmentalized protease was important because of all the immediate functional information that could be inferred about the enzyme, based on the structural homology to other proteases sharing this architecture.
FIRST CRYSTAL STRUCTURE OF DegP: THE HEXAMERIC FORM
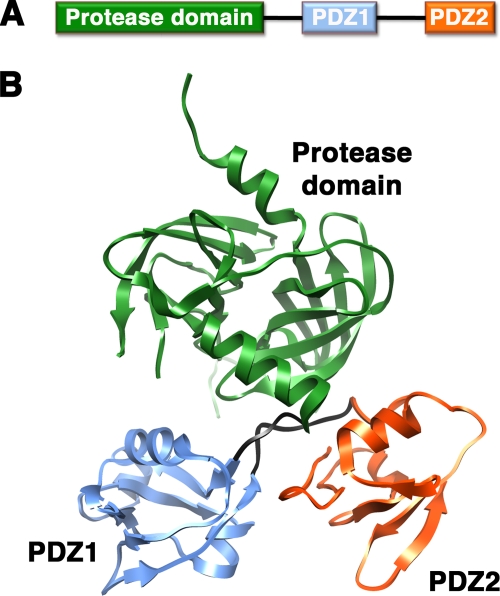
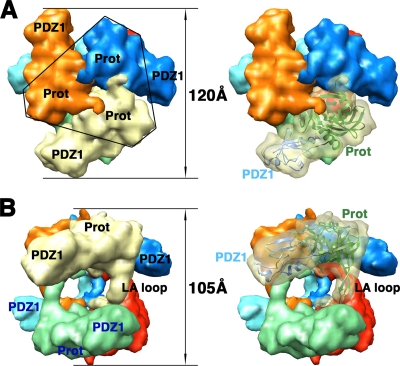
In 2002, Krojer et al. presented the first crystal structure of a hexameric form of DegP (20). The DegP monomer has modular architecture formed by a trypsin-like protease domain and two PDZ domains (PDZ1 and PDZ2) (Fig. 1A) containing the conserved structural features (8) that allow these domains to interact with the C-terminal residues of their substrates. Two flexible loops link the protease domain to the PDZ1 domain and PDZ1 to the PDZ2 domain (Fig. 1B). The X-ray structure revealed that DegP oligomerizes into a hexamer that encloses its proteolytic sites into a central chamber (Fig. 2 and 3). The hexamer is formed by the staggered association of two trimeric rings. Residues exclusively from the protease domains stabilize each trimer (Fig. 2A and 3A). Every monomer extends a loop (the LA loop) from its protease domain that winds around the LA loop of the opposite monomer, defining three spacing pillars between the two opposite trimers (Fig. 2B). The DegP hexamer was stabilized in the crystal form in two different conformations, open (Fig. 2) and closed (Fig. 3), which suggests movement (or flexibility) of the PDZ domains. In the open conformation, the PDZ domains are in an extended position that allows free access between the spacing pillars to the inner chamber (Fig. 2B), but access is blocked by the PDZ domains in the closed position (20) (Fig. 3B).
FIG. 1.
Structure of the DegP monomer. (A) Linear diagram of the DegP protein. (B) Ribbon representation of the X-ray structure of the DegP monomer. The protease domain is colored in green, the PDZ1 domain is in blue, and the PDZ2 domain is in orange. The loops between the protease domain and the PDZ1 domain and between both PDZ domains are shown in black. This image of DegP was prepared from PDB file 1KY9 by using the Chimera program (28).
FIG. 2.
Structure of the DegP hexamer in the open conformation. (A) Surface representations of the DegP hexameric cage in the open conformation, viewed along the threefold axis. Prot, protease domain. (B) View of the same structure after rotation of the view in panel A by ∼90° along an axis orthogonal to the threefold symmetry axis. In the open conformation, the PDZ domains do not cover the side entrances to the inner chamber. Each monomer is represented in a different color. One LA loop and the protease and PDZ1 domains within the same trimer are labeled in black. Domains of the opposite trimer are labeled in blue. The structure of the PDZ2 domain in the open conformation was not resolved. In the images on the right, one monomer is shown as a semitransparent surface and a ribbon representation shows the DegP monomer, with its protease and PDZ1 domains colored in green and blue, respectively, docked into the volume. Labels for both domains follow the color code described above. These images were prepared from PDB file 1KY9. An electron density map limited at a resolution of 10 Å was first obtained from the X-ray structure by using the EMAN program (25), and the Chimera program (28) was used to produce surface representations from this map.
FIG. 3.
Structure of the DegP hexamer in the closed conformation. Surface representations are shown as viewed along the threefold axis (A) and from the side (B). The color and label schemes are identical to those in Fig. 2. In this conformation, the PDZ domains from each monomer within a trimer (labeled in black) interact with the PDZ domains of the monomers in the opposite trimer (labeled in blue), closing the side entrances to the inner chamber. The surface representations on the right each show one of the monomers as a ribbon docked into the semitransparent volume. These images were prepared from PDB file 1KY9 by using the EMAN (25) and Chimera (28) programs as described in the legend to Fig. 2. Prot, protease domain.
The hexameric DegP represented by the X-ray structure was immediately accepted as the physiologically functional form of the protein, and the structure provided the basis for the first comprehensive functional model of DegP. The restricted access to the catalytic site confirmed the self-compartmentalized architecture of the protease and also led to the proposal that DegP could work in a way similar to that of other proteases with an enclosed catalytic site, such as the Clp proteases and the proteasome. In these enzymes, the unfolded substrate is translocated into the inner chamber, where degradation occurs. However, the DegP crystal structure also suggested several differences from these other, well-characterized systems. For instance, because the axial pores of the hexameric cage are completely blocked by the protease domain (Fig. 2A and 3A), substrate access to the inner chamber was proposed to be lateral, and the movement of the PDZ domains suggests that they act as gates to the entrances. One of the attractive aspects of this functional model was that it coupled the potential role of the PDZ domains in substrate-specific recognition with their potential roles in feeding the substrate into the chamber and acting as gatekeepers.
UNRESOLVED FUNCTIONAL ASPECTS OF THE DegP HEXAMERIC FORM
In the years following the publication of the DegP hexameric structure, several studies probed the different aspects of the proposed functional model. For instance, experimental support for the role of the PDZ domains in the recognition of substrates targeted for degradation was provided (11, 21).
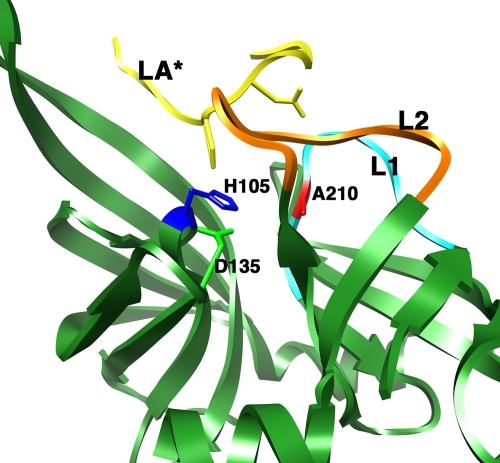
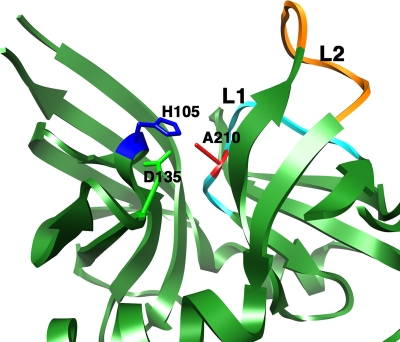
However, a number of functional aspects could not be explained from the existing X-ray structure. The most intriguing unresolved issue was the regulation of the protease/chaperone switch. How the fate of the substrate for this dual-function protease/chaperone enzyme is determined and what governs whether the substrate will be refolded or degraded have been long-standing questions in the field. Initially, it was suggested that temperature is an important factor in this process. By using the E. coli periplasmic MalS protein, a natural substrate for DegP, it was observed that the chaperone function of DegP dominates at temperatures below 28°C and that the proteolytic activity increases dramatically at temperatures above 28°C (36). These results, interpreted in the context of the X-ray structure of hexameric DegP, led to the hypothesis that it is the temperature-regulated conformation of the active-site residue Ser-210 that determines whether the substrate will be refolded or degraded. At low temperatures, the serine is in an inactive conformation away from the other residues of the catalytic triad and only the chaperone activity is carried out. However, it was proposed that elevated temperatures may induce a conformational change in Ser-210, assembling a functional catalytic triad and causing increased proteolytic activity to dominate over chaperone activity. The X-ray structure of the DegP hexamer (20) was consistent with this model, as the protein was crystallized at a low temperature and captured in the chaperone conformation. In this structure, the LA loop of each monomer protrudes into the active site of the subunit in the opposite trimer, forcing the L1 and L2 loops into a twisted, inactive conformation (Fig. 4). Although the model of a temperature-induced protease/chaperone switch was not proven by the resolution of a structure for DegP at a high temperature, the findings of a recent study performed with Thermotoga maritima HtrA (17) provided experimental support for the hypothesis of a temperature-induced conformational change in the catalytic site. Spin-labeling electron paramagnetic resonance and fluorescence spectroscopy experiments showed that a helical lid covering the T. maritima HtrA active site is lifted up at elevated temperatures, allowing the substrate access to the catalytic triad.
FIG. 4.
Context of the catalytic triad in the DegP hexamer. The ribbon representation depicts the area of the protease domain containing the catalytic triad in one of the monomers within the hexameric DegP structure. Mechanistically important loops L1 (cyan) and L2 (orange) are highlighted. The LA* loop shown in the figure is from the opposite monomer in the hexameric structure. Residues of the catalytic triad (D135, H105, and A210) are shown in stick mode.
Subsequent studies also suggested that DegP works as a chaperone under heat shock conditions (31) or exerts its proteolytic activity at low temperatures (33). Although the in vivo relevance of the DegP chaperone and protease activities at high and low temperatures, respectively, is unclear, these two studies certainly raise the possibility that other factors besides temperature control the protease/chaperone switch in the cell. In this context, it was found that neither of the PDZ domains is required for DegP chaperone activity but that the PDZ1 domain is essential for protease activity, as it permits the recognition of the substrate molecule (11, 36). Therefore, it is also possible that DegP may recognize substrate molecules targeted for degradation and refolding in different manners and that the mechanism through which DegP recognizes the substrate may also play a role in the protease/chaperone switch.
Certainly, the X-ray model of the DegP hexameric form showed that, structurally, the enzyme is a self-compartmentalized protease, as the catalytic triads of the individual monomers are sequestered within an inner chamber. However, there was no experimental evidence showing whether substrates are channeled into the chamber for degradation or refolding. Some of the structural features of the DegP hexamer were, in fact, difficult to reconcile with this working model. In particular, the inner chamber housing the proteolytic active sites is large enough to accommodate little more than individual secondary structure elements (Fig. 2B), and it was difficult to imagine how DegP could accommodate large, unfolded substrates or aggregates in the process of being refolded. This model was a snapshot of an isolated enzyme, and it is clear now that the presence of a substrate is required to see the functionally active structure of DegP.
A mutational study (14) challenged the idea that substrates were threaded into the interior of the hexameric cage for degradation or refolding. In this study, several DegP mutants were constructed with LA loops shorter than that of the wild type and therefore predicted to have reduced cage dimensions. Although the actual sizes of the cages were not directly confirmed by structural methods, surprisingly, these mutants maintained both chaperone and protease activities. This study raised, for the first time, the possibilities that substrates could gain access to the catalytic sites through the transient disassembly of the hexameric cage and that the interaction of DegP with a specific substrate could be the initial event promoting the disassembly of the hexamer. In this context, the hexameric form of DegP was proposed to constitute a double-layered safety mechanism (14), firstly protecting the cell from uncontrolled proteolysis by physically limiting the access of folded proteins to the catalytic sites and secondly maintaining the catalytic triad in an inactive conformation. Although this study suggested already that the DegP trimer was the functional unit of the enzyme, it did not provide experimental evidence regarding whether trimeric DegP or other oligomeric forms carry out the degradation or refolding of the substrate.
STRUCTURES OF DegP FUNCTIONAL FORMS: THE LARGE DegP24 AND DegP12 OLIGOMERS
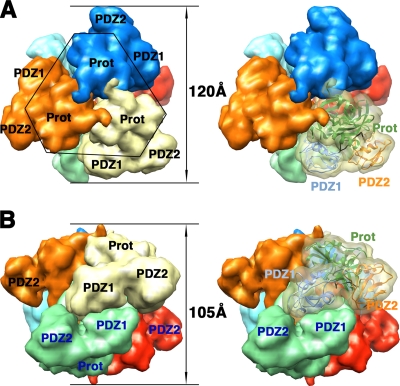
Recently, the structures of novel oligomeric forms of the DegP protein have been elucidated by X-ray crystallography and cryo-electron microscopy (cryo-EM). Two independent groups (13, 22) reported the structures of large 12-mer (DegP12) and 24-mer (DegP24) oligomeric cages. Both oligomers are induced in the presence of protein substrates and are made of identical trimeric units (Fig. 5 and 6) that are structurally very similar to the trimers forming the DegP hexameric form (Fig. 2B and 3B). Probably, the requirement for the presence of the substrate to trigger the reorganization of hexameric DegP into these larger cages explains why the 12-mer and 24-mer oligomeric forms were described just recently and not at the time the structure of the hexameric form was first obtained.
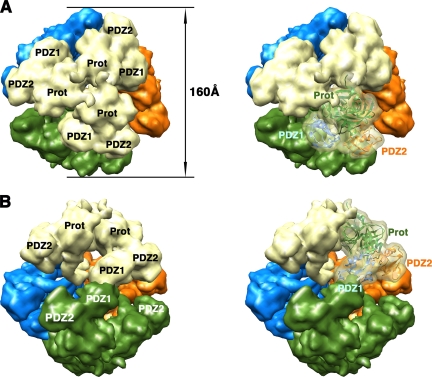
FIG. 5.
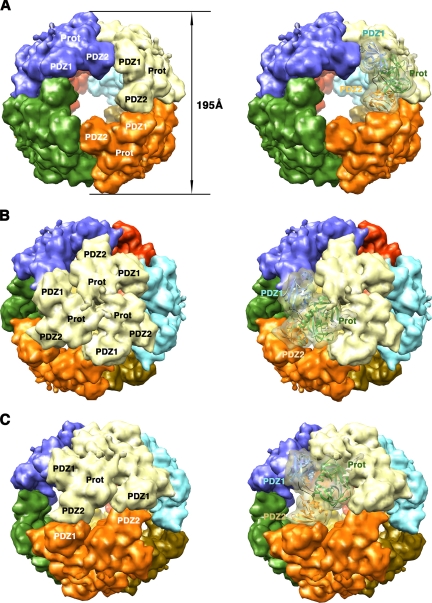
Structure of the DegP24 oligomer. Surface representations of the 24-mer DegP cage are shown as viewed along the fourfold (A), threefold (B), and twofold (C) axes. Each DegP trimer is represented in a different color. In the images on the right, one of the monomers is shown as a semitransparent volume containing the ribbon representation of the DegP monomer, color coded as described in the legend to Fig. 1. These images were prepared from PDB file 3CS0 by using the EMAN (25) and Chimera (28) programs as described in the legend to Fig. 2. Prot, protease domain.
FIG. 6.
Structure of the DegP12 oligomer viewed along the threefold (A) and approximate twofold (B) axes. Each trimer is represented in a different color. Images on the right show one of the DegP monomers as a ribbon representation docked into a semitransparent volume. Labels for this monomer follow the same color code used in Fig. 1. These images were prepared from PDB file 2ZLE by using the EMAN (25) and Chimera (28) programs as described in the legend to Fig. 2. Prot, protease domain.
The DegP24 cage is 195 Å in diameter, and the eight trimers in the structure are located at the vertices of an octahedron, forming a shell that encloses a large cavity 110 Å in diameter (Fig. 5A). The four trimeric units in the DegP12 oligomer form a tetrahedral shell about 160 Å in diameter surrounding a central cavity 78 Å in diameter (Fig. 6) (22). In both oligomeric forms, interactions between neighboring trimers occur side by side through PDZ domains, leaving pores in between wide enough to allow small folded proteins to diffuse in and out of the inner cavity (Fig. 5A, 5C, and 6B). The DegP24 structure obtained by X-ray crystallography (22) and cryo-EM (13) showed that all these PDZ domain interactions are identical and occur between the PDZ1 domain of one trimeric unit and the PDZ2 domain of the contiguous trimeric unit in a pattern that allows each trimer to interact with three other trimers in the oligomer (Fig. 5B). For the DegP12 oligomer, the available structures show discrepancies regarding how the interaction between trimers occurs. One of the cryo-EM structures (22) showed that the intertrimer contacts are made by adjacent PDZ1 domains and do not seem to involve the PDZ2 domain (Fig. 6B). However, a second, higher-resolution cryo-EM reconstruction (13) revealed that these contacts in the DegP12 cages are almost identical to the ones observed in the DegP24 structure and also involve the PDZ1 and PDZ2 domains (data not shown; the structure is not available in the Protein Data Bank [PDB]).
Both oligomeric states follow the architecture of a self-compartmentalized protease because the catalytic sites are enclosed within the inner cavity (Fig. 5 and 6). However, different from the DegP hexamer, in which the LA loop protrudes into the active site, the DegP24 and DegP12 oligomers have the LA loop displaced and the L1 and L2 loops set up as a functional proteolytic site (22) (Fig. 7).
FIG. 7.
Catalytic triad in the DegP24 oligomer. The view of the catalytic site of one DegP monomer within the structure of the 24-mer cage is analogous to the one shown in Fig. 4. The assembly of DegP into a large 24-mer cage removes the LA loop from an opposing monomer (not shown) and aligns the residues of the catalytic triad (D135, H105, and A210) into a functional conformation. L1 and L2 loops are shown in cyan and orange, respectively, and the residues of the catalytic triad are shown in stick mode.
FUNCTIONAL IMPLICATIONS FROM THE STRUCTURES OF THE LARGE DegP24 AND DegP12 OLIGOMERS
Important observations of the two studies (13, 22) describing the new DegP multimeric structures were that these large 12-mer and 24-mer spherical cages are induced by the presence of protein substrates and that once substrate degradation is completed, DegP reverts to the hexameric form. This finding, along with the new structures, redefined the functional model for DegP with the proposal that the DegP hexameric form is in fact the resting state of the enzyme and that the binding of substrate molecules triggers DegP reorganization into large oligomeric cages that are the functional forms for both the protease and the chaperone activities (13, 22).
Interestingly, the new structural work also provided, for the first time, experimental evidence for the encapsulation of the substrate into the internal cavity for degradation or refolding. The authors of one study (22) detected a density in the inner cavity of one of the cryo-EM DegP12 structures that could be assigned to a mostly folded substrate molecule. The internal density was not present in all the other structures obtained by either cryo-EM or X-ray crystallography (13, 22). This discrepancy is explained by the limitations of both techniques. Most likely, substrate molecules in the DegP internal cavity are in different conformational states, and probably their locations are also variable, causing the densities representing substrate molecules in the inner cavity to fade out during the averaging process inherent in structure determination (5, 6, 10).
Regarding the mechanism of enclosing substrate molecules into the inner cavity, the existing data (13, 22) are consistent with a model in which, upon substrate binding, DegP hexamers dissociate and reassociate as large cages around the substrate. Conversion between oligomeric states occurs probably via the dissociation and reassociation of trimers, based on the structural similarity of the trimeric units in the different DegP oligomeric forms. However, the possibility that the substrate can also be threaded through the large pores in the shells of the cages is not completely ruled out by the existing experimental evidence.
Another relevant question with respect to this functional model is whether refolding and degradation always occur after the substrate has been internalized into the inner chamber of one of the large cages. Alternatively, it is possible that under specific conditions DegP may act as a trimer without previously enclosing the substrate. Regarding this possibility, Jiang et al. (13) observed a positive effect on DegP-specific activity when increasing concentrations of proteolytically inactive DegPS210A were added. Such a concentration effect was not detected when a non-cage-forming DegP mutant (DegP ΔPDZ2) was added. Similarly, the degradation of a chromogenic peptide substrate that does not induce the formation of large cages was significantly accelerated in the presence of lysozyme, a protein substrate that induces the reorganization of DegP into large cages (22). These results suggest that substrate degradation occurs more efficiently in the context of the large cages. However, the DegP ΔPDZ2 mutant that is unable to form the large 12-mer and 24-mer cages is fully active, and this finding argues against the necessity of enclosing the substrate in a cavity as the only possible mechanism for DegP to perform its degradation or refolding activity. Nevertheless, it is likely that in vivo, native HtrA degrades substrates within the larger cages.
In addition, if conversion between oligomeric states occurs via the dissociation and reassociation of trimers and DegP trimeric mutants are active (13), how does the enzyme avoid performing any activity as a trimer during the oligomeric state transition? Jiang et al. (13) found that the PDZ2 domain exerts an inhibitory role and prevents DegP activity during the oligomeric state transition. A similar inhibition of activity mediated by the PDZ domain has been described in detail for the proteolytic activity of the human homolog of DegP, the HtrA2 protein (7). The assembly of the large cages in DegP releases the inhibitory effect of the PDZ2 domain, as this domain becomes involved in interaction with the PDZ1 domain of a neighboring trimer to stabilize the cage structure.
The structures of the large cages also provided new insights into the temperature protease/chaperone switch in DegP, showing that the LA loop that protrudes into the active site in the hexameric structure (Fig. 4) is displaced in the larger cages and that the L1 and L2 loops are set up into a functional proteolytic site (22) (Fig. 7). Therefore, it is clear from these structures that conversion from hexamers into the 12-mer and 24-mer structures is a crucial step for initiating the protease activity. However, the existence of an active catalytic triad does not imply that every substrate molecule internalized in the inner cavities of the large cages is degraded. In fact, Krojer et al. (22) determined that the folding state of the enclosed substrate is an important factor in deciding the substrate fate. The authors found that DegP degrades unfolded outer membrane proteins (OMPs) but stabilizes folded OMP protomers, concluding that the fate of the encapsulated protein depends on the substrate folding state and the ability of the substrate to adopt its native conformation fast enough to escape the cleaving machinery of DegP. Temperature is certainly a key factor regulating the folding state and the kinetics of a specific protein. Therefore, this result partially explains the proposed role of temperature in the protease/chaperone switch (36). Temperatures lower than 28°C may allow substrate molecules to readily fold into their native conformations, escaping DegP degradation activity and making DegP exhibit mostly chaperone activity. However, incubation at higher temperatures may induce protein misfolding and expose protease cleavage sites to the extent that substrate degradation occurs before the native state is reached. Alternatively, some proteins may adopt a partially unfolded state at high temperatures, which leaves them susceptible to degradation as well. Nevertheless, the structures available have not been resolved for a temperature range wide enough to rule out any effect of the temperature on the structures of the cages themselves. High-resolution structural analyses of the 12-mer and 24-mer cages at temperatures near 42°C should clarify whether temperature also affects DegP structure in a way that promotes its chaperone activity at temperatures below 28°C and its protease activity at higher temperatures.
ROLE OF MEMBRANE LIPIDS IN REGULATING THE LOCATION, OLIGOMERIC STATE, AND ACTIVITY OF DegP
DegP is located in the bacterial periplasmic space, and one important aspect of the functional model for DegP is whether the enzyme is attached to the membrane by interaction with the lipids. An earlier study suggested that DegP has affinity for the periplasmic side of the inner membrane and that it interacts in vitro with negatively charged head groups of phosphatidylglycerol and cardiolipin (32). The crystal structure of DegP24 (22), in fact, indicates that the enzyme contains clusters of lysine and arginine residues located in the PDZ domains, producing a positive charge around the entrances of the large pores of the cage that seems to be important to target cellular membranes. Accordingly, the authors of the study describing the structure (22) proposed that the large 24-mer cage may be wedged between the inner and outer bacterial membranes, acting as a macropore that allows the diffusion of OMP precursors to the outer membrane. However, no in vivo evidence of these macropores is available yet.
A very recent study (30) has also investigated whether membrane lipids have any effect on the activity and oligomeric state of DegP. The authors found that DegP forms bowl-shaped structures on lipid monolayers in the absence of any substrate protein. Electron microscopy images revealed three types of oligomeric bowl-shaped structures, with four-, five-, and sixfold symmetry. The symmetry axis of each structure is always perpendicular to the membrane plane. Negatively stained electron micrographs were used to create 3D reconstructions showing that the three structures are made of the same building block, which is a DegP trimer. In every structure, the trimeric units are positioned around the symmetry axis and associate with one another through lateral interactions mediated by the PDZ domains, similar to the contacts depicted in the DegP24 crystal structure (22). Interaction between trimers is flexible and allows for the assembly of all the different oligomers with their different curvatures: from the most concave structure, which is the one with fourfold symmetry that fits the structure of exactly one-half of the 24-mer cage, to the almost flat plane adopted by the oligomer with sixfold symmetry.
Interestingly, in the presence of the liposomes that induce the formation of the bowl-shaped structures, DegP shows dramatically higher proteolytic activity than that of lipid-free DegP when protein substrates are provided (30). These results suggest the possibility that in the cell, membrane lipids may induce the formation of bowl-shaped structures. These oligomers may constitute a reservoir of partially assembled 12-mer and 24-mer cages that quickly assemble around the substrate into functionally active large cages that cleave unfolded protein. Therefore, it is tempting to speculate that by having DegP organized into the bowl-shaped structures rather than in the hexameric form, the assembly of the large cages and the subsequent degradation of the substrate occur faster. Conversely, the chaperone activity decreases when the reaction is initiated with a solution containing lipid-induced bowl-shaped structures (30). This result indicates that the bowl-shaped structures may constitute the reservoir for the protease form but not for the chaperone form of DegP and suggests one additional factor influencing the fate of the substrate. Multiple investigations summarized in this review have considered the roles of several factors, such as temperature (36), the mechanism of substrate recognition (11), and the folding state of the substrate (22), in regulating the protease/chaperone switch. The recent results (30) are consistent with a model in which the fate of the substrate is also determined by regulating the DegP reservoir of bowl-shaped structures versus DegP hexamers. The question to be addressed in future studies is how the influence from all the described factors is integrated by DegP to decide whether the substrate will be refolded or degraded.
TRANSITION BETWEEN OLIGOMERIC STATES ALSO PROVIDES OPPORTUNITY FOR SELF-REGULATION OF THE LEVEL OF DegP
The structural information about DegP has also provided a framework for understanding how the cell eliminates excess DegP once the stress conditions that induced its overexpression and activity are overcome.
DegP is upregulated to quickly eliminate unfolded and damaged proteins during cellular stress (3, 29). Also, its proteolytic activity is allosterically enhanced in the presence of small peptides that mimic either unfolded substrates or hydrolysis products (26). Interestingly, peptides produced during substrate hydrolysis also induce self-cleavage of DegP (15), eventually leading to the elimination of the enzyme. These findings suggest that the autocleavage process of DegP, first described by Skorko-Glonek et al. (35), aims to eliminate the excess DegP produced under stress conditions once its enzymatic activities are no longer needed (15).
Two important observations (15) are that the hexameric cage structure is required for the autocleavage of DegP and that the cleaved DegP forms appear late in the reaction, once the full-length substrate is degraded. In the context of the available structural information about DegP, these findings suggest that while the full-length substrate is present, DegP remains mostly in the forms of 12-mer and 24-mer oligomeric cages and limited amounts of self-degradation occur. However, once substrate degradation is completed, DegP reassembles into the hexameric form and cleaves itself.
The X-ray structure of the 24-mer cage (22) is consistent with this hypothesis. In the large cages, the LA loop is displaced from the neighboring L1 and L2 loops to allow the formation of a properly functional active site (Fig. 7). However, in the hexameric form (20), the LA loop protrudes into the active site of the subunit in the opposite trimer, forcing the L1 and L2 loops into an inactive conformation (Fig. 4). Upon the completion of substrate degradation, the protease returns to its hexameric form, but peptides resulting from substrate hydrolysis allosterically stimulate HtrA proteolytic activity. It is likely that peptides stimulating HtrA may keep the catalytic L1 and L2 loops in an active conformation. Consequently, the LA loop may be cleaved because it is placed back into close proximity to the catalytic site, which may now be in an active conformation. Membrane lipids may also play a role in preventing the reassembly of DegP into hexamers by maintaining the enzyme in bowl-shaped oligomers and consequently delaying the autocleavage process. Additional structures and studies are required to provide experimental evidence supporting the proposal of this regulatory mechanism.
OUTLOOK
The functional model of DegP has evolved as the structures of its different oligomeric states have been elucidated. It is now clear that DegP adopts various forms, depending on the surrounding environment and the availability of a substrate. However, as described in this minireview, there are still a number of areas that require additional work. A very intriguing aspect of the DegP mechanism is how the protein integrates the information from the substrate and the environment and decides whether the substrate will be refolded or degraded. The recently observed bowl-shaped structures formed by DegP on lipid monolayers in the absence of any substrate protein are also an exciting discovery that speaks about the potential roles of membrane lipids in regulating DegP activity. Additional research is needed to elucidate how these specific structures fit into the functional model for the protein. Other pending questions are, for instance, whether different oligomeric forms target certain substrates or are dedicated to specific functions within the periplasmic space and to what degree this mechanism of transitioning between different oligomeric states extends to other members of the HtrA family. Certainly, additional structures will explain these and other aspects of the molecular mechanisms of DegP that are still unclear.
Acknowledgments
Images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH grant P41 RR-01081). J.O. acknowledges support from the Canadian Institutes of Health Research (CIHR) and from an Early Researcher Award from the Ministry of Research and Innovation.
Footnotes
Published ahead of print on 22 May 2009.
REFERENCES
- 1.Beal, M. F. 2005. Mitochondria take center stage in aging and neurodegeneration. Ann. Neurol. 58495-505. [DOI] [PubMed] [Google Scholar]
- 2.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10443-455. [DOI] [PubMed] [Google Scholar]
- 3.Duguay, A. R., and T. J. Silhavy. 2004. Quality control in the bacterial periplasm. Biochim. Biophys. Acta 1694121-134. [DOI] [PubMed] [Google Scholar]
- 4.Falkevall, A., N. Alikhani, S. Bhushan, P. F. Pavlov, K. Busch, K. A. Johnson, T. Eneqvist, L. Tjernberg, M. Ankarcrona, and E. Glaser. 2006. Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J. Biol. Chem. 28129096-29104. [DOI] [PubMed] [Google Scholar]
- 5.Frank, J. 1989. Image analysis of single macromolecules. Electron Microsc. Rev. 253-74. [DOI] [PubMed] [Google Scholar]
- 6.Frank, J. 1989. Three-dimensional imaging techniques in electron microscopy. BioTechniques 7164-173. [PubMed] [Google Scholar]
- 7.Gupta, S., R. Singh, P. Datta, Z. Zhang, C. Orr, Z. Lu, G. Dubois, A. S. Zervos, M. H. Meisler, S. M. Srinivasula, T. Fernandes-Alnemri, and E. S. Alnemri. 2004. The C-terminal tail of presenilin regulates Omi/HtrA2 protease activity. J. Biol. Chem. 27945844-45854. [DOI] [PubMed] [Google Scholar]
- 8.Harris, B. Z., and W. A. Lim. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 1143219-3231. [DOI] [PubMed] [Google Scholar]
- 9.Hegde, R., S. M. Srinivasula, Z. Zhang, R. Wassell, R. Mukattash, L. Cilenti, G. DuBois, Y. Lazebnik, A. S. Zervos, T. Fernandes-Alnemri, and E. S. Alnemri. 2002. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 277432-438. [DOI] [PubMed] [Google Scholar]
- 10.Ilari, A., and C. Savino. 2008. Protein structure determination by x-ray crystallography. Methods Mol. Biol. 45263-87. [DOI] [PubMed] [Google Scholar]
- 11.Iwanczyk, J., D. Damjanovic, J. Kooistra, V. Leong, A. Jomaa, R. Ghirlando, and J. Ortega. 2007. The role of the PDZ domains in Escherichia coli DegP protein. J. Bacteriol. 1893176-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffery, C. J. 2004. Molecular mechanisms for multitasking: recent crystal structures of moonlighting proteins. Curr. Opin. Struct. Biol. 14663-668. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, J., X. Zhang, Y. Chen, Y. Wu, Z. H. Zhou, Z. Chang, and S. F. Sui. 2008. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl. Acad. Sci. USA 10511939-11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jomaa, A., D. Damjanovic, V. Leong, R. Ghirlando, J. Iwanczyk, and J. Ortega. 2007. The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity. J. Bacteriol. 189706-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jomaa, A., J. Iwanczyk, J. Tran, and J. Ortega. 2009. Characterization of the autocleavage process of the Escherichia coli HtrA protein: implications for its physiological role. J. Bacteriol. 1911924-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, D. Y., and K. K. Kim. 2005. Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38266-274. [DOI] [PubMed] [Google Scholar]
- 17.Kim, D. Y., E. Kwon, Y. K. Shin, D. H. Kweon, and K. K. Kim. 2008. The mechanism of temperature-induced bacterial HtrA activation. J. Mol. Biol. 377410-420. [DOI] [PubMed] [Google Scholar]
- 18.Kim, K. I., S. C. Park, S. H. Kang, G. W. Cheong, and C. H. Chung. 1999. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli. J. Mol. Biol. 2941363-1374. [DOI] [PubMed] [Google Scholar]
- 19.Kooistra, J., J. Milojevic, G. Melacini, and J. Ortega. A new function of human HtrA2 as an amyloid-β oligomerization inhibitor. J. Alzheimer's Dis., in press. [DOI] [PMC free article] [PubMed]
- 20.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416455-459. [DOI] [PubMed] [Google Scholar]
- 21.Krojer, T., K. Pangerl, J. Kurt, J. Sawa, C. Stingl, K. Mechtler, R. Huber, M. Ehrmann, and T. Clausen. 2008. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc. Natl. Acad. Sci. USA 1057702-7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krojer, T., J. Sawa, E. Schafer, H. R. Saibil, M. Ehrmann, and T. Clausen. 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453885-890. [DOI] [PubMed] [Google Scholar]
- 23.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 1711574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipinska, B., S. Sharma, and C. Georgopoulos. 1988. Sequence analysis and regulation of the htrA gene of Escherichia coli: a sigma 32-independent mechanism of heat-inducible transcription. Nucleic Acids Res. 1610053-10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 12882-97. [DOI] [PubMed] [Google Scholar]
- 26.Meltzer, M., S. Hasenbein, P. Hauske, N. Kucz, M. Merdanovic, S. Grau, A. Beil, D. Jones, T. Krojer, T. Clausen, M. Ehrmann, and M. Kaiser. 2008. Allosteric activation of HtrA protease DegP by stress signals during bacterial protein quality control. Angew. Chem. (International ed. in English) 471332-1334. [DOI] [PubMed] [Google Scholar]
- 27.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26209-221. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz, N., and T. J. Silhavy. 2005. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 8122-126. [DOI] [PubMed] [Google Scholar]
- 30.Shen, Q. T., X. C. Bai, L. F. Chang, Y. Wu, H. W. Wang, and S. F. Sui. 2009. Bowl-shaped oligomeric structures on membranes as DegP's new functional forms in protein quality control. Proc. Natl. Acad. Sci. USA 1064858-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skorko-Glonek, J., E. Laskowska, A. Sobiecka-Szkatula, and B. Lipinska. 2007. Characterization of the chaperone-like activity of HtrA (DegP) protein from Escherichia coli under the conditions of heat shock. Arch. Biochem. Biophys. 46480-89. [DOI] [PubMed] [Google Scholar]
- 32.Skorko-Glonek, J., B. Lipinska, K. Krzewski, G. Zolese, E. Bertoli, and F. Tanfani. 1997. HtrA heat shock protease interacts with phospholipid membranes and undergoes conformational changes. J. Biol. Chem. 2728974-8982. [DOI] [PubMed] [Google Scholar]
- 33.Skorko-Glonek, J., A. Sobiecka-Szkatula, J. Narkiewicz, and B. Lipinska. 2008. The proteolytic activity of the HtrA (DegP) protein from Escherichia coli at low temperatures. Microbiology 1543649-3658. [DOI] [PubMed] [Google Scholar]
- 34.Skorko-Glonek, J., A. Wawrzynow, K. Krzewski, K. Kurpierz, and B. Lipinska. 1995. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene 16347-52. [DOI] [PubMed] [Google Scholar]
- 35.Skorko-Glonek, J., D. Zurawa, F. Tanfani, A. Scire, A. Wawrzynow, J. Narkiewicz, E. Bertoli, and B. Lipinska. 2003. The N-terminal region of HtrA heat shock protease from Escherichia coli is essential for stabilization of HtrA primary structure and maintaining of its oligomeric structure. Biochim. Biophys. Acta 1649171-182. [DOI] [PubMed] [Google Scholar]
- 36.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97339-347. [DOI] [PubMed] [Google Scholar]
- 37.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 1712689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swamy, K. H., C. H. Chung, and A. L. Goldberg. 1983. Isolation and characterization of protease do from Escherichia coli, a large serine protease containing multiple subunits. Arch. Biochem. Biophys. 224543-554. [DOI] [PubMed] [Google Scholar]