Abstract
The citrulline ureidase (CTU) activity has been shown to be associated with highly virulent Francisella tularensis strains, including Schu S4, while it is absent in avirulent or less virulent strains. A definitive role of the ctu gene in virulence and pathogenesis of F. tularensis Schu S4 has not been assessed; thus, an understanding of the significance of this phenotype is long overdue. CTU is a carbon-nitrogen hydrolase encoded by the citrulline ureidase (ctu) gene (FTT0435) on the F. tularensis Schu S4 genome. In the present study, we evaluated the contribution of the ctu gene in the virulence of category A agent F. tularensis Schu S4 by generating a nonpolar deletion mutant, the Δctu mutant. The deletion of the ctu gene resulted in loss of CTU activity, which was restored by transcomplementing the ctu gene. The Δctu mutant did not exhibit any growth defect under acellular growth conditions; however, it was impaired for intramacrophage growth in resting as well as gamma interferon-stimulated macrophages. The Δctu mutant was further tested for its virulence attributes in a mouse model of respiratory tularemia. Mice infected intranasally with the Δctu mutant showed significantly reduced bacterial burden in the lungs, liver, and spleen compared to wild-type (WT) Schu S4-infected mice. The reduced bacterial burden in mice infected with the Δctu mutant was also associated with significantly lower histopathological scores in the lungs. Mice infected with the Δctu mutant succumbed to infection, but they survived longer and showed significantly extended median time to death compared to that shown by WT Schu S4-infected mice. To conclude, this study demonstrates that ctu contributes to intracellular survival, in vivo growth, and pathogenesis. However, ctu is not an absolute requirement for the virulence of F. tularensis Schu S4 in mice.
Francisella tularensis, the etiological agent of tularemia, is a category A bioterrorism agent. High infectivity, ease of intentional aerosol dissemination, and lack of a licensed vaccine have made Francisella a potential biowarfare agent (5, 12, 34). The two major subspecies of Francisella have been divided on the basis of virulence, epidemiological distribution, and biochemical reactions (51). F. tularensis subspecies tularensis (type A strain) is highly virulent and the major cause of tularemia in North America, whereas F. tularensis subspecies holarctica (type B strain), prevalent in Europe and Asia, is less virulent. Biochemically, type A strains produce acid from glycerol and exhibit citrulline ureidase (CTU) activity, while type B strains do not exhibit these activities (21). In contrast to these biochemical differences, very limited variation is seen at the genetic level (25, 41), suggesting that differences in virulence between type A and B strains may arise from differential gene expression by nearly homologous genomes. The highly virulent Schu S4 strain represents type A F. tularensis subspecies tularensis and was originally isolated from a clinical case of tularemia in Ohio in 1941. To date, only a few virulence-associated genes have been characterized in this strain (22, 36, 37, 48), and its virulence determinants still remain poorly understood.
CTU, a member of the carbon-nitrogen hydrolase family protein encoded by the F. tularensis genome (FTT0435), degrades citrulline into ornithine, carbon dioxide, and ammonia (10). Citrulline is generated during the catabolism of arginine by bacterial arginine deiminase (ADI) (40, 47). Ornithine generated by citrulline degradation is either exchanged for arginine by an arginine-ornithine transporter or utilized for the generation of polyamines and energy in the form of ATP (40). Citrulline is also produced by macrophages during conversion of l-arginine and oxygen to nitric oxide (NO) by inducible NO synthase (iNOS). Citrulline thus formed can be recycled to l-arginine through an arginine-citrulline cycle, which not only regulates intracellular availability of l-arginine but, in turn, maintains a sustained production of NO by macrophages (19). However, unlike citrulline, macrophages have little or no capacity to convert ornithine, the breakdown product of citrulline into l-arginine (4). Recent reports have demonstrated that reactive nitrogen species derived from NO are critical for clearance of F. tularensis (27, 29). In addition, ammonia generated by degradation of citrulline has been proposed to play a role in alkalization of endosomal pH leading to phagosomal maturation arrest (25). Thus, interruption of the arginine-citrulline cycle through the degradation of citrulline into ornithine, CO2, and ammonia by CTU may assume an important role in the virulence of F. tularensis.
Until recently, CTU activity has been used to differentiate strains of F. tularensis with high virulence from strains with low virulence or avirulent strains (45). Previous studies have shown that the majority of virulent F. tularensis type A strains exhibit high CTU activity while strains lacking this enzyme activity are either less virulent or avirulent (10, 11). However, a direct relationship between CTU activity and virulence of F. tularensis could not be established. A majority of these previous studies were based on comparisons of CTU activity in naturally occurring wild-type (WT) virulent type A strains with that in less virulent or avirulent type B variants of F. tularensis. In the current study, a genetic approach was used to directly assess the role of CTU activity in the pathogenesis and virulence of the F. tularensis Schu S4 strain.
MATERIALS AND METHODS
Bacterial strains.
F. tularensis Schu S4, originally isolated from a human case of tularemia, was obtained from the U.S. Army Medical Research Institute for Infectious Diseases (USAMRIID; Frederick, MD). F. tularensis live vaccine strain (LVS; ATCC 29684; American Type Culture Collection, Rockville, MD) was kindly provided by Karen Elkins (U.S. Food and Drug Administration, Bethesda, MD). The bacteria were cultured on modified Mueller-Hinton (MH) chocolate agar plates (2, 13) or in MH broth (Difco Laboratories, Lawrence, KS) supplemented with ferric pyrophosphate and Iso-Vitalex (BD Biosciences, San Jose, CA). Active mid-log-phase bacteria were harvested and stored in liquid nitrogen; 1-ml aliquots were thawed periodically for use.
Generation of the Δctu mutant and transcomplementation.
F. tularensis Schu S4 was used for the generation of an in-frame gene deletion mutant of the ctu gene (Δctu). All genetic manipulations of the Schu S4 strain conformed to Centers for Disease Control guidelines and were performed in a biosafety level 3/animal biosafety level 3 facility at Albany Medical College. The sequences and locations of the primers, bacterial strains used, and plasmid constructs generated in this study are shown in Table 1. An allelic replacement method was adapted for the generation of the Δctu mutant of F. tularensis Schu S4 (15). A suicide plasmid vector, pDMK, kindly provided by Anders Sjostedt (University of Umeå, Sweden) was used for mutagenesis (27). A previously described splicing by overlap extension PCR method was used to generate a deletion within the coding region of the ctu gene in such a way that only the flanking regions of the gene remained (26). The PCR-amplified fragment containing up- and downstream regions minus the coding region of the ctu gene was cleaved with SalI/SpeI restriction enzymes and ligated into a similarly digested pDMK vector. The resultant pDMK::Δctu was transformed into chemically competent Escherichia coli S17-1 cells to yield E. coli pDMK::Δctu, and the colonies were selected on Luria-Bertani (LB) plates containing kanamycin (20 μg/ml). Early-log-phase cultures of E. coli pDMK::Δctu (∼107 CFU/ml) and F. tularensis Schu S4 (∼109 CFU/ml) grown in Chamberlain's chemically defined medium (7) were prepared for conjugation according to the method described earlier (2, 13). The transconjugants were selected on modified chocolate agar plates containing kanamycin (10 μg/ml) and polymyxin B (100 μg/ml) (2). The resultant colonies were screened for loss of both resistance to kanamycin and sensitivity to sucrose. The mutant colonies exhibiting such a phenotype were selected, and deletion of ctu was confirmed by PCR by using flanking primers and DNA sequencing. The Δctu mutant was further characterized for its virulence attributes by macrophage invasion assays and mouse survival studies.
TABLE 1.
Primer sequences, bacterial strains, and plasmids used and generated in this study
| Primer, strain, or plasmid | Primer sequence | Source or reference |
|---|---|---|
| Primers | ||
| Up ctuF (A) | 5′ CACGCGTCGACACATTCAAATGGTCAAGGAGTTTT 3′ | |
| Up ctuR (B) | 5′ ATCAAATCTCCTTTATAGCCGGT 3′ | |
| Dn ctuF (C) | 5′ CGGCTATAAAGGAGATTTGATATTGATAAAAATGTTTTTAGTTTAGAG 3′ | |
| Dn ctuR (D) | 5′ GGGACTAGTTCATCTAAACGCTAATCATGCTG 3′ | |
| CTUpKKF | 5′ AAAACTGCAGATGGCGAATATAAAAGTTGCAG 3′ | |
| CTUpKKR | 5′ TTAATACTTTCTAACAATTTCTTCA 3′ | |
| pKK kan CTU-F | 5′ TGAAGAAATTGTTAGAAAGTATTAAGTCGACTATTAAAAAAATTCATCAAG 3′ | |
| pKK kan CTU-R PstI | 5′ GGAGTAACTGCAGTATGTCACAT 3′ | |
| Strains or plasmidsa | ||
| F. tularensis Schu S4 | USAMRIID | |
| pDMK | 27 | |
| pDMK::Δctu | This study | |
| S17-1::pDMK::Δctu | This study | |
| F. tularensis Schu S4::Δctu (Δctu) | This study | |
| F. tularensis Schu S4::Δctu+pctu | This study |
E. coli S17-1 available in the Center for Immunology and Microbial Disease, Albany Medical College, was used for conjugation experiments.
For transcomplementation of the Δctu mutant, a pKK214::gfp vector expressing green fluorescent protein kindly provided by T. Kawula (University of North Carolina, Chapel Hill, NC) was used. The ctu gene was cloned downstream of the F. tularensis GroEL promoter by replacing gfp in the pKK214::gfp vector. Briefly, the ctu gene was amplified using Schu S4 genomic DNA as a template employing primers CTUpkkF and CTUpkkR (Table 1). Simultaneously, a kanamycin cassette was amplified from plasmid pkk214::gfp using the pkk kan CTU-F and pkk kan CTU-R PstI primers (Table 1). Both the ctu gene and kanamycin cassette PCR products were fused together by overlap extension. The final PCR product was digested with PstI and ligated into the similarly digested pKK214. This method allowed us to replace gfp with the ctu gene while keeping the kanamycin gene in frame. This construct was termed pctu and checked for the orientation of the ctu gene by PCR. The pctu containing the cloned ctu gene in the correct orientation was electroporated into the Δctu mutant as described earlier (3) to generate the transcomplemented Δctu+pctu strain. The expression of CTU in Δctu+pctu was confirmed by reverse transcriptase PCR (RT-PCR) and a citrulline ureidase activity assay.
For RT-PCR, RNA was isolated from overnight bacterial cultures by using the Trizol reagent (Invitrogen, Carlsbad, CA), and 1 μg of RNA was reverse transcribed using ctu gene-specific primers (CTUpkkR) by SuperScript II RT kit (Invitrogen, Carlsbad, CA). The cDNA was amplified using the CTUpkkF and CTUpkkR primers. The amplified products were electrophoresed on a 1.5% agarose gel and visualized on a UV transilluminator after staining with ethidium bromide.
CTU activity assay.
For assessment of CTU activity in the WT F. tularensis Schu S4, Δctu, and Δctu+pctu strains, a thin-layer chromatography (TLC)-based approach as described earlier was used (20, 24). The bacterial cultures were resuspended in 25 μl of 0.1 M phosphate-buffered saline (PBS; pH 6.5) to yield a concentration of 1 × 1010 CFU/ml and lysed by ultrasonication. The lysates were filtered using a 0.22-μm filter. Forty microliters of the filtrate was incubated with an equal volume of 0.7% (wt/vol) citrulline (Sigma, St. Louis, MO) at 30°C for 20 h. Three microliters of the reaction mix was spotted onto silica gel TLC plates (Partisil Diamond K6F; Schleicher & Schuell, Keene, NH). The spots were dried, and the TLC was carried out using n-butanol-acetic acid-water solvents mixed at a ratio of 40:10:17 in a glass chamber until the liquid front reached the top of the plates. The plates were removed from the glass chamber, dried, and sprayed with 0.5% ninhydrin dissolved in n-butanol. The plates were dried again in a fume hood and developed at 60°C for 30 min to visualize the colored spots. F. tularensis LVS, which does not exhibit CTU activity (38, 39), and bacterial lysates not treated with citrulline were used as negative controls. Citrulline (0.7% [wt/vol]) and ornithine (100 mM) were spotted as positive controls.
Growth curves.
WT Schu S4 and the Δctu and the Δctu+pctu strains were cultured in MH broth for 48 h at 37οC in a shaking incubator. Aliquots were withdrawn at 4-h intervals, and absorbance was recorded at 600 nm. To enumerate CFU, the aliquots were serially diluted in sterile PBS and plated onto MH chocolate agar plates. The plates were incubated for 48 h, and the colonies were counted and expressed as log10 CFU/ml.
Macrophage invasion assay.
To address the effect of ctu gene deletion on intramacrophage survival, a macrophage cell culture invasion assay was performed as described earlier (23, 31, 32). Bone marrow-derived macrophages (BMDMs) isolated from WT and inos−/− C57BL/6 mice and the MH-S cell line, a murine alveolar macrophage cell line (33), were used in these assays. MH-S cells or BMDMs were either left untreated or treated with recombinant gamma interferon (IFN-γ; 100 ng/ml; Sigma, St. Louis, MO) for 16 h prior to infection, and thereafter. The macrophages were infected with WT Schu S4 or the Δctu or Δctu+pctu strain at a multiplicity of infection of 100. The infection was synchronized by centrifuging the plates at 1,000 × g for 5 min at 4οC. Two hours after infection, the growth medium was replaced with medium containing gentamicin (100 μg/ml) to kill all adherent and extracellular bacteria. One hour later, the medium containing gentamicin was replaced with growth medium without any antibiotics, and the cells were incubated at 37οC in the presence of 5% CO2. The cells were lysed with 0.1% sodium deoxycholate 24 and 48 h later, diluted 10-fold in sterile PBS and spread onto chocolate agar plates (BD Biosciences, San Jose, CA) to quantitate the number of bacteria that replicated intracellularly. The Δctu+pctu strain was plated onto chocolate agar plates containing kanamycin (10 μg/ml) to ensure that recovered bacteria still carried the pctu plasmid. The results were expressed as log10 CFU/ml.
Measurement of NO.
The concentration of nitrite (NO2−), the oxidized metabolite of NO, was assessed by the Griess reaction. The culture supernatants of BMDMs unstimulated or stimulated with 100 ng/ml of IFN-γ and infected with the Δctu mutant or WT Schu S4 were collected at 24 and 48 h and analyzed for NO2− levels. One hundred microliters of each culture supernatant was mixed with an equal volume of Griess reagent (Promega, Madison, WI) and incubated at room temperature for 10 min in the dark. The optical density readings were recorded at 545 nm. A standard curve generated with various concentrations (2.5 to 10 μM) of sodium nitrite (NaNO2) was used for determining NO2− concentrations in culture supernatants. The data were expressed as μM concentrations of NO2−.
Mice experiments.
All experiments were conducted using 6- to 8-week-old BALB/c mice (Taconic, Germantown, NY) of both sexes. The mice were maintained in a specific-pathogen-free environment in the Animal Resource Facility at Albany Medical College. All Schu S4 challenge experiments were performed in a CDC-approved animal biosafety level 3 facility at Albany Medical College and conformed to the Institutional Animal Care and Use Committee guidelines.
Kinetics of bacterial clearance.
The effect of the ctu mutation on bacterial survival under in vivo conditions was determined by performing a kinetic experiment in mice. Six- to eight-week-old BALB/c mice were infected intranasally with 25 CFU of WT Schu S4 or the Δctu or Δctu+pctu strain. Mice were sacrificed on day 1, 3, or 5 postinfection (PI), and bacterial numbers were quantified in the lung, liver, and spleen of the infected mice. Briefly, the organs were subjected to mechanical homogenization using a Mini-BeadBeater-8 (BioSpec Products Inc. Bartlesville, OK). The tissue homogenates were spun at 1,000 × g for 10 s in a microcentrifuge to pellet tissue debris. The supernatants were diluted 10-fold in sterile PBS, and 10 μl of each dilution was spotted onto MH chocolate agar plates in duplicate and incubated at 37οC for 48 to 72 h in the presence of 5% CO2. The colonies on the plates were counted and expressed as CFU per organ as reported earlier (1, 49).
Histopathology.
The lungs from WT Schu S4-, Δctu strain-, or Δctu+pctu strain-infected BALB/c mice were excised and fixed in 10% neutral buffered formalin for histological evaluation. The lungs were collected on days 1, 3, and 5 PI. The lungs were inflated via instillation of PBS into the trachea prior to fixation and processed using standard histological procedures. The paraffin-embedded sections were stained with hematoxylin-eosin and examined by light microscopy. The hematoxylin-eosin-stained sections were analyzed in a blind fashion using a histopathological scoring system described earlier (1).
Survival experiments.
To analyze the role of ctu in virulence, time-to-death experiments were performed. BALB/c mice were deeply anesthetized via intraperitoneal injection of a cocktail of ketamine (Fort Dodge Animal Health, Fort Dodge, IA) and xylazine (Phoenix Scientific, St. Joseph, MO). Mice were challenged intranasally with 25 CFU of WT Schu S4 or the Δctu or Δctu+pctu strain in a volume of 20 μl PBS (10 μl/nare). The mice were monitored for a period of 21 days for morbidity and mortality. The survival results were plotted as Kaplan-Meier curves, and the statistical significance was determined by log-rank test.
Statistical analysis.
All results were expressed as means ± standard errors of the means, and comparisons between the groups were made using one-way analysis of variance (ANOVA) followed by Bonferroni's correction, the nonparametric Mann-Whitney test, or Student's t test. The survival data were analyzed using log-rank test, and P values were determined. Differences between the experimental groups were considered significant at a P value of <0.05.
RESULTS
The ctu gene is interrupted in avirulent or less virulent strains of F. tularensis.
The CTU protein in strain Schu S4 is 286 amino acids long and has a molecular mass of 32.29 kDa. Computer-based comparative analysis showed that the amino acid sequences of CTU from Schu S4 and those from virulent type A strains FSC198 and WY96-3418 were 100% identical. The CTU sequence of type A strains also exhibited 97% sequence homology with the attenuated F. tularensis subspecies holarctica type B LVS, FTA, and OSU18. However, unlike the single open reading frame (ORF) of type A strains, the CTU ORF in type B strains, including LVS, was found to be interrupted by stop codons at amino acids 54 and 141, resulting in a truncated protein product. In addition, the CTU sequence in LVS and other type B strains revealed amino acid changes/substitutions at positions 113, 136, 183, 204, 222, and 248. Thus, sequence analysis revealed that while the virulent type A strains of F. tularensis are all strongly CTU positive owing to the presence of an uninterrupted ORF, the avirulent or less virulent type B strains are CTU negative due to mutations in the ctu gene. The findings indicate that ctu gene sequence analysis may form a strong basis for the rapid differentiation of type A and B strains of F. tularensis.
Verification of ctu gene deletion in F. tularensis Schu S4.
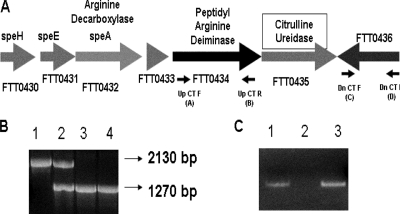
The genomic organization of the ctu gene is shown in Fig. 1A. Deletion of the ctu gene in the Δctu mutant was confirmed by PCR, DNA sequencing, and RT-PCR. A colony PCR using primer pairs located up- and downstream of the ctu gene resulted in a smaller fragment (∼1.27 kb) in the Δctu mutant than in the WT Schu S4 (∼2.13 kb), confirming the gene deletion (Fig. 1B). DNA sequencing of the regions flanking the deleted ctu gene revealed that deletion of the ctu gene was in frame and ORFs up- and downstream of the ctu gene were unaltered (data not shown). RT-PCR was performed using ctu gene-specific primers. This analysis also confirmed deletion of the ctu gene, as no transcripts were amplified in the Δctu mutant; however, ctu-specific transcripts were observed in WT Schu S4 and the transcomplemented strain (Fig. 1C).
FIG. 1.
Verification of ctu gene deletion in F. tularensis Schu S4. (A) Genomic organization of the ctu gene (FTT0435) of Schu S4. Small arrows indicate primer locations. (B) Confirmation of the ctu gene deletion by PCR. Flanking primers A plus D were used for confirmation of the ctu gene deletion. Lane 1, WT F. tularensis Schu S4; lane 2, a merodiploid stage indicating integration of pDMK::Δctu in the Schu S4 genome; lanes 3 and 4, the Δctu mutant. An amplification product of ∼1.27 kbp (lanes 3 and 4) compared to 2.13 kbp in WT Schu S4 (lane 1) confirmed the ctu gene deletion. (C) RT-PCR analysis. Lane 1, WT Schu S4; lane 2, the Δctu mutant; lane 3, the Δctu+pctu transcomplemented strain.
Deletion of the ctu gene results in loss of CTU activity.
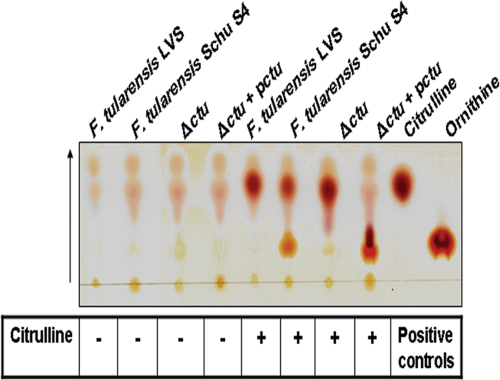
We further characterized the Δctu mutant for CTU activity by TLC. The lysates from WT Schu S4 degraded citrulline into ornithine, whereas the Δctu mutant, similar to F. tularensis LVS, lost its citrulline degrading capability. CTU activity was restored by complementing the ctu gene in trans in the Δctu+pctu strain (Fig. 2). The results demonstrate that the ctu gene in Schu S4 is required for degradation of citrulline into ornithine and that this function is specific to the ctu gene. The results also confirm findings from sequence analysis that LVS does not have a functional ctu gene.
FIG. 2.
CTU activity assay. The CTU activity assay was performed as described in Materials and Methods. Citrulline and ornithine were run as positive controls. The arrow indicates the direction of run.
Loss of CTU does not affect acellular growth.
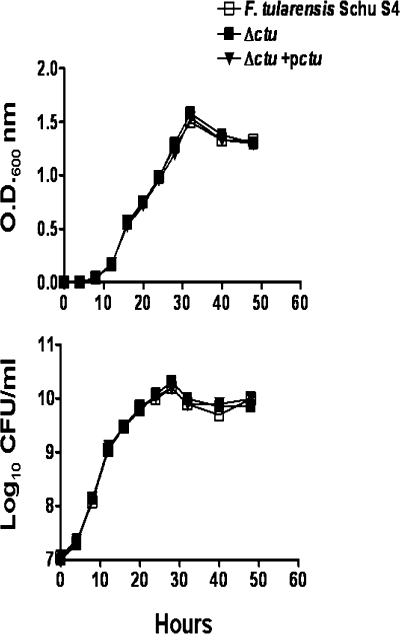
The role of the ctu gene under acellular growth conditions was assessed by comparing the growth curve of the Δctu mutant with those of WT Schu S4 and the Δctu+pctu transcomplemented strain. The Δctu mutant did not exhibit any growth defect, and its growth rate was similar to those of WT Schu S4 and the transcomplemented strain (Fig. 3). This result suggests that CTU activity is not required for growth under normal, acellular growth conditions.
FIG. 3.
In vitro growth analysis of the Δctu mutant of F. tularensis Schu S4. The growth curve for the Δctu mutant was generated and compared with those of WT Schu S4 and the Δctu+pctu transcomplemented bacteria. The optical density at 600 nm (O.D.600nm) (upper panel) and the corresponding CFU (lower panel) were recorded at the indicated times.
Loss of ctu attenuates intramacrophage survival.
On the basis of the association of CTU activity with highly virulent type A strains of F. tularensis, we hypothesized that deletion of the ctu gene would lead to attenuation of intramacrophage growth. We performed macrophage cell culture invasion assay in BMDMs and the MH-S cell line, using WT Schu S4, the Δctu mutant, and the Δctu+pctu transcomplemented strain for the quantitation of intramacrophage survival and replication. Despite equal numbers of bacteria recovered at 3 h PI, significantly lower numbers (five- to sevenfold) of Δctu mutants, relative to the number of WT Schu S4 and the transcomplemented strain, were recovered in BMDMs and the MH-S cells at 24 and 48 h PI (Fig. 4, left panels). Significantly reduced numbers of the Δctu mutant compared to the WT and transcomplemented strain were also recovered from IFN-γ-treated BMDMs and MH-S cells (Fig. 4, right panels). Transcomplementation restored growth of the Δctu mutant within BMDMs and MH-S cells to levels intermediate between the Δctu mutant and the WT F. tularensis Schu S4 strain. These results demonstrate that CTU contributes to the intramacrophage survival of F. tularensis Schu S4. However, in the absence of a complete clearance of the Δctu mutant by infected macrophages, our results indicate that ctu is not the sole factor responsible for the intracellular lifestyle of F. tularensis Schu S4 and that other factors in conjunction with ctu contribute to its intramacrophage survival and replication.
FIG. 4.
The Δctu mutant of Schu S4 is deficient for intramacrophage survival. A macrophage cell culture invasion assay was performed on BMDMs and the MH-S cells untreated (left panels) or treated with IFN-γ (right panels). The intracellular replication was quantitated at the indicated times and expressed as CFU/ml. The values represent the means ± standard errors (SE) of quadruplicate samples and are cumulative of the results of three experiments conducted. P values were determined using one-way ANOVA. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Enhanced killing of the Δctu mutant may be attributed to NO production in infected macrophages.
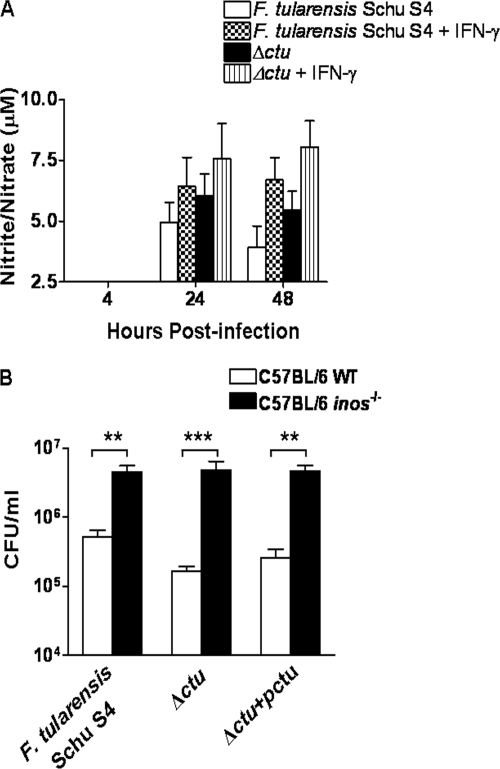
We hypothesized that suppression of NO production by CTU via interruption of the arginine-citrulline cycle would enhance intramacrophage survival of F. tularensis Schu S4. Since reactive nitrogen species are instrumental in the intramacrophage killing of F. tularensis (27-29), to examine whether the enhanced killing of the Δctu mutant relative to that of the WT Schu S4 strain was due to differences in the levels of NO produced by infected macrophages, levels of nitrite/nitrate, the stable oxidative product of NO, were measured by the Griess reaction and used as an indicator of NO production in culture supernatants of infected macrophages with or without IFN-γ treatment. Elevated nitrite levels were observed in culture supernatants of the Δctu strain-infected BMDMs compared to those observed in WT Schu S4-infected cells (Fig. 5A). Conversely, in BMDMs deficient for iNOS, intramacrophage survival of the Δctu mutant was restored, similar to that of WT Schu S4 (Fig. 5B). These results demonstrate that NO contributes to the enhanced intramacrophage killing of the Δctu mutant.
FIG. 5.
Enhanced intramacrophage killing of the Δctu mutant is NO dependent. (A) Culture supernatants from the invasion assay were analyzed for nitrite levels, using a commercial Griess reagent. The detection limit of the Griess reaction was 2.5 μM. The results are expressed as the means ± SEs from four replicate samples tested in triplicate and are representative of the results of two independent experiments conducted. (B) BMDMs derived from C57BL/6 WT and inos−/− mice were stimulated with IFN-γ (100 ng/ml) for 16 h prior to and after infection with the F. tularensis Schu S4, Δctu, or Δctu+pctu strain at a multiplicity of infection of 100. The infected BMDMs were lysed 24 h PI, and the bacterial numbers were quantitated. The results are expressed as the means ± standard deviations. The statistical analysis was performed using unpaired Student's t test. **, P < 0.01; and ***, P < 0.001.
Δctu mutant-infected mice exhibit significantly reduced bacterial burden and histopathology.
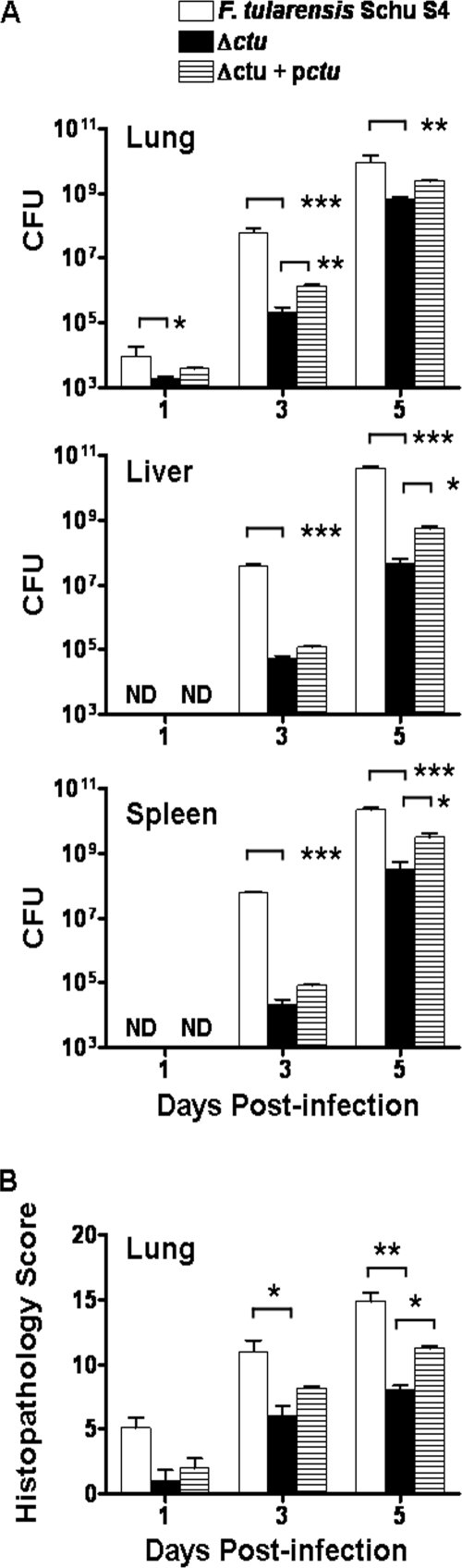
It was next investigated whether the enhanced killing of the Δctu mutant in macrophages could be replicated under in vivo conditions. BALB/c mice were infected intranasally with 25 CFU of WT Schu S4, Δctu mutant, or Δctu+pctu transcomplemented bacteria. Mice were sacrificed at the indicated times above, and bacterial burdens were quantitated in the lung, liver and spleen. At days 3 and 5 PI, at which 100% of Schu S4-infected mice succumbed to infection, significantly lower bacterial loads were observed in the lung, liver, and spleen of the Δctu mutant-infected mice than in the Schu S4-infected counterparts (Fig. 6A). The transcomplemented strain, the Δctu+pctu strain, exhibited partial restoration of the parental Schu S4 phenotype and was recovered in nearly 10-fold-higher numbers in the lungs at day 3 PI, and in the liver and spleen at day 5 PI, than was the Δctu mutant strain. These results demonstrate that ctu, in addition to its role in intramacrophage survival, is required for in vivo replication of F. tularensis Schu S4.
FIG. 6.
Quantitation of bacterial burden and histopathological lesions. Mice (n = 4 to 6 per group), infected with 25 CFU of WT Schu S4, the Δctu mutant, and the transcomplemented Δctu+pctu strain were sacrificed at the indicated times. The bacterial burden in the lungs, liver, and spleen (A) and the histopathological lesions in the lungs (B) were quantitated. The results are expressed as the means ± SEs and are cumulative of the results of two independent experiments conducted. P values were determined using one-way ANOVA. ND, not detected; *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Histological lesions in the lungs of mice infected with the Δctu mutant were quantitated using a previously described histopathological scoring (HPS) system (1, 31), and the scores were compared with those in the WT Schu S4- and Δctu+pctu strain-infected mice. Consistent with the reduced bacterial burdens, histopathological scores observed in the lungs at days 3 and 5 PI (Fig. 6B), and in the liver and spleen at day 3 PI, of mice infected with the Δctu mutant were significantly lower than those observed in Schu S4-infected mice (data not shown). Transcomplementation increased the severity of the lung lesions in the Δctu+pctu strain-infected mice compared to that in the Δctu mutant-infected mice. However, the severity of the lesions in the Δctu+pctu strain-infected mice never reached the extent observed in the WT Schu S4-infected mice. Collectively, these results demonstrate that ctu participates in the pathogenesis of F. tularensis Schu S4 strain.
The Δctu mutant of Schu S4 is partially attenuated for virulence in mice.
An attenuation in intramacrophage survival and reduced bacterial burden in mice infected with the Δctu mutant prompted us to further investigate the effect of ctu gene deletion on virulence in mice, using an intranasal challenge protocol (1, 2, 31, 32). BALB/c mice are extremely susceptible to Schu S4 infection, and a dose as low as 1 CFU administered intranasally can cause death in the infected mice (22; our unpublished data). Groups of 15 mice were each inoculated intranasally with 25 CFU of WT Schu S4, the Δctu strain, or the transcomplemented strain. The mice were monitored twice daily for morbidity and mortality for a period of 21 days. All mice inoculated with 25 CFU of Schu S4 succumbed to infection by day 5 PI. Although 100% of the mice infected with 25 CFU of the Δctu strain also succumbed to infection, a significantly extended median time to death compared to that of Schu S4-infected mice was observed (Fig. 7). Mice infected with the Δctu+pctu transcomplemented strain had an intermediate virulence phenotype. The results suggest that the Δctu strain undergoes slow replication in infected mice compared to WT Schu S4 strain; however, given the extremely high virulence and very low 100% lethal dose of Schu S4, subsequent increases in bacterial numbers are sufficient to cause death in the infected mice. These results also indicate that the ctu gene deletion causes only a partial attenuation of virulence in mice.
FIG. 7.
Mouse survival studies. Six- to eight-week old BALB/c mice (n = 15 per group) were infected intranasally with 25 CFU of WT Schu S4, the Δctu mutant, and the Δctu+pctu transcomplemented strain. The mice were monitored for morbidity and mortality for a period of 21 days. The results are expressed as Kaplan-Meier survival curves. The median survival times are shown in the table. The P values were determined using log-rank test. *, P < 0.006.
DISCUSSION
CTU activity has essentially been used as a marker to differentiate highly virulent strains of F. tularensis from less virulent or avirulent strains (10, 11, 38, 39, 42). Despite this exclusive association with a highly virulent phenotype, the actual contribution of CTU to virulence and pathogenesis of F. tularensis is not known. We attempted to address this important issue by generating a nonpolar ctu deletion mutant of the highly virulent Schu S4 strain of F. tularensis and further characterized this mutant for its virulence attributes in macrophages and mice. DNA sequence analysis of the Δctu mutant revealed that deletion of the ctu gene was in frame and did not alter the transcription of upstream genes (data not shown), as ctu is the last gene of the operon. Our in vitro analysis confirmed that the Δctu mutant was not growth defective under acellular conditions. However, the Δctu mutant was attenuated for intramacrophage survival and showed reduced virulence in intranasally infected mice.
Francisella utilizes l-arginine as a carbon and/or nitrogen source (7). The ctu gene of F. tularensis Schu S4 is carried on an operon that resembles the ADI system required for arginine utilization in several bacterial pathogens (6, 9, 18). The ctu (FTT0435), arginine deiminase (FTT0434), and arginine decarboxylase (speA [FTT0432]) genes similar to those found on other bacterial ADI operons may serve to carry out arginine catabolism in Francisella, whereas spermidine synthase (speE [FTT0431]) and S-adenosylmethionine decarboxylase (speH [FTT0430]) genes are required for polyamine biosynthesis (Fig. 1A). A recent report has shown that transcription of all these genes, including the ctu gene, is significantly upregulated following infection of macrophages with F. tularensis Schu S4 (50). The genomic organization of ctu with genes involved in arginine utilization and their transcriptional upregulation following macrophage infection (50) raise the possibility that deletion of ctu diminishes the ability of the Δctu mutant to grow in a nutrient-limiting macrophage environment. However, arginine decarboxylase (FTT0432), an enzyme that degrades arginine into agmatine, provides an additional arginine metabolism mechanism in Francisella that may compensate for the loss of CTU. Our laboratory is now in the process of creating deletion mutants of additional genes involved in arginine utilization in the virulent Schu S4 strain. These mutant strains will allow us to explore further whether arginine is a major substrate that is required for intramacrophage survival of Francisella.
The NO produced by IFN-γ-activated murine macrophages reduces infectivity of F. tularensis LVS and Schu S4 (17, 27, 30, 35). Similarly, iNOS is required to resolve LVS infection in mouse models (29). Our results have shown elevated NO levels in culture supernatants from the Δctu mutant-infected macrophages (Fig. 5A). Additionally, the Δctu mutant survived similarly to the WT Schu S4 in inos−/− macrophages (Fig. 5B), suggesting an NO-dependent mechanism for killing of the Δctu mutant. In activated macrophages, increased NO levels are associated with increased citrulline generated as a result of breakdown of arginine by iNOS. The citrulline is recycled to generate arginine via an arginine-citrulline cycle in the macrophages (19). The CTU of F. tularensis Schu S4 degrades citrulline to ornithine and ammonia and, thus, may inhibit arginine resynthesis in the infected macrophages. However, this process might require secretion of CTU by F. tularensis Schu S4. The PsortB software analysis of CTU did not predict its exact subcellular localization but provided identical scores for all possible locations, including that of the secreted form (data not shown). In the absence of concrete evidence on the secretory nature of CTU, we speculate that Francisella depletes the arginine pool in macrophages by an active uptake and metabolism of arginine via CTU and arginine decarboxylase, thereby reducing the substrate for iNOS and subsequent NO production. Inhibiting this aspect of the innate immune response could help Francisella resist killing by macrophages. Chlamydophila pneumoniae and Helicobacter pylori also use a similar strategy and deplete arginine to reduce iNOS activity and NO abundance (14, 46).
It has been shown that Francisella, when grown in an acidic medium, causes alkalization of the pH due to generation of ammonia (7). The ammonia produced via deamination of amino acids also serves to stabilize bacterial cytoplasmic pH upon exposure to an acidic environment, such as in the phagosomal vacuoles (40). The ammonia generated by CTU has been proposed to play a role in neutralization of endosomal pH that leads to phagosomal maturation arrest (25). It has also been reported that inhibition of acidification and phagosomal maturation enhances intramacrophage survival of Francisella (8, 43), Helicobacter pylori (44) and Mycobacterium tuberculosis (16). On the other hand, neutralization of phagosomal pH by ammonium chloride (NH4Cl) treatment of macrophages restores intramacrophage survival of H. pylori urease mutants, which are deficient for ammonia production (44). Thus, an inability to modulate the phagosomal environment or to maintain the bacterial pH homeostasis in the absence of ctu may also offer an explanation for attenuated intramacrophage survival of the Δctu mutant. However, NH4Cl treatment of macrophages resulted in a modest two- to threefold increase in the survival of ingested Δctu mutant at 24 h PI (data not shown). This small improvement in survival of the Δctu mutant following NH4Cl treatment of infected macrophages suggests that CTU alone may not cause a significant change in the phagosomal environment. Other genes like asparaginase, glutaminase, and arginine deiminase genes may still produce ammonia in the absence of CTU. The presence of these multifactorial and redundant mechanisms potentially argues in favor of our observation that the Δctu mutant was not cleared completely by the infected macrophages. It is possible that in the absence of ctu, these redundant mechanisms compensate for its loss.
The transcomplementation studies provided the evidence that the Δctu mutation itself is responsible for the reduced-virulence phenotype. Transcomplementation of Δctu restored virulence to the levels intermediate between the WT and the Δctu mutant phenotype in cell culture-based assays and a mouse model of respiratory tularemia. This partial, rather than full, restoration of the mutant to the WT phenotype could be attributed to plasmid loss in the absence of kanamycin selection in cellular and mouse infection models. The loss of pctu in the absence of antibiotic selection pressure may have compromised the growth of the complemented mutant, resulting in an intermediate phenotype. Similar observations have been reported earlier for the transcomplemented Schu S4 mutant strain of F. tularensis (37).
Mice infected with the Δctu mutant showed a significantly extended median time to death compared to that of the WT Schu S4-infected mice, but all mice eventually succumbed to infection. The results are not unexpected, as several other mutants of Schu S4 that are defective for intracellular survival have also been shown to retain virulence in mice (27, 36, 37). A possible explanation could be the existence of unidentified redundant virulence mechanisms in Schu S4 that mask the effect of a single gene deletion. In addition, due to the extremely high virulence of Schu S4, even small increases in bacterial numbers are sufficient to cause death in infected mice.
To conclude, this study provides definitive evidence that CTU activity contributes to intramacrophage survival and tularemia pathogenesis but is not the primary virulence factor of F. tularensis Schu S4. However, the association of the ctu gene with virulence may constitute a strong and rapid method for differentiation of highly virulent type A strains from less virulent or avirulent type B strains of F. tularensis.
Acknowledgments
Excellent technical support was provided by Michelle Wyland-O'Brien and Sherie O'Connell. We also thank Anders Sjöstedt, Umea University, Umea, Sweden, for providing the pDMK vector.
This work was supported by NIH grant P01 AI056320.
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Bakshi, C. S., M. Malik, M. Mahawar, G. S. Kirimanjeswara, K. R. Hazlett, L. E. Palmer, M. B. Furie, R. Singh, J. A. Melendez, T. J. Sellati, and D. W. Metzger. 2008. An improved vaccine for prevention of respiratory tularemia caused by Francisella tularensis SchuS4 strain. Vaccine 265276-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakshi, C. S., M. Malik, K. Regan, J. A. Melendez, D. W. Metzger, V. M. Pavlov, and T. J. Sellati. 2006. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 1886443-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, G. S., S. V. Myltseva, and F. E. Nano. 1995. Electroporation of Francisella tularensis. Methods Mol. Biol. 47149-154. [DOI] [PubMed] [Google Scholar]
- 4.Benninghoff, B., V. Lehmann, H. P. Eck, and W. Droge. 1991. Production of citrulline and ornithine by interferon-gamma treated macrophages. Int. Immunol. 3413-417. [DOI] [PubMed] [Google Scholar]
- 5.Bossi, P., and F. Bricaire. 2003. Tularemia, a potential bioterrorism weapon. Presse Med. 321126-1130. [PubMed] [Google Scholar]
- 6.Casiano-Colón, A., and R. E. Marquis. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 541318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamberlain, R. E. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13232-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 723204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Mol. Biol. Rev. 50314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming, D. E., and L. Foshay. 1955. Studies on the physiology of virulence of Pasteurella tularensis. I. Citrulline ureidase and deamidase activity. J. Bacteriol. 70345-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming, D. E., and L. Foshay. 1956. Studies on the physiology of virulence of Pasteurella tularensis. II. Serine deaminase and transaminase activity. J. Bacteriol. 71324-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher-Smith, M., J. Kim, R. Al-Bawardy, and D. Josko. 2004. Francisella tularensis: possible agent in bioterrorism. Clin. Lab. Sci. 1735-39. [PubMed] [Google Scholar]
- 13.Gil, H., G. J. Platz, C. A. Forestal, M. Monfett, C. S. Bakshi, T. J. Sellati, M. B. Furie, J. L. Benach, and D. G. Thanassi. 2006. Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc. Natl. Acad. Sci. USA 10312897-12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gobert, A. P., D. J. McGee, M. Akhtar, G. L. Mendz, J. C. Newton, Y. Cheng, H. L. T. Mobley, and K. T. Wilson. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 9813844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovliov, I., A. Sjostedt, A. Mokrievich, and V. Pavlov. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222273-280. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, A. H., P. D. Hart, and M. R. Young. 1980. Ammonia inhibits phagosome-lysosome fusion in macrophages. Nature 28679-80. [DOI] [PubMed] [Google Scholar]
- 17.Green, S. J., C. A. Nacy, R. D. Schreiber, D. L. Granger, R. M. Crawford, M. S. Meltzer, and A. H. Fortier. 1993. Neutralization of gamma interferon and tumor necrosis factor alpha blocks in vivo synthesis of nitrogen oxides from l-arginine and protection against Francisella tularensis infection in Mycobacterium bovis BCG-treated mice. Infect. Immun. 61689-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruening, P., M. Fulde, P. Valentin-Weigand, and R. Goethe. 2006. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 188361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guoyao, W. U., and J. T. Brosnon. 1992. Macrophages can convert citrulline into arginine. Biochemistry 28145-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, D. L., and J. Van Eys. 1965. The relationship between arginine, citrulline and ornithine in Tetrahymena pyriformis. J. Protozool. 12259-265. [DOI] [PubMed] [Google Scholar]
- 21.Jellison, W. L. 1980. Tularemia in North America, p. 161-193. In J. H. Steel (ed.), CRC handbook series in zoonosis, vol. 2. CRC Press, Boca Raton, FL. [Google Scholar]
- 22.Kadzhaev, K., C. Zingmark, I. Golovliov, M. Bolanowski, H. Shen, W. Conlan, and A. Sjöstedt. 2009. Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS ONE 4e5463. [DOI] [PMC free article] [PubMed]
- 23.Kirimanjeswara, G. S., J. M. Golden, C. S. Bakshi, and D. W. Metzger. 2007. Prophylactic and therapeutic use of antibodies for protection against respiratory infection with Francisella tularensis. J. Immunol. 179532-539. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, V. B., A. E. Bernardo, M. M. Alshaher, M. Buddhiraju, R. Purushothaman, and J. E. Morley. 1999. Rapid assay for nitric oxide synthase using thin-layer chromatography. Anal. Biochem. 26917-20. [DOI] [PubMed] [Google Scholar]
- 25.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37153-159. [DOI] [PubMed] [Google Scholar]
- 26.Lauriano, C. M., J. R. Barker, F. E. Nano, B. P. Arulanandam, and K. E. Klose. 2003. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229195-202. [DOI] [PubMed] [Google Scholar]
- 27.Lindgren, H., H. Shen, C. Zingmark, I. Golovliov, W. Conlan, and A. Sjostedt. 2007. Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immun. 751303-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindgren, H., L. Stenman, A. Tarnvik, and A. Sjostedt. 2005. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect. 7467-475. [DOI] [PubMed] [Google Scholar]
- 29.Lindgren, H., S. Stenmark, W. Chen, A. Tarnvik, and A. Sjostedt. 2004. Distinct roles of reactive nitrogen and oxygen species to control infection with the facultative intracellular bacterium Francisella tularensis. Infect. Immun. 727172-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loegering, D. J., J. R. Drake, J. A. Banas, T. L. McNealy, D. G. McArthur, L. M. Webster, and M. R. Lennartz. 2006. Francisella tularensis LVS grown in macrophages has reduced ability to stimulate the secretion of inflammatory cytokines by macrophages in vitro. Microb. Pathog. 41218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik, M., C. S. Bakshi, K. McCabe, S. V. Catlett, A. Shah, R. Singh, P. L. Jackson, A. Gaggar, D. W. Metzger, J. A. Melendez, J. E. Blalock, and T. J. Sellati. 2007. Matrix metalloproteinase 9 activity enhances host susceptibility to pulmonary infection with type A and B strains of Francisella tularensis. J. Immunol. 1781013-1020. [DOI] [PubMed] [Google Scholar]
- 32.Malik, M., C. S. Bakshi, B. Sahay, A. Shah, S. A. Lotz, and T. J. Sellati. 2006. Toll-like receptor 2 is required for control of pulmonary infection with Francisella tularensis. Infect. Immun. 743657-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mbawuike, I. N., and H. B. Herscowitz. 1989. MH-S, a murine alveolar macrophage cell line: morphological, cytochemical, and functional characteristics. J. Leukoc. Biol. 46119-127. [DOI] [PubMed] [Google Scholar]
- 34.Oyston, P. C., A. Sjostedt, and R. W. Titball. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2967-978. [DOI] [PubMed] [Google Scholar]
- 35.Polsinelli, T., M. S. Meltzer, and A. H. Fortier. 1994. Nitric oxide-independent killing of Francisella tularensis by IFN-gamma-stimulated murine alveolar macrophages. J. Immunol. 1531238-1245. [PubMed] [Google Scholar]
- 36.Qin, A., and B. J. Mann. 2006. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin, A., D. W. Scott, and B. J. Mann. 2008. Francisella tularensis subsp. tularensis Schu S4 disulfide bond formation protein B, but not an RND-type efflux pump, is required for virulence. Infect. Immun. 763086-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodionova, I. V. 1968. Citrulline ureidase activity in geographical races of Francisella tularensis. Dokl. Akad. Nauk SSSR 179457-460. [PubMed] [Google Scholar]
- 39.Rodionova, I. V. 1970. Differentiation of geographic races of Francisella tularensis on the basis of citrulline ureidase activity. Lab. Delo 142-43. [PubMed] [Google Scholar]
- 40.Ryan, S., M. Begley, C. G. Gahan, and C. Hill. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11432-445. [DOI] [PubMed] [Google Scholar]
- 41.Samrakandi, M. M., C. Zhang, M. Zhang, J. Nietfeldt, J. Kim, P. C. Iwen, M. E. Olson, P. D. Fey, G. E. Duhamel, S. H. Hinrichs, J. D. Cirillo, and A. K. Benson. 2004. Genome diversity among regional populations of Francisella tularensis subspecies tularensis and Francisella tularensis subspecies holarctica isolated from the US. FEMS Microbiol. Lett. 2379-17. [DOI] [PubMed] [Google Scholar]
- 42.Sandström, G., A. Sjostedt, M. Forsman, N. V. Pavlovich, and B. N. Mishankin. 1992. Characterization and classification of strains of Francisella tularensis isolated in the central Asian focus of the Soviet Union and in Japan. J. Clin. Microbiol. 30172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santic, M., M. Molmeret, K. E. Klose, and K. Y. Abu. 2006. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 1437-44. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz, J. T., and L. A. Allen. 2006. Role of urease in megasome formation and Helicobacter pylori survival in macrophages. J. Leukoc. Biol. 791214-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sjostedt, A. 2004. Gram-negative aerobic cocci. Family XVII. Francisellae, p. 200-210. In D. J. Brenner (ed.), Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 46.Smith, C. B., and D. E. Graham. 2008. Outer and inner membrane proteins compose an arginine-agmatine exchange system in Chlamydophila pneumoniae. J. Bacteriol. 1907431-7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonck, K. A., G. Kint, G. Schoofs, W. C. Vander, J. Vanderleyden, and S. C. De Keersmaecker. 2009. The proteome of Salmonella typhimurium grown under in vivo-mimicking conditions. Proteomics 9565-579. [DOI] [PubMed] [Google Scholar]
- 48.Twine, S., M. Bystrom, W. Chen, M. Forsman, I. Golovliov, A. Johansson, J. Kelly, H. Lindgren, K. Svensson, C. Zingmark, W. Conlan, and A. Sjostedt. 2005. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 738345-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wayne Conlan, J., H. Shen, R. Kuolee, X. Zhao, and W. Chen. 2005. Aerosol-, but not intradermal-immunization with the live vaccine strain of Francisella tularensis protects mice against subsequent aerosol challenge with a highly virulent type A strain of the pathogen by an alphabeta T cell- and interferon gamma-dependent mechanism. Vaccine 232477-2485. [DOI] [PubMed] [Google Scholar]
- 50.Wehrly, T. D., A. Chong, K. Virtaneva, D. E. Sturdevant, R. Child, J. A. Edwards, D. Brouwer, V. Nair, E. R. Fischer, L. Wicke, A. J. Curda, J. J. Kupko III, C. Martens, D. D. Crane, C. M. Bosio, S. F. Porcella, and J. Celli. 2009. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed]
- 51.Wong, J. D., and S. Shapiro. 1999. Francisella, p. 647-651. In P. R. Murray (ed.), Manual of clinical microbiology. ASM Press, Washington, DC.