Abstract
The outer membrane plasminogen activator Pla of Yersinia pestis is a central virulence factor in plague. The primary structure of the Pla β-barrel is conserved in Y. pestis biovars Antiqua, Medievalis, and Orientalis, which are associated with pandemics of plague. The Pla molecule of the ancestral Y. pestis lineages Microtus and Angola carries the single amino acid change T259I located in surface loop 5 of the β-barrel. Recombinant Y. pestis KIM D34 or Escherichia coli XL1 expressing Pla T259I was impaired in fibrinolysis and in plasminogen activation. Lack of detectable generation of the catalytic light chain of plasmin and inactivation of plasmin enzymatic activity by the Pla T259I construct indicated that Microtus Pla cleaved the plasminogen molecule more unspecifically than did common Pla. The isoform pattern of the Pla T259I molecule was different from that of the common Pla molecule. Microtus Pla was more efficient than wild-type Pla in α2-antiplasmin inactivation. Pla of Y. pestis and PgtE of Salmonella enterica have evolved from the same omptin ancestor, and their comparison showed that PgtE was poor in plasminogen activation but exhibited efficient antiprotease inactivation. The substitution 259IIDKT/TIDKN in PgtE, constructed to mimic the L5 region in Pla, altered proteolysis in favor of plasmin formation, whereas the reverse substitution 259TIDKN/IIDKT in Pla altered proteolysis in favor of α2-antiplasmin inactivation. The results suggest that Microtus Pla represents an ancestral form of Pla that has evolved into a more efficient plasminogen activator in the pandemic Y. pestis lineages.
Since the year 540, plague has killed some 200 million humans in three pandemics, i.e., the Justinian plague, the Black Death, and the modern plague (36). Genomic studies have estimated that the etiological agent, Yersinia pestis, evolved from the oral-fecal pathogen Yersinia pseudotuberculosis serotype O1b only shortly before the first pandemic, i.e., 5,000 to 20,000 years ago (1, 2, 46), which has made the bacterium a paradigm of the rapid evolution of a severe bacterial pathogen (57). At least four biovars of Y. pestis have been identified through metabolic and genomic studies; of these biovars, Antiqua, Medievalis, and Orientalis may be associated with the three plague pandemics, whereas the fourth biovar, Microtus, is associated with human-attenuated Y. pestis strains from two geographically distant infection foci in China (36, 59-61). A recent molecular analysis indicated that the biovars are not monophyletic and proposed the subdivision of Y. pestis into eight molecular groupings, which represent different evolutionary branches and histories and are only partially compatible with the biovars (1). Y. pestis evolved from Y. pseudotuberculosis along branch 0, which consists of “atypical” Y. pestis strains designated Angola, Microtus, and Pestoides; these are phylogenetically ancestral to the Antiqua, Medievalis, and Orientalis branches (1).
As a disease, plague exhibits various pathologies. Bubonic plague is the zoonotic form of the disease, which is usually acquired by humans from the bite of a flea that has been infected through a blood meal on a diseased rodent (36). The bacteria invade at the intradermal flea bite site and migrate to lymphatic vessels and then to regional draining lymph nodes, where they multiply and cause the development of buboes (44). Without early treatment, bubonic plague progresses to life-threatening septicemic plague, and hematogenous spread of the bacterium to lungs leads to pneumonic plague, a rapidly fatal and highly contagious airborne disease. Occasional injection of Y. pestis cells by the flea directly into the circulatory system leads to primary septicemic plague (43).
The plasminogen activator Pla is a cell surface protease encoded by the Y. pestis-specific plasmid pPCP1 (10, 48). Pla is essential in the pathogenesis of bubonic (43, 49) and pneumonic plague (28), whereas it has less of a role in primary septicemic plague (43, 49). The pla gene is highly transcribed in buboes of Y. pestis-infected mice (45), and Pla specifically potentiates migration of the bacteria to lymphatic tissue (43). Pla seems to have a different role in pneumonic plague, where it allows Y. pestis to replicate rapidly in the lungs, causing lethal fulminant pneumonia (28). Virulent Y. pestis strains lacking the Pla-encoding plasmid pPCP1 have been isolated in Asia (3), and they can be associated with primary septicemic plague (43).
Pla is an aspartic protease (22, 55) that activates human plasminogen (Plg) to the serine protease plasmin (47) and inactivates the plasmin inhibitor α2-antiplasmin (α2AP), thus affecting the main control system for plasmin activity (22). Plg is an abundant circulating zymogen, and its activation is central in the pathogenesis of plague (13, 28, 43), and plasmin is a powerful serine protease associated with cell migration and degradation of fibrin clots (29, 32, 37). In accordance with this, Pla-mediated bacterial adherence directs uncontrolled plasmin proteolysis onto basement membranes to enhance bacterial metastasis through tissue barriers (25, 27), and fibrinolysis by Pla-generated plasmin activity plays a role in the pathogenesis of bubonic plague (8).
Compared to those of other Y. pestis biovars, Microtus isolates have several unique genomic features that may be involved in their inherent inability to attack the human host, and specific losses of genes or gene functions are thought to be responsible for the human attenuation (59). Interestingly, the attenuation does not apply to the murine host. The predicted amino acid sequence of the Pla polypeptide is remarkably conserved: in the branches Antiqua, Medievalis, and Orientalis, the Pla sequences are completely identical, whereas a single amino acid substitution, T259I, has been detected in atypical Angola and Microtus strains (6, 38, 50). A genetic analysis of 260 isolates of Y. pestis showed that the T259I substitution in Pla is shared by all isolates of biovar Microtus but absent in those of other biovars (59). Many of the Pestoides strains lack the pPCP1 plasmid and hence also the pla gene (12), and pla sequences from Pestoides are not available.
Pla is a member of the omptin family of conserved outer membrane proteases/adhesins detected in several gram-negative bacterial pathogens (15, 17, 21). The omptins have the same molecular size, a β-barrel fold of 10 transmembrane β strands, and five surface-exposed loops, L1 to L5 (Fig. 1). The catalytic residues and the residues interacting with lipid A in the outer membrane are completely conserved (17, 21-23, 41, 55). The omptins cleave peptide substrates at basic residues (17) but show dramatic heterogeneity in the recognition of biologically important polypeptides, such as Plg, the antiprotease α2AP, gelatin, and progelatinases. Analyses of hybrid proteins created between Pla and the omptins PgtE of Salmonella enterica and OmpT of Escherichia coli have indicated that the differing polypeptide substrate selectivity of omptins is dictated by sequence variation in the mobile loop structures of the β-barrel (22, 40). Residue T259 in Pla is located at surface loop 5 and oriented inward in the active-site groove of the Pla barrel, close to residue K262, where Pla is autoprocessed (22, 23) (Fig. 1).
FIG. 1.
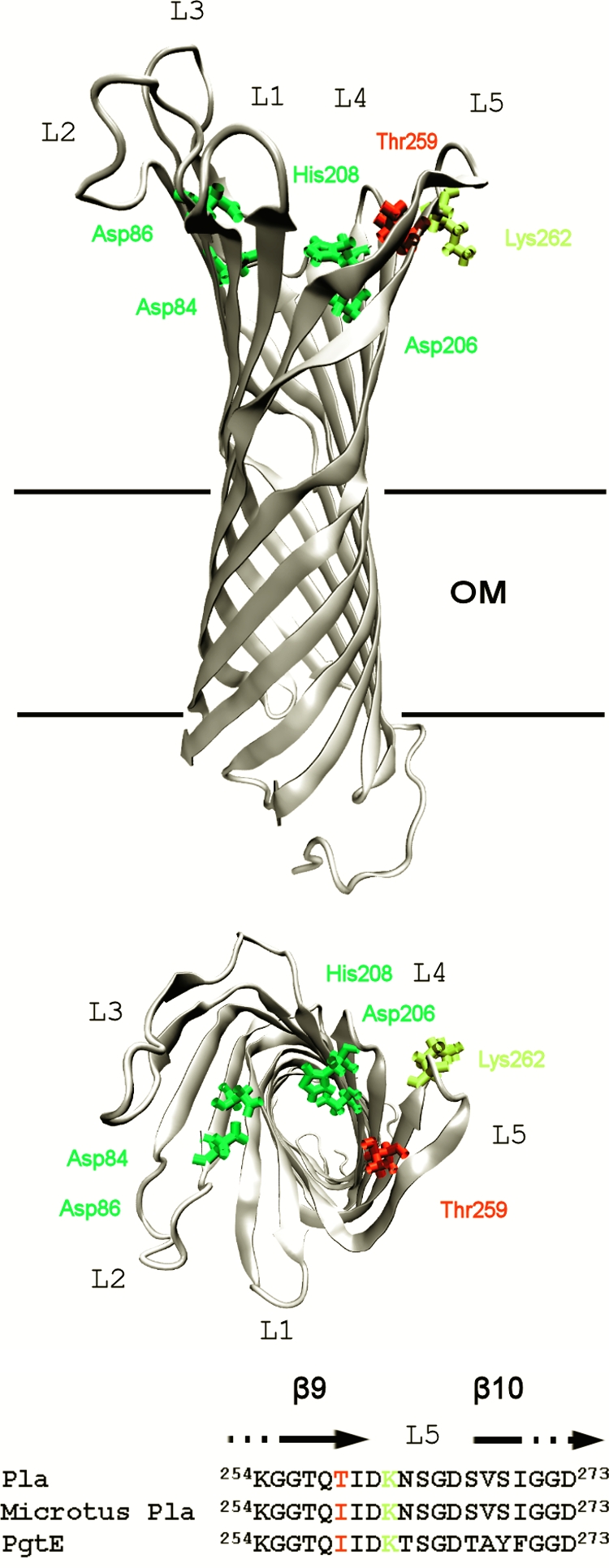
Model of Pla structure (23) and location of residue Thr259. Side (top drawing) and top (bottom drawing) views of the transmembrane β-barrel are shown. L1 to L5 are the surface loops. Catalytic residues Asp84, Asp86, Asp206, and His208 are indicated in green, Thr259 is in red, and the autoprocessing site Lys262 is in yellow. OM is the outer membrane. (C) Amino acid sequence of residues 254 to 273 at L5 and the termini of β-strands 9 and 10 in Pla, Microtus Pla, and PgtE are shown.
The omptin β-barrel has spread by horizontal gene transfer in gram-negative bacteria and adapted to the life-styles of host bacteria (15, 17, 21, 22, 40). Overall, the omptins give an example of an evolvable, robust enzyme fold (34) that easily acquires novel or improved functions. The fact that the single substitution T259I associates with ancestral Y. pestis Microtus and Angola populations suggests that Microtus Pla represents a form of the protein that preceded the common Pla protein. The central role of Plg activation in the pathogenesis of plague led us to analyze whether the single substitution T259I affects the fibrinolytic activities of the Pla molecule.
MATERIALS AND METHODS
Bacteria and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. The pla derivatives were expressed in the inducible pSE380 vector in E. coli XL1-Blue MRF′ essentially as described earlier (22, 23). Plasmid constructs pPlaT259I and pMRK3.51 were made by recombinant PCR as described earlier for the mutagenesis of pla (22) and pgtE (23), and the nucleotide sequences were verified by sequencing. The recombinant E. coli strains were cultivated overnight at 37°C in Luria broth (10 ml) supplemented with glucose (0.2% wt/vol), ampicillin (100 μg/ml), and tetracycline (12.5 μg/ml). For Pla protein expression, the culture was pelleted, suspended in 150 μl of phosphate-buffered saline (PBS, pH 7.1), and plated on Luria plates containing 5 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and antibiotics as described above. Recombinant Y. pestis strains were cultivated over 2 nights at 37°C in 10 ml of brain heart infusion broth supplemented with glucose (0.2% wt/vol), hemin (40 μg/ml), and ampicillin (100 μg/ml), and for expression of Pla, the bacteria were collected as described above and plated on brain heart infusion plates containing 5 μM IPTG, hemin (40 μg/ml), and ampicillin (100 μg/ml) and grown over 2 nights. For the assays, bacteria were collected in PBS, pelleted, and adjusted to an optical density at 600 nm of 1.2 (corresponding to ca. 109 cells/ml) or 2.0 (2 × 109 cells/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Reference(s) |
|---|---|---|
| Strains | ||
| E. coli XL1 Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Y. pestis KIM D34 | pPCP1− Δpgm pYV+ derivative of Y. pestis KIM-10 | 11, 54 |
| Plasmids | ||
| pSE380 | Expression vector; trc promoter lacO operator lacI bla | Invitrogen |
| pMRK1 | pla in pSE380 | 22 |
| pPlaT259I | Microtus pla in pSE380 | This study |
| pMRK1.51 | pla, 259TIDKN→IIDKT | 40 |
| pMRK3 | pgtE in pSE380 | 23 |
| pMRK3.51 | pgtE, 259IIDKT→TIDKN | This study |
Measurement of Pla protein expression.
Expression levels and isoforms of the Pla protein were assessed from cell envelope preparations (E. coli) (22) or from whole-cell samples (Y. pestis). The cell envelopes were prepared by sonicating 3 ml of the cell suspension (109 cells/ml) in PBS containing 2.5 mM EDTA four times for 30 s (17% amplitude, 3-s pulse, 1-s break) over crushed ice. The cells were pelleted by centrifugation (10 min, 3,000 rpm), and the supernatant was further centrifuged for 2 min at 6,000 rpm. The cell envelopes in the supernatant were pelleted (10 min, 13,000 rpm, 4°C) and suspended in PBS and then in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Half of the sample volume was boiled for 10 min. Whole-cell samples were prepared by mixing bacteria (109 cells/ml) with half of the volume of SDS-PAGE loading buffer and boiling them for 10 min. Samples were loaded into a 12% (wt/vol) SDS-PAGE gel, electrophoresed, and transferred onto a nitrocellulose membrane for detection with anti-His6-Pla antiserum (22), alkaline-phosphatase-conjugated anti-rabbit immunoglobulin G (IgG; Dako), and phosphatase substrate.
Fibrinolysis.
The fibrinolysis plate assay was performed basically as described previously (4), with minor modifications. Human fibrinogen (1.73 mg/ml; depleted of Plg, von Willebrand factor, and fibronectin; Kordia Life Sciences) was suspended in 2 ml water containing 5 μg/ml human Glu-Plg (American Diagnostica); 100 μl of bovine thrombin (5 NIH units; MP Biomedicals) in 100 mM sodium borate buffer (pH 7.74) was then added by gentle mixing, and the solution was immediately poured onto a six-well plate (Nunc). Five-microliter samples of bacterial suspensions (107 bacteria) in PBS were applied to the solidified plate, which was then incubated at 37°C for 20 h (E. coli) or 48 h (Y. pestis). Fibrinolysis was visually assessed as a clear zone around the bacteria.
Activation and cleavage of plasminogen and cleavage of α2AP.
For activity measurements, 4 or 0.4 μg human Glu-Plg was mixed with 8 × 107 bacteria in PBS in a total volume of 174 μl in microtiter plate (Nunc) wells. For cumulative analysis of plasmin formation, 30 μl of the chromogenic plasmin substrate Val-Leu-Lys-p-nitroaniline dihydrochloride (S-2251, 2.5 mg/ml; Chromogenix/DiaPharma) was immediately added and plasmin formation at 37°C was measured every 15 or 30 min for several hours in a microtiter plate reader at 405 nm (22). To analyze the time course of plasmin formation, 4 μg human Glu-Plg was mixed with bacteria at 37°C and the plasmin substrate was added at time points of 0, 1, 2, 5, 10, and 22 h; plasmin activity was then measured after 15 min. In Plg cleavage assays (22), 2.5 μg of human Glu-Plg and 4 × 107 bacteria (in PBS) were incubated in a total volume of 25 μl while shaking for 7 h at 37°C. After 2 and 7 h, the cells were pelleted, half the volume of SDS-PAGE loading buffer was added and the supernatant samples were boiled for 10 min. Ten-microliter samples were subjected to 12% (wt/vol) SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (GE Healthcare), and Plg polypeptides were detected by Western blotting with polyclonal anti-human Plg antibody (1 mg/ml, diluted 1:1,000; American Diagnostica), peroxidase-conjugated anti-rabbit IgG (1:2,500; GE Healthcare), and enhanced chemiluminescence detection reagents (GE Healthcare) according to the manufacturer's instructions. The membrane was exposed to X-ray film (Agfa) for 15 s. In assays where Plg cleavage was detected with monoclonal anti-human Plg catalytic domain antibody (R&D Systems), the following minor modifications were made. Three micrograms of Glu-Plg and 4.8 × 107 bacteria were incubated for 2 h in a total volume of 30 μl. For comparison, Plg cleavage by urokinase (uPA) at a concentration of 2.5 μg/ml was assessed. After incubation, supernatant samples of 15 μl were electrophoresed and detection was done with the monoclonal antibody (500 μg/ml, diluted 1:500) and peroxidase-conjugated anti-mouse IgG (GE Healthcare; 1:1,000). The exposure time was 2 min. Cleavage of human α2AP (0.8 μg; Calbiochem) was tested as detailed earlier (22, 26), by using 3 × 107 bacteria and incubation for 2 h under shaking (1,100 rpm) at 37°C.
Inactivation of plasmin and α2AP.
Inactivation of plasmin was measured by incubating 0.5 μg human plasmin (Sigma) with bacteria (8 × 107) in 170 μl of PBS for 30 min at 37°C in a microtiter plate. The chromogenic plasmin substrate (30 μl) was then added, and plasmin activity was measured at 405 nm after 90 min of incubation at 37°C. Inactivation of α2AP was measured basically as described earlier (26). Human α2AP (5.75 μg/ml) was incubated with bacteria (8 × 107) in PBS for 2 h at 37°C in a microtiter plate. Human plasmin (0.5 μg) was added, and the plate was incubated for 30 min at 37°C. Thirty microliters of the chromogenic plasmin substrate was added to make the final volume 200 μl, and plasmin activity was measured at 405 nm after 90 min of incubation at 37°C.
Sequence alignment and Pla structure model.
Y. pestis Pla amino acid sequences were obtained from the NCBI GenBank protein database and aligned by using the ClustalW program (7). Pairwise alignment of Pla (31) and PgtE (14) sequences was done with the needle program from the EMBOSS package (42). The Pla model was built with the MODELLER program (30) by using the existing OmpT structure (Protein Data Bank identifier: 1i78) as the template. Visualization was done with the VMD program (18).
RESULTS
Autoprocessed β-Pla is absent in Pla T259I.
To assess functional properties of the Microtus Pla variant, the substitution T259I was created in the Pla molecule originating from biovar Medievalis, and Pla and Pla T259I were expressed in Y. pestis KIM D34, which lacks the pPCP1 plasmid. Western blotting with polyclonal anti-Pla antiserum indicated that Pla and Pla T259I were expressed in similar amounts in KIM D34 cells (Fig. 2A). In SDS-PAGE, denatured Pla exhibits four isoforms (pre-Pla, α-Pla, β-Pla, and γ-Pla) (24, 47, 48), of which α-Pla and γ-Pla are different conformations of the mature Pla protein and β-Pla is the autoprocessed form (20, 22). Pla and Pla T259I showed different isoform patterns in the KIM D34 host; all isoforms were detected with Pla, whereas Pla T259I lacked the autoprocessed isoform β-Pla (Fig. 2A).
FIG. 2.
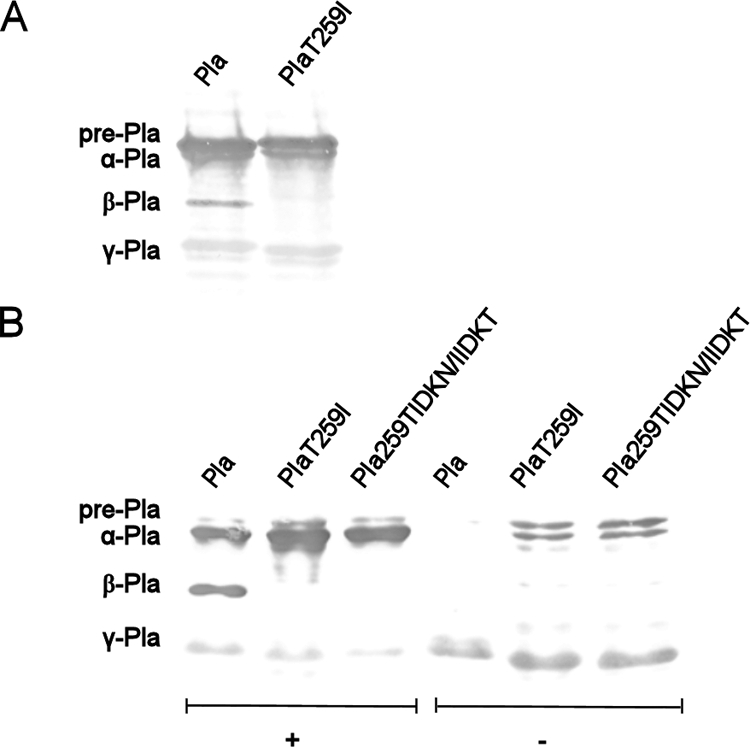
Expression and isoform patterns of Pla and the Pla derivatives in recombinant bacteria. (A) Pla and Pla T259I were expressed in Y. pestis KIMD34, and the Western blot assay shows the reactivity of whole-cell samples with anti-Pla antiserum. (B) Expression and isoforms of the Pla proteins in cell envelope preparations from recombinant E. coli. The protein constructs are indicated above the lanes, and the migration distances of the α, β, and γ isoforms of the mature Pla molecule and of nonprocessed pre-Pla are indicated on the left. The plus sign denotes samples boiled for 10 min before electrophoresis, and the minus sign denotes unboiled samples.
PgtE of S. enterica belongs to the same omptin subfamily and shares 75% sequence identity with Pla (15, 17, 21); in the L5 region, the sequences differ in 6/20 residues (Fig. 1C). We included in the assays the Pla derivative carrying the substitution 259TIDKN/IIDKT, which was available from previous work (40) and at L5 mimics the sequence of PgtE. To analyze the Pla derivatives in more detail and to compare Pla and PgtE (see below), we expressed the omptin variants in recombinant E. coli XL1-Blue, which has been successfully used to characterize the functions of both Pla and PgtE (22-24, 27, 40, 47). Pla and the Pla variants were expressed in similar amounts in cell envelope preparations from recombinant E. coli, as detected by Western blotting with anti-Pla antiserum (Fig. 2B). Differences in the Pla isoforms were detected in boiled samples of the three Pla variants also in the E. coli XL1 host. Wild-type Pla had all four forms, whereas Pla T259I and Pla 259TIDKN/IIDKT completely lacked the β-Pla form. In unboiled cell envelope samples, where the native-like folding of the β-barrel is preserved (33), wild-type Pla was detected almost completely in the γ-Pla form whereas the substituted Pla molecules also exhibited the pre-Pla and α-Pla forms of the molecule. Moreover, the γ-Pla form of Pla T259I and Pla 259TIDKN/IIDKT migrated faster than did wild-type Pla (Fig. 2B).
Microtus Pla is reduced in fibrinolysis.
Formation of fibrin clots prevents bacterial migration in infected tissues (5), and the plasmin-mediated degradation of fibrin is a possible pathogenetic mechanism of Pla (8, 15, 28). Strain KIM D34(pPlaT259I) was less efficient than KIM D34(pMRK1) in a fibrinolysis plate assay (Fig. 3A). The same result was obtained when the two plasmids were expressed in E. coli XL1 (Fig. 3B). Fibrinolysis was observed only when Plg was added to the plates.
FIG. 3.
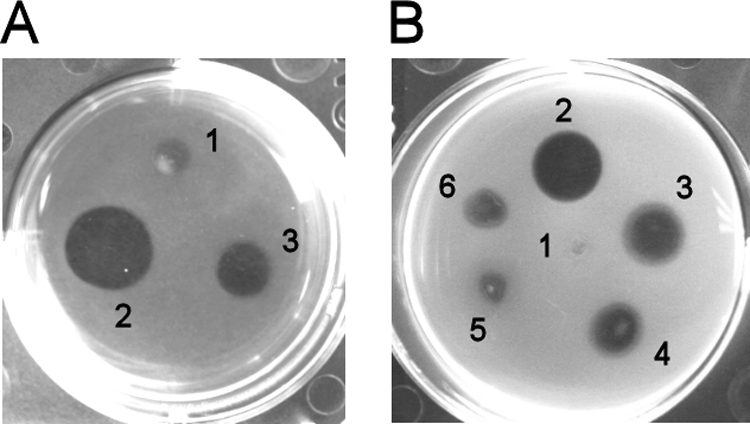
Fibrinolysis by recombinant Y. pestis KIM D34 and E. coli XL1. Bacteria (107 cells) in buffer were pipetted onto fibrin plates containing 5 μg/ml Plg, and dissolution of the clot was assessed after incubation for 20 h (E. coli) or 48 h (Y. pestis) at 37°C. (A) Bacterial numbering: 1, Y. pestis KIM D34(pSE380); 2, Y. pestis KIM D34(pMRK1); 3, Y. pestis KIM D34(pPlaT259I). (B) Bacterial numbering: 1, E. coli XL1(pSE380); 2, E. coli XL1(pMRK1); 3, E. coli XL1(pPlaT259I); 4, E. coli XL1(pMRK1.51); 5, E. coli XL1(pMRK3); 6, E. coli XL1(pMRK3.51).
E. coli XL1(pMRK3) with PgtE exhibited weak fibrinolytic activity, and E. coli XL1(pMRK1.51) expressing Pla 259TIDKN/IIDKT, which mimics L5 of PgtE, was less effective than the bacteria with the common Pla protein (Fig. 3B). The 259IIDKT/TIDKN substitution in PgtE (pMRK3.51) was therefore constructed to compare it to the reverse substitution in Pla; this construct showed slightly higher fibrinolytic activity than did PgtE (Fig. 3B). We observed no differences in isoform formation between PgtE and PgtE 259IIDKT/TIDKN (data not shown).
Formation of plasmin is reduced in recombinant Y. pestis and E. coli expressing Microtus Pla.
The findings above indicated that the T259I substitution in Pla affects Plg activation. In measurements of cumulative plasmin formation in Y. pestis, strains KIM D34(pMRK1), with the common Pla protein, and KIM D34(pPlaT259I) initiated plasmin formation equally effectively at a high Plg concentration (20 μg/ml), whereas reduced plasmin formation by KIM D34(pPlaT259I) was evident in assays with a lower concentration of Plg (2 μg/ml; Fig. 4A). Similarly, slower Plg activation by Pla T259I and by Pla 259TIDKN/IIDKT was observed at the lower Plg concentration in the E. coli XL1 background (Fig. 4B). In contrast, E. coli XL1(pMRK3) with PgtE was poor in a cumulative Plg activation assay, and its activity was improved by the 259IIDKT/TIDKN substitution in pMRK3.51.
FIG. 4.
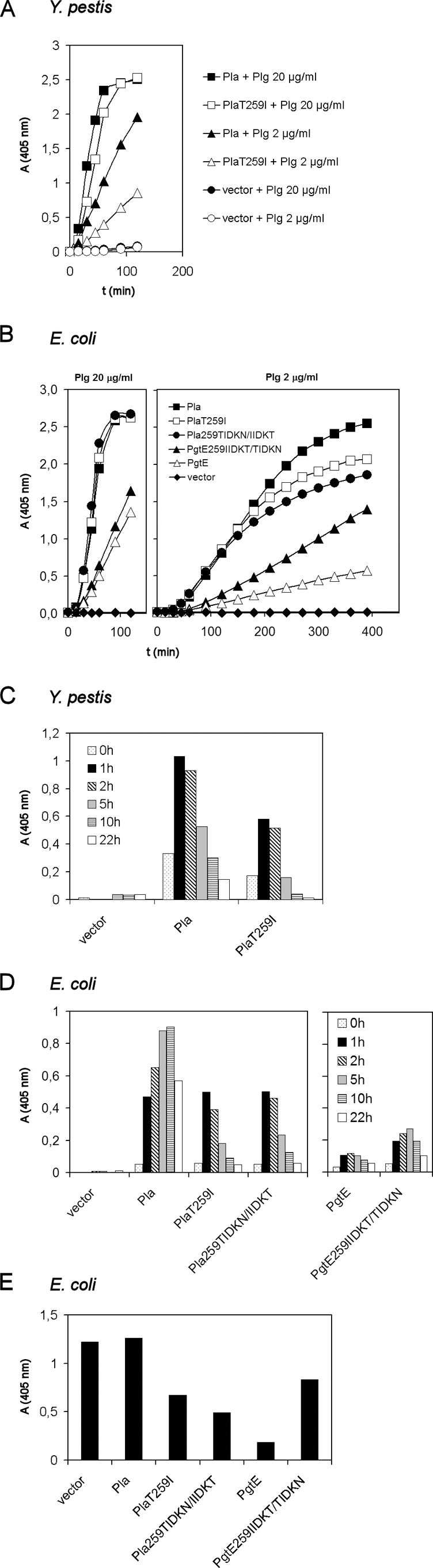
Plasminogen activation by recombinant Y. pestis KIM D34 and E. coli XL1 and plasmin inactivation by recombinant E. coli XL1. (A and B) Cumulative, initial plasmin formation by bacteria tested at two Plg concentrations (20 and 2 μg/ml). The expression host, protein constructs, and Plg concentrations are indicated. (C and D) Plasmin formation by bacteria after incubation for up to 22 h with Plg. Plasmin formation was analyzed by adding a chromogenic plasmin substrate at the indicated time points and measuring absorbance after 15 min. (E) Inhibition of plasmin activity by recombinant E. coli. Plasmin was incubated for 30 min with the bacteria, after which the plasmin substrate was added and the absorbance was measured after 90 min. The assays were repeated at least three times, and results of a representative assay with duplicate samples are shown.
We next measured plasmin activity during a 22-h incubation of bacteria with Plg. With Y. pestis KIM D34(pMRK1), the plasmin activity was highest after 1 to 2 h of incubation and then declined, whereas with KIM D34(pPlaT259I), the plasmin activity remained lower and was more transient (Fig. 4C). Plasmin activity formed by E. coli XL1(pMRK1) with wild-type Pla increased for up to 10 h but was decreased after a 22-h incubation, whereas the plasmin activity formed by E. coli with Pla T259I or Pla 259TIDKN/IIDKT was already decreased after 1 h of incubation and close to the background level at 22 h (Fig. 4D). A clear difference between Pla and PgtE was seen in the time course of plasmin formation: plasmin activity did not rise after 1 h of incubation of Plg with E. coli(pMRK3) (Fig. 4D). Also, in incubations for 1 to 10 h, E. coli XL1 with PgtE 259IIDKT/TIDKN showed an increase in the level of PgtE-mediated plasmin formation.
We also tested the proteinases in the Y. pseudotuberculosis background by using as a host strain Y. pseudotuberculosis PB1 Δwb, which represents the probable ancestor of Y. pestis (46). The T259I substitution in Pla destabilized the formation of plasmin activity also in this strain (data not shown).
Microtus Pla inactivates plasmin.
The reduction in plasmin levels observed after a longer incubation of Plg with PgtE-, Pla T259I-, or Pla 259IIDKT/TIDKN-expressing bacteria suggested that these omptin variants degrade the Plg substrate into proteolytically inactive fragments and into plasmin, which is then further degraded by the proteases. We therefore assessed whether the omptin proteases indeed inactivate plasmin; this was done by incubating human plasmin with the E. coli derivatives and then analyzing the remaining levels of plasmin activity (Fig. 4E). The level of plasmin activity remained similar with the vector strain E. coli(pSE380) and with E. coli(pMRK1), indicating that Pla did not inactivate plasmin during incubation. In contrast, the activity of plasmin was lowered after incubation with E. coli(pPlaT259I) and E. coli(pMRK1.51). Plasmin activity was also diminished by E. coli(pMRK3) with PgtE, whereas E. coli(pMRK3.51) with PgtE 259IIDKT/TIDKN caused only a partial reduction (Fig. 4E).
The I259T substitution is critical for generation of the catalytic plasmin light chain.
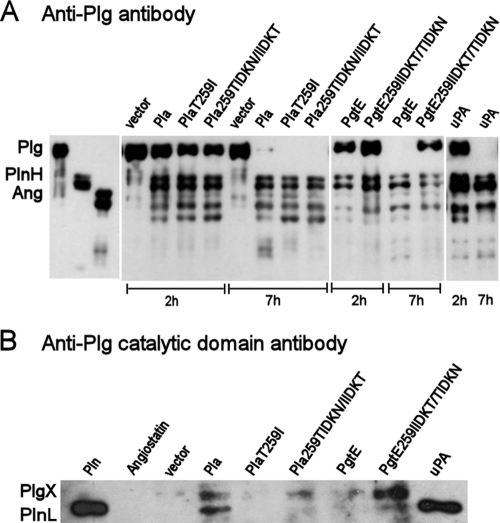
We assessed Plg cleavage by the recombinant E. coli strains by Western blotting with polyclonal anti-Plg antibodies which mainly recognize the noncatalytic, lysine-binding kringle domains in Plg and with a monoclonal anti-Plg catalytic domain antibody which recognizes the 25-kDa light chain of plasmin (PlnL) that contains the protease domain. The fragmentation of Plg by the bacteria was compared to Plg fragments obtained in incubation with the human Plg activator uPA. Western blotting with the anti-Plg antibody showed similar patterns of Plg cleavage by Pla, Pla T259I, and Pla 259TIDKN/IIDKT expressed in recombinant E. coli XL1 (Fig. 5A). The Pla constructs completely cleaved the Plg molecule within 7 h into peptides similar in size to the plasmin heavy chain (PlnH) and angiostatins. The latter are composed of kringle domains of Plg and lack the protease domain, and they are also contained in the PlnH chains (35). Formation of PlnH- and angiostatin-like fragments was detectable in uPA-mediated cleavage of Plg as well (Fig. 5A).
FIG. 5.
Degradation of plasminogen by Pla and PgtE variants expressed in recombinant E. coli XL1. Plg degradation by the bacteria was analyzed by Western blotting with anti-Plg antibody (A) and anti-Plg catalytic domain antibody (B). The migration distances of Plg, plasmin heavy chain (PlnH, apparent size of 70 kDa), plasmin light chain (PlnL, 25 kDa), and angiostatins (Ang) are shown on the left. PlgX indicates a fragment of Plg produced by the bacteria. The Pla and PgtE constructs expressed in E. coli XL1 and uPA-treated Plg are indicated above the lanes, incubation times in panel A are indicated below the lanes. In panel B, the incubation time was 2 h.
Recombinant E. coli XL1(pMRK3) with PgtE cleaved the Plg molecule as efficiently as did Pla-expressing E. coli XL1(pMRK1), whereas E. coli with the mutated PgtE 259IIDKT/TIDKN protein exhibited slower cleavage (Fig. 5A).
Immunoblotting with the anti-Plg catalytic domain antibody revealed the presence of PlnL in Plg preparations incubated for 2 h with E. coli XL1 Pla or with uPA (Fig. 5B). A peptide corresponding to PlnL was weakly detectable with PgtE 259IIDKT/TIDKN, whereas we failed to detect the PlnL chain in samples treated with XL1(pPlaT259I), XL1(pMRK1.51), XL1(pMRK3), or the vector strain (Fig. 5B). PlnL contains the catalytic domain of plasmin (51). An immunoreactive peptide (here named PlgX) was observed in the degradation of Plg by the omptins Pla, Pla 259TIDKN/IIDKT, and PgtE 259IIDKT/TIDKN; notably, this peptide was lacking in uPA-treated Plg samples (Fig. 5B).
Microtus Pla is efficient in α2AP inactivation.
Pla and PgtE are multifunctional and cleave and inactivate the major circulating plasmin inhibitor α2AP (22, 26, 52), which is important for the in vivo activity of plasmin. To assess whether the I259T substitution affects the activity of omptins toward another biologically important polypeptide substrate, we compared the mutated constructs for cleavage of the antiprotease (Fig. 6A). The Pla derivatives and PgtE cleaved α2AP, whereas E. coli XL1(pMRK3.51) with PgtE 259IIDKT/TIDKN exhibited slower proteolysis than did PgtE-expressing cells. In inhibition assays, E. coli strains expressing PgtE, Pla T259I, or Pla 259TIDKN/IIDKT abolished the antiprotease activity of α2AP. In contrast, E. coli with Pla or PgtE 259IIDKT/TIDKN only partially affected the antiproteolytic activity (Fig. 6B), indicating that although these two proteases cleaved the antiprotease, they were less effective in its inactivation.
FIG. 6.
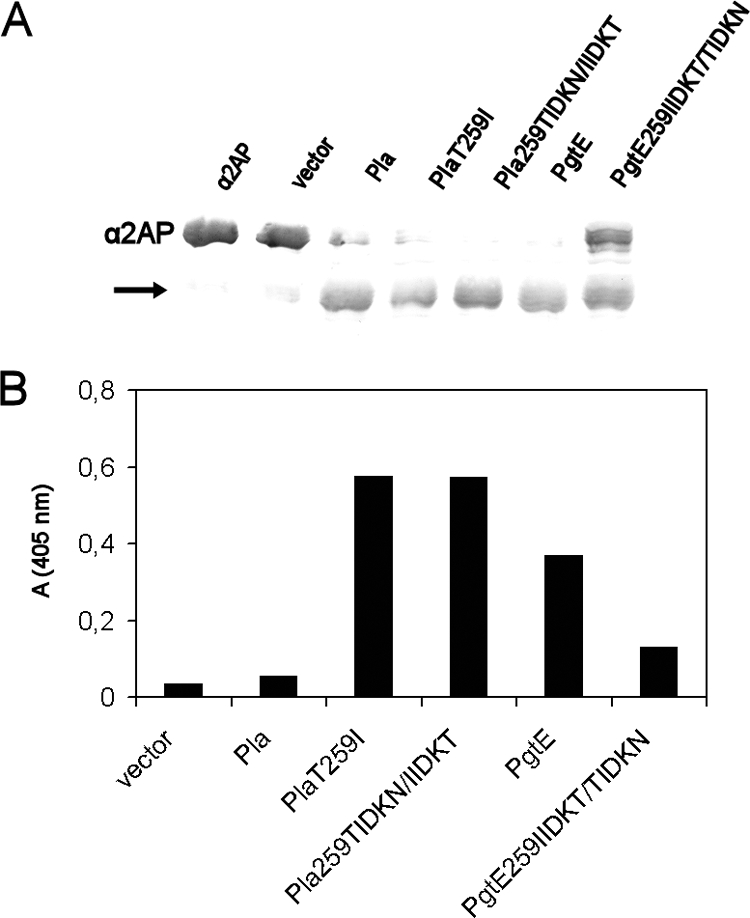
Degradation and inactivation of α2AP by recombinant E. coli XL1. (A) Degradation of α2AP after a 2-h incubation with bacteria, as analyzed by Western blotting. The protein constructs expressed in E. coli XL1 are indicated above the lanes. The migration distance of α2AP is shown on the left, and the arrow indicates a cleavage product of α2AP. (B) Inactivation of α2AP by bacteria. The Pla and PgtE constructs expressed in E. coli XL1 are indicated. Bacteria were incubated with α2AP for 2 h, after which plasmin and its chromogenic substrate were added and plasmin activity was measured after 15 min. The assay was repeated three times, and results of a representative assay with duplicate samples are shown.
DISCUSSION
Horizontal gene transfer, loss of gene functions, genomic rearrangements, and single-nucleotide polymorphism have been important in the evolution of Y. pestis (1, 57). Single-nucleotide polymorphisms have served as useful markers in the construction of phylogenetic patterns, but their biological effects have been only partially characterized. Here we describe a nonsynonymous nucleotide change that increases the fibrinolytic potential of a major bacterial virulence factor. Pla is essential for the virulence and epidemic spread of Y. pestis (28, 43, 45), and the pla gene in plasmid pPCP1 has been acquired by horizontal gene transfer after Y. pestis and Y. pseudotuberculosis diverged. The single substitution I259T present in the Pla protein of the pandemic or “typical” lineages of Y. pestis directs the proteolytic activity of Pla toward higher fibrinolysis and plasmin formation but lower antiprotease inactivation. The reverse substrate selectivity was seen with the close ortholog PgtE of the intracellular pathogen S. enterica, which has an ancestral form in common with Pla (15, 47). Plg activation by Pla is essential in plague (28, 43, 49), and the I259T substitution in Pla has likely been important in the evolution of plague pathogenesis. Plasmin and plasminogen bound to fibrin are resistant to regulation by α2AP (29), and the change in the selectivity of the common Pla protein toward plasmin generation is in accord with the observed importance of fibrinolysis in plague (8, 28).
The eukaryotic Plg activators uPA and tissue type plasminogen activator activate Plg through a single cleavage of the peptide bond between Arg560 and Val561, which is also cleaved by Pla (49). In addition to generating the PlnL chain, which carries the catalytic domain of plasmin, the Pla-expressing bacteria also generated an immunoreactive Plg fragment, here termed PlgX, which was not detected with uPA-cleaved Plg. This indicates that Pla is less specific than uPA in Plg activation and cleavage. With Pla T259I-expressing bacteria, Plg activation was slower than with Pla-expressing bacteria and we did not detect generation of the PlnL chain by Western blotting. As an enzyme detection method, immunoblotting is less sensitive than measurement of plasmin enzymatic activity and may thus fail to detect small amounts of plasmin or PlnL. The bacteria expressing Microtus Pla also inactivated plasmin more efficiently than did those expressing the common Pla protein. Taken together, the results strongly suggest that the low plasmin activity levels formed by Microtus Pla result from transient formation of active plasmin, subsequent or simultaneous cleavage of the PlnL chain, and inactivation of the formed plasmin activity.
The omptins in enteric bacteria share ca. 50% sequence identity, and Pla of human-pathogenic Y. pestis is the only omptin protease that has Thr at position 259 (41). The substitution T259I in Pla lowered plasmin formation but did not grossly change the amount of Pla peptides in the outer membrane, and our hypothesis is that the observed change in target selectivity results from a conformational change in Pla surface loop 5. Residue T259 and the autoprocessing site K262 are spatially located near the catalytic groove of the Pla protease, and it is likely that the lack of autoprocessing in Microtus Pla results from steric inhibition of peptide bond cleavage. Prevention of autoprocessing in the Microtus Pla molecule will restrict the mobility and flexibility of L5 residues and thus may alter interactions of Pla with polypeptide substrates. A conformational effect is also suggested by the finding that in unboiled E. coli cell envelope preparations, where the β-barrel fold is not destroyed (33), Pla was nearly completely in the γ-Pla form, whereas pre-Pla and α-Pla were detected in the substituted Pla proteins. Also, under nondenaturing conditions, the γ-Pla form migrated differently from γ forms of the substituted Pla molecules. The important role of the L5 region in correct substrate recognition by both Pla and PgtE is underlined by the findings that the mutated PgtE 259IIDKT/TIDKN protein showed an increase in plasmin formation and a decrease in α2AP inactivation and that the reverse change in substrate selectivity was observed with the reverse substitution, Pla 259TIDKN/IIDKT. We have earlier found that this region in L5 is important for the gelatinase activity that is expressed by PgtE but not by Pla (40).
PgtE contributes to the virulence of S. enterica by promoting bacterial survival in mouse macrophages and bacterial spread in infected mice (26, 40). The proteolytic activities of PgtE include degradation of cationic antimicrobial peptides (14) and complement proteins (39), highly active degradation and inactivation of α2AP in S. enterica cells from infected macrophages (26), and degradation of gelatin and activation of mammalian gelatinases (40). Among the omptins, the latter function appears unique to PgtE (15), and it is involved in the pathogenesis of salmonellosis (16).
In contrast to Pla, PgtE appears to cleave Plg/plasmin polypeptides mainly in an inactivating manner. Several lines of evidence support this conclusion. First, PgtE-expressing bacteria were as effective as Pla-expressing bacteria in cleaving the Plg molecule but produced very low levels of plasmin activity and fibrinolysis. These results are in line with previous observations showing poor plasmin formation by the PgtE omptin of S. enterica (47, 52). Second, the low level of plasmin activity did not increase during a 2- to 5-h incubation with Plg, whereas with Pla-expressing bacteria the plasmin activity levels were increased. Third, we detected generation of the PlnH chain and kringle domains but did not detect formation of the PlnL chain by E. coli(pMRK3). This suggests that the proteolytically active PlnL chain was rapidly degraded by PgtE. In the same assay, we detected the PlnL chain after incubation with Pla-expressing bacteria, as well as with uPA. Finally, we indeed observed that incubation of human plasmin with PgtE-expressing bacteria lowered the enzymatic activity of plasmin. The opposite situation was observed when α2AP was used as a substrate; Pla-expressing bacteria cleaved the α2AP polypeptide into a smaller fragment similarly as did PgtE-expressing bacteria but were less effective in abolishing the antiproteolytic activity of α2AP. Measurement of α2AP antiprotease activity relies on determining the change in apparent plasmin activity, and it should be noted that although PgtE-expressing bacteria directly inactivated plasmin in 30 min of incubation, we could observe clear plasmin activity compared to the vector strain in the antiprotease assay. Our results support the notion that PgtE should be functionally classified as an antiprotease inactivator rather than as a Plg activator (26).
Directed evolution of enzymes has shown that while most single amino acid substitutions are either neutral or deleterious, some single substitutions increase the enzyme's fitness (53). Such amino acid substitutions can be located in or be distant from the active site, and the distant mutations affect catalysis by slightly altering the geometry, electrostatic properties, or dynamics of amino acids in the active site. The β-barrel is a sturdy membrane-embedded structure that allows large changes in the surface loops without disturbing the β-barrel folding (19), and the omptin β-barrels fulfill the criteria of an evolvable and mutationally robust protein superfamily that is undergoing adaptive molecular evolution (34). The individual omptins in enteric bacteria exhibit differing functions (15), and substitution of nonconserved amino acid residues at the L1 to L5 structures broadens the substrate selectivity of Pla of Y. pestis and OmpT of E. coli toward progelatinase or Plg activation, respectively (22, 40), which indicates that the mobile surface loops are important in polypeptide substrate recognition by omptins. Varadarajan and coworkers (56) described an S223R amino acid substitution in OmpT that changed the cleavage bond selectivity from Arg-Arg to Ala-Arg in short synthetic peptides. Residue S223 is located in the intimate vicinity of the catalytic residues of OmpT.
We describe here a single residue change in Pla that has occurred naturally and has persevered in the evolution of pandemic Y. pestis branches. Plg is synthesized mainly in the liver and is abundant in circulation, whereas only very low concentrations of Plg mRNA or Plg protein have been detected in extrahepatic tissue sites, such as adipose tissue (58) and extracellular matrices (9), which the plague bacterium must bypass to reach lymphatic tissue. Hence, the improved Plg utilization and enhanced fibrinolysis by the common Pla protein can be critical in the pathogenesis of plague.
Acknowledgments
We thank Raili Lameranta and Johanna Heikkinen for technical assistance.
This work was financially supported by the Research Foundation of the University of Helsinki, the European Union Network of Excellence EuroPathoGenomics program, the Viikki Graduate School in Molecular Biosciences, and the Academy of Finland (grants 116507 and 130202).
Footnotes
Published ahead of print on 22 May 2009.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 10117837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., K. Aurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 9614043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anisimov, A. P., L. E. Lindler, and G. B. Pier. 2004. Intraspecific diversity of Yersinia pestis. Clin. Microbiol. Rev. 17434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beesley, E. D., R. R. Brubaker, W. A. Janssen, and M. J. Surgalla. 1967. Pesticins. 3. Expression of coagulase and mechanism of fibrinolysis. J. Bacteriol. 9419-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann, S., and S. Hammerschmidt. 2007. Fibrinolysis and host response in bacterial infections. Thromb. Haemost. 98512-520. [PubMed] [Google Scholar]
- 6.Chain, P. S. G., P. Hu, S. A. Malfatti, L. Radnedge, F. Larimer, L. M. Vergez, P. Worsham, M. C. Chu, and G. L. Andersen. 2006. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. J. Bacteriol. 1884453-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 313497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degen, J. L., T. H. Bugge, and J. D. Goguen. 2007. Fibrin and fibrinolysis in infection and host defense. J. Thromb. Haemost. 5(Suppl. 1)24-31. [DOI] [PubMed] [Google Scholar]
- 9.Farina, A. R., A. Tiberio, A. Tacconelli, L. Cappabianca, A. Gulino, and A. R. Mackay. 1996. Identification of plasminogen in Matrigel and its activation by reconstitution of this basement membrane extract. BioTechniques 21904-909. [DOI] [PubMed] [Google Scholar]
- 10.Ferber, D. M., and R. R. Brubaker. 1981. Plasmids in Yersinia pestis. Infect. Immun. 31839-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finegold, M. J., J. J. Petery, R. F. Berendt, and H. R. Adams. 1968. Studies on the pathogenesis of plague. Blood coagulation and tissue responses of Macaca mulatta following exposure to aerosols of Pasteurella pestis. Am. J. Pathol. 5399-114. [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia, E., P. Worsham, S. Bearden, S. Malfatti, D. Lang, F. Larimer, L. Lindler, and P. Chain. 2007. Pestoides F, an atypical Yersinia pestis strain from the former Soviet Union. Adv. Exp. Med. Biol. 60317-22. [DOI] [PubMed] [Google Scholar]
- 13.Goguen, J. D., T. Bugge, and J. L. Degen. 2000. Role of the pleiotropic effects of plasminogen deficiency in infection experiments with plasminogen-deficient mice. Methods 21179-183. [DOI] [PubMed] [Google Scholar]
- 14.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 1824077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haiko, J., M. Suomalainen, T. Ojala, K. Lähteenmäki, and T. K. Korhonen. 2009. Breaking barriers—attack on innate immune defences by omptin surface proteases of enterobacterial pathogens. Innate Immun. 1567-80. [DOI] [PubMed] [Google Scholar]
- 16.Handley, S. A., and V. L. Miller. 2007. General and specific host responses to bacterial infection in Peyer's patches: a role for stromelysin-1 (matrix metalloproteinase-3) during Salmonella enterica infection. Mol. Microbiol. 6494-110. [DOI] [PubMed] [Google Scholar]
- 17.Hritonenko, V., and C. Stathopoulos. 2007. Omptin proteins: an expanding family of outer membrane proteases in gram-negative Enterobacteriaceae. Mol. Membr. Biol. 24395-406. [DOI] [PubMed] [Google Scholar]
- 18.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—Visual Molecular Dynamics. J. Mol. Graphics 1433-38. [DOI] [PubMed] [Google Scholar]
- 19.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37239-253. [DOI] [PubMed] [Google Scholar]
- 20.Kramer, R. A., D. Zandwijken, M. R. Egmond, and N. Dekker. 2000. In vitro folding, purification and characterization of Escherichia coli outer membrane protease OmpT. Eur. J. Biochem. 267885-893. [DOI] [PubMed] [Google Scholar]
- 21.Kukkonen, M., and T. K. Korhonen. 2004. The omptin family of enterobacterial surface proteases/adhesins: from housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol. 2947-14. [DOI] [PubMed] [Google Scholar]
- 22.Kukkonen, M., K. Lähteenmäki, M. Suomalainen, N. Kalkkinen, L. Emödy, H. Lång, and T. K. Korhonen. 2001. Protein regions important for plasminogen activation and inactivation of α2-antiplasmin in the surface protease Pla of Yersinia pestis. Mol. Microbiol. 401097-1111. [DOI] [PubMed] [Google Scholar]
- 23.Kukkonen, M., M. Suomalainen, P. Kyllönen, K. Lähteenmäki, H. Lång, R. Virkola, I. M. Helander, O. Holst, and T. K. Korhonen. 2004. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol. Microbiol. 51215-225. [DOI] [PubMed] [Google Scholar]
- 24.Kutyrev, V., R. J. Mehigh, V. L. Motin, M. S. Pokrovskaya, G. B. Smirnov, and R. R. Brubaker. 1999. Expression of the plague plasminogen activator in Yersinia pseudotuberculosis and Escherichia coli. Infect. Immun. 671359-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lähteenmäki, K., S. Edelman, and T. K. Korhonen. 2005. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 1379-85. [DOI] [PubMed] [Google Scholar]
- 26.Lähteenmäki, K., P. Kyllönen, L. Partanen, and T. K. Korhonen. 2005. Antiprotease inactivation by Salmonella enterica released from infected macrophages. Cell. Microbiol. 7529-538. [DOI] [PubMed] [Google Scholar]
- 27.Lähteenmäki, K., R. Virkola, A. Saren, L. Emody, and T. K. Korhonen. 1998. Expression of plasminogen activator Pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infect. Immun. 665755-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lathem, W. W., P. A. Price, V. L. Miller, and W. E. Goldman. 2007. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science 315509-513. [DOI] [PubMed] [Google Scholar]
- 29.Lijnen, H. R., and D. Collen. 1995. Mechanisms of physiological fibrinolysis. Baillieres Clin. Haematol. 8277-290. [DOI] [PubMed] [Google Scholar]
- 30.Martí-Renom, M. A., A. C. Stuart, A. Fiser, R. Sanchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29291-325. [DOI] [PubMed] [Google Scholar]
- 31.McDonough, K. A., and S. Falkow. 1989. A Yersinia pestis-specific DNA fragment encodes temperature-dependent coagulase and fibrinolysin-associated phenotypes. Mol. Microbiol. 3767-775. [DOI] [PubMed] [Google Scholar]
- 32.Mignatti, P., and D. B. Rifkin. 1993. Biology and biochemistry of proteinases in tumor invasion. Physiol. Rev. 73161-195. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, K., and S. Mizushima. 1976. Effects of heating in dodecyl sulfate solution on the conformation and electrophoretic mobility of isolated major outer membrane proteins from Escherichia coli K-12. J. Biochem. 801411-1422. [DOI] [PubMed] [Google Scholar]
- 34.O'Loughlin, T. L., W. M. Patrick, and I. Matsumura. 2006. Natural history as a predictor of protein evolvability. Protein Eng. Des. Sel. 19439-442. [DOI] [PubMed] [Google Scholar]
- 35.O'Reilly, M. S., L. Holmgren, Y. Shing, C. Chen, R. A. Rosenthal, M. Moses, W. S. Lane, Y. Cao, E. H. Sage, and J. Folkman. 1994. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell 79315-328. [DOI] [PubMed] [Google Scholar]
- 36.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 1035-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plow, E. F., T. Herren, A. Redlitz, L. A. Miles, and J. L. Hoover-Plow. 1995. The cell biology of the plasminogen system. FASEB J. 9939-945. [DOI] [PubMed] [Google Scholar]
- 38.Prentice, M. B., K. D. James, J. Parkhill, S. G. Baker, K. Stevens, M. N. Simmonds, K. L. Mungall, C. Churcher, P. C. Oyston, R. W. Titball, B. W. Wren, J. Wain, D. Pickard, T. T. Hien, J. J. Farrar, and G. Dougan. 2001. Yersinia pestis pFra shows biovar-specific differences and recent common ancestry with a Salmonella enterica serovar Typhi plasmid. J. Bacteriol. 1832586-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramu, P., R. Tanskanen, M. Holmberg, K. Lähteenmäki, T. K. Korhonen, and S. Meri. 2007. The surface protease PgtE of Salmonella enterica affects complement activity by proteolytically cleaving C3b, C4b and C5. FEBS Lett. 5811716-1720. [DOI] [PubMed] [Google Scholar]
- 40.Ramu, P., L. A. Lobo, M. Kukkonen, E. Bjur, M. Suomalainen, H. Raukola, M. Miettinen, I. Julkunen, O. Holst, M. Rhen, T. K. Korhonen, and K. Lähteenmäki. 2008. Activation of pro-matrix metalloproteinase-9 and degradation of gelatin by the surface protease PgtE of Salmonella enterica serovar Typhimurium. Int. J. Med. Microbiol. 298263-278. [DOI] [PubMed] [Google Scholar]
- 41.Rawlings, N. D., F. R. Morton, C. Y. Kok, J. Kong, and A. J. Barrett. 2008. MEROPS: the peptidase database. Nucleic Acids Res. 36D320-D325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16276-277. [DOI] [PubMed] [Google Scholar]
- 43.Sebbane, F., C. O. Jarrett, D. Gardner, D. Long, and B. J. Hinnebusch. 2006. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. USA 1035526-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sebbane, F., D. Gardner, D. Long, B. B. Gowen, and B. J. Hinnebusch. 2005. Kinetics of disease progression and host response in a rat model of bubonic plague. Am. J. Pathol. 1661427-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sebbane, F., N. Lemaitre, D. E. Sturdevant, R. Rebeil, K. Virtaneva, S. F. Porcella, and B. J. Hinnebusch. 2006. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. USA 10311766-11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skurnik, M., A. Peippo, and E. Ervela. 2000. Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. Mol. Microbiol. 37316-330. [DOI] [PubMed] [Google Scholar]
- 47.Sodeinde, O. A., and J. D. Goguen. 1989. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonella typhimurium. Infect. Immun. 571517-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sodeinde, O. A., and J. D. Goguen. 1988. Genetic analysis of the 9.5-kilobase virulence plasmid of Yersinia pestis. Infect. Immun. 562743-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sodeinde, O. A., Y. V. Subrahmanyam, K. Stark, T. Quan, Y. Bao, and J. D. Goguen. 1992. A surface protease and the invasive character of plague. Science 2581004-1007. [DOI] [PubMed] [Google Scholar]
- 50.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, Y. Han, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Wang, J. Yu, H. Yang, J. Wang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11179-197. [DOI] [PubMed] [Google Scholar]
- 51.Summaria, L., B. Hsieh, W. R. Groskopf, and K. C. Robbins. 1967. The isolation and characterization of the S-carboxymethyl β (light) chain derivative of human plasmin. The localization of the active site on the β (light) chain. J. Biol. Chem. 2425046-5052. [PubMed] [Google Scholar]
- 52.Suomalainen, M., J. Haiko, P. Ramu, L. Lobo, M. Kukkonen, B. Westerlund-Wikström, R. Virkola, K. Lähteenmäki, and T. K. Korhonen. 2007. Using every trick in the book: the Pla surface protease of Yersinia pestis. Adv. Exp. Med. Biol. 603268-278. [DOI] [PubMed] [Google Scholar]
- 53.Tracewell, C. A., and F. H. Arnold. 2009. Directed enzyme evolution: climbing fitness peaks one amino acid at a time. Curr. Opin. Chem. Biol. 133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect. Immun. 43895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandeputte-Rutten, L., R. A. Kramer, J. Kroon, N. Dekker, M. R. Egmond, and P. Gros. 2001. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. EMBO J. 205033-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varadarajan, N., J. Gam, M. J. Olsen, G. Georgiou, and B. L. Iverson. 2005. Engineering of protease variants exhibiting high catalytic activity and exquisite substrate selectivity. Proc. Natl. Acad. Sci. USA 1026855-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wren, B. W. 2003. The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 155-64. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, L., D. Seiffert, B. J. Fowler, G. R. Jenkins, T. C. Thinnes, D. J. Loskutoff, R. J. Parmer, and L. A. Miles. 2002. Plasminogen has a broad extrahepatic distribution. Thromb. Haemost. 87493-501. [PubMed] [Google Scholar]
- 59.Zhou, D., Y. Han, Y. Song, P. Huang, and R. Yang. 2004. Comparative and evolutionary genomics of Yersinia pestis. Microbes Infect. 61226-1234. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, D., Z. Tong, Y. Song, Y. Han, D. Pei, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, Z. Du, J. Wang, Z. Guo, J. Wang, P. Huang, and R. Yang. 2004. Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, microtus. J. Bacteriol. 1865147-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, D., Y. Han, Y. Song, Z. Tong, J. Wang, Z. Guo, D. Pei, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, Z. Du, J. Bao, X. Zhang, J. Yu, J. Wang, P. Huang, and R. Yang. 2004. DNA microarray analysis of genome dynamics in Yersinia pestis: insights into bacterial genome microevolution and niche adaptation. J. Bacteriol. 1865138-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]