Abstract
The application of mitomycin C induction to 114 genetically diverse Streptococcus agalactiae strains generated 36 phage suspensions. On electron microscopy of the phage suspensions, it was possible to assign the phages to the Siphoviridae family, with three different morphotypes (A, B, and C). Phage genetic diversity was evaluated by a PCR-based multilocus typing method targeting key modules located in the packaging, structural, host lysis, lysogeny, replication, and transcriptional regulation clusters and in the integrase genes and by DNA digestion with EcoRI, HindIII, and ClaI. Thirty-three phages clustering in six distantly related molecular phage groups (I to VI) were identified. Each molecular group was morphotype specific except for morphotype A phages, which were found in five of the six phage groups. The various phage groups defined on the basis of molecular group and morphotype had specific lytic activities, suggesting that each recognized particular host cell targets and had particular lytic mechanisms. Comparison of the characteristics of lysogenic and propagating strains showed no difference in the serotype or clonal complex (CC) identified by multilocus sequence typing. However, all the lysogenic CC17 and CC19 strains presented catabolic losses due to a lack of catabolic decay of dl-alpha-glycerol-phosphate substrates (CC17) and of alpha-d-glucose-1-phosphate (CC19). Moreover, the phages from CC17 lysogenic strains displayed lytic replication in bacterial hosts from all S. agalactiae phylogenetic lineages other than CC23, whereas phages obtained from non-CC17 lysogenic strains lysed bacteria of similar evolutionary origin. Our findings suggest that the adaptive evolution of S. agalactiae exposed the bacteria of this species to various phage-mediated horizontal gene transfers, which may have affected the fitness of the more virulent clones.
The Lancefield group B Streptococcus, Streptococcus agalactiae—a major cause of neonatal infections—has increasingly been reported as a common pathogen in nonpregnant adults since the 1970s (40). The proportion of neonatal infections caused by serotype III multilocus sequence type 17 (ST-17) strains is higher than would be expected on the basis of the proportion of women and infants colonized by ST-17 strains in control populations (3, 5, 24, 26, 30, 33). The major clonal complexes (CC) 1, 12, 17, 19, and 23 have been associated with infections in adults (4, 5, 14, 21). The presence in the S. agalactiae genome of particular insertion sequences, a group II intron, and prophage DNA fragments (17, 36, 49) suggests that horizontal genetic transfer may play an important role in genome diversification and the emergence of virulent clones in S. agalactiae.
Temperate phages affect bacterial fitness by modifying anchor points for genome rearrangements, by disrupting genes, by protecting against lytic infection, by lysing competing strains through prophage induction, and by introducing new fitness factors (8, 18). Little is currently known about these phages. S. agalactiae phages were first isolated in 1969 (37). A phage-typing method has since been proposed for epidemiological investigations (1, 7, 45, 46). More recent analysis of sequenced S. agalactiae strains has revealed the presence of abundant regions resembling prophages (47, 48) although there is currently no evidence to suggest that these regions correspond to functional bacteriophages or remnants. We recently identified, cloned, and sequenced three prophage DNA fragments from S. agalactiae genomes that displayed significant alignment with previously identified prophage sequences from S. agalactiae and Streptococcus pyogenes. We found that these prophage DNA fragments were more frequent in the genomes of strains from a particular phylogenetic lineage isolated from cases of neonatal meningitis. Thus, genetic events coinciding with lysogeny may have increased the ability of strains to invade the brain endothelium (49).
We investigated lysogeny in S. agalactiae species by carrying out phage induction in 114 S. agalactiae strains representative of the genetic diversity of strains isolated from the genital tract and neonates. In accordance with the recommendations of the International Committee on Taxonomy of Viruses (11), the isolated phages were characterized by classical methods: electron microscopy analysis of virions, restriction endonuclease analysis (REA), and determination of the lytic spectrum of phages. In addition, a PCR-based method targeting phage sequences located in the packaging cluster, the structural cluster, the host lysis cluster, the integrase genes, the lysogeny cluster, the replication cluster, and the transcriptional regulation cluster was developed for multilocus phage characterization. Molecular features were used to assess the genetic diversity of S. agalactiae bacteriophages. We characterized the phage donors and propagating strains by serotyping, multilocus sequence typing (MLST), and the assessment of carbon source oxidation to identify particular features of S. agalactiae lysogenic strains.
MATERIALS AND METHODS
Bacterial strains.
Phage induction was carried out in 114 S. agalactiae strains; 113 S. agalactiae strains were selected on the basis of previously established characteristics (33, 36) as representative of the diversity of strains colonizing the vagina or responsible for neonatal infections. The remaining isolate was strain 2603V/R, which has already been sequenced (48). Fifty-nine strains were colonizing strains isolated from the vaginas of asymptomatic pregnant women (n = 37) and asymptomatic neonates (n = 22), and 55 strains originated from the cerebrospinal fluid samples of neonates suffering from meningitis. Twenty-eight strains were of serotype I, 19 were of serotype II, 55 were of serotype III, 3 were of serotype IV, and 4 were of serotype V, including the 2603V/R strain.
Isolation and propagation of S. agalactiae phages.
S. agalactiae phages were isolated in a four-step process (Fig. 1), as follows.
FIG. 1.
The various steps in phage isolation.
Step 1: phage induction.
Each of the 114 S. agalactiae strains was assessed as a potential donor strain and subjected to mitomycin C induction (45). Each strain was plated on Trypticase soy agar containing 5% (vol/vol) sheep blood and incubated overnight at 37°C. A single colony was then added in 4.5 ml of modified Todd Hewitt broth (mTHB) consisting of 30 g of THB base (Becton Dickinson, le Pont de Claix, France), 2 g of yeast extract (Oxoid LTD, Basingstoke Hampshire, England), 12 mg of CaCl2 (VWR Prolabo, Fontenay-sous-Bois, France), and 10 mg of l-tryptophan (Merck, Darmstadt, Germany) per liter; the culture was then incubated for 2 h at 30°C. Mitomycin C (Sigma-Aldrich, St.-Quentin-Fallavier, France) was added to the mTHB at a final concentration of 1 μg per ml. Incubation was continued for 2 h. The culture was then centrifuged at 1,500 × g for 15 min. The supernatant was filtered through a 0.45-μm-pore-size filter (Millipore, Bedford, MA). The 114 filtered supernatants, corresponding to putative phage filtrates, were stored at −80°C.
Step 2: phage isolation.
We assessed the lytic activity of each of the 114 putative phage filtrates against the 114 S. agalactiae strains used as indicator strains. Each phage filtrate was tested with each indicator strain, using a multiloop applicator: 10 μl of each filtrate was poured onto a base layer (20 ml) of mTHB containing 0.7% agar (wt/vol) and inoculated with an overnight culture of an indicator strain in mTHB. Plates were incubated overnight at 30°C and examined for the presence of lytic plaques.
Step 3: phage selection.
A putative phage filtrate was considered to be a phage suspension if it provided at least 20 clear plaques with at least two different indicator strains. A clear plaque was then picked and dissolved in 1 ml of mTHB. The phage suspensions thus obtained were stored at −80°C.
Step 4: phage propagation.
The indicator strain yielding the largest number of plaques for a given phage suspension was defined as the propagating strain for that phage suspension. Each phage was then propagated in its propagating strain. We added 1 ml of phage suspension to 10 ml of semisolid medium (mTHB plus 0.4% agar [wt/vol]) and 800 μl of an overnight culture of the propagating strain in mTHB. The mixture was then poured onto a base layer (20 ml) of mTHB containing 0.7% agar (wt/vol) and incubated for 24 h at 30°C. Phage propagation resulted in visible plaques, which were recovered in 10 ml of mTHB. The semisolid medium was then taken up and centrifuged at 1,500 × g for 15 min. The supernatant was passed through a filter with 0.45-μm pores and titrated by plating serial dilutions on a lawn of the propagating strain on modified Todd-Hewitt agar. The titrated induced phage preparations were kept at −80°C and named ϕn.
Phage characterization. (i) Electron microscopy analysis of phages.
Based on the guidelines of the International Committee on Taxonomy of Viruses (19), the induced phage preparations were centrifuged on a sucrose density gradient. Phage particles were negatively stained with 2% uranyl acetate, examined in a JEOL 1230 transmission electron microscope at an accelerating voltage of 120 kV, and photographed. Morphological types were defined on the basis of phage tail length.
(ii) Molecular characterization of isolated phages. (a) REA.
Phage DNA isolation and purification were carried out with a Lambda Minikit (Qiagen, Valencia, CA). The phage DNA was digested separately with EcoRI, HindIII, and ClaI (Roche Diagnostic, Meylan, France) by adding 15 μl of DNA to 15 IU of each restriction endonuclease. After 3 h of incubation at 37°C, the DNA fragments were separated by electrophoresis in 0.8% agarose gels using a voltage gradient of 50 V over 16 h. Digestion patterns were examined under UV transillumination after staining with ethidium bromide and manually compared.
(b) Multilocus characterization of phages by a PCR-based method.
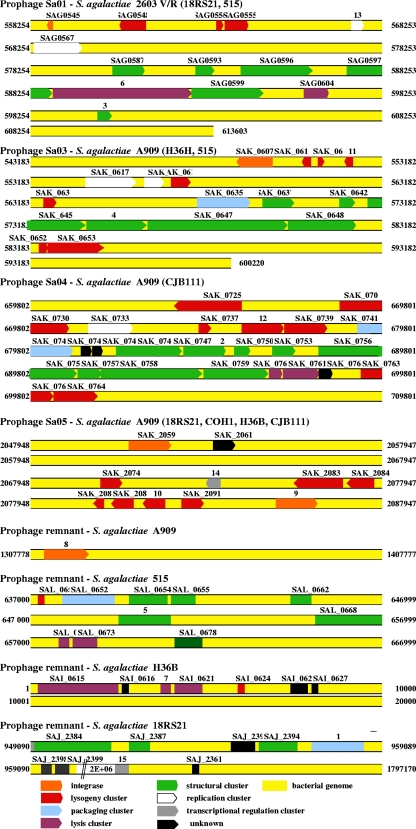
A PCR-based method was used to study the genetic diversity of phages and to assess the level of similarity between isolated phages. As S. agalactiae phages have yet to be sequenced, we performed an in silico analysis of the genomes of eight sequenced S. agalactiae strains: strains 515, A909, 18RS21, COH1, H36B, and CJB111 (47); strain 2603V/R (48); and strain NEM316 (16). We identified 15 target genes, numbered 1 to 15, encoding phage proteins (Fig. 2 and Table 1). These 15 target genes are located in four long sequences corresponding to four prophages—Sa1 (strains 2603V/R, 18RS21, and 515), Sa3 (strains A909, H36B, and 515), Sa4 (strains A909 and CJB111), and Sa5 (strains A909, 18RS21, COH1, H36B, and CJB111)—or in prophage remnants in the S. agalactiae genomes of strains A909, 515, H36B, and 18RS21. Using BLAST software, we aligned the genome sequences obtained, with a view to identifying sequences specific to each of the target genes. Primers for amplifying these specific sequences were designed with primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) (Table 1). PCR was performed on each of the isolated phages with the 15 primer pairs. The sequences amplified were located in the packaging cluster (PCR 1), the structural cluster (PCRs 2, 3, 4, and 5), the host lysis cluster (PCRs 6 and 7), the integrase genes (PCRs 8 and 9), the lysogeny cluster (PCRs 10, 11, and 12), the replication cluster (PCR 13), and the transcriptional regulation cluster (PCRs 14 and 15). The PCR assay was performed on a Chromo 4 System instrument (Bio-Rad, Hercules, CA) in a final volume of 25 μl containing 5 μl of extracted DNA, the necessary primers (each at a concentration of 0.5 μM), and 1× iQ SYBR Green Supermix (Qiagen SA, Courtaboeuf, France), including 3 mM MgCl2. Amplification was performed over 40 cycles of 10 s at 94°C, 10 s at a hybridization temperature specific for each primer set (Table 1), and 30 s at 72°C. The reaction products were cooled to 35°C and subjected to a post-PCR melting cycle by increasing the temperature by 0.2°C for each 10-s cycle, up to 95°C. PCR amplifications were validated by sequencing one of each of the amplicons obtained with each formulated primer pair. For all phages, PCR amplification was considered positive if an amplicon of the expected size was generated.
FIG. 2.
Identification and localization of the 15 target genes used for multilocus characterization of phages by a PCR method and identified by an in silico analysis of eight sequenced S. agalactiae genomes. The target genes numbered 1 to 15 were located in the packaging cluster (1), the structural cluster (2, 3, 4, and 5), the host lysis cluster (6 and 7), the integrase genes (8 and 9), the lysogeny cluster (10, 11 and 12), the replication cluster (13), and the transcriptional regulation cluster (14 and 15).
TABLE 1.
Multilocus PCR-based molecular characterization of phage: primers used and amplicon size
| Prophage | PCR no.a | Target gene
|
Reference strain(s) | Primer
|
Hybridization temp (°C) | Amplicon size (bp) | ||
|---|---|---|---|---|---|---|---|---|
| Locus | Description | Direction | Sequence (5′ → 3′) | |||||
| Sa01 | 13 | SAG0566 | Single-strand binding protein prophage lambda Sa1 | 2603 V/R, 18RS21 | Forward | GTGCTTTGGTTGGAATTAC | 54 | 132 |
| Reverse | TCTGTTGTTGGCTATTGC | |||||||
| 6 | SAG0598 | Prophage lambda Sa1; N-acetylmuramoyl-l-alanine amidase, family 4 | 18RS21, 2603V/R | Forward | ACAAATATCACGCACTAAAC | 54 | 289 | |
| Reverse | TCTTGACCAGTCCATTCC | |||||||
| 3 | SAG0610 | Hypothetical protein Sa1 | 2603V/R, H36B, COH1, 515, A909, CJB111 | Forward | TTGATATACTCCACATTAGC | 49 | 192 | |
| Reverse | CCTTCCTTGTTTCATACG | |||||||
| Sa03 | 4 | SAK_0646 | Prophage lambda Sa03; tail component; putative | A909, 2603V/R | Forward | AATACAATACAATAGAAGATTAC | 54 | 233 |
| Reverse | TCCGTCTTGAGTATAGTC | |||||||
| 11 | SAK_0613 | Prophage ps2; protein 07; excisionase | H36B, A909 | Forward | GACTATGGCGATTATGTG | 49 | 128 | |
| Reverse | CTATGCGTTGGATTATTG | |||||||
| Sa04 | 12 | SAK_0738 | DNA methylase prophage lambda W4 | CJB111, A909 | Forward | GGGATAAGAAAGCCAATC | 54 | 172 |
| Reverse | ACATAGATAGACGCATCG | |||||||
| 2 | SAK_0748 | Phage major capsid protein; HK97 family | CJB111, A909 | Forward | TGATTTCTCTTACTACTGGATTG | 52 | 136 | |
| Reverse | CGCTTCTGGTAGAACGAG | |||||||
| Sa05 | 14 | SAK_2079 | Prophage Sa05; ArpU family transcriptional regulator | A909, COH1 | Forward | GCGTTACCCAGTTTGATATAG | 47 | 67 |
| CJB111, 18RS21 | Reverse | GAAATAAGCCGAGAATGC | ||||||
| 10 | SAK_2090 | BRO domain protein, prophage antirepressor prophage Sa05 | A909, H36B | Forward | TAGAGCACCAAGGCGAATG | 54 | 102 | |
| CJB111 | Reverse | AAACGACCTCATCAACTAAACG | ||||||
| 9 | SAK_2094 | Prophage Sa05 site-specific recombinase; phage integrase family | A909, H36B | Forward | AAAGAGTAAAGCATTTCG | 49 | 526 | |
| CJB111, 18RS21 COH1 | Reverse | CCTAATCTATATTGGAGTTC | ||||||
| 8 | SAK_1326 | Site-specific recombinase, phage intergrase family | A909, H36B | Forward | TTTGACCTACGGGATTATG | 50.5 | 261 | |
| CJB111 | Reverse | TGAACGCCATCTTAGAAG | ||||||
| 5 | SAL_0666 | Putative prophage lambda Sa1; minor structural protein | 515, 18RS21 | Forward | CATAGAGATACACGACATC | 50.5 | 289 | |
| Reverse | TGAACGCTTGATAACATC | |||||||
| 7 | SAI_0620 | Phage holing; LL-H family | H36B, 515 | Forward | CTGTGGAAGTTGGTATTAAG | 47 | 94 | |
| 2603V/R | Reverse | TCTGTTAAACTGATATTATATTGC | ||||||
| 1 | SAJ_2395 | Phage terminase-like protein, large subunit | 18RS21, 515 | Forward | TGATAGATAAGTATGTGAGATTC | 50.5 | 251 | |
| Reverse | TTGTCTTTCCGAGTTAGC | |||||||
| 15 | SAJ_2357 | gp31 | 18RS21, 515 | Forward | ACTATTATATCATACGAGGAG | 50.5 | 247 | |
| Reverse | ATTGCTTCTAATTCTTGTTC | |||||||
PCRs were numbered as defined in Fig. 2.
We investigated the genetic relationship between the genomes of isolated phages by carrying out a hierarchical analysis by the Jaccard dichotomy coefficient method with SYSTAT12 (LogiLabo, Paris, France). The genetic features used for analysis were the amplification by PCR of the 15 phage gene sequences studied and of each of the DNA fragments obtained by REA with the three enzymes used in this study. An absence of gene amplification was not considered to indicate similarity between the phages studied.
(iii) Determination of phage lytic spectrum.
The lytic activity of the induced phage preparations was assessed against the 114 S. agalactiae strains. Each strain was used to inoculate 4.5 ml of mTHB, which was then incubated at 30°C for 2 h. For each strain, two petri dishes (90 mm) containing modified Todd-Hewitt agar were flooded with fresh cultures. Excess fluid was removed with a pipette, and the plates were dried. In accordance with the phage typing procedure (46), we used induced phage preparation dilutions corresponding to 106 and 107 phages per ml. The induced phage preparations were applied to the dried bacterial lawns with a multiloop applicator (Biddulf and Co., Manchester, United Kingdom). Plates were incubated overnight at 30°C and examined with oblique transmitted light against a dark background. Using a previously published method (27), we subjected the lytic reactions observed, characterized by >50 plaques or confluent lysis, to hierarchical analysis by the Jaccard dichotomy coefficient method, with SYSTAT12 (LogiLabo, Paris, France). A representation of the phages clustered as a function of their reactions with strains is shown as a dendrogram.
Characterization of donor and propagating strains.
Donor and propagating strains were serotyped and analyzed by MLST, as described by Jones et al. (21). Using the unweighted-pair group method with arithmetic mean, a tree was generated from allelic profile data, using Phylodendron and the entire group B streptococcus MLST database (http://pubmlst.org/sagalactiae/). CCs were designated on the basis of reported findings (21). We also assessed the ability of these strains to oxidize carbon sources, which is known to be variable in this species, with the Biolog system as previously described (13). Differences in catabolic functions between the donor and propagating strains were studied by chi square tests or Fisher-Yates tests. A P value of <0.05 was considered to indicate statistical significance.
RESULTS
Mitomycin C induction, according to the procedure summarized in Fig. 1, generated 36 induced phage suspensions from 24 different donor strains. We selected 21 indicator strains.
Electron microscopy analysis of phages isolated from the 36 S. agalactiae phage suspensions.
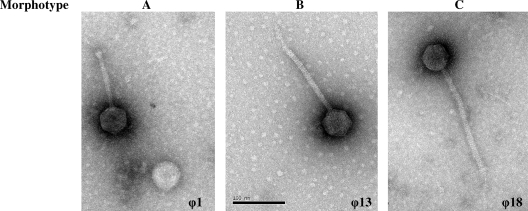
Examination of the 36 induced phage preparations (Fig. 3) showed that the phages belonged to the Siphoviridae family, characterized by isometric, nonenveloped, 54-nm-diameter heads and filamentous cross-banded tails with short terminal fibers. Three morphotypes were defined on the basis of tail length (34): morphotypes A (108 nm ± 10 nm), B (145 nm ± 10 nm), and C (215 nm ± 10 nm). Eight of the 36 phages observed were of morphotype A (22%), 17 were of morphotype B (47%), and 11 were of morphotype C (31%) (Fig. 3).
FIG. 3.
Electron micrograph of S. agalactiae phages. Three morphotypes were found (A, B, and C).
Molecular characterization of phage DNA. (i) REA.
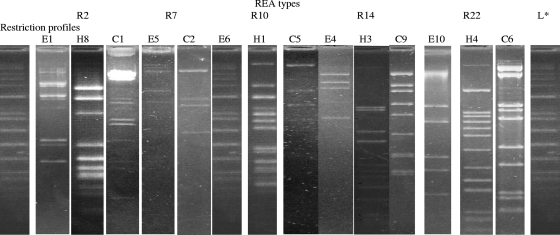
DNA extracted from the 36 induced phage preparations was digested with EcoRI, HindIII, and ClaI. EcoRI restriction provided 12 profiles (E1 to E12), HindIII restriction provided 12 profiles (H1 to H12), and ClaI restriction provided 13 profiles (C1 to C13). Restriction profiles were not obtained in seven cases. Twenty-three REA types were recovered by combining the REA profiles obtained with the three enzymes. Five of these REA types contained more than one phage (Fig. 4).
FIG. 4.
EcoRI, HindIII, and ClaI profiles of the five major REA types observed for phages from 36 phages suspensions. L*, Raoul ladder (MWRAU300; Q-Biogene, Strasbourg, France).
(ii) Multilocus phage characterization.
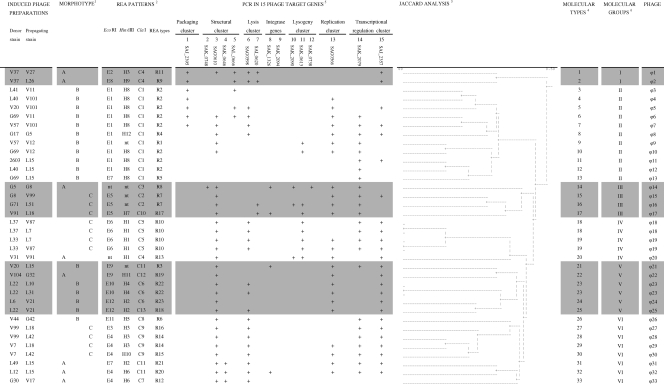
In cases of positive PCR amplification, the amplicons obtained with primer pairs targeting the 15 studied phage gene sequences were all of the expected sizes. The selected primers gave a high rate of amplification of phage gene sequences from the transcriptional regulation cluster (33/36, or 92%), the replication cluster (24/36, or 67%), the host lysis cluster (23/36, or 64%), and the structural cluster (32/36, or 89%) (Fig. 5). Amplification with the two primer pairs targeting gene sequences from the transcriptional regulation cluster resulted in the identification of four different genetic patterns. The two primer pairs targeting gene sequences in the host lysis cluster identified four genetic patterns, and the four primer pairs targeting gene sequences in the structural cluster identified six genetic patterns. By contrast, the selected primers gave low rates of amplification for sequences from the lysogeny cluster (10/36, or 28%; four genetic patterns), the integrase genes (4/36, or 11%; two genetic patterns), and the packaging cluster (7/36, or 19%) (Fig. 5).
FIG. 5.
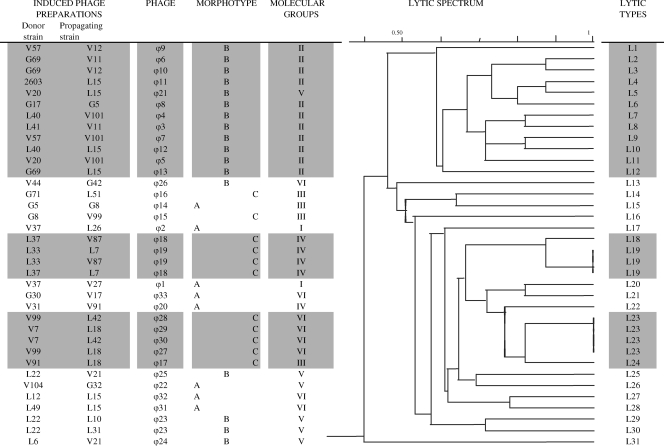
Characteristics of 36 S. agalactiae phages. Morphotypes were defined as a function of tail length (1). REA types were obtained by combining the restriction profiles obtained with the three restriction enzymes (2). Jaccard analysis shows a dendrogram of similarity values for the molecular characteristics of S. agalactiae phages, combining REA type and results for the 15 prophage sequences (3). Molecular types were obtained by combining REA type and results for the 15 prophage sequences (4). PCR results with screening for the 15 prophage sequences found in published S. agalactiae genome sequences are shown (5). Molecular groups were defined at a level of 42% dissimilarity (6).
Using the 15 PCR targeting phage genes, we obtained 25 different genetic patterns for the 36 phage suspensions studied.
(iii) Genetic relationship between phages.
Hierarchical analysis using the molecular phage characteristics established by REA and multilocus characterization was used to estimate the genetic relationship between the isolated phages (Fig. 5, dendrogram). Among the 36 induced phage preparations, 33 phages displayed genetic dissimilarity (ϕ1 to ϕ33) (Fig. 5) but clustered in six distantly related molecular phage groups (I to VI), with a level of 42% dissimilarity (Fig. 5). Each molecular phage group was characterized by a specific phage morphotype (Fig. 5). Phage group I clustered two phages (ϕ1 and ϕ2) of morphotype A; phage group II clustered 11 phages (ϕ3 to ϕ13) of morphotype B. Phage groups III and IV contained mostly (five of seven phages) morphotype phages C (ϕ15 to ϕ19), and phage group V contained mostly (four of five phages) morphotype B phages (ϕ21 and ϕ23 to ϕ25). Phage group VI contained two major lineages, one of morphotype C (ϕ27 to ϕ30) and the other of morphotype A (ϕ31 to ϕ33). Thus, morphotype A phages were found in five of the six phage molecular groups and may therefore be considered the most diverse genetically.
Lytic activities of S. agalactiae phages.
We observed 340 lytic reactions in tests of the lytic activity of the 36 induced phage preparations against the collection of 114 S. agalactiae strains. Hierarchical analysis of the lytic reactions observed for each phage distinguished 31 different lytic types (Fig. 6, dendrogram); 30 of these 31 lytic types were distributed in two major branches of the dendrogram, including 12 in branch A and 18 in branch B (Fig. 6). The diversity of phage groups, as defined by their genetic characteristics and morphotypes, was therefore correlated with their lytic activity. The 11 molecular group II/morphotype B phages had diverse lytic types (L1 to L12) (Fig. 6), but all theses lytic types were related and clustered in major branch A of the dendrogram, suggesting similarities in terms of the host cell targets recognized or lytic mechanisms involved. The molecular group IV/morphotype C phages (ϕ18 and 19) had lytic activities L18 and L19, displaying 87% similarity (Fig. 6, branch C), and the four molecular group VI/morphotype C phages (ϕ27 to ϕ30) had similar activities of the L23 type (Fig. 6, branch D), indicating strong similarities in terms of the host cell targets recognized or the lytic mechanisms involved. By contrast, the four molecular group V/morphotype B phages (ϕ21, the pair ϕ23 and ϕ24, and 25) and the three molecular group III/morphotype C phages (ϕ15 to ϕ17) had diverse lytic activities. The group of phages of morphotype A (ϕ1, ϕ2, ϕ14, ϕ20, ϕ22, and ϕ31 to 33), which was the most genetically diverse, also displayed the highest degree of diversity in lytic activities. Nevertheless, the lytic types generated by these morphotype A phages were all related and located in branch B of the dendrogram (Fig. 6).
FIG. 6.
Lytic activities of S. agalactiae phages. The dendrogram representing the lytic spectrum was generated by the Jaccard dichotomy coefficient method using the lytic reactions against the 114 S. agalactiae strains. Phages were defined by their morphotypes and molecular characteristics (molecular groups). Thirty-three lytic types were found.
Characteristics of donor and propagating strains.
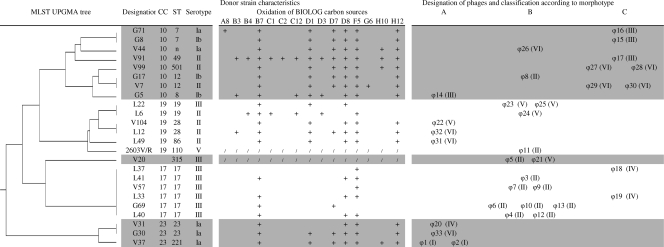
Determination of the serotypes and phylogenetic origins of the 24 donor and 21 propagating strains by MLST indicated that the distribution of strains within serotypes and within the S. agalactiae lineages of the studied population—CC10, CC17, CC19, and CC23—did not differ between donor and propagating strains (Fig. 7 and 8). By contrast, studies of catabolic functions showed a loss of catabolic function in the lysogenic strains of CC17 and CC19. Indeed, all five CC19 donor strains (100%) and only one of the six CC19 propagating strains (17%) showed no decay of alpha-d-glucose-1 phosphate substrate (H10) (P = 0.013). All six CC17 donor strains (100%) and only one of the three CC17 propagating strains (33%) showed no decay of dl-alpha-glycerol phosphate substrate (H12) (P = 0.083) (Fig. 7 and 8). All nine CC17 strains (100%) showed no decay of the d-ribose substrate (D7), whereas this was the case for only a few of the strains in the other CCs (3/27, or 11%; P < 0.001).
FIG. 7.
Phylogenetic origin, molecular characteristics, and catabolic functions of donor strains with morphotypes and molecular groups of phages induced by strains of each major phylogenetic lineage (CC). The molecular groups of each phage are specified in parentheses. UPGMA, unweighted-pair group method with arithmetic mean. A8, Tween 40; B3, arbutin; B4, d-cellobiose; B7, d-galactose; C1, alpha-d-lactose; C2, lactulose; C12, alpha-methyl-d-glucoside; D1, beta-methyl-d-glucoside; D3, palatinose; D7, d-ribose; D8, salicine; F5, l-malic acid; G6, l-glutamic acid; H10, alpha-d-glucose-1-phosphate; H12, dl-alpha-glycerol-phosphate; C11, 3-methyl-glucose.
FIG. 8.
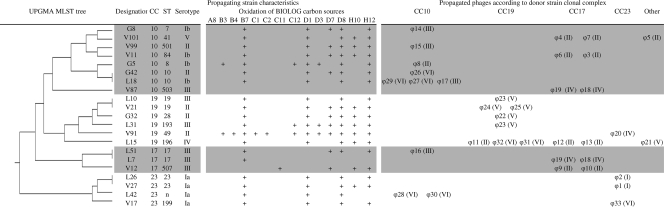
Phylogenetic origin, molecular characteristics and catabolic functions of propagating strains. The phages propagated by strains of the major phylogenetic lineage (CC) of S. agalactiae species were reported as a function of the phylogenetic position of donor strains. For each phage, the molecular group is specified in parentheses. UPGMA, unweighted-pair group method with arithmetic mean. A8, Tween 40; B3, arbutin; B4, d-cellobiose; B7, d-galactose; C1, alpha-d-lactose; C2, lactulose; C12, alpha-methyl-d-glucoside; D1, beta-methyl-d-glucoside; D3, palatinose; D7, d-ribose; D8, salicine; F5, l-malic acid; G6, l-glutamic acid; H10, alpha-d-glucose-1-phosphate; H12, dl-alpha-glycerol-phosphate; C11, 3-methyl-glucose.
Characteristics of phages as a function of the phylogenetic position of donor and propagating strains.
Isolated phages were phylogenetic lineage specific within S. agalactiae (Fig. 7). Indeed, the lysogenic CC23 donor strains provided phages only of the genetically diverse morphotype A group. CC17 strains produced phages of molecular group II/morphotype B or of molecular group IV/morphotype C. In addition, phages of these two groups were rarely supplied by strains of phylogenetic lineages other than CC17: only 2 of the 11 molecular group II/morphotype B phages and none of the molecular group IV/morphotype C phages were produced by strains of other lineages. The phages of molecular group V appeared to be more specifically responsible for the lysogeny of CC19 strains, given that only one of the five phages (ϕ21) of this molecular group was generated by a non-CC19 strain. Similarly, the phages of molecular group III and of molecular group VI/morphotype C appear to be more specifically able to integrate into CC10 strain genomes as all the phages displaying these characteristics were produced by strains of this phylogenetic lineage.
Phages obtained from lysogenic strains of each of the S. agalactiae phylogenetic lineages other than CC17 were able to infect and induce a viral lytic cycle more specifically in bacteria of a similar evolutionary origin (Fig. 8). Indeed, 7 of the 10 phages isolated from CC10 donor strains (70%) were propagated by CC10 strains. All of the phages isolated from CC19 donor strains (100%) were propagated by CC19 strains, and three of the four phages isolated from CC23 donor strains (75%) were propagated by CC23 strains. By contrast, the phages isolated from CC17 donor strains (eight molecular group II/morphotype B phages and two molecular group IV/morphotype C) were able to mediate lytic replication in bacterial hosts of each of the phylogenetic lineages of S. agalactiae other than CC23.
DISCUSSION
Phages have made an important contribution to bacterial evolution but have been little studied in the S. agalactiae species. S. agalactiae phages of bovine origin, assigned to the Siphoviridae family, were first isolated in 1969 (37), and a phage-typing method was proposed for investigations related to S. agalactiae infections in the early 1980s (45). Prophage sequences are abundant in the sequenced genome of S. agalactiae (47, 48), but little is known about the functionality of S. agalactiae bacteriophages. The mitomycin C prophage induction observed in S. agalactiae strains of four major lineages (CC10, CC17, CC19, and CC23) responsible for genital and neonatal colonization and infections indicates that S. agalactiae serves as a host for several temperate phages. Serotype V strains of CC1, which emerged in adults in the 1990s and in neonates more recently (5, 26), were not included in our collection, and their study is required. Although most of the donor strains (23/24) (Fig. 7) provided phages of the same molecular group and morphotype, polylysogeny could not be excluded in S. agalactiae species. Indeed, the method used here identified only intact phages, with defective phages or remnants entirely ignored.
The classical methods of phage characterization (11) and the result of our in silico analysis suggested that the phages integrated into the S. agalactiae genome were diverse. We further estimated the level of genetic diversity of the induced S. agalactiae phages by developing a multilocus typing system for characterization. For cellular organisms, genetic diversity and genetic relationships between individuals can be deduced by studying universally conserved genes. Phage genomes contain no ubiquitously present sequences and display high levels of recombination. It is therefore not possible to apply this technique to phage sequences although a similar approach, using loci corresponding to key modules of phages involved in infection and propagation, could be developed (8, 9, 35). For example, a PCR-based multilocus typing scheme was recently developed for studies of the genetic diversity of Shiga toxin-encoding bacteriophages (44). We developed a PCR-based multilocus typing method targeting key modules within the packaging cluster, the structural cluster, the host lysis cluster, the integrase genes, the lysogeny cluster, the replication cluster, and the transcriptional regulation cluster to provide an indication of the level of genetic diversity and relationship between phages induced within S. agalactiae species.
Comparison of the data obtained with classical and molecular methods for analyzing S. agalactiae phages led to the identification of molecular groups of phages, with specific morphotype features and lytic activities, each with its own ability to infect, replicate within, or integrate into host cells from well-defined intraspecies phylogenetic lineages (Fig. 7 and 8). Over the last 60 years, a marked change has occurred in the habitat of S. agalactiae, from predominantly bovine infections to several ecological niches in the environment, animals, and humans, with each colonizing population displaying specific features with respect to the various S. agalactiae lineages (23, 29, 39, 41, 50). As shown for other species (38), such changes in habitat expose bacteria to a wide range of environmental and nutritive constraints, imposing stressful conditions that lead to the induction of mutation and contribute to adaptive evolution. Our data suggest that these stress-induced conditions concomitantly expose the bacteria, which share their new habitat with specifically adapted bacteriophages, to different phage-mediated horizontal gene transfers.
Phages originating from the genetically homogeneous CC17 (ST-17) presented a wide spectrum of lytic activities against strains of all lineages (except CC23), whereas—as shown previously for a phage isolated from a CC19 strain (32)—phages originating from CC19, CC10, and CC23 presented a narrow range of lytic activity, principally against strains of the same CC (Fig. 8) (32). In recent years, S. agalactiae ST-17 strains have emerged as a major cause of S. agalactiae early-onset diseases (24). Our findings raise questions concerning the role of the lysogenic features observed here for ST-17 strains in the emergence of this pathogen. As suggested by experiments with cocultures of Salmonella strains carrying or lacking specific prophages (6), prophages may modify the competitive fitness of host strains through their ability to grow and, for a small proportion of the population, to annihilate rival bacteria through cell lysis. Phages active against isolates other than the strain causing disease were released from most of the bacterial isolates from septicemic patients, suggesting that the prophages present in sepsis-causing bacterial clones play a role in clonal selection during bacterial invasion (15). Given the particular ability of the phages of ST-17 strains to destroy extraclonal strains, such phenomena may confer a selective advantage for vaginal colonization or for invasive disease in neonates.
We found that lysogenic S. agalactiae strains of CC17 and CC19 had lost some catabolic functions (Fig. 7 and 8). Several studies in other species have demonstrated that genetic events causing a loss of catabolic function frequently induce changes in the expression of virulence (8, 12, 13, 28, 31, 43). In many species, lysogenic phages affect virulence because they carry genes encoding toxins, such as diphtheria toxin in Corynebacterium diphtheriae (10), Shiga toxin and type III secretion effectors in enterohemorrhagic Escherichia coli (42), staphylococcal enterotoxin A and Panton-Valentine leukocidin in Staphylococcus aureus (2, 22), streptococcal pyrogenic exotoxins (20), and cholera toxin in Vibrio cholerae (51). No such virulence factor associated with phages has been found in S. agalactiae CC17 and CC19, but lysogeny may affect the virulence of these two highly virulent clones by modifying bacterial fitness. For example, in vitro lysogenization of pneumococcal strains enhances bacterial adhesion to inert surfaces and pharyngeal cells (25). Localization of insertion sites of phages in the S. agalactiae genome and a greater understanding of temperate phage morons, particularly in phages isolated from ST-17 strains, may provide insight into why these strains are more likely than others to invade the central nervous system of neonates.
Acknowledgments
This study was supported by l'Université François-Rabelais and le Centre Hospitalier Universitaire of Tours, France.
N.V.D.M-M., A.S.D., and R.Q. conceived and designed the experiments. A.S.D. and N.V.D.M-M. performed isolation, propagation, and characterization of phages; A.S.D. and P.Y.S. performed electron microscopy analysis; and G.H.A., M.F.L., L.M. (MLST), A.S.D., N.V.D.M.-M., and R.Q. performed experiments for serotyping and metabolic properties of strains. A.S.D., N.V.D.M.-M., and R.Q. analyzed the data and wrote the paper.
Footnotes
Published ahead of print on 22 May 2009.
REFERENCES
- 1.Band, J. D., H. W. Clegg II, P. S. Hayes, R. R. Facklam, J. Stringer, and R. E. Dixon. 1981. Transmission of group B streptococci. Traced by use of multiple epidemiologic markers. Am. J. Dis. Child. 135355-358. [DOI] [PubMed] [Google Scholar]
- 2.Betley, M. J. J., and J. J. Mekalanos. 1985. Staphylococcal enterotoxin A is encoded by phage. Science 229185-187. [DOI] [PubMed] [Google Scholar]
- 3.Bidet, P., N. Brahimi, C. Chalas, Y. Aujard, and E. Bingen. 2003. Molecular characterization of serotype III group B-Streptococcus isolates causing neonatal meningitis. J. Infect. Dis. 1881132-1137. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg, H. M., D. S. Stephens, M. Modansky, M. Erwin, J. Elliot, R. R. Facklam, A. Schuchat, W. Baughman, and M. M. Farley. 1996. Invasive group B streptococcal disease: the emergence of serotype V. J. Infect. Dis. 173365-373. [DOI] [PubMed] [Google Scholar]
- 5.Bohnsack, J. F., A. Whiting, M. Gottschalk, D. M. Dunn, R. Weiss, P. H. Azimi, J. B. Philips III, L. E. Weisman, G. G. Rhoads, and F-Y. C. Lin. 2008. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. academic centers from 1995 to 1999. J. Clin. Microbiol. 461285-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossi, J., J. A. Fuentes, G. Mora, and N. Figueroa-Bossi. 2003. Prophage contribution to bacterial population dynamics. J. Bacteriol. 1856467-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, K. M., L. C. Vogel, S. P. Gotoff, C. A. Gadzala, J. Stringer, and W. R. Maxted. 1980. Nosocomial transmission of bacteriophage type 7/11/12 group B streptococci in a special care nursery. Am. J. Dis. Child. 134964-966. [DOI] [PubMed] [Google Scholar]
- 8.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomics rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Büchen-Osmond, C. 2006. Index to ICTVdb virus descriptions. ICTVdb Management, Mailman School of Public Health, Columbia University, New York, NY.
- 10.Buck, G. A., and N. B. Groman. 1981. Identification of deoxyribonucleic acid restriction fragments of beta-converting corynebacteriophages that carry the gene for diphtheria toxin. J. Bacteriol. 148153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canchaya, C. A., M. Ventura, and D. van Sinderen. 2007. Bacteriophage bioinformatics and genomics, p. 43-59. In S. McGrath and D. van Sinderen (ed.), Bacteriophage genetics and molecular biology. Caister Academic Press, Norfolk, United Kingdom.
- 12.Day, W. A., R. E. Fernandez, and A. T. Maurelli. 2001. Pathoadaptative mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect. Immun. 697471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domelier, A. S., N. van der Mee-Marquet, A. Grandet, L. Mereghetti, A. Rosenau, and R. Quentin. 2006. Loss of catabolic function in Streptococcus agalactiae strains and its association with neonatal meningitis. J. Clin. Microbiol. 443245-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farley, M. M., R. C. Harvey, T. Stull, J. D. Smith, A. Schuchat, J. D. Wenger, and D. S. Stephens. 1993. A population-based assessment of invasive disease due to group B Streptococcus in nonpregnant adults. N. Engl. J. Med. 3281807-1811. [DOI] [PubMed] [Google Scholar]
- 15.Gaidelyte, A., M. Vaara, and D. H. Bamford. 2007. Bacteria, phages and septicemia. PLoS ONE 2e1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Madsek, M. Zouine, E. Couvé, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 451499-1523. [DOI] [PubMed] [Google Scholar]
- 17.Granlund, M., F. Michel, and M. Norgren. 2001. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol. 1832560-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA. 962192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Committee on Taxonomy of Viruses. 1995. Virus taxonomy, 6th report of the International Committee on Taxonomy of Viruses. Arch. Virol. Suppl. 101-586. [PubMed] [Google Scholar]
- 20.Johnson, L. P., and P. M. Schlievert. 1983. A physical map of the group A streptococcal pyrogenic exotoxin bacteriophage T12 genome. Mol. Gen. Genet. 189251-255. [DOI] [PubMed] [Google Scholar]
- 21.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B Streptococcus. J. Clin. Microbiol. 412530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko, J., T. Kimura, S. Narita, T. Tomita, and Y. Kamio. 1998. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene 21557-67. [DOI] [PubMed] [Google Scholar]
- 23.Keefe, G. P., I. R. Dohoo, and E. Spangler. 1997. Herd prevalence and incidence of Streptococcus agalactiae in the dairy industry of Prince Edward Island. J. Dairy Sci. 80464-470. [DOI] [PubMed] [Google Scholar]
- 24.Lin, F-Y. C., A. Whiting, E. Adderson, S. Takahashi, D. M. Dunn, R. Weiss, P. H. Azimi, J. B. Philips III, L. E. Weisman, J. Regan, P. Clark, G. G. Rhoads, C. E. Frasch, J. Troendle, P. Moyer, and J. F. Bohnsack. 2006. Phylogenetic lineages of invasive and colonizing strains of serotype III group B streptococci from neonates: a multicenter prospective study. J. Clin. Microbiol. 441257-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeffler, J. M., and V. A. Fischetti. 2006. Lysogeny of Streptococcus pneumoniae with MM1 phage: improved adherence and other phenotypic changes. Infect. Immun. 744486-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manning, S. D., A. C. Springman, E. Lehotzky, M. A. Lewis, T. S. Whittam, and H. D. Davies. 2009. Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 471143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquet-Van der Mee, N., and A. Audurier. 1995. Proposals for optimization of the international phage typing system for Listeria monocytogenes: combined analysis of phage lytic spectrum and variability of typing results. Appl. Environ. Microbiol. 61303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 953943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel, C., C. Pelletier, M. Boussaha, D. G. Douet, A. Lautraite, and P. Tailliez. 2007. Diversity of lactic acid bacteria associated with fish and the fish farm environment, established by amplified rRNA gene restriction analysis. Appl. Env. Microbiol. 732947-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 864731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallen, M. J., and B. W. Wren. 2007. Bacterial pathogenomics. Nature 449835-842. [DOI] [PubMed] [Google Scholar]
- 32.Pritchard, D. G., S. Dong, J. R. Baker, and J. A. Engler. 2004. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology 1502079-2087. [DOI] [PubMed] [Google Scholar]
- 33.Quentin, R., H. Huet, F. S. Wang, P. Geslin, A. Goudeau, and R. K. Selander. 1995. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J. Clin. Microbiol. 332576-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quiberoni, A., D. Guglielmotti, A. Binetti, and J. Reinheimer. 2004. Characterization of three Lactobacillus delbrueckii subsp. bulgaricus phages and the physicochemical analysis of phage adsorption. J. Appl. Microbiol. 96340-351. [DOI] [PubMed] [Google Scholar]
- 35.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 1844529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolland, K., C. Marois, V. Siquier, B. Cattier, and R. Quentin. 1999. Genetic features of Streptococcus agalactiae strains causing neonatal infections, as revealed by pulsed-field gel electrophoresis and hylB gene analysis. J. Clin. Microbiol. 371892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell, H., N. L. Norcross, and D. E. Kahn. 1969. Isolation and characterization of Streptococcus agalactiae bacteriophage. J. Gen. Virol. 5315-317. [DOI] [PubMed] [Google Scholar]
- 38.Saint-Ruf, C., and I. Matic. 2006. Environmental tuning of mutation rates. Environ. Microbiol. 8193-199. [DOI] [PubMed] [Google Scholar]
- 39.Schrag, S. J. 2004. The past and future of perinatal group B streptococcal disease prevention. Clin. Infect. Dis. 391136-1138. [DOI] [PubMed] [Google Scholar]
- 40.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuchat, A. 2001. Group B streptococcal disease: from trials and tribulations to triumph and trepidation. Clin. Infect. Dis. 33751-756. [DOI] [PubMed] [Google Scholar]
- 42.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 1853596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shelburne, S. A., D. Keith, N. Horstmann, P. Sumby, M. T. Davenport, E. A. Graviss, R. G. Brennan, and J. M. Musser. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 1051698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, D. L., B. M. Wareing, P. C. Fogg, L. M. Riley, M. Spencer, M. J. Cox, J. R. Saunders, A. J. McCarthy, and H. E. Allison. 2007. Multilocus characterization scheme for Shiga toxin-encoding bacteriophages. Appl. Environ. Microbiol. 738032-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stringer, J. 1980. The development of a phage-typing system for group-B streptococci. J. Med. Microbiol. 13133-143. [DOI] [PubMed] [Google Scholar]
- 46.Stringer, J., and W. R. Maxted. 1979. Phage typing of group B streptococci. Lancet 10328. [DOI] [PubMed] [Google Scholar]
- 47.Tettelin, H., V. Masignani, M. J. Cieslewicz, C. Donati, D. Medini, N. L. Ward, S. V. Angiuoli, J. Crabtree, A. L. Jones, A. S. Durkin, R. T. Deboy, T. M. Davidsen, M. Mora, M. Scarselli, I. Margarity Ros, J. D. Peterson, C. R. Hauser, J. P. Sundaram, W. C. Nelson, R. Madupu, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, S. A. Sullivan, S. C. Daugherty, D. H. Haft, J. Selengut, M. L. Gwinn, L. Zhou, N. Zafar, H. Khouri, D. Radune, G. Dimitrov, K. Watkins, K. J. O'Connor, S. Smith, T. R. Utterback, O. White, C. E. Rubens, G. Grandi, L. C. Madoff, D. L. Kasper, J. L. Telford, M. R. Wessels, R. Rappuoli, and C. M. Fraser. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. USA 10213950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 9912391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Mee-Marquet, N., A. S. Domelier, L. Mereghetti, P. Lanotte, A. Rosenau, W. van Leeuwen, and R. Quentin. 2006. Prophagic DNA fragments in Streptococcus agalactiae strains and association with neonatal meningitis. J. Clin. Microbiol. 441049-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Mee-Marquet, N., L. Fourny, L. Arnault, A. S. Domelier, M. Salloum, M. F. Lartigue, and R. Quentin. 2008. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin and genital body sites. J. Clin. Microbiol. 462906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 2721910-1914. [DOI] [PubMed] [Google Scholar]