Abstract
A transposon insertion mutant has been identified in a Desulfovibrio desulfuricans G20 mutant library that does not grow in the presence of 2 mM U(VI) in lactate-sulfate medium. This mutant has also been shown to be deficient in the ability to grow with 100 μM Cr(VI) and 20 mM As(V). Experiments with washed cells showed that this mutant had lost the ability to reduce U(VI) or Cr(VI), providing an explanation for the lower tolerance. A gene encoding a cyclic AMP (cAMP) receptor protein (CRP) was identified as the site of the transposon insertion. The remainder of the mre operon (metal reduction) contains genes encoding a thioredoxin, thioredoxin reductase, and an additional oxidoreductase whose substrate has not been predicted. Expression studies showed that in the mutant, the entire operon is downregulated, suggesting that the CRP may be involved in regulating expression of the whole operon. Exposure of the cells to U(VI) resulted in upregulation of the entire operon. CdCl2, a specific inhibitor of thioredoxin activity, inhibits U(VI) reduction by washed cells and inhibits growth of cells in culture when U(VI) is present, confirming a role for thioredoxin in U(VI) reduction. The entire mre operon was cloned into Escherichia coli JM109 and the transformant developed increased U(VI) resistance and the ability to reduce U(VI) to U(IV). The oxidoreductase protein (MreG) from this operon was expressed and purified from E. coli. In the presence of thioredoxin, thioredoxin reductase, and NADPH, this protein was shown to reduce both U(VI) and Cr(VI), providing a mechanism for the cytoplasmic reduction of these metals.
Previous studies have shown that soluble U(VI) can be reduced to the less-soluble U(IV) by pure cultures of bacteria (19, 20, 25). This process can be useful for in situ reduction, which results in uranium precipitation and therefore decreased mobility in groundwater (8, 33). Desulfovibrio desulfuricans G20 and Desfulovibrio vulgaris, neither of which can use U(VI) as a respiratory electron acceptor, have been shown to directly reduce U(VI) (19, 24), and the mechanism for U(VI) reduction has been addressed. A purified hydrogenase and periplasmic cytochrome c3 from cell extracts of D. vulgaris will reduce U(VI) to U(IV) with hydrogen as the electron donor (19), suggesting that cytochrome c3 of D. vulgaris may be directly involved in U(VI) reduction. When a cytochrome c3 mutant of D. desulfuricans G20 was generated, it would not reduce U(VI) with H2 as the electron donor (25); however, growth and U(VI) reduction occurred with lactate as the electron donor, although at lower rates than the wild type. Cytochrome c3 was also found to be bound to insoluble U(IV), providing further evidence that this protein may be involved in U(VI) reduction (24). Electron microscopic images showed that reduced U(IV) was not only present in the periplasm but also in the cytoplasm (28), indicating that the periplasmic cytochrome c3 may be only partially responsible for the in vivo U(VI) reduction process, with an additional pathway in the cytoplasm.
In order to identify this additional mechanism, transposon insertion mutants were generated. This mutant library has also been used to identify genes involved in sediment fitness (10, 21) and syntrophic growth (16). In this study, the mutants were screened for loss of U(VI) resistance. A mutant was identified that was sensitive to U(VI) and would not grow with 2 mM U(VI) or reduce it in suspensions of washed cells. This was the only mutant identified that would not reduce U(VI) in both tests. The disrupted operon (named mre, for metal reduction) was characterized, and it is shown here that the mechanism for the U(VI) reduction process involves at least three genes, including thioredoxin, thioredoxin reductase, and an additional metal oxidoreductase. Some or all of these components are likely also responsible for Cr(VI) and As(V) reduction by this organism.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
D. desulfuricans G20 (34) was obtained from the lab of Judy Wall, University of Missouri, Columbia. A mutant library of strain G20 containing 5,760 mutants was previously generated (10) and grown in lactate-sulfate medium (21) prepared under N2 and containing sodium lactate (62 mM), Na2SO4 (50 mM), MgSO4 (8 mM), NH4Cl (5 mM), HEPES (25 mM), K2HPO4 (2.2 mM), CaCl2 · 2H2O (0.6 mM), 0.1% yeast exact, and trace amounts of other minerals and vitamins. The pH was adjusted to 7.2 prior to autoclaving, and NaHCO3 (8 mM), cysteine HCl (0.025%) as reductant, and 1,050 mg/liter kanamycin to select for the insertion were added to the medium after autoclaving. To avoid metal precipitation, a modified lactate-sulfate medium was used for all the U(VI) and Cr(VI) incubations. This modified medium contained sodium lactate (62 mM), NaHCO3 (40 mM), Na2SO4 (20 mM), MgSO4 (4 mM), NH4Cl (5 mM), K2HPO4 (0.2 mM), CaCl2 · 2H2O (0.2 mM), and trace amounts of other minerals and vitamins at 25% of the level in the previously described medium. The liquid was flushed with N2 (80%)-CO2 (20%) before autoclaving, and cysteine HCl (0.025%) was added after autoclaving. All G20 cultures were incubated anaerobically, and all cultures were incubated at 37°C.
A mutant named Thio1 was obtained that did not grow in the presence of U(VI). This mutant was obtained by screening a previously generated (10) Tn10 mutant library (containing 5,760 mutants) for the ability to grow in the above medium with 2 mM uranyl acetate added. Individual frozen cultures were thawed and inoculated into deep-well microtiter plates, incubated, and then scored for growth. When Thio1 was grown with uranyl acetate, kanamycin was not added to the culture, as kanamycin forms a precipitate with U(VI) in lactate sulfate medium (this study).
Escherichia coli was grown aerobically in Luria-Bertani broth. For anaerobic growth, a medium was prepared which contained glycerol (20 mM), fumarate (20 mM), NaHCO3 (10 mM), and Casamino Acids (0.005%) (31). The medium was flushed under N2-CO2 and dispensed in serum tubes in 10-ml volumes. Kanamycin (100 μg/ml) was added to the medium after autoclaving to select for transformants.
Viable plate counts.
A viable count method was used to monitor growth in liquid cultures with U(VI) addition, as U(IV) precipitates interfere with optical density (OD) readings. The solid medium included agar (1.5%), 0.005% PdCl2 added after autoclaving as a substitute for cysteine HCl, and kanamycin (175 μg/ml) to select for maintenance of the transposon. Note that less kanamycin is needed for solid than liquid media. Solid media were poured into plates after addition of antibiotics. Plates were cooled, dried overnight in a laminar flow hood, and reduced for 12 to 24 h in an anaerobic glove box (Coy Laboratory Products, Inc., Grass Lake, MI). Subsequent manipulations and incubations were also done in the glove box. Samples (0.1 ml) of the cultures were taken every 3 hours during growth and suspended in 0.9 ml NaHCO3 buffer (30 mM) to dissolve any U(VI) precipitates that may have been binding cells together. The cell suspensions were serially diluted to 10−8, and 0.1 ml of each dilution was spread onto a lactate-sulfate agar plate. Plates were removed from the incubator after 2 to 3 days of incubation, and those with between 50 and 500 colonies were chosen for colony counts.
Washed-cell experiments.
Cells were grown in 1-liter volumes in lactate-sulfate medium. After 48 h growth, cells were collected by centrifugation and resuspended in 100 ml of buffer containing 30 mM NaHCO3 under N2/CO2 (4:1). Cells were washed twice and finally resuspended in 10 ml of buffer. The washed cell assay mixtures were incubated at 37°C and contained 9.9 ml of bicarbonate buffer under N2/CO2 with sodium lactate (62 mM), Na2SO4 (20 mM), cysteine HCl (0.025%), 0.1 ml of cells, and either uranyl acetate (1 mM) or K2CrO4 (0.5 mM).
Genome sequence and phylogenetic analyses.
The D. desulfuricans G20 genomic sequence was obtained from http://www.jgi.doe.gov. BLAST, alignment, and other bioinformatic analyses were carried out with CLUSTALW 1.82, NNPP promoter finder 2.2, through the NCBI website (http://www.ncbi.nlm.nih.gov), the BCM searchlauncher website (http://searchlauncher.bcm.tmc.edu), genomix (http://www.genomatix.de), and VIMSS computational genomics (http://www.vimss.org).
General molecular methods.
Plasmids were isolated with the Qiaprep miniprep kit (Qiagen Inc., Valencia, CA), and chromosomal DNA was isolated with the Easy DNA kit (Invitrogen Corp., Carlsbad, CA). PCRs were performed with the Taq DNA polymerase system (Invitrogen Corp.). When the template was larger than 3 kb, the Long Template PCR system (Roche, Mannheim, Germany) was used. T4 DNA ligase (Invitrogen Corporation, Carlsbad, CA) was used for ligations (incubated 1 h), in order to clone PCR products into pGEM4Z and pET15b (Table 1) (100 ng of each). Plasmid constructs were transformed into Escherichia coli strains JM109 and BL21 (Table 1) using chemically competent cells (5). DNA sequencing was carried out with the dideoxynucleotide chain termination method at the Oklahoma Medical Research Foundation (Oklahoma City, OK). The location of the transposon insertion was determined using arbitrary PCR (10).
TABLE 1.
Escherichia coli strains, plasmids, and primers used in this study
| Plasmid, strain, or primer | Genotype, description, or sequence |
|---|---|
| Plasmids | |
| pGem4Z (Promega) | 2.7-kb plasmid carries the lacZ α-peptide and multiple cloning region arrangement from pUC18 |
| pET15b (Novagen) | 5.7-kb plasmid carries an N-terminal His tag sequence followed by a thrombin site and three cloning sites |
| E. coli strains | |
| JM109 | F′ traD36 proA+B+lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV44 e14−gyrA96 recA1 relA1 endA1 thi hsdR17 |
| BL21 | F′ ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) |
| Primers | |
| Poperon | 5′-GCTCTAGAGCCCTTGTGCGGAATATAAA-3′/5′-TCCCCCCGGTACCCCGCAACACACTGTAA-3′ |
| Poxido | 5′-TCCCCCCGGACACCCAGTTCCGAAGTGAT-3′/5′-GCTCTAGATTTACACAGCATGCCACTCC-3′ |
| PthioGSP1 | 5′-ATCACTTCGGAACTGGGTGT-3′ |
| PthioGSP2 | 5′-TACCCCGCAACACACTGTAA-3′ |
| PthioGSP3 | 5′-CTGCTGGCACGGAGTTAGC-3′ |
| PdTanchor | 5′-GACCACGCGTATCGATGTCGACTTTTTTTTTTTTV-3′ |
| Panchor | 5′-GACCACGCGTATCGATGTCGAC-3′ |
| Plink12 | 5′-GCCGGATTCGAGATAAAACA-3′/5′-ATGACGGCAAGGAGCTGTAT-3′ |
| Plink23 | 5′-GCCCTTGTGCGGAATATAAA-3′/5′-TTTACACAGCATGCCACTCC-3′ |
| Plink34 | 5′-TTGCAGAACCGCACATAGTC-3′/5′-AAAAAGCGAAAGAGCTGCTG-3′ |
| Plink45 | 5′-ATCGCCATCGGCTTGATTC-3′/5′-GACAATGCGAGCACCTTGAA-3′ |
| Plink56 | 5′-GCCGGATTCGAGATAAAACA-3′/5′-ATGACGGCAAGGAGCTGTAT-3′ |
| Plink67 | 5′-GGGTGAGTAACGCGTGGATT-3′/5′-AGCAGAGGCCCCCTTTACC-3′ |
| Plink78 | 5′-GTCAAAGCTGGTTTCCGAACTG-3′/5′-GGCTTTCGGCAGGAAAAAA-3′ |
| PnhaCRT | 5′-TAAAGCCTGGTGCCTAATGG-3′/5′-TGAATCGGCCAACGCGCGGG-3′ |
| ParylRT | 5′-CTACGGCTACACTAGAAGAA-3′/5′-AAAAGGATCTTCACCTAGAT-3′ |
| PcAMPRT | 5′-CCTTTTCTACGGGGTCTGACG-3′/5′-TAAAAATGAAGTTTTAAATCGG-3′ |
| PthioRT | 5′-CCAGCCGGAAGGGCCGAG-3′/5′-CATCCAGTCTATTAATTGTTG-3′ |
| PthioRRT | 5′-CGTTAAGGGATTTTGGTCATG-3′/5′-GAGATTATCAAAAAGGATCTTC-3′ |
| PhypoRT | 5′-CCATTATTATCATGACATTAG-3′/5′-TTTCGTCTCGCGCGTTTCGGA-3′ |
| PoxidoRT | 5′-GTTATGGCAGCACTGCATAACG-3′/5′-TTCTCTTACTGTCATGCCATCC-3′ |
| PacrRT | 5′-TACCGCTGTTGAGATCCAG-3′/5′-TGTCTCATGAGCGGATACAT-3′ |
| PapsBRT | 5′-AGTGCTCATCATTGGAAAACG-3′/5′-GCCGGGAAGCTAGAGTAAGT-3′ |
Analysis of the mre operon.
In order to determine the transcriptional start of the mre operon, amplification of the 5′ end was performed with the 5′/3′ random amplification of cDNA ends (RACE) kit (Roche, Mannheim, Germany). Cells (10 ml) were grown in lactate-sulfate medium to an OD at 600 nm (OD600) of 0.2. Then, the cells were treated with 2 mM U(VI) to increase expression of the mre operon. Total RNA was isolated after 4 h of U(VI) treatment with the RNeasy minikit (Qiagen Inc., Valencia, CA) following the kit's manual. RNA (1 μg) was used as a template to synthesize the single-stranded cDNA with PthioGSP1 primer (Table 1) and reverse transcriptase. This generated a cDNA with the 3′ end at the 5′ end of the mRNA. The single-stranded cDNAs were cleaned with the High Pure PCR purification kit (included in the 5′/3′ kit), and a poly(A) tail was added at the 3′ end using terminal transferase. Then, two rounds of PCR were performed with the cDNA using Taq polymerase and PthioGSP2, PdTanchor (Table 1, first round) and PthioGSP3, Panchor (Table 1, second round). PCR products were purified with the High Pure PCR purification kit and sequenced with primers PthioGSP3 and Panchor (Table 1). The 3′ end of the cDNA was identified as it is adjacent to the adapter tail. This is equivalent to the 5′ end of the mRNA.
In order to determine whether all eight genes in the mre operon are transcribed together, seven sets of primers were designed to amplify the seven gaps of the eight genes. The RNA samples described above (1 μg) were used to synthesize a double-stranded cDNA library with SuperScript reverse transcriptase II and random primers (Roche, Germany). PCR was then performed using the seven sets of Plink primers (Plink12, Plink23, Plink34, Plink45, Plink56, Plink67, and Plink78 [Table 1]) and Taq polymerase, and the PCR products were visualized by gel electrophoresis.
The entire mre operon was cloned into E. coli as follows. The operon was amplified using Pfu polymerase (Takara, Japan) with the primer set Poperon (Table 1). This amplified DNA starting at 130 bases upstream of the ATG of the first open reading frame (ORF) in the operon to the end of the last ORF. The blunt end PCR product was then ligated into pGem4Z (Table 1) that had been cut with SmaI. The ligated construct pXLthio1 was then transformed into E. coli JM109.
Real-time PCR for quantification of mRNAs.
The cDNA library described above was used for real-time PCR, carried out with the Bio-Rad (CA) MyIQ cycler based on the instrument's manual. Primer sequences were designed with the Invitrogen oligo designer to generate 100-bp amplicons and amplification was as previously described (15). Relative quantification of mRNA expression was calculated using the PFAFFL method (15) and the following equation: ratio = (Etarget)ΔCt_target (control − treated)/(Ereference)ΔCt_reference (control − treated). The apsB gene was used as a reference.
U(VI) concentration determination.
Culture samples were diluted to U(VI) concentrations between 1 μM and 100 μM with nanopure water. A 1-ml sample at the appropriate U(VI) concentration was added to a quartz cuvette with 0.5 ml of Uraplex complexant (Chemchek Instruments, WA), and the U(VI) concentration was determined with a kinetic phosphorescence analyzer (KPA-10; Chemchek).
Purification of the oxidoreductase.
The oxidoreductase gene (mreG) was cloned by amplification of the gene using the primer set Poxido (Table 1). This amplified the complete ORF. The PCR product was ligated into pET15b as described above and the recombinant pXLox1 was transformed into E. coli BL21. The transformant was grown aerobically in 100 ml Magic medium (Invitrogen, CA) at 37°C and cells were collected after 24 h of incubation by centrifugation. All subsequent steps were carried out in an anaerobic glove box using the His•Bind purification kit (Novagen, Darmstadt, Germany). Reagents were flushed with N2 before use. Cells were disrupted by bead beating (Biospec Products Inc.) for 5 min. The purity of the protein was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the protein concentration was determined with the bicinchoninic acid protein assay (Pierce, PA).
U(VI) and Cr(VI) reduction by the purified oxidoreductase.
U(VI) and Cr(VI) reductase activities were assayed at room temperature under anaerobic conditions in an assay mixture containing U(VI) or Cr(VI) (1 mM) in 30 mM NaHCO3 buffer flushed with N2-CO2 (4:1) and dispensed into a sealed cuvette in a total volume of 2 ml. The purified oxidoreductase (100 ng), E. coli thioredoxin (100 ng; Sigma, CA), E. coli thioredoxin reductase (10 ng; Sigma, CA), and NADPH (1 mM; Sigma, CA) were added to the cuvette with a syringe. The NADPH concentration was measured at 340 nm for 5 minutes. After 1 h, the final U(VI) concentration was determined with the KPA, or the Cr(VI) concentration was measured colorimetrically (1).
RESULTS
Response of the mutant to other metals.
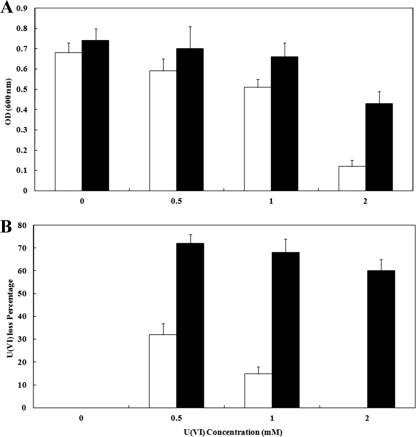
A transposon mutant library has been generated (10) and a mutant (Thio1) was identified that was deficient in the ability to grow with U(VI). The initial experiments showed that this mutant would not grow in the presence of 2 mM U(VI) and would not reduce U(VI) during growth or in washed cell experiments (Fig. 1). The ability of this mutant to tolerate other metals, including As(V) and Cr(VI), was then tested. Thio1 was completely inhibited by 20 mM As(V) and by 100 μM Cr(VI) in lactate sulfate medium (Fig. 2A). As well, washed cells would not reduce Cr(VI) (Fig. 2B).
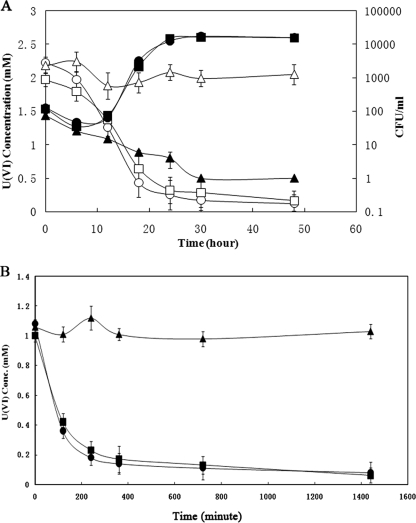
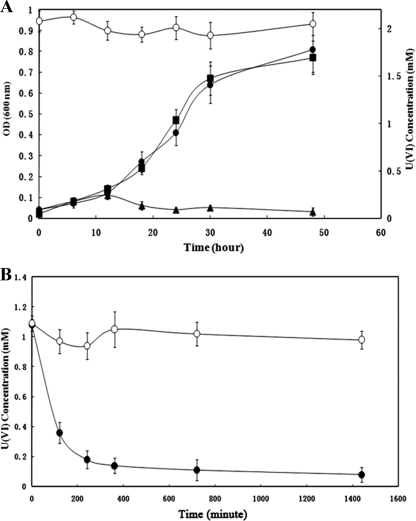
FIG. 1.
(A) Cell concentration (CFU; filled) and U(VI) concentration (open) during growth of the parental strain of G20 (circles), Thio1 mutant (triangles), and Thio2 mutant (squares). (B) U(VI) reduction by washed cells of the parental strain (•), Thio2 (▪), and Thiol (▴).
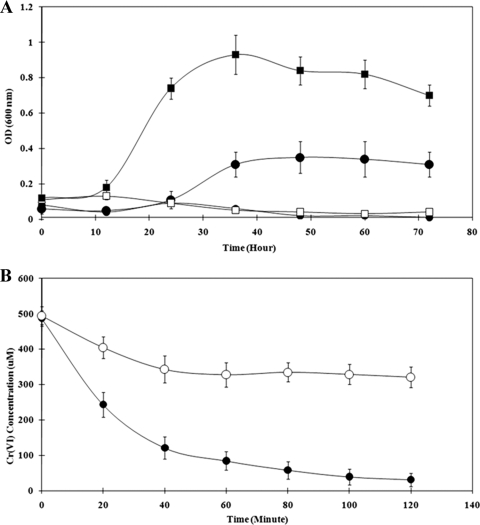
FIG. 2.
(A) Growth curve of G20 (filled) and Thio1 mutant (open) incubated with As(V) (squares) or Cr(VI) (circles). (B) Washed cell experiment showing Cr(VI) reduction over time by G20 (•) and the Thio1 mutant (○).
Analysis of the mre operon.
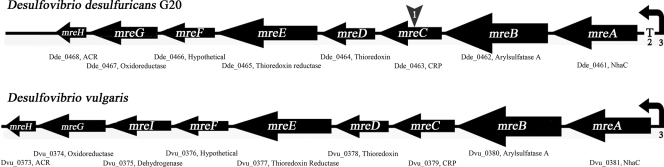
The mre operon disrupted in the Thio1 mutant contains eight genes. According to the NCBI annotation, they encode a Na+/H+ antiporter (mreA) most likely involved in pH homeostasis and sodium extrusion; an arylsulfatase (mreB) thought to catalyze the hydrolysis of sugar-sulfate (sulfatide) bonds and release inorganic sulfate; a cyclic AMP (cAMP) receptor protein (CRP; mreC) which is a transcription factor that binds to CRP binding sites in DNA, likely causing upregulation or downregulation of the downstream genes (4); thioredoxin (mreD); thioredoxin reductase (mreE); a hypothetical protein (mreF); an oxidoreductase (mreG) annotated as pyruvate ferredoxin/flavodoxin oxidoreductase family protein; and an acyl coenzyme A synthetase (mreH), which is homologous to the first enzyme in β-oxidation (Fig. 3).
FIG. 3.
Diagram showing the mre operon of G20 with a comparison to the orthologous operon of D. vulgaris. Gene annotation and reference numbers from NCBI are given below each gene. The alignment is from http://www.microbesonline.org. 1, transposon insertion; 2, base “T” at 470800 of the G20 genome (NCBI) is the transcriptional start; 3, promoters predicted with NNPP2.2.4.
The link PCR test showed that all eight genes in the putative operon are transcribed as a single mRNA (Fig. 3; see also Fig. S1 in the supplemental material). The transcriptional start is the base T at position 470800. Upstream of the start codon of the first gene in the operon (mreA) at bp −56, a CRP binding site was identified using Genomix. It is therefore likely that the CRP encoded by this operon may also regulate expression of the operon.
Desulfovibrio vulgaris reduces U(VI) in a manner similar to strain G20. It is the only bacterium with a sequenced genome that has a very similar operon. However, the D. vulgaris operon includes an additional gene (mreI) that encodes a putative dehydrogenase (Fig. 3).
Expression of genes in the mre operon.
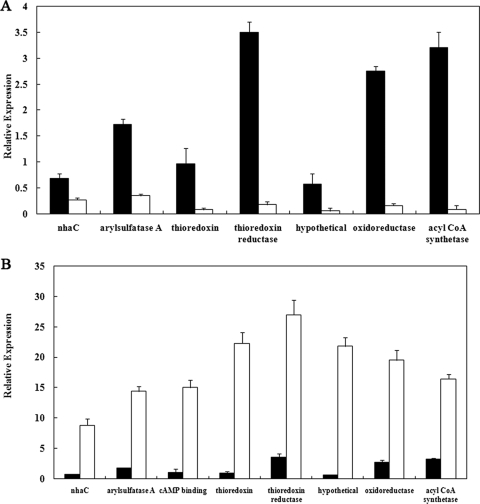
It was critical to determine whether the transposon had disrupted mreC or whether it inactivated the whole operon. It was hypothesized that mreC could be the potential activator of the mre operon and that inactivation of mreC would have an effect similar to inactivation of the entire operon. To test this hypothesis, expression of all genes in the mre operon except mreC were compared during growth in lactate-sulfate medium. Expression of genes downstream of mreC was dramatically decreased (90% to 95% decrease) in the mutant. However, expression of the two genes upstream of mreC was also decreased by 50% to 70% (Fig. 4A), indicating that the mreC may be involved in controlling the expression of the whole operon.
FIG. 4.
(A) Relative expression of genes in the thioredoxin operon of G20 (▪) and in the Thio1 mutant (□). Expression of the CRP gene (not shown) was given a relative value of 1. (B) The influence U(VI) in the growth medium on expression of genes in the thioredoxin operon. Relative expression is shown in the presence (□) or absence (▪) of U(VI). CRP expression in the absence of U(VI) was given a relative value of 1.
In order to further explore the relationship between the mre operon and U(VI) reduction, expression of each gene in the presence and absence of U(VI) was determined. All eight genes were shown to be upregulated 10- to 80-fold in the presence of U(VI) (Fig. 4B).
Role of thioredoxin.
Thioredoxin is believed to be a cytoplasmic electron donor for many biological reactions (11). Whether the thioredoxin and the thioredoxin reductase are involved in the U(VI) reduction process and serve as the source of electrons is an important question. Cadmium is a known inhibitor of thioredoxin and has been shown to bind at the Cys32 and the Asp26 residues of the E. coli thioredoxin (27). The addition of 20 μM CdCl2 had no effect on the growth of strain G20 in lactate-sulfate medium (Fig. 5A). However, when 2 mM U(VI) was added to cells incubated with CdCl2, neither growth nor U(VI) reduction occurred. CdCl2 was also shown to inhibit U(VI) reduction by washed cells (Fig. 5B), indicating that cadmium acts directly on the U(VI) reduction process. These results clearly show that thioredoxin is required for U(VI) reduction and suggest that thioredoxin may be the cytoplasmic electron donor for U(VI) reduction.
FIG. 5.
(A) Growth (OD600) curve for G20 with no addition (▪), with 20 μM CdCl2 (•), and with CdCl2 and 2 mM U(VI) (▴) added to the medium. The graph also shows the effect of CdCl2 on U(VI) reduction (○). (B) Plot showing the effect of the presence (○) or absence (•) of 20 μM CdCl2 on U(VI) reduction by washed cells of G20.
There are eight thioredoxin and thioredoxin reductase genes in the D. desulfuricans G20 genome (see Table S1 in the supplemental material). Aside from Thio1, another mutant (Thio2) is also available from a previous study and has been shown to be deficient in sediment fitness (21). Thio2 can grow and reduce U(VI) at rates similar to strain G20 (Fig. 1A), indicating that the thioredoxin reductase whose function has been destroyed in Thio2 is not involved in U(VI) reduction.
Cloning the mre operon into E. coli.
To determine whether genes in the mre operon could allow for the direct reduction of U(VI), the whole operon was cloned, generating pXLthio1, which was transformed into E. coli strain JM109. Transformants had a fourfold-higher growth yield than the plasmid-only control when grown in anaerobic medium with 2 mM U(VI) (Fig. 6). Under anaerobic conditions in the presence of 2 mM U(VI), 60% of U(VI) in the culture was reduced by the transformant with no apparent reduction by the untransformed strain. These results clearly point to a role for this operon during in vivo U(VI) reduction.
FIG. 6.
Effect of the recombinant thioredoxin operon on growth of E. coli in the presence of U(VI) (A) and U(VI) reduction by growing cultures sampled after 48 h (B). Results are shown for E. coli transformed with pXLthio1 (▪) or the untransformed strain (□).
Metal reduction by the purified oxidoreductase.
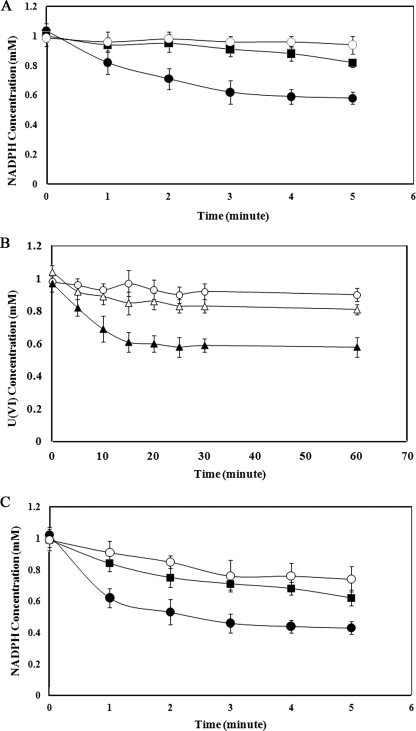
Since reduction of U(VI) appeared to be carried out by a product of the operon, mreG annotated to encode an oxidoreductase was cloned to determine whether it was involved in the process. The oxidoreductase was expressed and purified from E. coli transformed with pXLox1. A U(VI) reduction assay was carried out with the purified oxidoreductase, NADPH, thioredoxin, and thioredoxin reductase. In this system NADPH is oxidized (Fig. 7A), with electrons transferred to thioredoxin in a reaction involving the thioredoxin reductase. The U(VI) is then reduced in a reaction most likely involving the oxidoreductase (Fig. 7B). As several chromium reduction systems are capable of reducing U(VI), we tested the ability of the oxidoreductase to catalyze the reduction of Cr(VI) and showed that the system was also capable of Cr(VI) reduction (Fig. 7C).
FIG. 7.
Time courses of enzyme assays with the purified oxidoreductase. Assays were carried out independently to measure different components. (A) NADPH oxidation in an assay containing 1 mM U(VI), the purified oxidoreductase, thioredoxin reductase, thioredoxin, and NADPH (•). The final U(VI) concentration was 0.61 mM. Controls included only U(VI) in buffer (○) or U(VI) together with thioredoxin and thioredoxin reductase (▪). (B) U(VI) reduction in an assay containing 1 mM U(VI), the purified oxidoreductase, thioredoxin reductase, thioredoxin, and NADPH (▴), or in assays lacking the oxidoreductase (Δ) or thioredoxin (○). (C) NADPH oxidation in an assay with 1 mM Cr(VI), the purified oxidoreductase, thioredoxin reductase, thioredoxin, and NADPH (•) or in an assay mixture lacking thioredoxin and thioredoxin reductase (○) or Cr(VI) (▪).
DISCUSSION
A gene encoding a CRP within the mre operon has been disrupted by a transposon. CRPs are global transcriptional regulators that are broadly distributed in bacteria (4). When CRP binds cAMP, it may act as a repressor if it binds downstream of the RNA polymerase binding site. However, in most studied cases, the CRP-cAMP complex binds upstream of the RNA polymerase binding site and interacts with the α-subunit of the RNA polymerase (22), resulting in the activation of the gene or operon. In the D. desulfuricans G20 mre operon, a CRP binding site was detected at bp −56 upstream of the start codon of mreA. Both this information and the fact that reverse transcription-PCR data show induction of the operon in the presence of U(VI) indicate a likely role of CRP as an inducer. Although an induction of all of the genes in the operon was observed on exposure to U(VI), increases in expression were not uniform (Fig. 4B). This phenomenon has been previously observed (15, 18) and may be due to variability associated with the reverse transcription-PCR process.
CRP homologs have been shown to be involved in induction of genes used for various functions, including carbohydrate metabolism (7), modulation of virulence gene expression (26), biofilm formation (13), and others. The only other metal resistance gene that we are familiar with that is regulated by a CRP is the plasmid-encoded mercuric reductase (merA) (32), which is induced during catabolite repression (when cAMP levels go up). Several iron-related genes are also induced by the CRP-cAMP complex. These include the fur gene of E. coli, which is a repressor for various iron uptake systems (6). In addition, expression of a fecA homolog (ferric citrate uptake gene) in Stenotrophomonas maltophila is indirectly controlled by CRP, which can induce synthesis of a diffusible signal factor that activates fecA expression (12).
Strain G20 has been shown to grow with and reduce 20 mM arsenate using a modified ars reduction system (15). In bacteria, respiration-independent arsenate reduction is catalyzed by the gene product of arsC, an arsenate reductase, which either obtains electrons from thioredoxin or glutaredoxin, but never both (30). The system in D. desulfuricans G20 involves a monocistronic arsC gene and an arsRBCC operon (15). Two of the three arsC gene products have been shown to be most closely related to the arsC genes of other deltaproteobacteria and to those of strains of Bacillus (15). They both contain the three conserved cysteine residues typical of thioredoxin-dependent arsenate reductases. For these enzymes, electrons derived from NADPH are transferred to thioredoxin in a reaction catalyzed by thioredoxin reductase (17). Electrons can then be transferred to As(V) or other electron acceptors by the arsC gene product. The third arsC located in the ars operon is closely related to Wolinella succinogenes arsC gene, which has not been characterized. Since the CRP mutant cannot grow with As(V) (Fig. 2), this latter arsC gene most likely also uses thioredoxin as an electron donor.
Aside from their role in arsenic reduction, thioredoxins have only been shown to supply electrons for a small group of enzymes, including ribonucleotide reductase (23), phosphoadenosine phosphosulfate reductase, and methionine sulfoxide reductase (3). However, thioredoxins have been shown to be involved in the oxidative stress response in several organisms. In Rhodobacter spheroides and Rhodobacter capsulatus, expression of thioredoxin is regulated by the oxygen concentration of the medium (14) and survival of R. capsulatus in the presence of the oxidizers, H2O2, O2, and others was lower for a thioredoxin mutant. In Bacillus subtilis, a thioredoxin was shown to be induced by the presence of H2O2 as well as being induced during heat shock (29). This information indicates the importance of thioredoxins in dealing with oxidized compounds. As the thioredoxin studied here appears to provide electrons for reduction of several oxidized metals, one may conclude that the thioredoxin may protect cells against oxidation.
Previous work has suggested a mechanism for periplasmic reduction of U(VI) (24, 25). However, studies carried out in bicarbonate-buffered solutions have shown that the reduced U(IV) is not only in the periplasm but also present in the cytoplasm (28). The data presented here suggest an alternative mechanism for U(VI) reduction that is likely cytoplasmic. Experiments comparing U(VI) reduction of Thio1 to the parent suggest that for high concentrations of U(VI), the dominant mechanism for U(VI) reduction involves thioredoxin.
Cr(VI) and U(VI) are chemically quite different; however, several oxidoreductases have been shown to reduce both chemical compounds. These include ChrR from E. coli and Pseudomonas and NfsA from E. coli (2). These are proteins that obtain electrons from NADH and can carry out two-electron reduction of not only Cr(VI) and U(VI) but also quinones (9). We could not detect any significant homology between the Cr(VI)- and U(VI)-reducing oxidoreductase from G20 and the ChrR from Pseudomonas putida or E. coli.
In this work, a novel U(VI)/Cr(VI) reductase has been described in strain G20. We have proposed functions for the CRP, the oxidoreductase, thioredoxin, and thioredoxin reductase. There are still four other genes in the mre operon for which roles in this process are not clear. D. vulgaris has a very similar mre operon in its genome, containing similar genes, suggesting that this structure has been evolutionarily conserved. It is therefore possible that these four other genes may play a role in metal resistance in sulfate-reducing bacteria.
Supplementary Material
Acknowledgments
This research was supported a grant from the Environmental Remediation Science Program of the Office of Biological and Environmental Research of the U.S. Department of Energy, Office of Science.
Footnotes
Published ahead of print on 29 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ackerley, D. F., C. F. Gonzalez, M. Keyhan, R. Blake II, and A. Matin. 2004. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ. Microbiol. 6851-860. [DOI] [PubMed] [Google Scholar]
- 2.Barak, Y., D. F. Ackerley, C. J. Dodge, L. Banwari, C. Alex, A. J. Francis, and A. Matin. 2006. Analysis of novel soluble chromate and uranyl reductases and generation of an improved enzyme by directed evolution. Appl. Environ. Microbiol. 727074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boschi-Muller, S., S. Azza, and G. Branlant. 2001. E. coli methionine sulfoxide reductase with a truncated N terminus or C terminus, or both, retains the ability to reduce methionine sulfoxide. Protein Sci. 102272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Mol. Biol. Rev. 56100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. USA 692110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Lorenzo, V., M. Herrero, F. Giovannini, and J. B. Neilands. 1988. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur. J. Biochem. 173537-546. [DOI] [PubMed] [Google Scholar]
- 7.Emmer, M., B. deCrombrugghe, I. Pastan, and R. Perlman. 1970. Cyclic AMP receptor protein of E. coli: its role in the synthesis of inducible enzymes. Proc. Natl. Acad. Sci. USA 66480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganesh, R., K. G. Robinson, G. D. Reed, and G. S. Sayler. 1997. Reduction of hexavalent uranium from organic complexes by sulfate- and iron-reducing bacteria. Appl. Environ. Microbiol. 634385-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez, C. F., D. F. Ackerley, S. V. Lynch, and A. Matin. 2005. ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J. Biol. Chem. 28022590-22595. [DOI] [PubMed] [Google Scholar]
- 10.Groh, J. L., Q. Luo, J. D. Ballard, and L. R. Krumholz. 2005. Adaptation of microarray technology for signature tagged mutagenesis (STM) of Desulfovibrio desulfuricans G20 and Shewanella oneidensis MR-1 in anaerobic sediment survival experiments. Appl. Environ. Microbiol. 717064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54237-271. [DOI] [PubMed] [Google Scholar]
- 12.Huang, T.-P., and A. C. L. Wong. 2007. A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 735034-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalivoda, E. J., N. A. Stella, D. M. O'Dee, G. J. Nau, and R. Q. Shanks. 2008. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl. Environ. Microbiol. 743461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, K., E. HŠrtig, and G. Klug. 2003. Thioredoxin 2 is involved in oxidative stress defence and redox-dependent expression of photosynthesis genes in Rhodobacter capsulatus. Microbiology 149419-430. [DOI] [PubMed] [Google Scholar]
- 15.Li, X., and L. R. Krumholz. 2007. Regulation of arsenate resistance in Desulfovibrio desulfuricans G20 by an arsRBCC operon and an arsC gene. J. Bacteriol. 1893705-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, X., Q. Luo, N. Q. Wofford, K. L. Keller, M. M. McInerney, J. D. Wall, and L. R. Krumholz. 2009. A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J. Bacteriol. 1092675-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Y., Y. Hu, X. Zhang, H. Xu, E. Lescop, B. Xia, and C. Jin. 2007. Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis. J. Biol. Chem. 28211078-11083. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Maury, L., F. J. Florencio, and J. C. Reyes. 2003. Arsenic sensing and resistance system in the Cyanobacterium synechocystis sp. strain PCC 6803. J. Bacteriol. 1855363-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lovley, D. R., and E. J. Phillips. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovley, D. R., P. K. Widman, J. C. Woodward, and E. J. Phillips. 1993. Reduction of uranium by cytochrome c3 of Desulfovibrio vulgaris. Appl. Environ. Microbiol. 593572-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo, Q., J. L. Groh, J. D. Ballard, and L. R. Krumholz. 2007. Identification of genes that confer sediment fitness to Desulfovibrio desulfuricans G20. Appl. Environ. Microbiol. 736305-6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami, K., J. T. Owens, T. A. Belyaeva, C. F. Meares, S. J. Busby, and A. Ishihama. 1997. Positioning of two alpha subunit carboxy-terminal domains of RNA polymerase at promoters by two transcription factors. Proc. Natl. Acad. Sci. USA 9411274-11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr, M. D., and E. Vitols. 1966. Thioredoxin from Lactobacillus leichmannii and its role as hydrogen donor for ribonucleoside triphosphate reductase. Biochem. Biophys. Res. Commun. 25109-115. [DOI] [PubMed] [Google Scholar]
- 24.Payne, R. B., L. Casalot, T. Rivere, J. H. Terry, L. Larsen, B. J. Giles, and J. D. Wall. 2004. Interaction between uranium and the cytochrome c3 of Desulfovibrio desulfuricans strain G20. Arch. Microbiol. 181398-406. [DOI] [PubMed] [Google Scholar]
- 25.Payne, R. B., D. M. Gentry, B. J. Rapp-Giles, L. Casalot, and J. D. Wall. 2002. Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl. Environ. Microbiol. 683129-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rickman, L., C. Scott, D. M. Hunt, T. Hutchinson, M. C. MenŽndez, R. Whalan, J. Hinds, M. J. Colston, J. Green, and R. S. Buxton. 2005. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 561274-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rollin-Genetet, F., C. Berthomieu, A.-H. Davin, and E. Quemeneur. 2004. Escherichia coli thioredoxin inhibition by cadmium: two mutually exclusive binding sites involving Cys32 and Asp26. Eur. J. Biochem. 2711299-1309. [DOI] [PubMed] [Google Scholar]
- 28.Sani, R. K., B. M. Peyton, and A. Dohnalkova. 2006. Toxic effects of uranium on Desulfovibrio desulfuricans G20. Environ. Toxicol. Chem. 251231-1238. [DOI] [PubMed] [Google Scholar]
- 29.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Všlker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 1801867-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silver, S., and L. T. Phung. 2005. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl. Environ. Microbiol. 71599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spencer, M. E., and J. R. Guest. 1973. Isolation and properties of fumarate reductase mutants of Escherichia coli. J. Bacteriol. 114563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Summers, A. O., L. Knight-Olliff, and C. Slater. 1982. Effect of catabolite repression on the mer operon. J. Bacteriol. 149191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall, J. D., and L. R. Krumholz. 2006. Uranium reduction. Annu. Rev. Microbiol. 60149-166. [DOI] [PubMed] [Google Scholar]
- 34.Wall, J. D., T. Murnan, J. Argyle, R. S. English, and B. J. Rapp-Giles. 1996. Transposon mutagenesis in Desulfovibrio desulfuricans: development of a random mutagenesis tool from Tn7. Appl. Environ. Microbiol. 623762-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.