Abstract
Kingella kingae is a member of the Neisseriaceae and is being recognized increasingly as an important cause of serious disease in children. Recent work has demonstrated that K. kingae expresses type IV pili that mediate adherence to respiratory epithelial and synovial cells and are selected against during invasive disease. In the current study, we examined the genome of K. kingae strain 269-492 and identified homologs of the rpoN and the pilS and pilR genes that are essential for pilus expression in Pseudomonas aeruginosa but not in the pathogenic Neisseria species. The disruption of either rpoN or pilR in K. kingae resulted in a marked reduction in the level of transcript for the major pilus subunit (pilA1) and eliminated piliation. In contrast, the disruption of pilS resulted in only partial reduction in the level of pilA1 transcript and a partial decrease in piliation. Furthermore, the disruption of pilS in colony variants with high-density piliation resulted in variants with low-density piliation. Mutations in the promoter region of pilA1 and gel shift analysis demonstrated that both σ54 and PilR act directly at the pilA1 promoter, with PilR binding to two repetitive elements. These data suggest that the regulation of K. kingae type IV pilus expression is complex and multilayered, influenced by both the genetic state and environmental cues.
Kingella kingae is a gram-negative bacterium that belongs to the Neisseriaceae family. Improvements in diagnostics have led to the increased recognition of K. kingae as an important cause of a number of pediatric diseases, including septic arthritis, osteomyelitis, and endocarditis (10, 27, 32, 37, 39). Several studies have shown that K. kingae is a leading etiology of septic arthritis and osteomyelitis, with one study reporting that K. kingae accounts for the majority of osteoarticular infections in children younger than 36 months of age (5, 10, 27, 32, 38).
K. kingae is believed to initiate infection by colonizing the pharynx, a conclusion supported by the isolation of the same strain of K. kingae from both the respiratory tract and the blood of individuals with invasive disease (40). Following colonization, the organism breaches the epithelium, potentially by means of an RTX toxin (19), and then enters the blood and disseminates to deeper tissues, such as bones and joints. Recent work has shown that K. kingae expresses type IV pili that are essential for mediating adherence to respiratory epithelial and synovial cells (18), presumably facilitating the colonization of the respiratory tract and the seeding of joints. These fibers have been demonstrated previously to vary in density, with a correlation between the number of pili and the colony type (8, 13). Variants that express high levels of pili have a spreading/corroding colony type, while variants expressing low levels of pili have a nonspreading/noncorroding colony type (8). Interestingly, the likelihood of any level of piliation is much lower in invasive isolates than in respiratory tract isolates of K. kingae, suggesting a loss of piliation during the pathogenesis of invasive disease (T. E. Kehl-Fie et al., unpublished data).
Given the difference in piliation between respiratory tract isolates and invasive isolates, we set out to develop a better understanding of the factors controlling pilus expression. In this work, we provide evidence that the expression of K. kingae type IV pili is regulated by σ54 and by a two-component regulatory system with significant homology to the PilR/PilS system involved in Pseudomonas aeruginosa pilus regulation (3, 14, 16). The loss of σ54 or PilR resulted in the complete elimination of pilus expression, while the loss of PilS resulted in only a partial reduction in piliation. The introduction of the pilS mutation into variants with the spreading/corroding colony type converted them to the nonspreading/noncorroding colony type. The analysis of the pilA1 promoter revealed a σ54 binding domain that is upstream of the transcriptional start site and is required for pilus expression. Additional work established that PilR binds to two repetitive elements in the pilA1 promoter.
MATERIALS AND METHODS
Bacterial strains and eukaryotic cell lines.
Strains used in this study are listed in Table 1. K. kingae strains were stored at −80°C in brain heart infusion broth with 30% glycerol. Escherichia coli strains were stored at −80°C in Luria-Bertani (LB) broth with 15% glycerol. K. kingae strains were grown at 37°C with 5% CO2 on TSA II chocolate agar (BD Biosciences, San Jose, CA) supplemented with 50 μg/ml kanamycin as appropriate. E. coli strains were grown in LB broth or on LB agar at 37°C supplemented with 50 μg/ml kanamycin or 100 μg/ml ampicillin as appropriate. Chang cells (ATCC CCL-20.2; human conjunctiva) and Hig-82 cells (ATCC CRL-1832; rabbit synovial cells) were cultured at 37°C with 5% CO2 as previously described (19).
TABLE 1.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| 269-492 | K. kingae isolate from St. Louis Children's Hospital | 19 |
| KK01 | Nonspreading/noncorroding variant of strain 269-492 | 19 |
| KK03 | Spreading/corroding variant of strain 269-492 | 19 |
| 269-492 pilF | Strain 269-492 with an aphA3 insertion in pilF | 19 |
| 269-492 pilA1 | Strain 269-492 with an aphA3 insertion in pilA1 | 18 |
| 269-492 recJ | Strain 269-492 with an aphA3 insertion in recJ | 18 |
| KK01 recJ | Strain KK01with an aphA3 insertion in recJ | This work |
| KK03 recJ | Strain KK03 with an aphA3 insertion in recJ | This work |
| 269-492 rpoN | Strain 269-492 with an aphA3 insertion in rpoN | This work |
| 269-492 Δ−12 | Strain 269-492 with an aphA3 insertion in recJ and replacement of the σ54 −12 DNA GC binding region with AA | This work |
| 269-492 pilS | Strain 269-492 with an aphA3 insertion in pilS | This work |
| KK01 pilS | Strain KK01 with an aphA3 insertion in pilS | This work |
| KK03 pilS | Strain KK03 with an aphA3 insertion in pilS | This work |
| 269-492 pilR | Strain 269-492 with an aphA3 insertion in pilR | This work |
| KK01 pilR | Strain KK01 with an aphA3 insertion in pilR | This work |
| KK03 pilR | Strain KK03 with an aphA3 insertion in pilR | This work |
| 269-492/ACA#1− | Strain 269-492 with an aphA3 insertion in recJ and replacement of ACA#1 repetitive element with random DNA | This work |
| KK01 ACA#1− | Strain KK01 with an aphA3 insertion in recJ and replacement of ACA#1 repetitive element with random DNA | This work |
| KK03 ACA#1− | Strain KK03 with an aphA3 insertion in recJ and replacement of ACA#1 repetitive element with random DNA | This work |
| 269-492/ACA#2− | Strain 269-492 with an aphA3 insertion in recJ and replacement of ACA#2 repetitive element with random DNA | This work |
| KK01 ACA#2− | Strain KK01 with an aphA3 insertion in recJ and replacement of ACA#2 repetitive element with random DNA | This work |
| KK03 ACA#2− | Strain KK03 with an aphA3 insertion in recJ and replacement of ACA#2 repetitive element with random DNA | This work |
| 269-492/ACA#1−/ACA#2− | Strain 269-492 with an aphA3 insertion in recJ and replacement of ACA#1/ACA#2 repetitive elements with random DNA | This work |
| KK01 ACA#1−/ACA#2− | Strain KK01 with an aphA3 insertion in recJ and replacement of ACA#1/ACA#2 repetitive elements with random DNA | This work |
| KK03 ACA#1−/ACA#2− | Strain KK03 with an aphA3 insertion in recJ and replacement of ACA#1/ACA#2 repetitive elements with random DNA | This work |
| DH5α | E. coli F− φ80dlacZΔM15 Δ(lacZYA-argF)U169deoRrecA1endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 | 29 |
| DH5α MalC2 | DH5α containing pMalC2, which expresses MBP | This work |
| DH5α MAL-PilR | DH5α containing pMalC2:pilR, which expresses MBP-PilR | This work |
| DH5α pilA1:ΔSS | DH5α containing pHAT10/pilA1:ΔSS, which expresses HAT-PilA1ΔSS | This work |
Generation of K. kingae mutants.
To create gene disruptions in K. kingae, the relevant gene was cloned into pUC19 and then interrupted with an antibiotic cassette. The disrupted gene then was transformed into K. kingae by natural competence (19), and transformants were selected for by plating on agar supplemented with the appropriate antibiotic. To disrupt pilS, a fragment containing pilS was amplified from K. kingae strain 269-492 by PCR using the primers PilSk/ofwd and PilSk/orev and ligated into BamHI-digested pUC19, creating pUC19/pilS. The aphA3 cassette was released from pFalcon2 (12) by digestion with PuvII and was ligated into SpeI-digested/T4 polymerase-treated pUC19/pilS, creating pUC19/pilS:kan. To disrupt pilR, a fragment containing pilR was amplified using the primers pilRRev and pilRFwd and was ligated into EcoRI/SalI-digested pUC19, creating pUC19/pilR. An MluI site was introduced into pUC19/pilR using PCR primers pilRk/oFwd and pilRk/oRev, generating pUC19/pilR:MluI. The aphA3 cassette was released from pFalcon2 and ligated into MluI-digested pUC19/pilR:MluI, creating pUC19/pilR:kan. The K. kingae rpoN::kan mutant was isolated in a screen of a mariner transposon library for nonadherent mutants (T. E. Kehl-Fie, unpublished data).
To create mutations in the promoter region of pilA1, the plasmid pPRBK was created (18), containing pilA1 and the upstream recJ gene interrupted by the aphA3 cassette. To replace the σ54 −12 binding region with nonhomologous sequence, the QuikChange II XL kit (Stratagene, La Jolla, CA) and PCR primers ΔpilA −12 fwd and ΔpilA −12 rev were used, creating plasmid pPRBK:Δ-12. Following transformation into K. kingae, the presence of the mutation was confirmed by DNA sequencing. To replace the ACA#1 and ACA#2 repetitive elements with random DNA, pPRBK and derivatives were amplified with primers ΔACA#1Fwd and ΔACA#1Rev or primers ΔACA#2Fwd and ΔACA#2Rev, producing linear fragments lacking the ACA repeats and containing either a BglII site (ACA#1) or an SpeI site (ACA#2). The products then were treated with DpnI to remove parental DNA and digested with either BglII (ACA#1) or SpeI (ACA#2). The digested fragments were circularized by ligation, creating pPRBK:ΔACA#1, pPRBK:ΔACA#2, and pPRBK:ΔACA#1/#2. After transformation into K. kingae, the ΔACA#1, ΔACA#2, and ΔACA#1/#2 mutations were confirmed by PCR and restriction digestion.
Generation of anti-PilA1 antibody.
To purify PilA1 for antibody production, an N-terminal His affinity tag (HAT) fusion was created. A fragment containing pilA1 lacking the coding region for the predicted signal sequence was amplified by PCR from K. kingae strain 269-492 using primers pilinFΔSS and PilinR. The pilA1 fragment was ligated into BamHI/EcoRI-digested pHAT10, creating pHAT10/pilA1:ΔSS. The HAT-PilA1ΔSS fusion was purified by affinity chromatography using Talon resin (Clontech, Mountain View, CA) and then was resolved on an sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and excised. Antiserum GP65 was raised against HAT-PilA1ΔSS in guinea pigs (Cocalico Biologicals, Reamstown, PA).
Purification of type IV pili.
K. kingae pili were purified using a modification of the method described for the purification of Eikenella corrodens type IV pili (15). K. kingae strain 269-492 was grown for 18 h on 20 TSA II chocolate agar plates, and bacterial growth was scraped from the plates and suspended in 20 ml of 150 mM ethanolamine, pH 10.5 (EA buffer). Pili were sheared from the bacterial surface by subjecting the bacterial suspension to a handheld homogenizer for 2 min. To remove bacterial cells and debris, the suspension was centrifuged for 10 min at 10,000 × g and then for 30 min at 10,000 × g. The resulting supernatant was subjected to precipitation with 10% ammonium sulfate. The precipitated pili were resuspended in EA buffer to 1/10 of the original volume and then dialyzed overnight in 2 liters of EA buffer. The suspension again was subjected to precipitation with 10% ammonium sulfate, and the purified pili were examined by SDS-PAGE and by transmission electron microscopy.
Analysis of adherence, pilus expression, pilin production, and transcript levels.
The adherence of K. kingae to respiratory epithelial and synovial cells was assessed using quantitative adherence assays. Briefly, bacteria were incubated for 17 to 18 h overnight on chocolate agar and resuspended in brain heart infusion broth to an optical density at 600 nm of 0.8. Approximately 6.5 × 106 CFU were inoculated onto fixed confluent cell monolayers in 24-well plates. Monolayers were fixed with 2% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.4, for 2 h at 4°C with gentle rocking and then were washed three times with 1× Tris-buffered saline and replenished with fresh tissue culture media. Following the inoculation of bacteria, the 24-well plates were centrifuged for 5 min at 1,000 rpm and then were incubated for 25 min at 37°C. Monolayers were washed four times with phosphate-buffered saline (PBS) to remove nonadherent bacteria and then treated with 1× trypsin-EDTA (Sigma-Aldrich, St. Louis, MO) for 20 min at 37°C to release adherent bacteria. Appropriate dilutions were prepared and spread on agar plates, and the percent adherence was determined by dividing the number of adherent CFU by the number of CFU in the inoculum. Each sample was assayed in triplicate.
To assess PilA1 production, K. kingae strains were grown for 17 to 18 h on chocolate agar and resuspended to an optical density at 600 nm of 0.8 and then analyzed by Western blotting using antiserum GP65. To confirm equal loadings and uniform transfer, all membranes were stained with Ponceau S prior to Western blotting. The surface expression of pili was assessed by negative staining transmission electron microscopy as described previously (19), with the following modifications. Bacteria were resuspended in 0.2 M ammonium acetate, pH 7.4, instead of PBS, and the grids were not washed before examination using a Philips CM-12 electron microscope (FEI, Hillsboro, OR).
To perform quantitative real-time PCR, RNA and cDNA were prepared. Briefly, RNA was extracted using TRIreagent (Sigma, St. Louis, MO) and the RNeasy minikit by following the lipid-rich tissue protocol (Qiagen, Valencia, CA). Residual DNA was removed with RQ1 DNase (Fisher Scientific, Pittsburgh, PA), which was inactivated prior to generating cDNA with random hexamers and Superscript II (Invitrogen, Carlsbad, CA). The primer sets used for quantitative real-time PCR are listed in Table 2.
TABLE 2.
Primers used in this study
| Name | Sequencea |
|---|---|
| PilSk/orev | GCATGGATCCGTCGTTGGCTTGGCGCATGGC |
| PilSk/ofwd | GCATGGATCCGCAGCCTGCAACATTTGTTAGCCG |
| pilRRev | ACGTGTCGACCATATTGATTATTCAATATTCAAGCGTTCC |
| pilRFwd | ACGTGAATTCATGACCAAACACAGTTTGAATCGCC |
| pilRk/oFwd | ACGTACGCGTGCGAAAGCGGTTCGGGTAAAGAG |
| pilRk/oRev | ACGTACGCGTCCGAAATATAGACAGGCACAACGGC |
| ΔpilA-12fwd | CAAATGGCATGCACTCTAATACCAAGTAAGGCAAGTAACACAGC |
| ΔpilA-12rev | GCTGTGTTACTTGCCTTACTTGGTATTAGAGTGCATGCCATTTG |
| ΔACA#1Fwd | ATCGTGAGATCTGTGACTGAGGTCTTGGCATGCACTCTGCTACCAAGTAAGGCAAGTAAC |
| ΔACA#1Rev | ATCGTGAGATCTGTGGATCGGCTAGAATGGTATTTTATTAAACTGCCTTTTTTTGTCAGTCGCAC |
| ΔACA#2Fwd | ATCGTGACTAGTGCACTGCGTCGGCGGCACGTTGATAAGCGAATTGCACCCAAATTG |
| ΔACA#1Rev | ATCGTGACTAGTGGTCTAGCGTCGAAAATAGCGCGCTGATTGAAGCCTGCAATCTG |
| SRLFwd | ATGCATGGTACCAGAAAGCGAATTGAACGCGTTGC |
| SRLRev | ATGCATGTCGACGCTTTCACGTCTTCGGGCAGC |
| pilRDSBF | ATCGGGATCCTTTTTGATTTGGGTATAAAAGGGTATGCCGTAGG |
| pilRDSBR | ATCGGGATCCTTTGGACAAAAGGGCAAAAAGCAGCC |
| PilinFΔSS | TACGGGATCCAGATTACACCAAACGTTCACGCGTATCTGA |
| PilinR | TACGGAATTCTTAGCCACCATTGGTAGCGGTAGTAGTGCC |
| pilA1pdr-6fam | FAM-CTCAGTCAAAGCACTTTTCACG |
| pilA1pdr | CTCAGTCAAAGCACTTTTCACG |
| pilA1pdf | GAGCAGGGAACAGATTGCAGG |
| Pilin Region Fwd | ACGTGAATTCAAGCGCGTATGCCGTGCGAC |
| Pilin Region S3 | GCCGATTATGATGCCGATGGC |
| pilA1 RT-left | CAAACGTTCACGCGTATCTG |
| pilA1 RT-right | GGCCATGCATTGTTAGAAGC |
| ftsZ RT-left | CCAGAGCGAACCAAAGTCTC |
| ftsZ RT-right | AAGCTATACTCGCCCTGCTG |
| pillR RT-left | GTTGATGTCCGCATTGTGTC |
| pilR RT-right | GGCGATAGAACAGGTCTTGG |
| moxR RT-left | TTTATGTTGGCGGACGAAAT |
| moxR RT-right | CGTCCACCGATACTTGACCT |
| raiA-right | GCGGTTACCTTGTCGGTAGA |
| raiA-left | TCTTCCACTGCTTCAACGTG |
FAM, 6-carboxyfluorescein.
To perform primer runoff analysis, cDNA was created using the primer pilA1pdr-6fam, which anneals at the 5′ end of pilA1, and either Superscript II (Invitrogen, Carlsbad, CA) or Transcriptor (Roche Applied Sciences, Indianapolis, IN). Samples were electrophoresed on an Applied Biosystems 3100 genetic analyzer and analyzed using Applied Biosystems Genescan v3.7.1 software (Applied Biosystems, Foster City, CA).
Gel shift.
To assess the binding of PilR to the pilA1 promoter, we employed a maltose-binding protein-PilR (MBP-PilR) fusion protein and an ∼500-bp fragment containing the pilA1 promoter. To generate the MBP-PilR protein, the pilR gene was amplified from strain 269-492 using the primers pilRRev and pilRFwd and was ligated into EcoRI/SalI-digested pMalC2 (New England Biolabs, Ipswich, MA), creating pMalC2:pilR. To express MBP-PilR or MBP, overnight cultures were back diluted 1/50 into fresh LB and grown at 37°C for 2 h and then induced with 0.1 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for an additional 3 h. Bacteria were harvested by centrifugation, and the pellet then was resuspended in suspension buffer (20 mM HEPES, pH 7.5, 0.1 M NaCl, 1 mM EDTA) and lysed via sonication. Following sonication, the lysate was clarified by centrifugation and then incubated overnight at 4°C with amylose resin. The resin was washed four times with 10 bed volumes of suspension buffer. Protein was eluted from the resin in suspension buffer supplemented with 10 mM maltose. The pilA1 promoter was amplified from either pPRBK or pPRBK containing the ACA mutations using the primers pilA1pdf and either pilA1pdr or pilA1pdr-6fam. A nonspecific competitor fragment was amplified using the primers Pilin Region Fwd and Pilin Region S3. Binding reactions were performed as described previously (28), with the exception that the volume was increased to 30 μl. Samples were separated on a 4% native polyacrylamide gel prepared with 0.5× Tris-borate-EDTA (TBE) and were visualized using a Typhoon 9410 (GE Bioscience, Piscataway, NJ).
Transmission electron microscopy.
Piliated bacteria and dilutions of purified pili were examined by negative staining transmission electron microscopy using uranyl acetate.
MS analysis.
The gel slice corresponding to purified pili was subjected to in-gel trypsin digestion (the detailed protocol is at http://www.genome.duke.edu/cores/proteomics/sample-preparation/), followed by liquid chromatography-mass spectrometry-mass spectrometry (LC-MS/MS) analysis using a nanoAcquity liquid chromatograph (Waters Corp) coupled to an LTQ-Orbitrap XL (Thermo Scientific). The top three most intense multiply charged ions were interrogated by tandem MS with product ion detection in the Orbitrap. Raw data were processed using Mascot Distiller v2.0 and were searched using the Mascot v2.2 search engine against K. kingae protein sequences, with 10 ppm precursor and a 0.02-Da product ion tolerance.
Nucleotide sequence accession numbers.
The pilSR and rpoN loci were deposited in GenBank under accession numbers EU930332 and EU930331, respectively. The accession numbers for Pseudomonas aeruginosa PilR, PilS, and σ54 are AAP81269, NP_253236.1, and NP_253152.1, respectively.
RESULTS
Confirmation that Kingella kingae surface fibers are type IV pili.
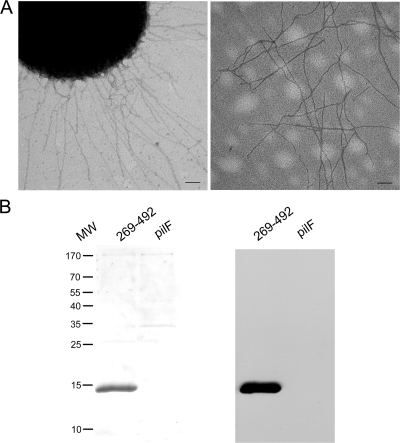
In initial experiments, we set out to confirm that K. kingae surface fibers are type IV pili and contain PilA1. Fibers were sheared from the surface of K. kingae strain 269-492 and then were precipitated with ammonium sulfate. K. kingae strain 269-492 pilF was used as a control and was subjected to the same protocol, taking advantage of the fact that this strain makes PilA1 but is unable to assemble surface fibers (PilA1 accumulates in the cell) (18). As shown in Fig. 1A, the examination of the purified material from strain 269-492 by negative staining transmission electron microscopy revealed abundant fibers that were identical to the fibers on the surface of whole bacteria. As shown in Fig. 1B, the examination of the purified fibers by SDS-PAGE revealed a major band ∼15 kDa in size, in agreement with the predicted molecular mass of PilA1, and examination by Western analysis with antiserum GP65 directed against PilA1 demonstrated strong reactivity with the ∼15-kDa band. The 15-kDa band was excised from the gel and subjected to LC-MS/MS, revealing the sequence for PilA1 and confirming that the surface fibers are type IV pili.
FIG. 1.
Evidence that K. kingae surface fibers are type IV pili. (A) Fibers on the surface of K. kingae strain 269-492 (left) and the preparation of purified pili (right) after staining with uranyl acetate and examining by transmission electron microscopy. Scale bars represent 100 nM. (B) Coomassie blue-stained SDS-PAGE gel of purified pili (left) and a Western blot of purified pili (right) using the antiserum GP65 directed against PilA. Strain K. kingae 269-492pilF (pilF) is unable to assemble pili and served as a control. MW, molecular size in kilodaltons.
σ54 regulates pilA1 transcription.
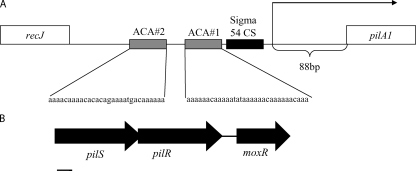
To develop a better understanding of the regulation of K. kingae type IV pili, we began by analyzing the promoter region of pilA1 (the gene encoding the major pilus subunit) (18) (Fig. 2). Western blotting and quantitative real-time PCR demonstrated that spreading/corroding and nonspreading/noncorroding colony types express pilA1 at different levels (Fig. 3A and B). The sequencing of the region between recJ and pilA1 revealed no differences between a spontaneous stable spreading/corroding colony variant of K. kingae strain 269-492 (strain KK03) and a spontaneous stable nonspreading/noncorroding colony variant of K. kingae strain 269-492 (strain KK01) (19), arguing that the pilA1 promoter region does not differ between spreading/corroding and nonspreading/noncorroding colony types. Primer runoff analysis indicated that the pilA1 transcriptional start site is 88 bp upstream of the putative pilA1 start codon in strains 269-492, KK03, and KK01 (Fig. 2). These data suggest that the conversion between spreading/corroding and nonspreading/noncorroding colony types is mediated by changes outside the pilA1 promoter region.
FIG. 2.
Diagram of the pilA1 promoter region and the PilS and PilR region. (A) pilA1 promoter region and the locations of the transcriptional start site, σ54 binding region, and ACA#1 and ACA#2 repeats. (B) pilS/pilR locus. The bar equals 200 bp.
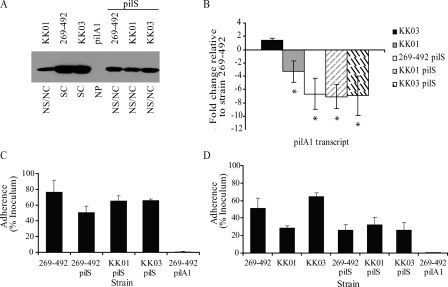
FIG. 3.
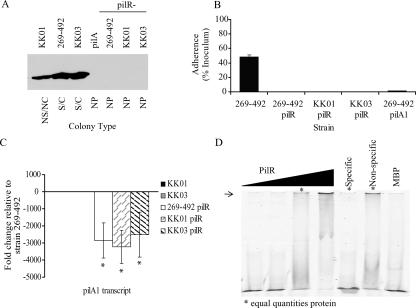
Contribution of PilS to K. kingae PilA1 expression and adherence. (A) Western blot for PilA1 in K. kingae strains KK01, 269-492, KK03, 269-492 pilA1, 269-492 pilS, KK01 pilS, and KK03 pilS. NS/NC, nonspreading/noncorroding; SC, spreading/corroding; NP, nonpiliated. (B) Levels of pilA1 transcript measured by quantitative real-time PCR in K. kingae strains KK03, KK01, 269-492 pilS, KK01 pilS, and KK03 pilS relative to that of strain 269-492 (*, P ≤ 0.05 using the unpaired t test compared to results for the parental strain). (C) Adherence to Chang respiratory epithelial cells by K. kingae strains 269-492, 269-492 pilS, KK01 pilS, KK03 pilS, and 269-492 pilA1. (D) Adherence to Hig-82 synovial cells by K. kingae strains 269-492, KK01, KK03, 269-492 pilS, KK01 pilS, KK03 pilS, and 269-492 pilA1.
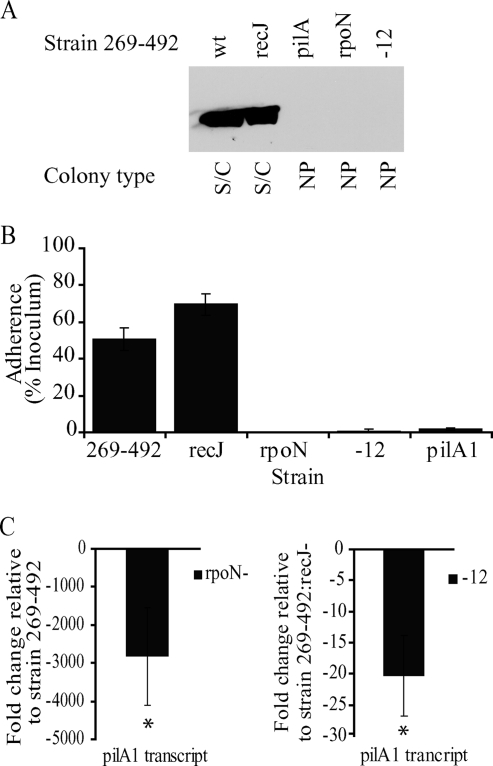
The examination of the pilA1 promoter region revealed the minimal consensus sequence for the alternative sigma factor σ54 (CAAATGGCATGCACTCTGCTACCAAGTA) (underlining indicates the conserved −12 and −24 bases) (1) upstream of the transcriptional start site (Fig. 1), and a review of the K. kingae genome (Kehl-Fie et al., unpublished) identified a gene that encodes a homolog of σ54, a gene that we have designated rpoN by analogy to other species. K. kingae σ54 is 45% similar and 30% identical to σ54 from Pseudomonas aeruginosa. To assess whether rpoN contributes to K. kingae type IV pilus expression or is a pseudogene, as it is in Neisseria species (20), we disrupted rpoN in strain 269-492 and assessed the resulting mutant for pilus expression and adherence. The examination of a whole-cell lysate of the rpoN mutant by Western blotting revealed no PilA1 (Fig. 4A). The colony morphology of the rpoN mutant changed from the spreading/corroding morphology of the parent strain to the morphology observed with nonpiliated clinical isolates (data not shown). Consistent with this observation, the rpoN mutant had no pili when examined by negative staining electron microscopy (data not shown) and was nonadherent in assays with Chang cells (Fig. 4B), similar to K. kingae strain 269-492 pilA1, a nonpiliated control (18). The further analysis of the rpoN mutant revealed that the level of pilA1 transcript was approximately 1/3,000 that of the parent strain (Fig. 4C). As a control, we performed quantitative real-time PCR on the gene downstream of rpoN and found no effect of the insertion in rpoN, arguing against a polar effect of the insertion (data not shown). Taken together, these data suggest that pilA1 expression is regulated by σ54.
FIG. 4.
Contribution of σ54 to K. kingae PilA1 expression and adherence. (A) Western blot for PilA1 in K. kingae strains 269-492, 269-492 recJ (recJ), 269-492 pilA1 (pilA1), 269-492 rpoN (rpoN), and 269-492 Δ−12 (-12). wt, wild type; S/C, spreading/corroding; NP, nonpiliated. (B) Adherence to Chang respiratory epithelial cells by K. kingae strains 269-492, 269-492 recJ, 269-492 rpoN, 269-492 Δ−12, and 269-492 pilA1 (pilA1). (C) Levels of pilA1 transcript measured by quantitative real-time PCR in K. kingae strains 269-492 rpoN and 269-492 Δ−12 relative to those of strains 269-492 and 269-492 recJ, respectively (*, P ≤ 0.05 using the unpaired t test compared to results for the parental strain).
To assess whether σ54 acts directly at the pilA1 promoter, the σ54 minimal consensus sequence was mutated by converting the GC nucleotides at the −12 position to AA, generating strain Δ−12. The characterization of the Δ−12 mutant revealed no detectable PilA1 by Western blotting (Fig. 4A), no visible pili by electron microscopy (data not shown), and no adherence to Chang cells (Fig. 4B). As shown in Fig. 4C, the Δ−12 mutant also had reduced levels of pilA1 transcript. These data suggest that σ54 acts directly on the pilA1 promoter.
PilS and PilR regulate pilA1 transcription.
In other organisms that express type IV pili and utilize σ54 to regulate pilin expression, regulation also requires a two-component regulatory system consisting of PilS, the sensor kinase, and PilR, the response regulator (14, 16, 22, 28, 36). Along these lines, the examination of the K. kingae genome identified tandem genes encoding proteins with homology to PilS and PilR (Fig. 2). K. kingae PilS is 42% similar and 26% identical to PilS from Pseudomonas aeruginosa and contains a conserved histidine kinase domain (residues 321 to 388) and a conserved ATP binding domain (residues 435 to 535). Interestingly, while the P. aeruginosa PilR protein and most PilR homologs are approximately 50 kDa in size, the K. kingae PilR protein has a predicted molecular mass of 61 kDa. The difference in molecular mass is due to an insertion in K. kingae PilR (residues 138 to 228) between the conserved receiver domain (residues 11 to 123, including a predicted phosphorylation site) and the conserved σ54-interacting domain (residues 231 to 399). Excluding the insertion between the receiver domain and the σ54-interacting domain, K. kingae PilR and P. aeruginosa PilR are 74% similar and 44% identical. To assess the roles of the PilS and PilR homologs in K. kingae pilus expression, we disrupted pilS and pilR in K. kingae strains 269-492, KK03, and KK01. Real-time quantitative PCR confirmed that the pilS disruption did not affect pilR or moxR (downstream of pilR) transcript levels, and that the pilR disruption did not affect moxR transcript levels (data not shown).
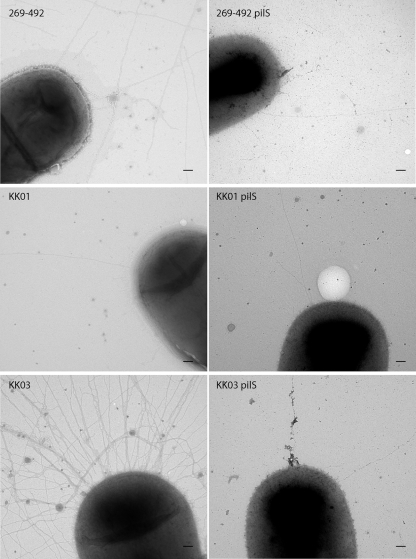
As shown in Fig. 3A, the examination of the pilS mutants by Western blotting revealed a decrease in the levels of PilA1 in the 269-492 and the KK03 derivatives comparable to the level of PilA1 in KK01. When assessed for adherence, the pilS mutants adhered to Chang respiratory epithelial cells at levels comparable to those of wild-type strain 269-492 (Fig. 3C). Previous work established that the level of adherence to synovial cells correlates with the density of piliation (18). To assess whether the reduction in PilA1 correlated with a reduction in piliation, we examined the pilS mutants for adherence to Hig-82 synovial cells. As shown in Fig. 3D, adherence by the three pilS mutants was comparable to adherence by KK01 (a nonspreading/noncorroding colony type). Consistent with this result, when the three pilS mutants were examined by electron microscopy, all three had levels of pili that were comparable to the levels in strain KK01 (Fig. 5). Interestingly, the mutation of pilS in strains 269-492 and KK03 also resulted in a shift from a spreading/corroding colony type to a nonspreading/noncorroding colony type (data not shown). As shown in Fig. 3B, all three pilS mutants had levels of pilA1 transcript that were slightly lower than the levels in KK01. These data suggest that PilS enhances PilA1 expression but is not absolutely required.
FIG. 5.
Contribution of PilS to K. kingae piliation. Negative staining transmission electron microscopy of K. kingae strains 269-492, KK01, KK03, 269-492 pilS, KK01 pilS, and KK03 pilS. The scale bar represents 100 nM.
As shown in Fig. 6A, the examination of the pilR mutants by Western blotting revealed no detectable PilA1. In all three strain backgrounds, the colony morphology of the pilR mutants shifted to the morphology observed with nonpiliated clinical isolates. Consistent with these results, the pilR mutants were nonpiliated when examined by negative staining electron microscopy (data not shown) and were nonadherent in assays with Chang cells (Fig. 6B). Further analysis revealed that the level of pilA1 transcript was approximately 1/3,000 that of strain 269-492 in all three mutants (Fig. 6C). Taken together, these data suggest that PilR is required for pilA1 transcription.
FIG. 6.
Contribution of PilR to K. kingae PilA1 expression and adherence. (A) Western blot for PilA1 in K. kingae strains KK01, 269-492, KK03, 269-492 pilA1 (pilA1), 269-492 pilR, KK01 pilR, and KK03 pilR. NS/NC, nonspreading/noncorroding; S/C, spreading/corroding; NP, nonpiliated. (B) Adherence to Chang respiratory epithelial cells by K. kingae strains 269-492, 269-492 pilR, KK01 pilR, KK03 pilR, and 269-492 pilA1. (C) Levels of pilA1 transcript measured by quantitative real-time PCR in K. kingae strains KK01, KK03, 269-492 pilR, KK01 pilR, and KK03 pilR relative to those of strain 269-492 (*, P ≤ 0.05 using the unpaired t test compared to results for parental strain). (D) Gel shifts with MBP-PilR. Lanes 1 to 4 contain increasing quantities of MBP-PilR (0, 0.3, 3, and 30 μg) incubated with 50 ng of labeled probe. Lanes 5 and 6 show 3 μg of MBP-PilR incubated with 50 ng of labeled probe and either 400× specific competitor (specific) or 400× nonspecific competitor (nonspecific). Lane 7 shows 30 μg of MPB with 50 ng of labeled probe. The arrow highlights the shifted DNA band at the top of the gel, indicating interaction with PilR.
To assess if PilR acts directly on the pilA1 promoter, PilR was expressed as an MBP fusion protein, and gel shift assays with the pilA1 promoter region were performed. As shown in Fig. 6D, PilR retarded the migration of an ∼500-bp fragment containing the intergenic region between pilA1 and recJ, supporting the conclusion that PilR acts at the pilA1 promoter and regulates pilA1 transcription.
Two repetitive elements facilitate binding of PilR to the pilA1 promoter.
Based on work in P. aeruginosa, a 5′-(N)4-6C/GTGTC-3 motif has been suggested to be a PilR binding sequence. This motif also is present upstream of the major pilin gene in other organisms that have been suggested to regulate pilus expression using the PilS/PilR two-component response regulator (11, 17). Interestingly, the examination of the K. kingae pilA1 promoter region revealed no evidence of the 5′-(N)4-6C/GTGTC-3 motif. However, as shown in Fig. 2, the pilA1 upstream sequence contains repetitive elements that we have designated ACA#1 and ACA#2 and are located 117 and 228 bp upstream of the predicted pilA1 start site. These repetitive elements contain three to five degenerate repetitive units consisting of 5′-(A)1-6C(A)1-6-3′ and are upstream of the σ54 DNA binding region. To assess whether these repetitive units contribute to pilA1 transcription, we replaced them individually and in combination with random DNA in K. kingae strains 269-492, KK01, and KK03.
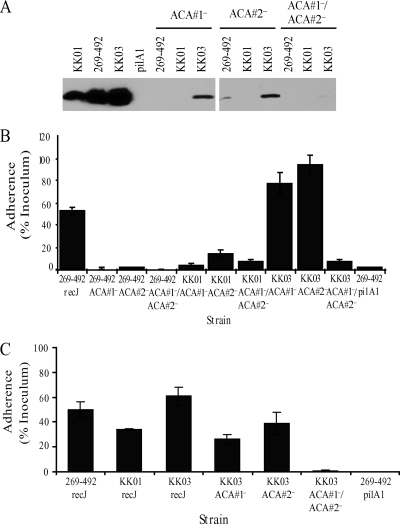
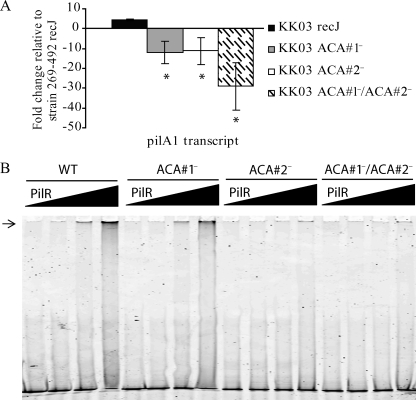
When the ACA mutants were examined by Western blotting, all were found to have substantially decreased levels of PilA1 (Fig. 7A). As show in Fig. 7B, in strains 269-492 and KK01, the loss of either ACA#1 or ACA#2 resulted in a substantial reduction in adherence to Chang epithelial cells. In contrast, in strain KK03 a reduction in adherence was observed only when both the ACA#1 and the ACA#2 repeats were replaced. To assess whether the decreased level of PilA1 correlated with a decrease in piliation in the KK03 ACA#1 and ACA#2 single mutants, we examined these mutants for adherence to Hig-82 cells and by electron microscopy. As shown in Fig. 7C, the KK03 ACA#1 and ACA#2 single mutants adhered at levels comparable to adherence by KK01 and decreased compared to adherence by the KK03 parent, suggesting a reduction in piliation. Consistent with the diminished adherence to Hig-82 cells, the examination of the KK03 ACA single mutants by electron microscopy revealed a reduction in the number of pili to levels slightly lower than those in strain KK01 (data not shown). The examination of the 269-492 and KK01 ACA#1 and ACA#2 single mutants and the 269-492, KK01, and KK03 ACA#1/ACA#2 double mutants revealed a majority of bacteria with no pili and rare bacteria with short pili (data not shown), in agreement with the low levels of PilA1 that were detected by Western blotting. The analysis of pilA1 transcript levels revealed decreased pilA1 transcript in all three KK03 ACA mutants, with the greatest decrease observed in the KK03 ACA double mutant (Fig. 8A). Similar results were obtained when the 269-492 and KK01 ACA mutants were examined for the level of pilA1 transcript (data not shown). These data suggest that both ACA repeats contribute to pilA1 transcription.
FIG. 7.
Contribution of ACA repetitive elements to K. kingae adherence. (A) Western blot for PilA1 in K. kingae strains KK01, 269-492, KK03, 269-492 pilA1 (pilA1), 269-492 ACA#1−, KK01 ACA#1−, KK03 ACA#1−, 269-492 ACA#2−, KK01 ACA#2−, KK03 ACA#2−, 269-492 ACA#1−/ACA#2−, KK01 ACA#1−/ACA#2−, and KK03 ACA#1−/ACA#2−. (B) Adherence to Chang respiratory epithelial cells by K. kingae strains 269-492 recJ, 269-492 ACA#1−, 269- 492 ACA#2−, 269-492 ACA#1−/ACA#2−, KK01 ACA#1−, KK01 ACA#2−, KK01 ACA#1−/ACA#2−, KK03 ACA#1−, KK03 ACA#2−, KK03 ACA#1−/ACA#2−, and 269-492 pilA1. (C) Adherence to Hig-82 synovial cells by K. kingae strains 269-492 recJ, KK01 recJ, KK03 recJ, KK03 ACA#1−, KK03 ACA#2−, KK03 ACA#1−/ACA#2−, and 269-492 pilA1.
FIG. 8.
Contribution of ACA repetitive elements to K. kingae pilus expression. (A) Levels of pilA1 transcript measured by quantitative real-time PCR in K. kingae strains KK03 recJ, KK03 ACA#1−, KK03 ACA#2−, and KK03 ACA#1−/ACA#2− relative to the levels for strain 269-492 recJ (*, P ≤ 0.05 using the unpaired t test compared to results for the parental strain). (B) Gel shifts with increasing concentrations of MBP-PilR (0, 0.03, 0.3, and 3 μg) with 50 ng of labeled wild-type probe (WT) or probe lacking repetitive element ACA#1, repetitive element ACA#2, or both ACA#1 and ACA#2. Lanes 1 to 4 contain wild-type probe. Lanes 5 to 8 contain probe lacking the ACA#1 element. Lanes 9 to 12 contain probe lacking the ACA#2 element. Lanes 13 to 16 contain probe lacking both the ACA#1 and ACA#2 elements. The arrow highlights the shifted DNA band at the top of the gel, indicating interaction with PilR.
Given the absence of the conserved PilR DNA binding sequence found in P. aeruginosa and other species upstream of pilA1, we examined whether the ACA repeats contribute to the binding of PilR to the pilA1 promoter. To perform this analysis, a 500-bp fragment corresponding to the native pilA1 promoter lacking ACA#1, ACA#2, or both ACA#1 and ACA#2 repetitive elements was amplified and assessed by gel shift. As shown in Fig. 8B, the loss of either ACA element resulted in a decreased affinity of PilR for the pilA1 promoter. Interestingly, the loss of the ACA#1 repetitive element resulted in only a small decrease in affinity. These results suggest that the ACA repetitive elements contribute to the expression of PilA1 by promoting PilR binding to the pilA1 promoter.
DISCUSSION
K. kingae is being recognized increasingly as a common cause of septic arthritis, bacteremia, and osteomyelitis in young children. Previous work demonstrated that type IV pili are essential for K. kingae adherence to respiratory epithelial cells (16) and generally are absent from isolates recovered from joint fluid or bone samples (Kehl-Fie et al., unpublished). To develop a better understanding of the clinically relevant transition between piliated and nonpiliated organisms, we undertook studies to identify the factors controlling pilus expression. Initial observations demonstrated that the highly piliated spreading/corroding colony type expresses PilA1 at higher levels than the sparsely piliated nonspreading/noncorroding colony type. Additional experiments established that the variation in piliation between spreading/corroding and nonspreading/noncorroding colony variants (KK01 and KK03,respectively) is not due to changes in the intergenic region between pilA1 and recJ. Further analysis revealed that PilA1 expression is regulated by the alternative sigma factor σ54 and the PilS/PilR two-component regulatory system, with both σ54 and PilR acting at the pilA1 promoter and being necessary for wild-type levels of pilA1 transcript. The involvement of σ54 and the PilS/PilR two-component system in K. kingae type IV pilus expression suggests that both genetic elements and environmental signals influence K. kingae piliation.
The analysis of the K. kingae genome identified genes encoding homologs of σ54, PilS, and PilR, which have been shown to be essential for type IV pilus expression in P. aeruginosa and a variety of other organisms (14, 16, 22, 28, 36). In contrast to P. aeruginosa, Neisseria gonorrhoeae and Neisseria meningitidis contain only remnants of the rpoN, pilS, and pilR genes and use σ70 to drive pilus expression (4, 9, 20). In our studies, we found that the disruption of rpoN and pilR in K. kingae resulted in the complete loss of PilA1 production, piliation, and adherence, indicating that σ54 and the PilS/PilR two-component regulatory system are required for K. kingae pilus expression. In addition, disruptions of rpoN and pilR resulted in a colony morphology that was similar to the colony morphology of nonpiliated clinical isolates, regardless of the colony morphology of the parental strain. Primer runoff analysis demonstrated that spreading/corroding and nonspreading/noncorroding colony types have the same transcriptional start site located 88 bp upstream of the predicted pilA1 start codon. The mutation of a conserved minimal σ54 DNA binding element (1) resulted in a reduction in PilA1 levels, the loss of piliation, and the loss of adherence, suggesting that σ54 acts directly at the pilA1 promoter.
The examination of the pilA1 promoter revealed no evidence of the motif corresponding to the reported DNA binding domain of P. aeruginosa PilR (17). Instead, the pilA1 promoter contains two ACA repetitive elements that are necessary for the efficient binding of PilR and the expression of pili. While the mutation of both ACA repetitive elements resulted in undetectable levels of PilA1 in the 269-492 and KK01 backgrounds and barely detectable levels in the KK03 background, as well as substantially reduced pilA1 transcript levels in all three backgrounds, the reduction in pilA1 transcript was not as great as that observed in the pilR mutants. Despite the fact that our gel shift assays demonstrated no clear PilR binding to the pilA1 promoter lacking both ACA#1 and ACA#2, we speculate that there is residual PilR binding to the mutated pilA1 promoter in whole bacteria. Interestingly, 27 additional ACA repetitive elements are scattered throughout the genome, suggesting that the K. kingae PilS/PilR system regulates additional factors involved in pathogenesis. Of note, the Dichelobacter nodosus PilS/PilR two-component system regulates type IV pili and several non-pilus genes, including an RTX-like gene (28).
In contrast to our findings with σ54 and PilR, the loss of PilS resulted in only a partial reduction in pilA1 transcript level, piliation, and adherence. Interestingly, when the pilS mutation was introduced into strain 269-492 (a strain with an intermediate number of pili), strain KK03 (an isogenic variant with a high number of pili), and strain KK01 (an isogenic variant with a low number of pili), the level of pilA1 transcript was reduced to the same level in all cases. In strains 269-492 and KK03, the pilS mutation resulted in a shift from spreading/corroding colonies to nonspreading/noncorroding colonies. These data suggest that spontaneous mutation in pilS is one mechanism that is responsible for the switch from spreading/corroding colonies to nonspreading/noncorroding colonies.
The transcription of pilA1 when PilS is lacking raises several interesting questions regarding the activity of PilR and suggests similarities to other systems. In Helicobacter pylori, the response regulator Hp166 has been shown to have activity in the absence of the cognate sensor kinase Hp165. The binding of Hp166 to essential genes in the absence of Hp165 and canonical phosphorylation has been suggested to be due to a higher affinity of Hp166 for promoters in front of essential genes than nonessential genes, allowing transcription in the absence of phosphorylation (2, 6, 7, 26, 30). In P. aeruginosa, the AlgR response regulator is capable of activating a subset of the genes it regulates in the absence of its cognate sensor, FimS, and canonical phosphorylation (24, 34). Similarly, the P. aeruginosa AlgB response regulator has been shown to have activity in the absence of phosphorylation (21, 24, 35). While K. kingae PilR clearly retains activity in the absence of PilS, it is not clear whether this activity is phosphorylation dependent or phosphorylation independent. If the residual PilR activity is phosphorylation dependent, another histidine kinase could be serving as a phosphodonor (31, 33), or PilR could be phosphorylated by a high-energy small molecule, such as acetyl phosphate (23, 25). If the residual K. kingae PilR activity is phosphorylation independent, the mechanism responsible for the activity is less apparent. Regardless of whether the residual activity of K. kingae PilR in the absence of PilS is phosphorylation dependent or phosphorylation independent, it is intriguing to speculate that the 91-amino-acid insertion between the receiver domain and the σ54 binding domain is responsible for the residual activity.
In this study, we demonstrate that the expression of K. kingae pilA1 is regulated by σ54 and a two-component response regulator. Our results indicate that K. kingae type IV pilus expression is complex and multilayered and involves both genetic and biochemical components, suggesting that K. kingae combines aspects of regulation from both P. aeruginosa and the pathogenic Neisseria species. The further study of K. kingae type IV pilus regulation provides an opportunity to develop a better understanding of a presumed key virulence factor and may lead to the discovery of additional factors that are involved in pathogenesis. Furthermore, the continued study of K. kingae PilS and PilR may provide insights into the growing number of species known to contain two-component response regulators with atypical activity, including H. pylori and P. aeruginosa.
Acknowledgments
This work was supported by NIH training grant T32-GM07067 to T.E.K.-F.
We thank the Duke Proteomics Core Facility and staff for assistance with mass spectrometry.
Footnotes
Published ahead of print on 22 May 2009.
REFERENCES
- 1.Barrios, H., B. Valderrama, and E. Morett. 1999. Compilation and analysis of sigma54-dependent promoter sequences. Nucleic Acids Res. 274305-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 1822068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, J. M., T. Koga, and S. Lory. 1994. Identification and characterization of PilS, an essential regulator of pilin expression in Pseudomonas aeruginosa. Mol. Gen. Genet. 243565-574. [DOI] [PubMed] [Google Scholar]
- 4.Carrick, C. S., J. A. Fyfe, and J. K. Davies. 2000. The genome of Neisseria gonorrhoeae retains the remnants of a two-component regulatory system that once controlled piliation. FEMS Microbiol. Lett. 186197-201. [DOI] [PubMed] [Google Scholar]
- 5.Chometon, S., Y. Benito, M. Chaker, S. Boisset, C. Ploton, J. Berard, F. Vandenesch, and A. M. Freydiere. 2007. Specific real-time polymerase chain reaction places Kingella kingae as the most common cause of osteoarticular infections in young children. Pediatr. Infect. Dis. J. 26377-381. [DOI] [PubMed] [Google Scholar]
- 6.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsyth, M. H., P. Cao, P. P. Garcia, J. D. Hall, and T. L. Cover. 2002. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. J. Bacteriol. 1844630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Froholm, L. O., and K. Bovre. 1972. Fimbriation associated with the spreading-corroding colony type in Moraxella kingii. Acta Pathol Microbiol. Scand. B Microbiol. Immunol. 80641-648. [PubMed] [Google Scholar]
- 9.Fyfe, J. A., C. S. Carrick, and J. K. Davies. 1995. The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a σ70 promoter during growth in vitro. J. Bacteriol. 1773781-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gené, A., J. J. Garcia-Garcia, P. Sala, M. Sierra, and R. Huguet. 2004. Enhanced culture detection of Kingella kingae, a pathogen of increasing clinical importance in pediatrics. Pediatr. Infect. Dis. J. 23886-888. [DOI] [PubMed] [Google Scholar]
- 11.Heinrich, D. W., and A. C. Glasgow. 1997. Transcriptional regulation of type 4 pilin genes and the site-specific recombinase gene, piv, in Moraxella lacunata and Moraxella bovis. J. Bacteriol. 1797298-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40214-224. [DOI] [PubMed] [Google Scholar]
- 13.Henriksen, S. D. 1969. Corroding bacteria from the respiratory tract. 1. Moraxella kingii. Acta Pathol. Microbiol. Scand. 7585-90. [PubMed] [Google Scholar]
- 14.Hobbs, M., E. S. Collie, P. D. Free, S. P. Livingston, and J. S. Mattick. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 7669-682. [DOI] [PubMed] [Google Scholar]
- 15.Hood, B. L., and R. Hirschberg. 1995. Purification and characterization of Eikenella corrodens type IV pilin. Infect. Immun. 633693-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishimoto, K. S., and S. Lory. 1992. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J. Bacteriol. 1743514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, S., K. S. Ishimoto, and S. Lory. 1994. PilR, a transcriptional regulator of piliation in Pseudomonas aeruginosa, binds to a cis-acting sequence upstream of the pilin gene promoter. Mol. Microbiol. 141049-1057. [DOI] [PubMed] [Google Scholar]
- 18.Kehl-Fie, T. E., S. E. Miller, and J. W. St. Geme III. 2008. Kingella kingae expresses type IV pili that mediate adherence to respiratory epithelial and synovial cells. J. Bacteriol. 1907157-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehl-Fie, T. E., and J. W. St. Geme III. 2007. Identification and characterization of an RTX toxin in the emerging pathogen Kingella kingae. J. Bacteriol. 189430-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskos, L., J. P. Dillard, H. S. Seifert, J. A. Fyfe, and J. K. Davies. 1998. The pathogenic neisseriae contain an inactive rpoN gene and do not utilize the pilE sigma54 promoter. Gene 20895-102. [DOI] [PubMed] [Google Scholar]
- 21.Leech, A. J., A. Sprinkle, L. Wood, D. J. Wozniak, and D. E. Ohman. 2008. The NtrC family regulator AlgB, which controls alginate biosynthesis in mucoid Pseudomonas aeruginosa, binds directly to the algD promoter. J. Bacteriol. 190581-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y., G. Hao, C. D. Galvani, Y. Meng, L. De La Fuente, H. C. Hoch, and T. J. Burr. 2007. Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell-cell aggregation. Microbiology 153719-726. [DOI] [PubMed] [Google Scholar]
- 23.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. USA 89718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, S., U. Selvaraj, D. E. Ohman, R. Quarless, D. J. Hassett, and D. J. Wozniak. 1998. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 180956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCleary, W. R., J. B. Stock, and A. J. Ninfa. 1993. Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 1752793-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDaniel, T. K., K. C. Dewalt, N. R. Salama, and S. Falkow. 2001. New approaches for validation of lethal phenotypes and genetic reversion in Helicobacter pylori. Helicobacter 615-23. [DOI] [PubMed] [Google Scholar]
- 27.Moumile, K., J. Merckx, C. Glorion, P. Berche, and A. Ferroni. 2003. Osteoarticular infections caused by Kingella kingae in children: contribution of polymerase chain reaction to the microbiologic diagnosis. Pediatr. Infect. Dis. J. 22837-839. [DOI] [PubMed] [Google Scholar]
- 28.Parker, D., R. M. Kennan, G. S. Myers, I. T. Paulsen, J. G. Songer, and J. I. Rood. 2006. Regulation of type IV fimbrial biogenesis in Dichelobacter nodosus. J. Bacteriol. 1884801-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor NY.
- 30.Schär, J., A. Sickmann, and D. Beier. 2005. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J. Bacteriol. 1873100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 32.Verdier, I., A. Gayet-Ageron, C. Ploton, P. Taylor, Y. Benito, A. M. Freydiere, F. Chotel, J. Berard, P. Vanhems, and F. Vandenesch. 2005. Contribution of a broad range polymerase chain reaction to the diagnosis of osteoarticular infections caused by Kingella kingae: description of twenty-four recent pediatric diagnoses. Pediatr. Infect. Dis. J. 24692-696. [DOI] [PubMed] [Google Scholar]
- 33.Wanner, B. L. 1992. Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J. Bacteriol. 1742053-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitchurch, C. B., T. E. Erova, J. A. Emery, J. L. Sargent, J. M. Harris, A. B. Semmler, M. D. Young, J. S. Mattick, and D. J. Wozniak. 2002. Phosphorylation of the Pseudomonas aeruginosa response regulator AlgR is essential for type IV fimbria-mediated twitching motility. J. Bacteriol. 1844544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wozniak, D. J., and D. E. Ohman. 1991. Pseudomonas aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J. Bacteriol. 1731406-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, S. S., and D. Kaiser. 1997. Regulation of expression of the pilA gene in Myxococcus xanthus. J. Bacteriol. 1797748-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yagupsky, P. 2004. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect. Dis. 4358-367. [DOI] [PubMed] [Google Scholar]
- 38.Yagupsky, P., R. Dagan, C. W. Howard, M. Einhorn, I. Kassis, and A. Simu. 1992. High prevalence of Kingella kingae in joint fluid from children with septic arthritis revealed by the BACTEC blood culture system. J. Clin. Microbiol. 301278-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yagupsky, P., N. Peled, and O. Katz. 2002. Epidemiological features of invasive Kingella kingae infections and respiratory carriage of the organism. J. Clin. Microbiol. 404180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yagupsky, P., N. Porat, and E. Pinco. 2009. Pharyngeal colonization by Kingella kingae in children with invasive disease. Pediatr. Infect. Dis. J. 28155-157. [DOI] [PubMed] [Google Scholar]