Abstract
Studies of the Staphylococcus aureus LytSR two-component regulatory system have led to the identification of the cid and lrg operons, which affect murein hydrolase activity, stationary-phase survival, antibiotic tolerance, and biofilm formation. The cid gene products enhance murein hydrolase activity and antibiotic tolerance whereas the lrg gene products inhibit these processes in a manner believed to be analogous to bacteriophage-encoded holins and antiholins, respectively. Importantly, these operons have been shown to play significant roles in biofilm development by controlling the release of genomic DNA, which then becomes an important structural component of the biofilm matrix. To determine the role of LytSR in biofilm development, a lytS knockout mutant was generated from a clinical S. aureus isolate (UAMS-1) and the effects on gene expression and biofilm formation were examined. As observed in laboratory isolates, LytSR was found to be required for lrgAB expression. Furthermore, the lytS mutant formed a more adherent biofilm than the wild-type and complemented strains. Consistent with previous findings, the increased adherence of the mutant was attributed to the increased prevalence of matrix-associated eDNA. Transcription profiling studies indicated that the lrgAB operon is the primary target of LytSR-mediated regulation but that this regulatory system also impacts expression of a wide variety of genes involved in basic metabolism. Overall, the results of these studies demonstrate that the LytSR two-component regulatory system plays an important role in S. aureus biofilm development, likely as a result of its direct influence on lrgAB expression.
The Staphylococcus aureus lytSR operon was shown more than a decade ago to encode a novel two-component regulatory system that affects murein hydrolase activity and autolysis (4). Disruption of lytSR caused altered murein hydrolase activity produced by the cells and spontaneous cell lysis. Upon stimulation, the LytS sensor component presumably interacts with its cognate response regulator, LytR, which then activates the transcription of genes under its control. One known target of this system is the lrgAB operon, which along with the related cidABC operon has been shown to be a regulator in the control of cell death and lysis (11, 23). The cidA gene encodes a putative holin protein that is an effector of murein hydrolase activity and cell lysis (23), while lrgA encodes a putative antiholin that is an inhibitor of these processes (11).
Recent studies indicate that a biological function of the cid and lrg operons is to control cell death and lysis during biofilm development (2, 22, 24), providing a source of extracellular genomic DNA (eDNA) for use as a biofilm matrix molecule. Indeed, eDNA has been shown to be an essential matrix molecule produced by many bacterial species during biofilm development (1, 22, 29, 32). The cidA gene product is a positive effector of cell lysis and DNA release in the biofilm (24), while the lrg operon is an inhibitor of cell lysis and inhibits the release of DNA in the biofilm (18a). Given the critical role of this system in the control of cell death and lysis, it is not surprising that the transcription of these genes is subject to complex regulatory control. Recent studies have demonstrated the presence of two overlapping regulatory networks involved in the regulation of the cidABC and lrgAB operons, one in response to carbohydrate metabolism and the other responding to changes in membrane potential (Δψ) (21, 35). The former signaling pathway is mediated by the product of the cidR gene, a LysR-type transcriptional regulator. During logarithmic growth in the presence of excess glucose and oxygen, S. aureus secretes high levels of acetic acid, which then induces cidABC and lrgAB transcription in a cidR-dependent manner (35). In contrast, the LytSR two-component system senses decreases in Δψ (21) and presumably initiates the transfer of a phosphoryl group to its cognate response regulator, LytR. Activated LytR is then hypothesized to induce lrgAB promoter activity.
To examine the potential role of the LytSR two-component regulatory system in controlling cell death and lysis during biofilm development, we disrupted the lytSR operon in a clinical S. aureus strain, UAMS-1, and examined the effect of this mutation on biofilm formation and on cid and lrg transcription. Consistent with previous results (21), this study demonstrates the role of the LytSR two-component regulatory system in Δψ-induced lrgAB transcription and reveals its role in the induction of lrgAB by glucose. Furthermore, the LytSR regulatory system was found to be necessary for normal biofilm development.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All S. aureus strains were grown in either tryptic soy broth (TSB) (Difco Laboratories, Detroit, MI) or filter-sterilized NZY broth (3% [wt/vol] N-Z Amine A [Sigma Chemical Co., St. Louis, MO], 1% [wt/vol] yeast extract [Fisher Scientific, Fair Lawn, NJ], pH 7.5), supplemented as necessary with 1.5% (wt/vol) granulated agar (Difco). Escherichia coli DH5α was grown in Luria-Bertani medium (Fisher Scientific) supplemented as necessary with 1.5% (wt/vol) granulated agar (Difco). Unless otherwise stated, liquid cultures were grown in Erlenmeyer flasks at 37°C with shaking at 250 rpm in a volume not greater than 10% of the flask volume. All antibiotics were purchased from either Sigma Chemical Co. or Fisher Scientific and were used at the following concentrations: ampicillin, 50 μg/ml; erythromycin (Em), 2.0 μg/ml; chloramphenicol (Cm), 5.0 μg/ml; and tetracycline (Tc), 5.0 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Highly transformable strain; restriction deficient | 16 |
| UAMS-1 | Clinical osteomyelitis isolate | 10 |
| KB999 | UAMS-1 lytS::Em; Emr | This study |
| KB1050 | UAMS-1 cidA::Em; Emr | 25 |
| E. coli | 13 | |
| DH5α | Host strain for construction of recombinant plasmid | |
| Plasmids | ||
| pCR2.1 | E. coli subcloning vector; Ampr | Invitrogen |
| pDG647 | Source of Emr cassette; Emr Ampr | 12 |
| pCL52.2 | Temperature-sensitive shuttle vector; Tcr Spr | 27 |
| pCN51 | Shuttle vector carrying Pcad promoter; Emr Ampr | 6 |
| pCN50 | Source of Cmr cassette; Cmr Ampr | 6 |
| pBK123 | Shuttle vector, pCN51ΔEm::CAT; Cmr | This study |
Abbreviations: Emr, erythromycin resistance; Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Spr, spectinomycin resistance; ORF, open reading frame.
Allele replacement of the lytS gene in S. aureus UAMS-1.
A lytS mutation in S. aureus UAMS-1 was generated, following the allele replacement strategy described previously (21), using the temperature-sensitive shuttle plasmid pCL52.2 (27). Briefly, a 542-bp DNA fragment originating from the 5′ end of lytS was amplified using the PCR with the upstream primer, 5′lytSR-F-EcoRI, and the downstream primer, 5′lytSR-R-BamHI, incorporating EcoRI and BamHI restriction endonuclease recognition sites near the DNA ends (Table 2). The PCR products were digested with EcoRI and BamHI and then ligated into the EcoRI and BamHI sites of the plasmid pDG647 (12) upstream of the Em cassette. Next, a 558-bp DNA fragment spanning a region 3′ to lytS was PCR amplified with the upstream primer, 3′lytSR-F-ClaI, and the downstream primer, 3′lytSR-R-PstI, incorporating ClaI and PstI restriction sites (Table 2). These DNA fragments were digested with ClaI and PstI and then ligated into the ClaI and PstI sites of pDG647 downstream of the Em cassette already containing the 5′ lytS fragment. The 1.3-kb Em cassette, flanked by lytS sequences, was excised from this plasmid by digestion with EcoRI and PstI and then ligated into pCL52.2 (27) to generate the final knockout construct, pTP200. This plasmid was then transformed into S. aureus strain RN4220 by electroporation (28), spread onto tryptic soy agar (TSA) plates containing Em, and then incubated at 37°C overnight. The plasmid was then reisolated and transformed into UAMS-1. To obtain the lytS mutant, the UAMS-1(pTP200) strain was grown at the nonpermissive temperature (45°C) on TSA plates containing Em to select for cells in which the plasmid had integrated into the chromosome via homologous recombination. To promote a second recombination event, a single colony was inoculated into antibiotic-free TSB and grown at 30°C for 5 days, with 1:1,000 dilutions into fresh antibiotic-free TSB each day. After the fifth day, dilutions of the culture were spread on TSA plates containing Em to yield isolated colonies. PCR and Southern blot analyses were performed with the Emr and Tcs colonies to verify that the lytS gene had been deleted in UAMS-1 (data not shown). The confirmed mutant strain was designated KB999.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′ → 3′) | Nucleotide positiona |
|---|---|---|
| 5′ lytSR-F-EcoRI | CCCGAATTCTGCAACGGGACAATTGTTAG | 299003-299022 |
| 5′ lytSR-R-BamHI | CCCGGATCCCAACGTGC TTTCCATGTACG | 299530-299549 |
| 3′ lytSR-F-ClaI | TCCACATTTTTTCTTCAAATCGATTAACACGATTTCAGC | 300588-300626 |
| 3′ lytSR-R-PstI | CCAAAAAGTCTGCAGGCTCGATGTCGATTCAAATTGTAATCG | 301102-301143 |
| lytSR-F-BamHI | CCCGGATCCAAACATAATTATAATTCACTGAGGTGCTATCGTGCTATCGC | 299394-299434 |
| lytSR-R-EcoRI | CCCGAATTCCGACCATTGCCTCCTACGTTTG | 302020-302041 |
| lrgA-F | CCCGTTAAATCAAACGTAGGAGG | 302013-302032 |
| lrgA-R | CCCGGTTAATCATGAGCTTGTGC | 302461-302480 |
| cidA-F | CCCCGTATTTAGAAAGGGATGGCGCC | 2706686-2706708 |
| cidA-R | CCCCAAGGCTTGCACGTAATCATTC | 2706274-2706295 |
| LytS-F | TCCAGCAGTTTTTCGCTATGTATAAAAGTC | 299212-299241 |
| LytS-R | TTCCAATGTTTCTTTTACATTTTCTGCCTC | 301272-301301 |
| LytR-F | CCCGGATCCGACAAGAGGAG GAATAAATATG | 301163-301184 |
| LytR-R | CCCGAATTCCGACCATTGCCTCCTACGTTTG | 302020-302041 |
Based on the MRSA-252 genome sequence.
Complementation of the lytS mutation in KB999 was achieved by PCR amplifying a DNA fragment encompassing the UAMS-1 lytSR operon using the primers lytSR-F-BamHI and lytSR-R-EcoRI (Table 2). The resulting PCR products were ligated into the BamHI and EcoRI sites of the gram-positive expression vector, pBK123, which was generated by replacing the Em resistance cassette in the plasmid, pCN51 (6), with the Cm resistance cassette from pCN50 (6). This placed the expression of lytSR under the control of the cadmium-inducible promoter derived from the cadC gene (8). This recombinant plasmid was designated pBK5.
Northern blot analysis.
Overnight cultures of S. aureus strains were used to inoculate NZY broth to an optical density at 600 nm (OD600) of 0.1. Cells were harvested 2, 4, and 6 h after inoculation, corresponding to the early exponential, late exponential, and early stationary phases, respectively. The cells were pelleted by centrifugation and resuspended in lysis buffer, and total RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) and the Fastprep FP120 instrument (Bio 101, Vista, CA) and analyzed by Northern blotting as described previously (21). Digoxigenin (DIG)-labeled lrgA, cidA, lytS, and lytR DNA probes were synthesized using a PCR-based DIG probe synthesis kit (Roche) using the primer pairs listed in Table 2 and UAMS-1 genomic DNA as the template.
Microarray analysis.
To identify the genes affected by the lytS mutation, RNA samples were isolated in triplicate from the parental S. aureus strain UAMS-1 and the lytS mutant (KB999) grown in NZY broth in the presence of 35 mM glucose and subjected to transcription profiling as described previously (34). The genes identified in the microarray analysis were categorized according to the classification used in the KEGG pathway database (http://www.kegg.com).
Growth and analysis of biofilm.
To grow static biofilm, isolated colonies of S. aureus strains were inoculated into 3.0 ml of TSB-NaCl-Glc (3% NaCl, 0.5% glucose) and incubated overnight at 37°C and 250 rpm. These overnight cultures were diluted to an OD600 of 0.05 in fresh TSB-NaCl-Glc, and 200 μl of each culture was inoculated into 96-well Costar 3596 plates (Corning Life Sciences, Acton, MA) and incubated statically for 24 h at 37°C. Prior to the inoculation, the wells were precoated overnight with 20% human plasma (Sigma Chemical Co., St. Louis, MO) in bicarbonate buffer at 4°C. Biofilm quantification was performed as described previously (3, 7, 24) by staining with crystal violet and observing the absorbance at 655 nm of each well with a model 680 microtiter plate reader (Bio-Rad, Hercules, CA). For biofilms grown in flow-cell chambers, overnight cultures of S. aureus strains UAMS-1, KB999, and KB999(pBK5) were diluted into fresh TSB to an OD600 of 0.002. Using a sterile syringe with a 22-gauge needle, 3.0 ml of diluted culture was inoculated into the injection port of a medium-filled BST FC 270 flow-cell apparatus containing a polycarbonate coupon (Biosurface Technologies, Bozeman, MT) and allowed to incubate for 2 h to provide ample time for the bacteria to attach to the surface of the coupon. After this incubation period, culture medium (0.15% TSB and 0.12% glucose) was pumped into the flow-cell chamber using a Rainin RP-1 peristaltic pump (Rainin Instrument LLC, Woburn, MA) with a flow rate of 0.35 ml/min, and the biofilm was allowed to grow for 72 h. Macroscopic images of the biofilms were captured using a Canon EOS 350D digital camera.
For confocal laser scanning microscopy (CLSM), the biofilm was stained with Syto-9 and Toto-3 (Invitrogen) and then imaged using a Zeiss 510 Meta CLSM with an Achroplan 10× (0.45 numerical aperture) water dipping objective. The Syto-9 fluorophore was exited with an argon laser at 488 nm, and the emission band-pass filter used was 515 ± 15 nm. Excitation of Toto-3 was achieved using a HeNe 633-nm laser, and emissions were collected using a 680- ± 30-nm filter. z-stacks were collected at 1.0-μm intervals, and the images were compiled to generate three-dimensional renderings. All confocal parameters were set using wild-type biofilm and were used as standard settings for comparison to the biofilms produced by the mutant and complementation strains. Regions of interest within the biofilms were selected from similar areas within each flow-cell chamber, and each confocal experiment was repeated a minimum of four times. CLSM z-stack processing was performed using both the Zeiss ZEN LE software package (Carl Zeiss, Jena, Germany) and Volocity software (Improvision, Lexington, MA). Measurements of the biofilms produced were performed using the COMSTAT software package (14), calculating the biomass, maximum thickness, and colony volume at the substratum.
Purification and quantification of eDNA.
eDNA associated with the static biofilms grown in this study was isolated and quantified as described previously (24). To account for potential differences in biofilm biomass between samples, identically inoculated Costar plates were used for calculating the relative OD600 of each biofilm compared to the OD600 of the untreated UAMS-1 biofilm. For this purpose, the biofilm from each well was dislodged from the substratum and the OD600 of the cell suspension was determined.
RESULTS
Generation of lytS mutation in UAMS-1.
Previous studies revealed that a lytS mutation in the laboratory S. aureus isolate, 8325-4, had a dramatic effect on murein hydrolase activity and cell lysis (4). Since cell lysis is important during biofilm development, we generated a lytS mutation in the S. aureus clinical isolate and well-characterized biofilm-producing strain, UAMS-1, and examined the ability of this strain (designated KB999) to form a biofilm. Previous analyses of the lytS and lytR genes in 8325-4 revealed that they are cotranscribed under the control of the lytS promoter (4). Thus, to determine the effect of the lytS mutation on lytS and lytR expression, RNA samples were isolated from UAMS-1, KB999 (lytS mutant), and KB999(pBK5) (lytS mutant containing the lytSR complementation plasmid) and a Northern blot analysis was performed. Similar to the case with 8325-4, the lytS mutation in KB999 eliminated expression of both the lytS and lytR genes, indicating that these genes form a dicistronic operon and that expression of these genes could be restored to wild-type levels in mutant cells containing the lytSR-expressing plasmid (see Fig. S1 in the supplemental material). Interestingly, several of the phenotypes associated with the original lytS mutation in the 8325-4 strain were not observed in KB999 (unpublished results), including the increased spontaneous and Triton X-100-induced lysis as was previously observed for strain 8325-4 during planktonic growth (4).
LytSR-mediated control of transcription.
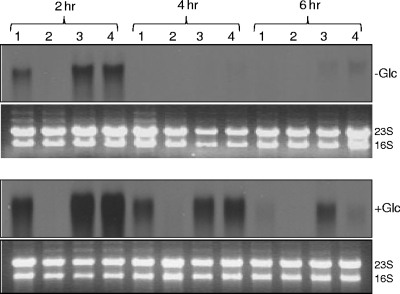
Despite the observation that the KB999 strain did not exhibit an altered cell lysis phenotype during planktonic growth, the lytS mutation still had a dramatic effect on lrgAB transcription. As shown in Fig. 1, the 1.2-kb lrgAB transcripts were undetectable in KB999 and were completely restored in the lytS mutant strain containing the lytSR complementation plasmid. Furthermore, the effect of the lytS mutation was observed at multiple time points in the growth cycle and in the presence and absence of 35 mM glucose (Fig. 1), indicating that both constitutive and glucose-inducible expression of lrgAB (25) is abolished by disruption of the lytS gene. We also performed Northern blot analyses to determine the effect of the lytS mutation on cidABC transcription, which is known to be induced by growth in the presence of 35 mM glucose in a cidR-dependent manner (34). These experiments, however, demonstrated normal cidABC expression, indicating that the LytSR two-component system is not involved in the control of this operon under these conditions (unpublished data).
FIG. 1.
The effect of lytS mutation on lrgAB expression. Total cellular RNAs from S. aureus UAMS-1 (lane 1), KB999 (lane 2), KB999(pBK5) (lane 3), or UAMS-1(pBK5) (lane 4) cells grown in either NZY broth (-Glc) or NZY broth with 35 mM glucose (+Glc) were isolated at 2, 4, and 6 h postinoculation. Ten micrograms of each RNA sample was separated in a 1% (wt/vol) agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to an lrgA-specific probe. The ethidium bromide-stained gels of the RNA used in these experiments are also shown.
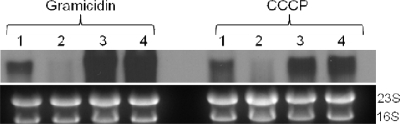
It has also been demonstrated previously that lrgAB transcription in the laboratory isolate RN6390 is induced by agents that dissipate the membrane potential (Δψ) associated with the cytoplasmic membrane (21). To examine the role of LytSR in sensing Δψ changes in UAMS-1, S. aureus cultures were grown to late exponential phase (4 h) and treated with either 25 μg/ml gramicidin or 10 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP), and RNA was collected and examined using Northern blot analyses. As demonstrated in Fig. 2, lrgAB transcription was induced by both of these agents in the wild-type strain but not in the lytS mutant. Importantly, Δψ-mediated induction of lrgAB expression in the lytS mutant was restored in cells containing the lytSR-expressing plasmid (Fig. 2, lane 3). These results indicate that Δψ-inducible lrgAB transcription in the UAMS-1 strain is dependent on the LytSR two-component regulatory system, as previously observed for the RN6390 strain (21).
FIG. 2.
Δψ induction of lrgAB expression is lytSR dependent. S. aureus UAMS-1 (lane 1), KB999 (lane 2), KB999(pBK5) (lane 3), and UAMS-1(pBK5) (lane 4) were incubated for 4 h and then treated with either gramicidin or CCCP. The cultures were then incubated for an additional 15 min and harvested by centrifugation, and total cellular RNA was isolated. Ten micrograms of each RNA sample was separated in a 1% (wt/vol) agarose-formaldehyde gel, transferred to a nylon membrane, and analyzed by Northern blotting using a DIG-labeled lrgA-specific probe as described in the legend for Fig. 1.
Effect of lytS mutation on biofilm formation.
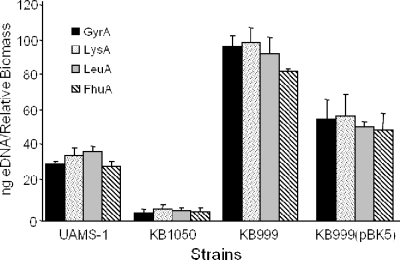
Studies from our laboratory have demonstrated the importance of S. aureus cidA in the control of cell lysis and DNA release during biofilm development (24). A cidA mutant produced lower levels of eDNA than the parental strain, resulting in an unstable biofilm, demonstrating the role of DNA as an important component of the S. aureus biofilm matrix. More recent studies have also shown that an lrgAB mutant produced increased levels of eDNA in the biofilm matrix, resulting in a more adherent biofilm (18a). As a positive effector of lrgAB transcription, we hypothesized, the lytS mutant would also exhibit a biofilm phenotype, possibly similar to that of the lrgAB mutant. As shown in Fig. 3, the lytS mutant was found to form a thicker, more adherent biofilm than the parental strain, UAMS-1. Furthermore, the KB999 strain harboring the lytSR expressing plasmid produced a biofilm whose adherence was similar to that of UAMS-1 (Fig. 3). These results are in contrast to those generated using the cidA mutant (KB1050), which produces a biofilm that is more easily disrupted (see reference 24) (Fig. 3). To determine whether the differences in biofilm adherence correspond with changes in the amount of eDNA in the matrices of these biofilms, we also quantified the levels of eDNA present in each of the biofilms examined. The total amount of eDNA present in 24-h static unwashed biofilm of S. aureus UAMS-1, KB1050, KB999, and KB999(pBK5) was quantified by real-time PCR as described previously using four different primer pairs specific for four randomly selected chromosomal genes (24). As shown in Fig. 4, the lytS mutant exhibited elevated eDNA levels associated with the biofilm cells compared to those of the UAMS-1 and complemented strains.
FIG. 3.
Static biofilm assay. Biofilms of either S. aureus UAMS-1, KB1050, KB999, or KB999(pBK5) were grown statically for 24 h in a plasma-coated 96-well microtiter plate and processed as described in Materials and Methods. The picture of the processed dry plate (left image) was taken using a digital camera and is representative of three independent experiments. The absorbance of the dried plate was read at 655 nm and plotted. These data are the averages for three independent experiments, each performed in triplicate, and the error bars correspond to the standard errors of means.
FIG. 4.
eDNA quantification in static biofilm. Biofilms of either S. aureus UAMS-1, KB1050, KB999, or KB999(pBK5) were grown, and total eDNA from 24-h unwashed biofilm was isolated as described in Materials and Methods. The total eDNA present in each biofilm was quantified by real-time PCR, using primer pairs specific for gyrA (gyrase A), leuA (2-isopropylmalate synthase), lysA (diaminopimelate decarboxylase A), and fhu (ferrichrome transport ATP-binding protein A). The values are expressed as nanograms of eDNA per relative biofilm biomass, as described in Materials and Methods. These data are the averages for three independent experiments, each performed in triplicate, and the error bars correspond to the standard errors of the means.
Effect of lytS mutation on biofilm maturation.
To determine the effect of the lytS mutation on biofilm maturation, we grew the UAMS-1, KB999, and KB999(pBK5) strains under flow-cell conditions. As shown in macro images of the biofilm formed (Fig. 5A), very little difference in overall structure was observed between these strains. Thus, we stained the biofilms with Syto 9 (a viable cell stain) and Toto-3 (stains dead cells and eDNA) and performed CLSM to examine the biofilm structures in more detail. Although no differences in staining patterns were observed, this analysis revealed that the lytS mutant was slightly thicker than the parental and complemented strains (Fig. 5B). COMSTAT analyses supported this observation, demonstrating that the lytS mutant biofilm had an average thickness of 50.1 μm, compared to average thicknesses of 37.0 μm for the parental strain and 42.4 μm for the complemented strain (Fig. 5D). Similar trends were also observed for total biomass, since the mutant had an average of 32.7 μm3/μm2 while the parental and complemented strains averaged 20.8 and 25.7 μm3/μm2, respectively (Fig. 5D). More strikingly, visualization of the interface between the biofilms and the substratum (Fig. 5C, bottom-up images) indicated that the lytS mutant exhibited a more intimate association with the substratum than the parental and complemented strains. Again, this was supported by COMSTAT analysis of multiple images (Fig. 5D), which indicated that the mutant had an average colony volume at the substratum of 1,220.5 nm3, compared to 531.1 nm3 and 308.4 nm3 for the parental and complemented strains, respectively. The results of these experiments are consistent with those of the static assays (see Fig. 3) and suggest that the signal transduction events mediated by the LytSR two-component regulatory system are important for early events involved in adherence.
FIG. 5.
Effect of lytSR on biofilm maturation. S. aureus UAMS-1 (parent), KB999 (lytS mutant), and KB999(pBK5) (complemented strain) biofilms were grown in flow-cell chambers and analyzed after 3 days of growth. (A) Representative biofilm macro images were taken using a Canon EOS 350D digital camera. (B) The samples were stained with Syto-9 (green) and Toto-3 (red) to indicate live and dead cell populations, respectively, and visualized by CLSM equipped with a Plan-Apochromat 10× objective. The images presented are top-down views of each biofilm and are accompanied by orthogonal views (above). (C) Representative z-stack images from each biofilm were compiled, and the biofilm adjacent to the substratum is visualized as a bottom-up view. (D) The biofilm images were analyzed using COMSTAT software, and total biomass (black bars; units = μm3/μm2), average thickness (white bars; units = μm), and average colony volume at the substratum (gray bars; unit = 10 nm3) for each strain are presented. The error bars correspond to the standard errors of the means.
Identification of additional LytSR-regulated genes.
To identify additional genes regulated by the LytSR two-component regulatory system and potentially important for biofilm development, RNA samples were isolated from logarithmically growing UAMS-1 and KB999 grown in NZY broth in the presence of 35 mM glucose and subjected to microarray analysis. Unlike the cidR mutation, which affected expression of only the cidABC, lrgAB, and alsSD operons (34), the lytS mutation affected expression of many genes. The lytS mutation had an overwhelmingly negative impact on gene expression by virtue of the fact that 460 genes were expressed at levels that were at least twofold lower in the lytS mutant than in UAMS-1 (see Table S1 in the supplemental material), versus only seven genes that were expressed at levels at least twofold higher in the mutant (see Table S2 in the supplemental material). Analysis of the genes which displayed threefold or higher expression in UAMS-1 than that for the lytS mutant (Table 3) revealed that most of the genes downregulated by the lytS mutation were involved in carbohydrate, energy, or nucleotide metabolism. Other downregulated genes included those involved in replication, transcription, and translation. Only one locus (SACOL2131), encoding a putative Dps family protein, was more than threefold upregulated in KB999 compared to UAMS-1, exhibiting 4.2-fold higher levels of these transcripts in the lytS mutant than in UAMS-1. Strikingly, expression of the lrgAB operon was by far the most dramatically impacted by the lytS mutation, with the lrgA- and lrgB-specific transcripts expressed at levels that were 11.3- and 7.4-fold lower in the lytS mutant than in UAMS-1.
TABLE 3.
Summary of genes downregulated by lytS mutationa
| Gene category (nb) | ORF no. | Common name | Description | Fold downregulation |
|---|---|---|---|---|
| Metabolism (249) | ||||
| Carbohydrate metabolism (16) | SACOL1741 | icd | Isocitrate dehydrogenase | 3.4 |
| SACOL1449 | sucA | 2-Oxoglutarate dehydrogenase, E1 component | 3.1 | |
| SACOL1448 | sucB | Dihydrolipoamide acetyltransferase | 3.0 | |
| SACOL1262 | sucC | Succinyl-CoA synthase, beta subunit | 3.5 | |
| SACOL1782 | fhs | Formate-tetrahydrofolate ligase | 3.7 | |
| SACOL2198 | alsD | Alpha-acetolactate decarboxylase | 3.0 | |
| SACOL2199 | alsS | Acetolactate synthase, catabolic | 3.4 | |
| SACOL2553 | cidC | Pyruvate oxidase | 3.1 | |
| SACOL2146 | PTS system, mannitol-specific IIBC components | 3.3 | ||
| SACOL2149 | mtlD | Mannitol-1-phosphate 5-dehydrogenase | 3.0 | |
| SACOL0175 | PTS system, IIABC components | 3.5 | ||
| SACOL0543 | glmU | UDP-N-acetylglucosamine pyrophosphorylase | 3.3 | |
| Energy metabolism (12) | SACOL1124 | ctaA | Cytochrome oxidase assembly protein | 3.3 |
| SACOL2097 | atpA | ATP synthase F1, alpha subunit | 3.4 | |
| SACOL2101 | atpB | ATP synthase F0, A subunit | 3.0 | |
| SACOL2094 | atpC | ATP synthase F1, epsilon subunit | 3.5 | |
| SACOL2095 | atpD | ATP synthase F1, beta subunit | 3.6 | |
| SACOL2100 | atpE | ATP synthase F0, C subunit | 3.5 | |
| SACOL2099 | atpF | ATP synthase F0, B subunit | 3.9 | |
| SACOL2096 | atpG | ATP synthase F1, gamma subunit | 3.5 | |
| SACOL2098 | atpH | ATP synthase F1, delta subunit | 3.2 | |
| SACOL0944 | NADH dehydrogenase, putative | 3.0 | ||
| SACOL2102 | Putative ATP synthase protein I | 4.0 | ||
| Nucleotide metabolism (19) | SACOL1078 | purL | Phosphoribosylformylglycinamidine synthase II | 4.1 |
| SACOL1080 | purM | Phosphoribosylaminoimidazole synthetase | 4.7 | |
| SACOL1081 | purN | Phosphoribosylglycinamide formyltransferase | 4.4 | |
| SACOL1077 | purQ | Phosphoribosylformylglycinamidine synthase I | 4.4 | |
| SACOL1076 | purS | Phosphoribosylformylglycinamidine synthase | 4.2 | |
| SACOL1075 | purC | Phosphoribosylaminoimidazole-succinocarboxamide synthase | 4.4 | |
| SACOL1083 | purD | Phosphoribosylamine-glycine ligase | 5.8 | |
| SACOL1073 | purE | Phosphoribosylaminoimidazole carboxylase, catalytic subunit | 5.5 | |
| SACOL1079 | purF | Amidophosphoribosyltransferase | 4.5 | |
| SACOL2130 | deoD2 | Purine nucleoside phosphorylase | 3.1 | |
| SA0925 | purH | Bifunctional purine biosynthesis protein (N315) | 4.7 | |
| SACOL2130 | deoD2 | Purine nucleoside phosphorylase | 3.1 | |
| Amino acid metabolism (11) | SACOL2047 | leuB | 3-Isopropylmalate dehydrogenase | 3.4 |
| SACOL0671 | Hydrolase, alpha/beta hydrolase fold family | 4.3 | ||
| SACOL2280 | ureA | Urease, gamma subunit | 3.2 | |
| SACOL2281 | ureB | Urease, beta subunit | 3.3 | |
| SACOL2282 | ureC | Urease, alpha subunit | 3.3 | |
| SACOL2327 | hutG | Formiminoglutamase | 3.1 | |
| Metabolism of cofactors and vitamins (7) | SACOL2265 | mobB | Molybdopterin-guanine dinucleotide biosynthesis protein B | 3.3 |
| SACOL2266 | moeA | Molybdopterin biosynthesis MoeA protein, putative | 3.1 | |
| Lipid metabolism (4) | SACOL1243 | plsX | Fatty acid/phospholipid synthesis protein PlsX | 3.0 |
| SACOL0637 | mvaD | Mevalonate diphosphate decarboxylase | 3.4 | |
| SACOL0636 | mvk | Mevalonate kinase | 4.1 | |
| SACOL0638 | Phosphomevalonate kinase | 3.3 | ||
| Genetic information processing (46) | ||||
| Replication and repair (1) | SACOL0438 | ssb2 | Single-stranded DNA-binding protein | 3.4 |
| Transcription (5) | SACOL0589 | rpoC | DNA-directed RNA polymerase, beta subunit | 3.1 |
| SACOL0588 | rpoB | DNA-directed RNA polymerase, beta subunit | 3.2 | |
| SACOL2113 | rho | Transcription termination factor Rho | 3.0 | |
| SACOL2147 | Transcriptional antiterminator, BglG family/DNA-binding protein | 3.6 | ||
| SACOL2517 | Transcriptional regulator, MerR family | 3.0 | ||
| Translation (36) | SACOL2236 | rplB | Ribosomal protein L2 | 3.5 |
| SACOL2238 | rplD | Ribosomal protein L4 | 3.4 | |
| SACOL2227 | rplE | Ribosomal protein L5 | 3.8 | |
| SACOL2224 | rplF | Ribosomal protein L6 | 3.1 | |
| SACOL0585 | rplJ | Ribosomal protein L10 | 3.2 | |
| SACOL2207 | rplM | Ribosomal protein L13 | 3.0 | |
| SACOL2229 | rplN | Ribosomal protein L14 | 3.5 | |
| SACOL2220 | rplO | Ribosomal protein L15 | 3.4 | |
| SACOL2232 | rplP | Ribosomal protein L16 | 3.5 | |
| SACOL2223 | rplR | Ribosomal protein L18 | 3.5 | |
| SACOL1725 | rplT | Ribosomal protein L20 | 3.3 | |
| SACOL1702 | rplU | Ribosomal protein L21 | 3.4 | |
| SACOL2234 | rplV | Ribosomal protein L22 | 4.2 | |
| SACOL2237 | rplW | Ribosomal protein L23 | 3.3 | |
| SACOL2228 | rplX | Ribosomal protein L24 | 3.6 | |
| SACOL2231 | rpmC | Ribosomal protein L29 | 4.1 | |
| SACOL2221 | rpmD | Ribosomal protein L30p/L7e | 3.1 | |
| SACOL1726 | rpmI | Ribosomal protein L35 | 4.4 | |
| SACOL1274 | rpsB | Ribosomal protein S2 | 3.6 | |
| SACOL2233 | rpsC | Ribosomal protein S3 | 4.2 | |
| SACOL2222 | rpsE | Ribosomal protein S5 | 3.6 | |
| SACOL0592 | rpsG | Ribosomal protein S7 | 3.9 | |
| SACOL2240 | rpsJ | Ribosomal protein S10 | 3.1 | |
| SACOL2214 | rpsK | Ribosomal protein S11 | 3.0 | |
| SACOL0591 | rpsL | Ribosomal protein S12 | 3.7 | |
| SACOL2226 | rpsN | Ribosomal protein S14 | 3.8 | |
| SACOL1292 | rpsO | Ribosomal protein S15 | 3.1 | |
| SACOL2230 | rpsQ | Ribosomal protein S17 | 3.3 | |
| SACOL2235 | rpsS | Ribosomal protein S19 | 3.9 | |
| SACOL1642 | rpsT | Ribosomal protein S20 | 3.0 | |
| SA0352 | rpsF | 30S ribosomal protein S6 (N315) | 3.1 | |
| SACOL2225 | rpsH | Ribosomal protein S8 | 3.7 | |
| SACOL0590 | 30S ribosomal protein L7 | 3.6 | ||
| SACOL0593 | fusA | Translation elongation factor G | 3.7 | |
| SACOL1727 | infC | Translation initiation factor IF-3 | 4.5 | |
| SACOL0958 | General stress protein 13 | 3.4 | ||
| Folding, sorting, and degradation (4) | SACOL0205 | pflA | Pyruvate formate-lyase-activating enzyme | 3.3 |
| SACOL2283 | ureE | Urease accessory protein UreE | 3.6 | |
| SACOL2284 | ureF | Urease accessory protein UreF | 3.3 | |
| SACOL2563 | ATP-dependent Clp protease, putative | 3.2 | ||
| Membrane transport (5) | SACOL0247 | lrgA | Holin-like protein LrgA | 11.3 |
| SACOL0248 | lrgB | Holin-like protein LrgB | 7.4 | |
| SACOL2554 | cidB | Putative membrane protein | 3.1 | |
| SACOL0175 | PTS system, IIABC components | 3.5 | ||
| SACOL2146 | PTS system, mannitol-specific IIBC components | 3.3 | ||
| Others (10) | ||||
| Conserved hypothetical proteins (5) | SACOL0218 | Conserved hypothetical protein | 3.0 | |
| SACOL1136 | Conserved hypothetical protein | 3.1 | ||
| SACOL1802 | Conserved hypothetical protein | 3.5 | ||
| SACOL2518 | Conserved hypothetical protein | 3.0 | ||
| SA1472 | Conserved hypothetical protein (N315) | 3.5 | ||
| Others not well characterized (5) | SACOL2534 | frp | NAD(P)H-flavin oxidoreductase | 3.3 |
| SACOL0220 | Flavohemoprotein, putative | 3.2 | ||
| SACOL0778 | Sulfatase family protein; similar to anion binding protein S. aureus Mu50 | 3.2 | ||
| SACOL1305 | HD/HDIG/KH domain protein; unclassified protein | 3.1 | ||
| SACOL2072 | ATP-dependent RNA helicase, DEAD box family | 3.0 | ||
| Intergenic region (10) |
The ORF number prefix SACOL indicates the S. aureus COL genome, and SA indicates the S. aureus N315 genome. CoA, coenzyme A; PTS, phosphotransferase system.
n, number of genes involved in that pathway, as suggested by the KEGG pathway.
DISCUSSION
The results of this study provide additional insight into the S. aureus LytSR two-component regulatory system and reveal the impact of this system on biofilm development. Initial studies of this system in the laboratory isolate 8325-4 demonstrated its role in the control of murein hydrolase activity and autolysis during planktonic growth (4) and ultimately led to the identification of the Cid/Lrg regulatory system involved in bacterial programmed cell death (2, 22). In contrast to 8325-4, however, the lytS mutant derivative of UAMS-1 generated in this study did not show a lysis defect under planktonic growth conditions. Indeed, spontaneous lysis of the mutant or that induced by Triton X-100 was similar to that of UAMS-1 (unpublished results). In addition, murein hydrolase activity produced by UAMS-1 and that of its lytS mutant derivative were comparable. These similarities were observed despite the fact that the UAMS-1 lytS mutant exhibited defective regulatory control of lrgAB transcription, as had been previously demonstrated with laboratory isolates (5, 21). Why the two strains exhibit different phenotypes during planktonic growth remains unknown; however, it has been speculated that high-passage isolates, such as 8325-4, accumulate defects in the regulation of cell death and lysis due to selection for mutations that enhance stationary-phase survival during repeated rounds of planktonic growth (26). Indeed, one such mutation may involve the rsbU gene, which spontaneously arose in the 8325-4 background (17) and was subsequently found to be important in the control of cid and lrg expression (26). Thus, the differences observed between these two strains may be a result of rsbU or other undefined regulatory mutations.
Notably, the strain-dependent effects of the lytS mutation are similar to the effects of the lrgAB mutation in laboratory and clinical isolates. Previous studies demonstrated that an lrgAB mutation in the laboratory isolate and 8325-4 derivative, RN6390, also resulted in defects in murein hydrolase activity and lysis during planktonic growth (11). However, recent studies of an lrgAB mutant derivative of UAMS-1 revealed that it did not produce a murein hydrolase or lysis phenotype during planktonic growth (unpublished results). In both cases, the lytS and lrgAB mutations produced a pronounced phenotype during biofilm growth. As demonstrated in this study, the lytS mutation resulted in a thicker, more adherent biofilm in the static assay (Fig. 3) and a thicker biofilm that exhibited more-intimate contact with the substratum in the flow-cell assay (Fig. 5). Analysis of the matrix produced in the static assay revealed that it contained increased levels of eDNA compared to those of the parental and complemented strains (Fig. 4). Consistent with LytSR being a positive effector of lrgAB transcription, the lrgAB mutant exhibited a phenotype similar to that of the lytS mutant under flow-cell conditions (18a). On the other hand, the cidA mutation caused a lysis defect during both planktonic and sessile growth. Overall, these results indicate that the UAMS-1 lytSR and lrgAB operons have biofilm-specific functions.
Recent studies have suggested the presence of two overlapping regulatory networks that control cidABC and lrgAB expression, one involving induction of expression as a consequence of carbohydrate metabolism (25) and the other being dependent on changes in Δψ (21). The CidR regulator is required for induction of cidABC and lrgAB expression in response to acetic acid accumulation in the culture supernatant resulting from the metabolism of excess glucose (35). In contrast, the LytSR two-component regulatory system is required for lrgAB expression under a variety of conditions, including the presence of glucose, gramicidin, and CCCP, as well as the constitutive expression of lrgAB during early exponential phase (see Fig. 1 and 2). In most two-component systems, external stimulation triggers a phosphorylation cascade that results in the phosphorylation and activation of the response regulator component of the system, which then induces transcription of genes under its control. In some cases, however, response regulators can be directly phosphorylated by small-molecule phospho donors, including acetyl phosphate and carbamoyl phosphate (18, 20, 33). The results of the current study leave open the possibility that two mechanisms of LytSR-mediated control of lrgAB transcription exist, one involving sensing of membrane potential and the other mediated by direct phosphorylation by acetyl phosphate as a result of the accumulation of acetate. However, how these signals are coordinated to induce lrgAB expression during biofilm formation remains unknown and is an important focus of our laboratory.
Another aspect of this study was to determine the impact of lytSR on global gene expression using transcription profiling studies. As indicated by comparison of the transcriptomes of the UAMS-1 and lytS mutant strains, the lytS mutation resulted in the downregulation of 460 genes and the upregulation of seven genes. Most of the genes downregulated in the lytS mutant included those involved in metabolism, particularly carbohydrate, energy, and nucleotide metabolism (Table 3; see also Tables S1 and S2 in the supplemental material). These results are in contrast to the cidR mutation, which affected transcription of only the cidABC, lrgAB, and alsSD operons (34), although the effect of the lytS mutation on transcription of most genes was much more subtle (between two- and fourfold changes in transcript levels) than that of the cidR mutation (ranging from 13.0- to 43.9-fold changes in gene expression). The most dramatic effect of the lytS mutation on transcription was on the lrgAB operon itself, causing an 11.3-fold decrease in transcript levels. Other genes downregulated by the lytS mutation included those involved in genetic information processing, such as replication, transcription, and translation (Table 3; see also Tables S1 and S2 in the supplemental material). Although one might predict that these changes would affect the growth rate of the lytS mutant, the growth rate of this strain was comparable to that of the wild type (data not shown). The many genes downregulated in the lytS mutant include the alsSD operon, which was previously shown to be regulated in a CidR-dependent manner (31, 34). In addition, levels of the cidB and cidC genes were shown to be moderately decreased in the lytS mutant compared to those in the parental strain, indicating that cidBC expression is affected by the lytS mutation and is independent of cidA. Indeed, previous studies demonstrate that the cid operon is transcribed by two transcripts, one spanning the cidA, cidB, and cidC genes (25) and the other spanning cidB and cidC only (26). Thus, only expression of the cidBC transcript appears to be affected by the lytS mutation. Based on the results of these microarray experiments, we propose that the LytSR two-component regulatory system directly affects lrgAB transcription and that the impact of the lytS mutation on other genes is indirect.
In conclusion, the results generated by this study demonstrate that the lytSR regulatory locus plays a significant role in the control of cell lysis during biofilm development, likely by controlling lrgAB expression and the regulation of eDNA release. Given the complex nature of biofilms, including the varied metabolic microenvironments that exist within these structures, continued studies of the LytSR regulatory system will be important for achieving a better understanding of the roles that carbohydrate metabolism and membrane potential play in the development of a maturing biofilm. One possibility is that the transition into anaerobic metabolism during biofilm maturation (9, 15, 19, 30) could play a key signaling role in the LytSR-mediated control of lrgAB expression. This and other possibilities are the focus of our current investigations.
Supplementary Material
Acknowledgments
This project was funded by NIH grant no. R01AI038901 and DOD grant no. DAAD 19-03-1-0191 to K.W.B.
Footnotes
Published ahead of print on 5 June 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allesen-Holm, M., K. B. Barken, L. Yang, M. Klausen, J. S. Webb, S. Kjelleberg, S. Molin, M. Givskov, and T. Tolker-Nielsen. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 591114-1128. [DOI] [PubMed] [Google Scholar]
- 2.Bayles, K. W. 2007. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 51721-726. [DOI] [PubMed] [Google Scholar]
- 3.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 714206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 1785810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charpentier, E., A. I. Anton, P. Barry, B. Alfonso, Y. Fang, and R. P. Novick. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 706076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbisier, P., G. Ji, G. Nuyts, M. Mergeay, and S. Silver. 1993. luxAB gene fusions with the arsenic and cadmium resistance operons of Staphylococcus aureus plasmid pI258. FEMS Microbiol. Lett. 110231-238. [DOI] [PubMed] [Google Scholar]
- 9.Gera, I. 2008. The bacterial biofilm and the possibilities of chemical plaque control. Literature review. Fogorv. Sz. 10191-99. (In Hungarian.) [PubMed] [Google Scholar]
- 10.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 633373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 1821794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167335-336. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 14.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 1462395-2407. [DOI] [PubMed] [Google Scholar]
- 15.Kong, K. F., C. Vuong, and M. Otto. 2006. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296133-139. [DOI] [PubMed] [Google Scholar]
- 16.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 17.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 1804814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Mann, E. E., K. C. Rice, B. R. Boles, J. L. Endres, D. Ranjit, L. Chandramohan, L. H. Tsang, M. S. Smeltzer, A. R. Horswill, and K. W. Bayles. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS one 4e5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquis, R. E. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 15198-207. [DOI] [PubMed] [Google Scholar]
- 20.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 1752793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patton, T. G., S. J. Yang, and K. W. Bayles. 2006. The role of proton motive force in expression of the Staphylococcus aureus cid and lrg operons. Mol. Microbiol. 591395-1404. [DOI] [PubMed] [Google Scholar]
- 22.Rice, K. C., and K. W. Bayles. 2008. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 7285-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice, K. C., B. A. Firek, J. B. Nelson, S. J. Yang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 1852635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice, K. C., E. E. Mann, J. L. Endres, E. C. Weiss, J. E. Cassat, M. S. Smeltzer, and K. W. Bayles. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 1048113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice, K. C., J. B. Nelson, T. G. Patton, S. J. Yang, and K. W. Bayles. 2005. Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J. Bacteriol. 187813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rice, K. C., T. Patton, S. J. Yang, A. Dumoulin, M. Bischoff, and K. W. Bayles. 2004. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 1863029-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sau, S., J. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 1791614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73133-138. [DOI] [PubMed] [Google Scholar]
- 29.Spoering, A. L., and M. S. Gilmore. 2006. Quorum sensing and DNA release in bacterial biofilms. Curr. Opin. Microbiol. 9133-137. [DOI] [PubMed] [Google Scholar]
- 30.Stewart, P. S., and M. J. Franklin. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6199-210. [DOI] [PubMed] [Google Scholar]
- 31.Tsang, L. H., J. E. Cassat, L. N. Shaw, K. E. Beenken, and M. S. Smeltzer. 2008. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE 3e3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 2951487. [DOI] [PubMed] [Google Scholar]
- 33.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 6912-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, S. J., P. M. Dunman, S. J. Projan, and K. W. Bayles. 2006. Characterization of the Staphylococcus aureus CidR regulon: elucidation of a novel role for acetoin metabolism in cell death and lysis. Mol. Microbiol. 60458-468. [DOI] [PubMed] [Google Scholar]
- 35.Yang, S. J., K. C. Rice, R. J. Brown, T. G. Patton, L. E. Liou, Y. H. Park, and K. W. Bayles. 2005. A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J. Bacteriol. 1875893-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.