Abstract
DnaA is a replication initiator protein that is conserved among bacteria. It plays a central role in the initiation of DNA replication. In order to monitor its behavior in living Escherichia coli cells, a nonessential portion of the protein was replaced by a fluorescent protein. Such a strain grew normally, and flow cytometry data suggested that the chimeric protein has no substantial loss of the initiator activity. The initiator was distributed all over the nucleoid. Furthermore, a majority of the cells exhibited certain distinct foci that emitted bright fluorescence. These foci colocalized with the replication origin (oriC) region and were brightest during the period spanning the initiation event. In cells that had undergone the initiation, the foci were enriched in less intense ones. In addition, a significant portion of the oriC regions at this cell cycle stage had no colocalized DnaA-enhanced yellow fluorescent protein (EYFP) focus point. It was difficult to distinguish the initiator titration locus (datA) from the oriC region. However, involvement of datA in the initiation control was suggested from the observation that, in ΔdatA cells, DnaA-EYFP maximally colocalized with the oriC region earlier in the cell cycle than it did in wild-type cells and oriC concentration was increased.
Initiation of DNA replication is highly regulated to coordinate with cell proliferation. It begins with a series of events in which the replication machinery is assembled at the replication origin of the chromosomal DNA (15, 26, 28, 38). Central to this process are the initiator proteins that bind to the origin of replication and eventually lead to the unwinding of the origin and to helicase loading on the unwound region. Previous biochemical studies and recent structural studies of the bacterial initiator protein DnaA have proposed the molecular mechanism of the action of ATP-DnaA in forming a large oligomeric complex to remodel the unique origin, oriC, and trigger duplex melting (12, 26). However, it is still not clear how the timing of initiation is controlled so that it takes place at a fixed time in the cell cycle. It has been reported that a basal level of DnaA molecules is bound by high-affinity DnaA binding sites (DnaA boxes R1, R2, and R4) at oriC throughout the cell cycle (9, 37). It is also suggested that noncanonical ATP-DnaA binding sites within oriC are occupied at elevated levels of the initiator molecules prior to the initiation event (18, 25). Thus, regulation of the activity and availability of DnaA is an important factor for the initiation control.
At least three schemes are known to prevent untimely initiations in Escherichia coli. First, oriC is subject to sequestration, a process that prevents reinitiation, possibly by blocking ATP-DnaA from binding to newly replicated oriC (8, 24). E. coli oriC contains 11 GATC sites that are normally methylated on both strands by Dam methyltransferase. Immediately after passage of the replication fork, GATC sites are in a hemimethylated state, with the newly synthesized strands remaining unmethylated. SeqA binds specifically to such sites and, at oriC, protects these regions from reinitiation for about one-third of the cell cycle (6, 39). Second, in a process termed regulatory inactivation of DNA (RIDA), ATP-DnaA molecules are converted to an inactive ADP-bound form after initiation by the combined action of a β subunit of DNA polymerase III holoenzyme and Hda (16, 17). Newly synthesized DnaA molecules are able to bind ATP for the next initiation event, since its cellular concentration is much higher than that of ADP. ATP-DnaA is also regenerated from the inactive ADP-DnaA later in the cell cycle (21). Finally, the chromosomal segment datA serves to reduce the level of free DnaA protein by titrating a large number of DnaA molecules after replication of the site close to oriC (20).
Cytological studies would be very useful for developing our understanding of the regulation mechanisms associated with the initiation step. In the present study, we tagged E. coli DnaA with a fluorescent protein in order to monitor its behavior in live cells. Microscopic observation revealed that DnaA is distributed all over the nucleoid. Remarkably, the majority of cells bore distinct foci that emitted brighter fluorescence against a weak fluorescent background on the nucleoid. We analyzed the behavior of these foci during the cell cycle with respect to oriC and datA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Table 1 lists the primer oligonucleotides that we used for PCR. In all PCRs, the high-fidelity KOD-plus DNA polymerase (Toyobo) was used.
TABLE 1.
Oligonucleotide primers used for PCR
| Name | Sequence (5′-3′) |
|---|---|
| PME | GTAGTAGCGTTTAAACTCTTCGATC |
| BGL | GAAACAGAAGATCTCTTGCGCAG |
| YFP-NU | CCGGTGACGCAAACGATGGTGAGCAAGGGC |
| YFP-NL | GCCCTTGCTCACCATCGTTTGCGTCACCGG |
| YFP-CU | GACGAGCTGTACAAGCAGCCGCAACGTGCT |
| YFP-CL | AGCACGTTGCGGCTGCTTGTACAGCTCGTC |
| attTn7LR | CAGTTCTCTAGACTTCAGCCCTGGAATGTT |
| attTn7LL | CTGCGTAAGCCCGGGGCATTTTTCTTCCTG |
| attTn7RR | GAAAAATGCCCCGGGCTTACGCAGGGCATC |
| attTn7RL | CACGCGGAATTCCTGGGCCGTGGCGATCAG |
| lacOterLR | CAAATCTAGAGTGCTAACGAAACACCTGGA |
| lacOterLL | CATACCCGGGTCCTGCATATCTCTTGTGCG |
| lacOterRR | AGGACCCGGGTATGCCGACTAAACGCTTTG |
| lacOterRL | GCTGTCTAGACCATGACCTGGTATGGACAC |
| lacOdatLR | GCGATCTAGAATGCACATCAACGCCATGTT |
| lacOdatLL | CTTGCCCGGGCTACAATTTTCACATATTTC |
| lacOdatRR | GTAGCCCGGGCAAGGCGCTCGCGCCGCATC |
| lacOdatRL | AGCGTCTAGATAGCGTAAGCACGATGGTTC |
| tetR-mCherry5 | ATTGATCATATGCGGATTAGAAAAACAACTTAAATGTGAAAGTGGGTCTGGCGGCGGTGGTAGCGGCGGTGGCGGTAGCGGTGGCGGTGGCAGCATGGTGAGCAAGGGCGAG |
| tetR-mCherry6 | AGCTCCAGCCTACACTTACTTGTACAGCTC |
| tetR-mCherry7 | GAGCTGTACAAGTAAGTGTAGGCTGGAGCT |
| tetR-mCherry4 | AGTCTTTGTCTTCTGCGTCGGGTTGATCGTGGTTGCGGAAATTGTGTTGTCATATGAATATCCTCCTTA |
| tetR-mCherry8 | TCTACTGTTTCTCCATACCCGTTTTTTTGGGCTAGCAGGAGGAATTCACCATGTCTAGATTAGATAAAA |
| tetR-mCherry9 | GCATCAGGCATCGGCACACAGTGCCGGATGCACCGCGCGACGCATCCGACCATATGAATATCCTCCTTA |
| del13mersR | ATCTCTGTCGACGATCGCACTGCCCTGTGG |
| del13mersL1 | AAATAAGTCGACAGATCGTGCGATCTACTG |
To create MG1655dnaA-eyfp, two portions of the dnaA gene on a plasmid were amplified by PCR using primer sets BGL plus YFP-NL and YFP-CU plus PME. PCR amplification of eyfp was carried out using pEYFP (Clontech) as a template and YFP-NU and YFP-CL as primers. The primers YFP-NL and YFP-NU as well as the primers YFP-CU and YFP-CL are complementary. Therefore, PCR in the presence of primers BGL and PME and the three fragments BGL to YFP-NL, YFP-NU to YFP-CL, and YFP-CU to PME yielded a dnaA gene segment from the BglII to PmeI sites, in which sequences corresponding to residues 87 to 104 of DnaA were replaced by eyfp. This fragment was used to replace the BglII-PmeI fragment of pKH5002SBdnaA and was subsequently introduced into the chromosomal dnaA locus by two successive homologous recombinations in vivo as described elsewhere (33).
To introduce a 240-copy lacO array at ori1 (15 kb counterclockwise from oriC) (42), two portions of the chromosomal attTn7 locus were amplified by PCR using primer sets attTn7LR plus attTn7LL and attTn7RR plus attTn7RL. The termini of the two fragments, attTn7LR to attTn7LL and attTn7RR to attTn7RL, overlap by 24 bp and carry a SmaI site in the overlapping sequence introduced by primers attTn7LL and attTn7RR. PCR in the presence of these two fragments and the primers attTn7LR and attTn7RL yielded one fused fragment. The fused fragment was cleaved at both ends with XbaI and EcoRI and introduced into the XbaI-EcoRI region of pUC18. The lacO array-kan fragment obtained from pLAU43 (22) as an XbaI-EcoRI fragment was then inserted by blunt-end ligation into the SmaI site of the resulting plasmid. From this plasmid, a portion of the attTn7 sequence carrying the lacO array-kan fragment in the middle was cut out with XbaI and EcoRI and introduced into MG1655dnaA-eyfp by the method of Datsenko and Wanner (11).
The lacO array-kan fragment of pLAU43 was introduced similarly at dat1 (20 kb clockwise from datA) and at ter2 (42), respectively, of MG1655dnaA-eyfp, except that pKH5002SB (20) was used instead of pUC18 and the XbaI site was created at both ends of the fused fragments for introduction into the XbaI site of pKH5002SB. For introduction into dat1, primer sets lacOdatLR plus lacOdatLL and lacOdatRR plus lacOdatRL were used, and for introduction into ter2, primer sets lacOterLR plus lacOterLL and lacOterRR plus lacOtertRL were used.
The inducible fluorescent repressor gene (PBAD-lacI-ecfp) was transferred by P1 transduction from TH130 [=AB1157Δ(353820-353822)::(araC PBAD-lacI-ecfp FRT-kan-FRT)] (13). The kan marker was removed during a final step using pCP20 as described elsewhere (11).
The strain carrying the inducible fluorescent repressor gene (PBAD-lacI-ecfp-tetR-mCherry) was created as follows. The mCherry sequence was amplified from pmCherry (Clontech) using primers tetR-mCherry5 and tetR-mCherry6. The FRT-cat-FRT sequence was amplified from pKD3 (11) by using primers tetR-mCherry7 and tetR-mCherry4. PCR in the presence of these two fragments as templates and tetR-mCherry5 plus tetR-mCherry4 as primers yielded a fragment consisting of (tetR)-mCherry-FRT-cat-FRT-(yajR). This fragment was introduced into YK1382 [ori1::tetO array ter2::lacO array ΔyajR::(araC PBAD-lacI-ecfp tetR-eyfp-FRT-cat-FRT)] following the method of Datsenko and Wanner (11). From the resulting strain, the ΔyajR::(araC PBAD-lacI-ecfp tetR-mCherry-FRT-cat-FRT) region was transferred to the MG1655dnaA-eyfp ori1::tetO array-dat1::lacO array by P1 transduction.
The host strain used in Fig. 2 features the inducible tetR-mCherry gene. To construct this strain, the chromosomal sequence from tetR through cat-FRT of the strain carrying ΔyajR::(araC PBAD-lacI-ecfp tetR-mCherry-FRT-cat-FRT) (described above) was amplified by PCR using primers tetR-mCherry8 and tetR-mCherry9. The amplified fragment was used to replace the lacI-ecfp sequence of MG1655dnaA-eyfp Δ(353820-353822)::(araC PBAD-lacI-ecfp FRT-cat-FRT) following the method of Datsenko and Wanner (11). The cat marker was removed using pCP20.
FIG. 2.
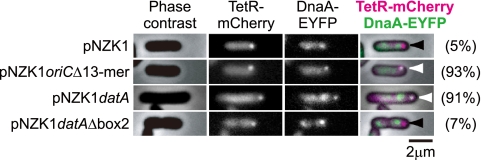
Visualization of DnaA-EYFP bound to oriC and datA on a low-copy-number plasmid. TetR-mCherry was temporarily induced from a host chromosome that also carries dnaA-eyfp in place of dnaA. In the merged picture at the far right, TetR-mCherry and DnaA-EYFP are shown in magenta and green, respectively. White arrowheads indicate colocalization of DnaA-EYFP and TetR-mCherry foci. Black arrowheads indicate TetR-mCherry foci that are not colocalized with DnaA-EYFP fluorescence. Percentages of the fluorescent spots of DnaA-EYFP that colocalized with the spots of TetR-mCherry out of 100 of the latter surveyed are indicated in parentheses.
To construct pNZK1, the tetO array-Gmr fragment was cut out from pLAU44 (22) using XbaI and XhoI and was introduced into pKV713 (19) by replacing the SnaBI-HpaI region in the sopABC region. pNZKdatA carries the chromosomal datA EcoNI-XhoI fragment that was inserted into the EcoRI site of pNZK1 by blunt-end ligation. The datAΔbox2 mutation is described elsewhere (34). pNZK1oriCΔ13-mer was made by inserting the chromosomal oriC region that lacked the 13-mer region at the SalI site of pNZK1. The defective oriC fragment was amplified using pCM959 (7) as a template and del13mersR and del13mersL1 as primers.
Cells were grown at 30°C in all experiments. In the experiments in Fig. 1, M9 medium containing 0.2% d-glucose was used. In the experiments in Fig. 3 to 5, M9 medium containing 0.2% l-arabinose was used. Overnight cultures were inoculated into fresh medium and grown until the optical density at 650 nm reached 0.2 to 0.3. In the experiments in Fig. 2, cells grown in M9 medium containing 0.2% d-glucose and 25 μg/ml ampicillin were harvested at room temperature by a brief centrifugation, suspended in M9 medium containing 0.2% l-arabinose, and grown for an additional 90 min to induce the fluorescent repressors.
FIG. 1.
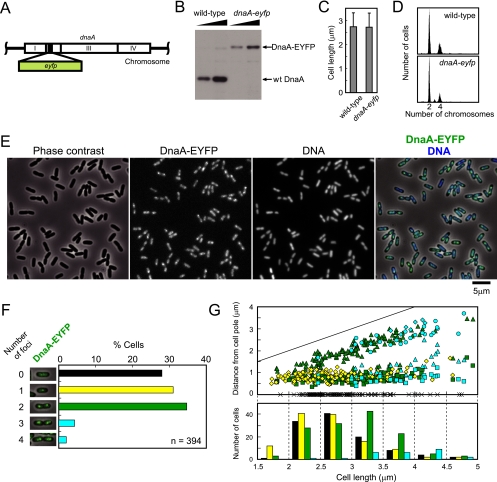
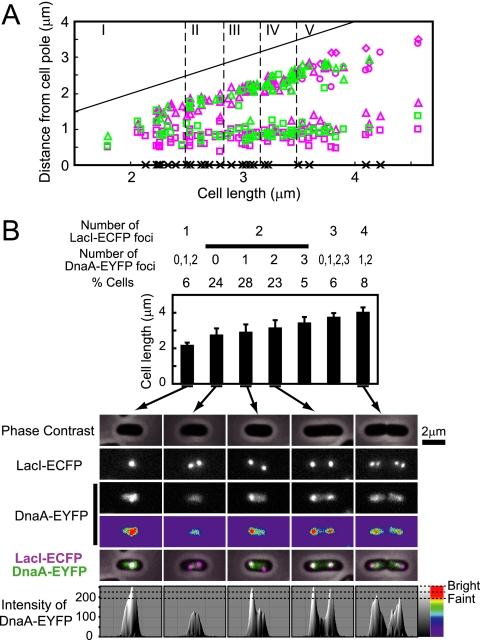
dnaA-eyfp strain. (A) Replacement of a portion of the domain II-encoding region of chromosomal dnaA by eyfp. (B) Immunoblotting using an antibody specific for DnaA. (C) Cell length distribution measured by microscopy. Three hundred cells were selected at random. Averages are shown with standard deviations. (D) Fluorescence histograms of DNA after replication runout. (E) Localization and focus formation of DnaA-EYFP at the nucleoid. In the merged picture at the far right, DnaA-EYFP and DNA are shown in green and blue, respectively. (F) Quantification of cells bearing each number of the DnaA-EYFP foci. Pictures of representative cells are presented. (G) Cellular positions of DnaA-EYFP foci. Positions of the foci scored in panel F were measured from a pole of each cell and plotted against the cell length. Cells with no focus are plotted as crosses on the abscissa. Colors of symbols represent cellular focus number as shown in panel F. In cells with two or more foci, the first, the second, the third, and the fourth focus from a cell pole are shown by a square, a triangle, a circle, and a diamond, respectively. The straight line indicates cell length. The lower panel shows the distribution of cells with each focus number in seven cell size classes. Focus numbers are represented by colors.
FIG. 3.
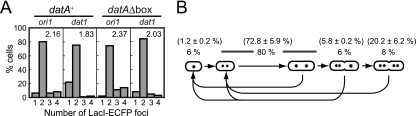
Characterization of strains used to observe the colocalization of DnaA-EYFP with chromosomal loci. (A) Proportions of cells that harbor each number of LacI-ECFP foci. Strains and positions of LacI-ECFP foci are indicated at the top. One hundred cells were chosen at random for the focus counting. Average focus number per cell is shown. (B) Schematic of the cell cycle of the dnaA-eyfp lacI-ecfp/lacO (ori1) strain in minimal medium at 30°C. LacI-ECFP foci are shown as dots in cells at different stages of the cell cycle. The percentages of cells with each focus number are shown. The percentages in parentheses indicate the proportion of the cells with one to four oriC copies per cell (from left to right) measured by flow cytometry after replication runout. Averages of five experiments are shown with standard deviations.
FIG. 5.
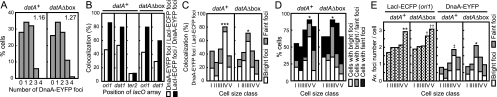
Quantification of fluorescent focus number and colocalization frequencies of the DnaA-EYFP foci with oriC and datA regions. Data were obtained from 100 cells that were examined in Fig. 4. (A) Quantification of cells (dnaA-eyfp lacI-ecfp/lacO array at ori1) carrying each observed number of DnaA-EYFP foci. The average number of foci per cell is shown in the graph. (B) Colocalization frequency of DnaA-EYFP foci with ori1, dat1, or ter2. White bars, number of LacI-ECFP foci colocalized with DnaA-EYFP foci/number of total LacI-ECFP foci; black bars, number of DnaA-EYFP foci colocalized with LacI-ECFP foci/number of total DnaA-EYFP foci. (C) Colocalization frequency (number of LacI-ECFP foci at ori1 colocalized with DnaA-EYFP foci/number of total LacI-ECFP foci at ori1) for cells in each size class. The five cell size classes are I (2.3 ± 0.2 μm), II (2.6 ± 0.1 μm), III (3.0 ± 0.1 μm), IV (3.3 ± 0.1 μm), and V (3.8 ± 0.3 μm) for datA+ and I (2.1 ± 0.2 μm), II (2.5 ± 0.1 μm), III (2.7 ± 0.1 μm), IV (2.9 ± 0.1 μm), and V (3.4 ± 0.2 μm) for datAΔbox. White and dark rectangles represent bright and faint foci, respectively. Significant differences of frequency of colocalization of DnaA-EYFP foci (bright or bright plus faint) at ori1 are indicated with asterisks (*, P < 0.05; ***, P < 0.001; comparison between two cell size classes of all combinations by Fisher's exact test). (D) Proportions of cells bearing DnaA-EYFP foci in each size class. Categories of cells are indicated at the right. Gray rectangles represent cells carrying both bright and faint foci. Significant differences are indicated with asterisks (*, P < 0.05; comparison of percentages of cells carrying at least one bright DnaA-EYFP focus between two cell size classes of all combinations by Fisher's exact test). (E) Average numbers of LacI-ECFP (lacO array at ori1) and DnaA-EYFP foci per cell in each cell size class. White and dark rectangles represent bright and faint foci of DnaA-EYFP, respectively. Significant differences from all other groups are indicated by asterisks (*, P < 0.05, and **, P < 0.01, by Mann-Whitney U test). †, P < 0.05 against cell size classes I to III but P > 0.05 against cell size class V; ††, P < 0.01 against cell size classes I to III but P > 0.05 against cell size class IV; ‡, P < 0.01 against cell size classes I to III but P > 0.05 against cell size class V as determined by Mann-Whitney U test. In the case of DnaA-EYFP, sums of bright and faint focus numbers were compared.
Fluorescence microscopy.
A portion of a culture was concentrated by brief centrifugation and was suspended in fresh medium at room temperature. An aliquot was transferred to a poly-l-lysine-coated glass slide. Microscopic images were obtained and analyzed using an Axiovert 200 M inverted microscope (Carl Zeiss) and MetaMorph software. Relative intensity of DnaA-enhanced yellow fluorescent protein (EYFP) fluorescence was measured using ImageJ software and the interactive three-dimensional surface plot plug-in. In this work, only the fluorescent spots with intensity levels from 192 to 255 in 0-to-255 grayscale levels, shown as the red color in Fig. 4B (DnaA-EYFP), were regarded as foci. Within this category, faint foci and bright foci were defined as those emitting fluorescence intensities of 192 to 223 and 224 to 255, respectively.
FIG. 4.
Subcellular localization of DnaA-EYFP and LacI-ECFP foci. (A) The positions of DnaA-EYFP and LacI-ECFP (ori1) foci in each cell. One hundred cells of the dnaA-eyfp lacI-ecfp/lacO (ori1) strain were examined. The ordinate represents the distance of the focus center as measured from a cell pole. Green square, DnaA-EYFP focus point in cells with a single DnaA-EYFP focus or the DnaA-EYFP focus point closest to the chosen cell pole in cells that exhibited more than one DnaA-EYFP focus point; green triangle, the second DnaA-EYFP focus area; green circle, the third DnaA-EYFP focus area; magenta square, ori1 focus point in cells with a single ori1 focus or the ori1 focus point closest to the chosen cell pole in cells that exhibited more than one ori1 focus; magenta triangle, the second ori1 focus area; magenta circle, the third ori1 focus area; magenta diamond, the fourth ori1 focus area. Cells with no focus are plotted as crosses on the abscissa. The straight line indicates cell length. (B) Classification according to cellular focus number. Cells analyzed in panel A were grouped according to the number of foci as shown. Proportions of cells and a graph of average cell length with standard deviation in each category are indicated. Pictures of representative cells in five categories are shown. DnaA-EYFP is presented by black and white and by colors. In the merged pictures, LacI-ECFP and DnaA-EYFP are shown by magenta and green, respectively. Fluorescence intensity of DnaA-EYFP is shown at the bottom. The intensity scale is shown at the right both by a linear gradation of black and white and by colors.
Other methods.
A Western blotting experiment using a polyclonal antibody against DnaA was carried out as described elsewhere (33). Flow cytometry was carried out as described elsewhere (32).
RESULTS
Creating a strain that expresses a functional fluorescent DnaA protein.
Thirty to 35 amino acid residues in domain II of E. coli DnaA have been shown to be dispensable for cell viability (33). Based on this finding, we replaced a portion of the chromosomal dnaA gene that codes for these nonessential residues with gfp, eyfp, ecfp, or mCherry. Viable mutant strains were obtained, and phenotypes of these constructs were assessed by measuring growth rate and by gathering data from flow cytometry, microscopy, and Western blotting. The strain expressing DnaA-EYFP, in which residues 87 to 104 of DnaA were replaced by EYFP (Fig. 1A and B), was chosen for further study since it exhibited phenotypes that seemed closest to those of the wild-type strain. The growth rate of this strain was indistinguishable from that of the wild-type strain in rich and in minimal media, at temperatures from 30 to 43°C (data not shown). Cell length was indistinguishable from that of the wild-type strain (Fig. 1C). Initiation frequency and synchrony were assessed by flow cytometry of cells treated with rifampin (rifampicin) and cephalexin (cefalexin) for several generation times to allow for the progression of replication forks to the termini while inhibiting new initiation and cell division, respectively. The number of chromosomes per cell measured in this experiment is equal to the number of origins per cell that were present at the time of addition of the drugs. The results shown in Fig. 1D indicate that replication disturbance in the dnaA-eyfp strain is small in terms of both frequency and synchrony of replication initiation.
Fluorescence microscopy revealed that DnaA-EYFP is located on the nucleoid (Fig. 1E). In addition, in many cells, bright fluorescent foci were observed against weak signals covering all the nucleoids. In Fig. 1F, randomly selected cells were classified according to the cellular number of fluorescent foci. Of 394 cells examined, ∼30% exhibited no distinct focus but the remainder presented one to four fluorescent foci. The majority of the latter group of cells had one focus point, mostly located either at the cell center or at the point one-quarter from a cell pole, or two foci, mostly located at positions approximately one-quarter and three-quarters away from a cell pole (Fig. 1G). On the average, cells with two foci were elongated relative to those with no or one focus point. A small portion of cells that were relatively elongated exhibited three to four foci.
oriC or datA is visible on a low-copy-number plasmid when bound by DnaA-EYFP.
From the observed locations of the DnaA-EYFP foci (Fig. 1G), we speculated that the fluorescent foci may have represented DnaA-EYFP molecules that were assembled at oriC or datA, located 467 kb apart on the chromosome, or at both of these locations. Therefore, we first examined whether oriC or datA could be visualized when they were present on a mini-F plasmid that is known to be present at one to two copies per cell. For this purpose, we constructed pNZK1, a mini-F-derived plasmid that carries a tetO array (22). To visualize the plasmid, the fluorescent repressor TetR-mCherry was expressed from the chromosomally integrated gene under the control of the araC-PBAD promoter in the dnaA-eyfp strain. The sop region was deleted in this plasmid since it is involved in the plasmid localization at the cell center (30), and this behavior would make it difficult to distinguish the plasmid from the host chromosome. Many plasmid-free segregants formed due to the absence of the sop region, but some cells presented pNZK1 that could be visualized by bound TetR-mCherry molecules in the cytoplasm. Only a small portion (5%) of pNZK1 was present with a detectable level of DnaA-EYFP fluorescence, despite the fact that it features two DnaA boxes at the plasmid origin of replication. It appeared that these rare cases were due to the fact that the fluorescence of DnaA-EYFP on the chromosome happened to colocalize with the plasmids. A portion of oriC that was devoid of the 13-mer region, and thus inactive for initiation origin, was ligated to this plasmid in order to assess the fluorescence intensity of DnaA-EYFP bound to the DnaA box-containing segment of oriC. As shown in Fig. 2 (pNZK1oriCΔ13-mer), most of the TetR-mCherry foci colocalized with the fluorescent foci of DnaA-EYFP.
In the case of datA on pNZK1, the fluorescence of DnaA-EYFP was also colocalized with most of the TetR-mCherry foci (Fig. 2, pNZK1datA). Furthermore, DnaA-EYFP was not visible on most of the datA plasmid when it carried the datAΔbox2 mutation, in accordance with the inability of the sequence to titrate a large number of DnaA molecules (32, 34).
Our results suggest that a single copy of oriC or datA on the chromosome should be detectable when DnaA-EYFP is assembled at the relevant locations.
Most fluorescent foci of DnaA-EYFP are colocalized with the oriC region.
In order to examine whether the fluorescent foci of DnaA-EYFP colocalize with oriC, the lacO array was inserted at the ori1 locus (16 kb counterclockwise of oriC) (42) on the chromosome of the dnaA-eyfp strain. We visualized its presence through the binding of LacI-enhanced cyan fluorescent protein (ECFP) as supplied by the chromosomally integrated gene under the control of the PBAD promoter. The strain grew normally like the wild-type strain with a generation time of ∼120 min at 30°C in M9-arabinose minimal medium. As measured by the number of LacI-ECFP foci, the cellular copy number of the ori1 locus under exponential growth conditions was one to four with an average of 2.16 (Fig. 3A, datA+). Figure 3B depicts the cell cycle of the datA+ strain as inferred from the data in Fig. 3A. In addition to the proportion of the cells with each ori1 focus number, the percentages of the cells with each oriC copy number that were measured by flow cytometry after replication runout are shown. The average cellular number of oriC (2.45) was ∼13% higher than that of ori1 foci (Fig. 3A). This may be due to the presence of two copies of newly replicated oriC in a single focus point. Significant variety appeared to exist with respect to initiation time, but a majority of the cells exhibited two ori1 foci and divided soon after the formation of four foci. From the data in Fig. 3B, it is estimated that, on average, replication commences at ∼27 min before cell division and that two copies of the newly replicated oriC remain associated at a single focus point for ∼17 min. This period is comparable to the time reported to be required for spatial separation of oriC after its replication (0.1- to 0.2-cell age for cells growing with the doubling time of 100 to 150 min) (1, 3, 35).
In Fig. 4A, 100 cells of the ori1-labeled dnaA-eyfp strain were chosen at random and positions of the fluorescent foci in each cell were plotted. Cells analyzed in Fig. 4A are classified in Fig. 4B according to the focus number, and pictures of representative cells in five classes are shown. The number of DnaA-EYFP foci was 0 to 3 with an average of 1.16 (Fig. 5A, datA+). Most of the DnaA-EYFP foci were colocalized with the LacI-ECFP foci at ori1 (Fig. 5B, datA+, LacI-ECFP foci/DnaA-EYFP foci). On the other hand, when the strain carrying the lacO array at the replication terminus (ter2) (42) was examined as a control, most of the DnaA-EYFP foci did not colocalize with the LacI-ECFP foci at ter2 (Fig. 5B). This result argues against a nonspecific distribution of the DnaA-EYFP foci.
DnaA-EYFP forms compact and bright foci in the oriC region during a period spanning the initiation event.
About half of the LacI-ECFP foci at ori1 did not colocalize with any foci of DnaA-EYFP fluorescence (Fig. 5B, datA+, DnaA-EYFP/LacI-ECFP). To investigate the relationship between colocalization frequencies and stage in the cell cycle, we divided 100 cells into five groups (I to V), each consisting of 20 cells, according to cell lengths reflecting stages in the cell cycle (Fig. 4A and 5C to E, datA+).
Remarkably, the localization frequency of the DnaA-EYFP foci at ori1 varied significantly depending on cell cycle stage. It was greatest in the size class IV cells (Fig. 5C, datA+). When the 100 sampled cells were classified according to the number of fluorescent foci, 23 cells had two well-separated ori1 foci, at which compact DnaA-EYFP foci were colocalized (Fig. 4B). These cells occupied the great majority of size class IV. Figure 5D (datA+) indicated that group IV was also highest in the number of cells carrying DnaA-EYFP foci. Moreover, bright foci were most abundant in this size group. Judging from the flow cytometry data (Fig. 3B), cells in size class IV are assumed to be at a stage around the initiation of replication and swiftly produce three to four ori1 foci or divide into two baby cells with one ori1 focus before forming two ori1 foci. The significantly increased number of ori1 foci per cell in class V cells (Fig. 5E, datA+) supports this assumption. Therefore, it is suggested that DnaA-EYFP accumulates in the oriC region to form bright foci during a period spanning the initiation event.
Figure 5C (datA+) also suggested that a significant portion of the accumulated DnaA-EYFP is released from the oriC region after initiation. In addition, DnaA-EYFP foci that still colocalized with ori1 after initiation appeared to be enriched in those with less-intense fluorescence. A possibility was ruled out that most of the released DnaA-EYFP foci from ori1 remained as foci at different positions on the nucleoid, since the focus number of DnaA-EYFP was decreased in cells in classes I to III and V (Fig. 5E, datA+). A reduction in the fluorescence intensity of the remaining foci after initiation was also suggested from the increased number of cells exhibiting only faint fluorescence of DnaA-EYFP in classes I to III and V (Fig. 5D, datA+).
Localization of DnaA-EYFP at datA and effect of datA on the behavior of DnaA-EYFP.
The initiator titration site datA is located about 467 kb away from oriC (20). In order to examine possible colocalization of the DnaA-EYFP focus with this site, the lacO array was inserted at the dat1 locus (20 kb clockwise of datA) on the chromosome of the dnaA-eyfp lacI-ecfp strain. It was revealed that the frequency of colocalization of DnaA-EYFP foci with dat1 was at a level comparable to that with ori1 (Fig. 5B, datA+). Measurements of cell length and distance of the fluorescent focus from a pole of each cell did not give any significant difference from the result obtained with ori1 (data not shown). Thus, it was not possible to discriminate between oriC and datA in datA+ cells.
On the other hand, in cells lacking the datA function, the frequency of colocalization of DnaA-EYFP foci with ori1 or dat1 (LacI-ECFP foci/DnaA-EYFP foci) was ∼80% or ∼60%, respectively (Fig. 5B, datAΔbox). It might be necessary to take into account some background level of colocalization like that observed at the ter2 locus. Furthermore, comparison of the localization frequencies of DnaA-EYFP on dat1 between datA+ and datAΔbox strains suggested that nearly half of the DnaA-EYFP foci at dat1 in the datA+ strain are formed in a manner dependent on the datA function (Fig. 5B, DnaA-EYFP foci/LacI-ECFP foci).
A notable feature observed in the datAΔbox strain was that there was an ∼10% increase in the average cellular number of DnaA-EYFP foci relative to the datA+ strain (Fig. 5A) and the population of cells that exhibited enriched DnaA-EYFP foci at ori1 shifted to smaller cell size classes (Fig. 5C). Results in Fig. 5D and E suggested that, as with datA+ cells, the reduction in frequency of colocalization of DnaA-EYFP foci at ori1 after initiation (Fig. 5C) is due to dissociation of the foci into smaller assemblies or monomers.
Consistent with the previous results obtained in different genetic backgrounds (20, 32), the cellular number of ori1 (and dat1 as well) was increased in the datAΔbox strain (Fig. 3A). These data support the idea that the absence of the datA function leads to the assembly of DnaA-EYFP foci at oriC earlier in the cell cycle and tends to trigger untimely initiations.
DISCUSSION
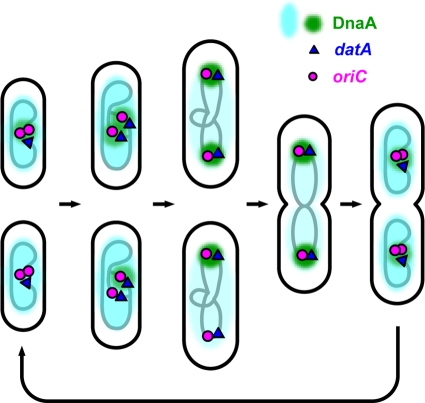
This study revealed that DnaA-EYFP of E. coli is located on the nucleoid and forms brilliant foci in many cells. Inspection of individual cells at different cell cycle stages suggested that the chimeric initiator dynamically changes its assembly form in the cell cycle (Fig. 6). During a period spanning the time of the initiation event, the brightest foci of DnaA-EYFP were colocalized with the oriC region. Shortly after the initiation, about half of the foci disappeared and a significant portion of the remaining foci were less intense than the foci at the time around the initiation. Although colocalization does not prove direct interaction, these observations may be a visual confirmation of the previous in vivo footprinting data (9, 37).
FIG. 6.
Model for dynamic DnaA localization during E. coli cell cycle. Shown is a cell cycle from two newly replicated origins to four newly replicated origins that was a majority of the results in this study. Clusters of DnaA molecules observed as DnaA-EYFP foci in this study are shown in green. DnaA molecules distributed on the entire nucleoid are shown in blue. The lower three cells at early to midcell stages represent cells that have the oriC-datA region free of DnaA-EYFP foci during a period after initiation.
The mechanism of dissociation of DnaA from oriC is not clear. RIDA is assumed to operate after initiation. Therefore, ATP-DnaA should be converted to inactive ADP-DnaA and may be released from oriC. Sequestration of oriC by SeqA seems to prevent association of DnaA with weaker binding sites in oriC (29). In addition, other factors such as integration host factor (IHF) and factor for inversion stimulation (FIS) may be involved in the release or prevention of binding of DnaA after the initiation (9). The fact that DnaA-EYFP foci did not completely disappear from all ori1 loci may indicate that a dynamic equilibrium of association and dissociation exists between DnaA-EYFP and the oriC-datA region at or near a critical level that is detectable as a focus. It appeared that many DnaA-EYFP foci are irregularly shaped after initiation. Although it was possible that they are in the course of dissociation or association, this was not taken into consideration in the present work, since it was difficult to classify the morphology of the foci accurately.
Accumulation of DnaA-EYFP to form a bright focus would be accelerated by desequestration of the promoter region of dnaA as well as of oriC. The dnaA gene is located close to oriC. It is also autoregulated (2, 5). Therefore, newly synthesized DnaA molecules, assumed to be in the ATP-bound form, may be rapidly synthesized and recruited into the DnaA foci. Rejuvenation of DnaA to the ATP-bound form with the help of acidic phospholipids (10) may take over RIDA during the reassembly process. Maximum binding to oriC at the time of initiation may be achieved by binding to weaker binding sites aided by architectural proteins such as HU and IHF (23).
A significant portion of cells carrying two ori1 foci had only one colocalized focus of DnaA-EYFP (Fig. 4B). They might result from the highly cooperative nature of intermolecular interaction of DnaA during the association and dissociation process. The possibility that the chimeric initiator has some defect in behaving synchronously at the two loci is not ruled out, however.
datA has a large DnaA titration capacity but is relatively weak in its affinity for DnaA due to rapid dissociation (32). These properties allow datA to serve as a temporary reservoir of DnaA for controlled accumulation of the initiator at oriC for the next initiation event. In addition to datA, there are ∼300 DnaA boxes on the chromosome (36). Although these sites appear to have little initiator-titration capacity (34), they may be involved in accommodating released DnaA-EYFP molecules, inasmuch as fluorescence was observed on the entire nucleoid in cells that exhibit no distinct focus (Fig. 1F and 4B).
We could not discriminate between oriC and datA in datA+ cells. Rather, it appeared that these two loci were colocalized with the DnaA-EYFP focus. However, the reduced localization frequency of DnaA-EYFP foci at dat1 in ΔdatA cells (Fig. 5B) seems to suggest that about half of the DnaA-EYFP foci colocalized with dat1 in datA+ cells are formed at datA. oriC may be also complexed with DnaA-EYFP in the same foci. It is tempting to speculate that the DnaA cluster is involved in formation of the Ori macrodomain (31, 41), which includes both oriC and datA.
It has been reported elsewhere that E. coli DnaA is located at the cell membrane (27). Furthermore, the same group reported recently that DnaA forms helical structures along the longitudinal cell axis by confocal fluorescence microscopy using DnaA-green fluorescent protein (GFP)-expressing cells and by immunofluorescence microscopy using wild-type cells and DnaA antibody (4). We have not observed any significant DnaA-EYFP fluorescence around the cell membrane (Fig. 1E and data not shown). Neither have we obtained fluorescent images suggesting helical structures of DnaA-EYFP assembly. If DnaA-EYFP adopts helical structures as reported, fluorescent foci should be visible near the inner cell membrane with certain intervals. Our study, however, cannot rule out a possibility that a small portion of DnaA-EYFP molecules adopt helical structures. Our trial of confocal microscopy has been inconclusive due to color bleaching. We have also attempted immunofluorescence microscopy using DnaA antibody. However, we have been unable to obtain reproducible results due to unknown reasons. We do not know the reason for the apparent discrepancy between the present results and the previous reports. It seems, however, that our DnaA-EYFP construct is much more active in the initiation than is the GFP-tagged DnaA described by Boeneman et al. (4), inasmuch as our strain does not exhibit significant differences in cell length and initiation frequency from the wild-type strain (Fig. 1C and D) and is not temperature sensitive.
Recently, it was reported that DnaA of Bacillus subtilis also forms fluorescent foci in live cells when tagged with GFP (40). Interestingly, the foci colocalize with oriC only at early or late stages of the cell cycle, when replication machinery is assembling or disassembling. During most of the cell cycle, DnaA molecules colocalize with the replication fork, tethered to the replication machinery by YabA, a unique protein that is absent in E. coli (14). Although DnaA serves as the ubiquitous initiator protein in bacteria, the regulation of initiation in gram-positive bacteria is very different from that in E. coli, inasmuch as several proteins known to be involved in this step have no known counterparts. Even in gram-negative proteobacteria, different mechanisms appear to be exploited by different bacterial species (43). In E. coli, it has been reported that replisome components assemble at oriC and, after initiation, behave independently from oriC (35). Further study will be required to adequately understand the diverse initiation control systems in prokaryotes.
Acknowledgments
We are grateful to T. Hatano for suggestions about microscopic analysis.
This work was supported in part by Grants-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (to T.O.) and by NIG Cooperative Research Program (2006-A49, 2007-A43, and 2008-B11).
Footnotes
Published ahead of print on 5 June 2009.
REFERENCES
- 1.Adachi, S., T. Fukushima, and S. Hiraga. 2008. Dynamic events of sister chromosomes in the cell cycle of Escherichia coli. Genes Cells 13181-197. [DOI] [PubMed] [Google Scholar]
- 2.Atlung, T., E. S. Clausen, and F. G. Hansen. 1985. Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet. 200442-450. [DOI] [PubMed] [Google Scholar]
- 3.Bates, D., and N. Kleckner. 2005. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121899-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeneman, K., S. Fossum, Y. Yang, N. Fingland, K. Skarstad, and E. Crooke. 2009. Escherichia coli DnaA forms helical structures along the longitudinal cell axis distinct from MreB filaments. Mol. Microbiol. 72645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, R. E., K. O'Day, and A. Wright. 1985. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40159-169. [DOI] [PubMed] [Google Scholar]
- 6.Brendler, T., and S. Austin. 1999. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 182304-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buhk, H.-J., and W. Messer. 1983. The replication origin region of Escherichia coli: nucleotide sequence and functional units. Gene 24265-279. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62967-979. [DOI] [PubMed] [Google Scholar]
- 9.Cassler, M. R., J. E. Grimwade, and A. C. Leonard. 1995. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 145833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crooke, E. 2001. Escherichia coli DnaA protein-phospholipid interactions: in vitro and in vivo. Biochimie 8319-23. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erzberger, J. P., M. L. Mott, and J. M. Berger. 2006. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13676-683. [DOI] [PubMed] [Google Scholar]
- 13.Hatano, T., Y. Yamaichi, and H. Niki. 2007. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol. Microbiol. 641198-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi, M., Y. Ogura, E. J. Harry, N. Ogasawara, and S. Moriya. 2005. Bacillus subtilis YabA is involved in determining the timing and synchrony of replication initiation. FEMS Microbiol. Lett. 24773-79. [DOI] [PubMed] [Google Scholar]
- 15.Kaguni, J. M. 2006. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 60351-375. [DOI] [PubMed] [Google Scholar]
- 16.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 9461-71. [DOI] [PubMed] [Google Scholar]
- 17.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 204253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawakami, H., K. Keyamura, and T. Katayama. 2005. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 28027420-27430. [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki, Y., C. Wada, and T. Yura. 1990. Roles of Escherichia coli heat shock proteins DnaK, DnaJ and GrpE in mini-F plasmid replication. Mol. Gen. Genet. 220277-282. [DOI] [PubMed] [Google Scholar]
- 20.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 123032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurokawa, K., S. Nishida, A. Emoto, K. Sekimizu, and T. Katayama. 1999. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 186642-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau, I. F., S. R. Filipe, B. Soballe, O. A. Okstad, F. X. Barre, and D. J. Sherratt. 2003. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 49731-743. [DOI] [PubMed] [Google Scholar]
- 23.Leonard, A. C., and J. E. Grimwade. 2005. Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol. Microbiol. 55978-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77413-426. [DOI] [PubMed] [Google Scholar]
- 25.McGarry, K. C., V. T. Ryan, J. E. Grimwade, and A. C. Leonard. 2004. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. USA 1012811-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mott, M. L., and J. M. Berger. 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5343-354. [DOI] [PubMed] [Google Scholar]
- 27.Newman, G., and E. Crooke. 2000. DnaA, the initiator of Escherichia coli chromosomal replication, is located at the cell membrane. J. Bacteriol. 1822604-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen, O., and A. Løbner-Olesen. 2008. Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells. EMBO Rep. 9151-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nievera, C., J. J. Torgue, J. E. Grimwade, and A. C. Leonard. 2006. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell 24581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90951-957. [DOI] [PubMed] [Google Scholar]
- 31.Niki, H., Y. Yamaichi, and S. Hiraga. 2000. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 14212-223. [PMC free article] [PubMed] [Google Scholar]
- 32.Nozaki, N., Y. Yamada, and T. Ogawa. 2009. Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells 14329-341. [DOI] [PubMed] [Google Scholar]
- 33.Nozaki, S., and T. Ogawa. 2008. Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 1543379-3384. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa, T., Y. Yamada, T. Kuroda, T. Kishi, and S. Moriya. 2002. The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 441367-1375. [DOI] [PubMed] [Google Scholar]
- 35.Reyes-Lamothe, R., C. Possoz, O. Danilova, and D. J. Sherratt. 2008. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 13390-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth, A., and W. Messer. 1998. High-affinity binding sites for the initiator protein DnaA on the chromosome of Escherichia coli. Mol. Microbiol. 28395-401. [DOI] [PubMed] [Google Scholar]
- 37.Samitt, C. E., F. G. Hansen, J. F. Miller, and M. Schaechter. 1989. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 8989-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sclafani, R. A., and T. M. Holzen. 2007. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 41237-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slater, S., S. Wold, M. Lu, E. Boye, K. Skarstad, and N. Kleckner. 1995. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82927-936. [DOI] [PubMed] [Google Scholar]
- 40.Soufo, C. D., H. J. D. Soufo, M.-F. Noirot-Gros, A. Steindorf, P. Noirot, and P. L. Graumann. 2008. Cell-cycle-dependent spatial sequestration of the DnaA replication initiator protein in Bacillus subtilis. Dev. Cell 15935-941. [DOI] [PubMed] [Google Scholar]
- 41.Valens, M., S. Penaud, M. Rossignol, F. Cornet, and F. Boccard. 2004. Macrodomain organization of the Escherichia coli chromosome. EMBO J. 234330-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, X., C. Possoz, and D. J. Sherratt. 2005. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 192367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zakrzewska-Czerwinska, J., D. Jakimowicz, A. Zawilak-Pawlik, and W. Messer. 2007. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 31378-387. [DOI] [PubMed] [Google Scholar]