Abstract
The hydration of oleic acid into 10-hydroxystearic acid was originally described for a Pseudomonas cell extract almost half a century ago. In the intervening years, the enzyme has never been characterized in any detail. We report here the isolation and characterization of oleate hydratase (EC 4.2.1.53) from Elizabethkingia meningoseptica.
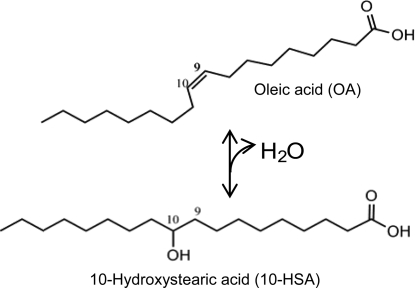
The ability of cells to convert oleic acid (OA) into 10-hydroxystearic acid (10-HSA) was discovered by Wallen et al. in Pseudomonas sp. strain 3266 in 1962 (Fig. 1) (12). In the following years, many other strains were identified that were also able to convert OA into 10-HSA or to further oxidize it to 10-ketostearic acid (2, 3, 5, 7). The Pseudomonas cells generally start to produce optically pure d-10-HSA in the stationary growth phase, and they do not seem to metabolize it any further, since levels of product accumulate in the fermentation broth. The putative enzyme for this conversion is referred to as oleate hydratase (EC 4.2.1.53); however, so far it has not been purified or characterized in any detail.
FIG. 1.
Reaction catalyzed by oleate hydratase; the conversion of OA into 10-HSA.
Kinetic studies have been performed with cell extracts, giving some insight into the stereospecificity and the possible mechanism of the reaction. Studies with 18O-labeled water reported the incorporation of 18O at the C-10 position of 10-HSA, confirming a hydration mechanism (7). The reaction was shown to be reversible; however, the detected concentration ratio at equilibrium was always in the range of 85:15 in favor of 10-HSA (9).
Here we report the isolation and first biochemical characterization of the oleate hydratase protein from Elizabethkingia meningoseptica (formerly known as Pseudomonas sp. strain 3266).
The primer set GM3 and GM4 (8) was used for PCR amplification of the Pseudomonas sp. strain 3266 16S-rRNA genes. The product (1,444 bp) was sequenced, and 16S phylogeny analysis resulted in a unanimous determination of the species as Elizabethkingia (Chryseobacterium) meningoseptica with a >99.8% resemblance.
Purification of the protein.
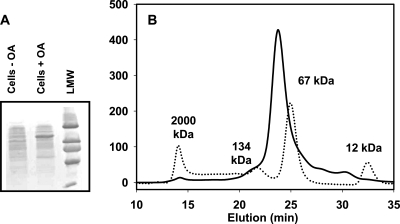
OA hydratase was purified from E. meningoseptica cells that were grown in the presence of 0.3% OA. The cell extract was separated by columns, and the OA conversion activity of the fractions was monitored with thin-layer chromatography (5). In parallel, sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis showed the coelution of a 70-kDa protein with the enzyme activity. The expression of this protein was also strongly upregulated in cells grown in the presence of OA (Fig. 2A) . Three column steps were required to purify oleate hydratase: (i) a column with DEAE fast-flow Sepharose (activity eluted between 300 and 450 mM NaCl, pH 8), (ii) a column with butyl Sepharose (5 to 400 mM potassium phosphate, pH 7.5), and (iii) a Superdex-200 column. Fractions containing oleate hydratase were combined and concentrated to 5 mg/ml.
FIG. 2.
(A) SDS-PAGE. Lane 1, cell extract from noninduced cells; lane 2, cell extract from OA-induced cells; lane 3, low-molecular-weight markers, from top: 94,000, 67,000, 43,000, 30,000, 20,000, and 14,000. (B) Size exclusion chromatography of oleate hydratase (solid line) and mass markers (dotted line), blue dextran (2,000 kDa), bovine serum albumin dimer (134 kDa), bovine serum albumin monomer (67 kDa), and cytochrome c (12 kDa).
Size exclusion chromatography of the purified protein (Fig. 2B) revealed a molecular mass of ±70 kDa, indicating a monomeric native conformation. Inductively coupled plasma optical emission spectrometry analysis (Optima 4300 DV; Perkin Elmer, Norwalk, CT) showed that the protein sample contained 1.2 g-atom of calcium per protein molecule. The sample did not contain a significant amount of copper, iron, magnesium, zinc, manganese, nickel, or cobalt.
Sequencing of the gene.
Protein digestion was performed essentially as described previously (11). Mass spectra were acquired on a hybrid quadrupole time-of-flight mass spectrometer (Waters QTof Premier), and tandem mass spectra were manually interrogated for peptide sequence information. Four peptide sequences were used to design degenerate primers (see Table S1 in the supplemental material). The primer combination 2F and 1R resulted in a PCR product of 150 bp that was used as a template to sequence the rest of the 1.94-kbp gene (ohyA) with a two-step primer walking method (10) (see Fig. S1 in the supplemental material). Subsequently the gene was cloned into the pBAD/His A vector (Invitrogen). Recombinant protein synthesis in Escherichia coli TOP10 cells was induced with 0.2% arabinose for 5 h. The recombinant oleate hydratase fused to an N-terminal His tag was purified with Ni-Sepharose 6 fast-flow resin (GE Healthcare).
Sequence analysis.
A BLAST search of the oleate hydratase amino acid sequence of 73,487.5 Da (see Fig. S2 in the supplemental material) against the nonredundant database afforded 165 homologues (1). The majority of these proteins were annotated as cross-myosin-reactive antigen (6), which originates from their homology to the Streptococcus pyogenes 67-kDa protein that was isolated and sequenced because of its ability to bind to myosin antigens (6). No further biochemical characterization of this protein was reported until a very recently published patent, which describes the cloning and expression of this S. pyogenes protein and its fatty acid hydratase activity (I. Feussner, E. Hornung, and A. Liavonchanka, Production of hydroxy fatty acids using fatty acid hydratase from Streptococcus pyogenes, WO patent application 2008119735, 2008).
Enzymatic activity.
In Table 1, we report apparent Km and Vmax values for oleate hydratase under different conditions. The parameters are labeled as “apparent” in view of the low solubility of the OA in Tris buffer. The real Vmax is most likely significantly higher, whereas the real Km is expected to be much lower. The recombinant enzyme was incubated with different concentrations of OA in 20 mM Tris (pH 8)-50 mM NaCl at various stirring speeds and different temperatures. Reactions were stopped by the addition of 50 μl 3 M HCl, and the conversion was analyzed with gas chromatography (GC). Fatty acids (with palmitic acid as an internal standard) were extracted from the aqueous layer with diethyl ether and converted to their n-propyl esters for GC analysis. Separations were effected with a Shimadzu GC2014 gas chromatograph on a CP-Sil 5CB column (length, 50 m; diameter, 0.53 mm; film thickness, 1 μm) with a temperature gradient of 5°C/min from 180 (holding for 2 min) to 250°C (holding for 10 min). The identities of the conversion products were confirmed by GC-mass spectrometry analysis with a Shimadzu GC MS-QP 2010 S gas chromatograph mass spectrometer (data not shown).
TABLE 1.
Kinetic parameters of oleate hydratase at different mixing speeds and temperaturesa
| Mixing speed (rpm) | Temp (°C) | Apparent Vmax (mU/mg) | Apparent Km (mM) |
|---|---|---|---|
| 1,000 | 30 | 170 ± 90 | 0.21 ± 0.99 |
| 1,400 | 30 | 140 ± 15 | 0.30 ± 0.26 |
| 1,400 | 22 | 160 ± 90 | 0.121 ± 1.0 |
| 2,200 | 22 | 390 ± 70 | 0.56 ± 0.54 |
| 2,200b | 22 | 340 ± 40 | 0.078 ± 2.2 |
Vmax and Km were determined from Michaelis-Menten fits of initial rate data points that were determined at three different apparent substrate concentrations (2 mM, 4 mM, and 8 mM), each in at least two independent experiments.
Data points obtained in a single experiment in the presence of 2.5% isopropyl alcohol.
The maximal specific activity of oleate hydratase is 390 ± 70 mU/mg, and the lowest, most accurate apparent Km is 0.3 ± 0.26 mM (Table 1). These values are within 10% identical to those for the wild-type protein.
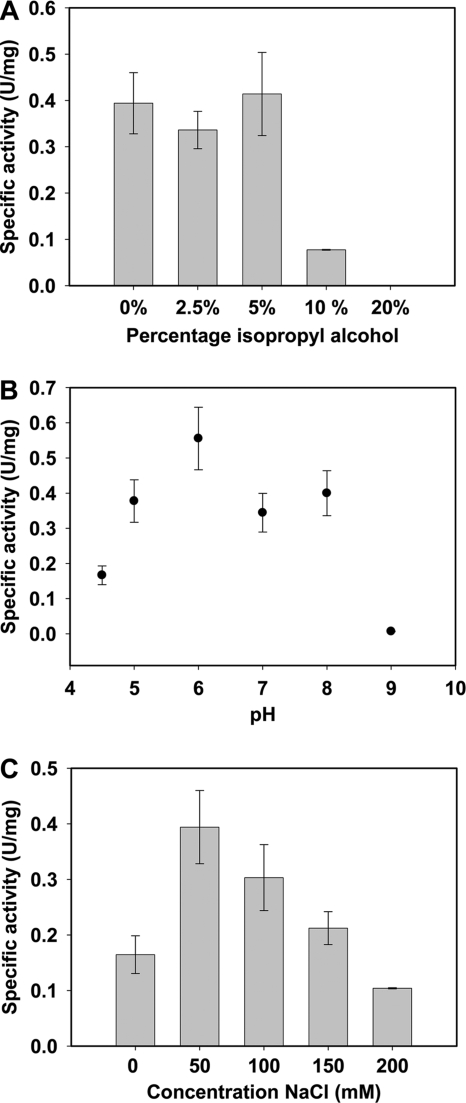
The specific activity of the enzyme did not change significantly in the presence of 2.5% or 5% isopropyl alcohol; however, it decreased at isopropyl alcohol concentrations higher than 10% (Fig. 3A). Furthermore, the addition of EDTA or EGTA (both at a 1,000-fold excess) had no effect on the activity of the enzyme. This suggests that calcium has no role in the catalytic mechanism of oleate hydratase.
FIG. 3.
Influence of different concentrations of isopropyl alcohol (A) or different pH values (B) on the specific activity of oleate hydratase with 4 mM OA at 2,200 rpm and 22°C. (C) Influence of the salt concentration on the specific activity at 2,200 rpm, pH 8, and 22°C.
The activity profile of the enzyme at different pH values is depicted in Fig. 3B. The protein and the OA were added to 20 mM sodium acetate (pH 4.5 and pH 5), 20 mM 2-(N-morpholino)ethanesulfonic acid (pH 6), 20 mM Tris-HCl (pH 7 and pH 8), or 20 mM bicine (pH 9). The maximum activity was detected around pH 6, which is in agreement with earlier measurements (4). The presence of salt also influenced the activity (Fig. 3C), with an optimal concentration of 50 mM.
The purification of oleate hydratase and sequencing of its functional gene have revealed a new type of hydratase. Further research is required to understand the mechanism of OA hydration and the physiological role of this new enzyme.
Nucleotide sequence accession number.
The nucleotide sequence of the 1.94-kbp open reading frame of OA has been deposited in GenBank under accession no. GQ144652.
Supplementary Material
Acknowledgments
We thank Harald Ruijssenaars (TNO location Delft-Julianalaan) for providing the Elizabethkingia meningoseptica strain and Ben Abbas (Department of Biotechnology, Delft University of Technology) for performing the 16S phylogeny analysis.
This project was financially supported by The Netherlands Ministry of Economic Affairs and the B-Basic partner organizations (www.b-basic.nl) through B-Basic, a public-private NWO-ACTS (Advanced Chemical Technologies for Sustainability) program, and by The Netherlands Proteomics Centre and the Kluyver Centre for Genomics of Industrial Fermentation.
Footnotes
Published ahead of print on 22 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.el-Sharkawy, S. H., W. Yang, L. Dostal, and J. P. Rosazza. 1992. Microbial oxidation of oleic acid. Appl. Environ. Microbiol. 582116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson, J. A., C. A. M. MacKenzie, and K. N. Joblin. 1995. Conversion of oleic acid to 10-hydroxystearic acid by two species of ruminal bacteria. Appl. Microbiol. Biotechnol. 441-6. [DOI] [PubMed] [Google Scholar]
- 4.Hudson, J. A., C. A. M. Mackenzie, and K. N. Joblin. 1996. Factors affecting the formation of 10-hydroxystearic acid from oleic acid by a ruminal strain of Enterococcus faecalis. Appl. Microbiol. Biotechnol. 45404-407. [DOI] [PubMed] [Google Scholar]
- 5.Kaneshiro, T., L. K. Nakamura, and M. O. Bagby. 1995. Oleic acid transformations by selected strains of Sphingobacterium thalpophilum and Bacillus cereus from composted manure. Curr. Microbiol. 3162-67. [DOI] [PubMed] [Google Scholar]
- 6.Kil, K. S., M. W. Cunningham, and L. A. Barnett. 1994. Cloning and sequence analysis of a gene encoding a 67-kilodalton myosin-cross-reactive antigen of Streptococcus pyogenes reveals its similarity with class II major histocompatibility antigens. Infect. Immun. 622440-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koritala, S., L. Hosie, C. T. Hou, C. W. Hesseltine, and M. O. Bagby. 1989. Microbial conversion of oleic acid to 10-hydroxystearic acid. Appl. Microbiol. Biotechnol. 32299-304. [Google Scholar]
- 8.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic-relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel-electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164165-172. [DOI] [PubMed] [Google Scholar]
- 9.Niehaus, W. G., Jr., and G. J. Schroepfer, Jr. 1965. The reversible hydration of oleic acid to 10D-hydroxystearic acid. Biochem. Biophys. Res. Commun. 21271-275. [DOI] [PubMed] [Google Scholar]
- 10.Pilhofer, M., A. P. Bauer, M. Schrallhammer, L. Richter, W. Ludwig, K.-H. Schleifer, and G. Petroni. 2007. Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res. 35e135/1-e135/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sevcenco, A.-M., G. C. Krijger, M. W. H. Pinkse, P. D. E. M. Verhaert, W. R. Hagen, and P.-L. Hagedoorn. 2009. Development of a generic approach to native metalloproteomics: application to the quantitative identification of soluble copper proteins in Escherichia coli. J. Biol. Inorg. Chem. 14631-640. [DOI] [PubMed] [Google Scholar]
- 12.Wallen, L. L., R. G. Benedict, and R. W. Jackson. 1962. The microbiological production of 10-hydroxystearic acid from oleic acid. Arch. Biochem. Biophys. 99249-253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.