Abstract
Multidrug resistance (MDR) plasmids belonging to the IncA/C plasmid family are widely distributed among Salmonella and other enterobacterial isolates from agricultural sources and have, at least once, also been identified in a drug-resistant Yersinia pestis isolate (IP275) from Madagascar. Here, we present the complete plasmid sequences of the IncA/C reference plasmid pRA1 (143,963 bp), isolated in 1971 from the fish pathogen Aeromonas hydrophila, and of the cryptic IncA/C plasmid pRAx (49,763 bp), isolated from Escherichia coli transconjugant D7-3, which was obtained through pRA1 transfer in 1980. Using comparative sequence analysis of pRA1 and pRAx with recent members of the IncA/C plasmid family, we show that both plasmids provide novel insights into the evolution of the IncA/C MDR plasmid family and the minimal machinery necessary for stable IncA/C plasmid maintenance. Our results indicate that recent members of the IncA/C plasmid family evolved from a common ancestor, similar in composition to pRA1, through stepwise integration of horizontally acquired resistance gene arrays into a conserved plasmid backbone. Phylogenetic comparisons predict type IV secretion-like conjugative transfer operons encoded on the shared plasmid backbones to be closely related to a group of integrating conjugative elements, which use conjugative transfer for horizontal propagation but stably integrate into the host chromosome during vegetative growth. A hipAB toxin-antitoxin gene cluster found on pRA1, which in Escherichia coli is involved in the formation of persister cell subpopulations, suggests persistence as an early broad-spectrum antimicrobial resistance mechanism in the evolution of IncA/C resistance plasmids.
Antimicrobial compounds have been used extensively in agriculture since the 1960s not only to treat and prevent disease in plants, fruits, vegetables, and animals but also to promote growth in fish, poultry, and other livestock (42). The risk of transferring antimicrobial drug resistance to nonresistant bacteria and the propagation of multidrug-resistant (MDR) bacteria from agricultural to clinical and/or community-associated settings are being debated by research, regulatory, and health authorities (27, 28). In this context, the recent discovery of a group of self-transferable IncA/C antimicrobial resistance plasmids, which are widely distributed among agricultural nontyphoidal Salmonella enterica isolates from the United States (24, 45) has caused considerable concern in the public health community. Similar IncA/C plasmids were identified in an MDR isolate from Madagascar of Yersinia pestis, the causative agent of the plague (16), and MDR strains of Vibrio cholerae O139 from China (34), as well as in MDR isolates of the fish pathogen Photobacterium damselae subsp. piscicida from the United States and Japan (21). While the IncA/C group of MDR plasmids seems to be efficient in collecting antimicrobial resistance traits and mobilizing them across geographical and taxonomical borders, little is known about the evolutionary origin of these plasmids or the genetic basis for their spread.
The IncA/C reference plasmid, pRA1, was isolated in 1971 from the fish pathogen Aeromonas liquefaciens, later renamed Aeromonas hydrophila, as a transferable antimicrobial resistance plasmid conferring resistance to sulfonamides and tetracyclines (2). The repA gene of pRA1, located at the origin of replication and responsible for encoding the replication initiation protein A, has been sequenced (25) and is used for PCR-based replicon typing of IncA/C plasmids (7). repA genes from all sequenced IncA/C plasmids to date share at least 98% nucleotide sequence identity.
To better understand the evolutionary origin of IncA/C plasmids, pRA1 was isolated, sequenced, and compared to all IncA/C plasmid sequences currently available. In addition to pRA1, a pRA1-derived cryptic IncA/C plasmid, designated pRAx, was also sequenced and included in the analysis. pRAx was isolated from Escherichia coli D7-3, a strain that was obtained through the conjugative transfer of pRA1 from A. hydrophila in 1980 (30). While the laboratory history of the pRAx-carrying strain E. coli D7-3 since the conjugative plasmid acquisition is unknown, pRAx was included in this study as it tested positive for the repA reference gene from pRA1 (100% nucleotide sequence identity) but negative for 11 out of 12 additional IncA/C marker genes that were shown to be part of a conserved plasmid backbone shared by recently isolated IncA/C plasmids (45).
MATERIALS AND METHODS
Strain sources.
The original pRA1-carrying Aeromonas hydrophila strain (2) was obtained from Catherine Llanes (Laboratoire de Bactériologie, Hôpital Jean Minjoz, Besançon Cedex, France) who obtained it from Martine Couturier (Université Libre de Bruxelles, Belgium). The pRAx-carrying E. coli strain D7-3 was obtained from Stuart Levy (Tufts University, Boston, MA). The history of both bacterial strains prior to their arrival at the Institute for Genome Science is unknown, except for what has been published (2, 26, 30, 31). Both strains were received as agar slants and streaked on LB agar plates supplemented with tetracycline. A single colony from each of these plates was picked, grown overnight, and used to generate glycerol stock cultures.
Plasmid typing and DNA isolation.
PCR-based typing of the IncA/C-specific replication initiation protein RepA and of the conserved plasmid backbone identified in recent IncA/C plasmids was performed as described previously (7, 42). pRA1 plasmid DNA was isolated from the original host strain, Aeromonas hydrophila, using a protocol for the preparation of DNA from large, low-copy plasmids (46). This protocol utilizes the CosMCPrep kit (Agencourt Bioscience Corporation, Beverly, MA) for DNA enrichment, followed by DNA purification from agarose gels (1% low-melting point agarose; Sigma-Aldrich, St. Louis, MO) using β-agarase (US Biological, Swampscott, MA), followed by phenol-chloroform extractions. To increase the yield of plasmid DNA preparations from E. coli D7-3, the copy number of pRAx per cell was increased prior to the isolation, using the EZ-Tn5 <R6Kγori/KAN-2> Tnp transposome kit (Epicentre Biotechnologies, Madison, WI). Briefly, the E. coli D7-3 strain carrying pRAx was made electrocompetent and transformed with the EZ-Tn5 transposome according to the manufacturer's protocol, allowing for random insertion of the <R6Kγori/KAN-2> transposon. Plasmid DNA was isolated from the resulting pooled transformants, and the DNA was used to transform electrocompetent pir E. coli EC100D cells (Epicentre Biotechnologies, Madison, WI). Transformants from this second electroporation step were screened for resistance to tetracycline and kanamycin, thereby selecting for the presence of EZ-Tn5 transposons (kanamycin resistance) integrated into the pRAx plasmid backbone (tetracycline resistance). pRAx derivatives containing the R6Kγori insertion showed a considerable increase in copy number. Plasmid DNA was then purified using the Qiagen (Valencia, CA) QIAfilter plasmid Midi kit according to the manufacturer's protocols.
Plasmid sequencing.
pRA1 and pRAx were assembled from whole-plasmid shotgun Sanger sequence reads. Sequencing libraries were constructed as previously reported (37) and sequenced using 3730xl DNA analyzer (Applied Biosystems). Assembly and manual closure were followed by manual annotation as previously described (37).
Plasmid mating experiments.
Conjugative plasmid transfer systems were set up as described previously (45). Briefly, a rifampin-resistant recipient strain was generated from 1-ml overnight cultures of E. coli DH10B (Invitrogen) by streaking the concentrated cells from 1 ml onto LB supplemented with rifampin (100 μg/ml) and by picking resistant clones after 24 h of incubation. For mating reactions, donor and recipient strains were grown in an orbital shaker (225 rpm) in brain heart infusion (BHI) medium at 37°C to mid-log phase, except for the pYR1 donor strain Yersinia ruckeri YR71, which was grown at 30°C. Medium for all donor strains contained tetracycline (10 μg/ml), and medium for the recipient strain contained rifampin (100 μg/ml). Donor and recipient bacteria were harvested by centrifugation (16,100 × g, 1 min), and 200 μl of donor and 400 μl of recipient were mixed and spotted onto BHI agar plates. Conjugation mixtures were allowed to incubate for 2.5 h at 37°C, after which the cells were resuspended in phosphate-buffered saline, diluted, and plated onto BHI agar containing rifampin (100 μg/ml) and tetracycline (10 μg/ml) to counterselect for the donor and the resistance marker encoded on the plasmid.
Comparative sequence analysis.
Unless otherwise stated, sequence comparisons on the amino acid or nucleotide level were carried out with the BLAST algorithm. Shared genes found on all IncA/C plasmids were identified using the BLAST Score Ratio Tool (36). Single-nucleotide polymorphisms (SNPs) were identified with the “show-snps” utility that is part of the MUMmer whole-genome alignment tool (11). For the comparison of conjugative plasmid transfer operons, the protein sequences of TraD, TraB, and TraF were concatenated for each of the compared plasmids or integrating conjugative elements. GenBank accession numbers of all proteins can be found in Table S1 in the supplemental material. TraD protein sequences showed at least 64% amino acid identity and 98% sequence coverage compared to TraDpRA1 (621 amino acids), TraB protein sequences showed 49% identity and 49% coverage compared to TraBpRA1 (438 amino acids), and TraF protein sequences showed 41% identity and 41% coverage compared to TraFpRA1 (346 amino acids). Protein sequences were aligned using the MUSCLE software (14). Phylogenetic trees were created using the maximum likelihood estimations with PAUP* (41) and based on Bayesian inference with MrBayes (39).
Nucleotide sequence accession numbers.
The plasmid sequences pRA1 and pRAx have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/GenBank) under accession no. FJ705807 and FJ705806, respectively.
RESULTS
The IncA/C reference plasmid pRA1.
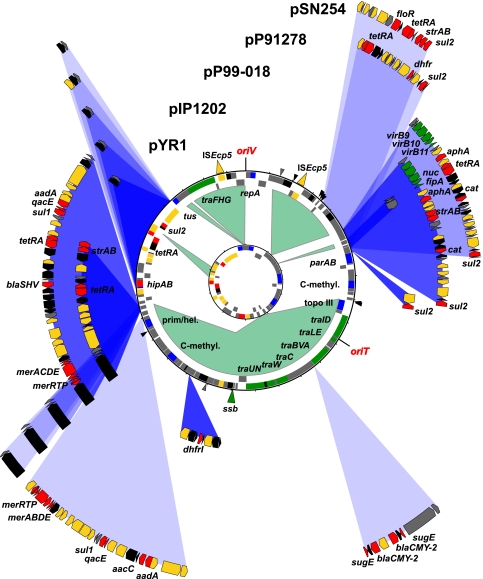
pRA1 is a circular plasmid of 143,963 bp containing 160 genes, 133 of which are encoded on the same strand as the IncA/C-specific replication initiation protein gene (repA) (Fig. 1). Some of the general features of pRA1 and all other IncA/C plasmids sequenced to date are summarized in Table 1. pRA1 and recent IncA/C plasmid isolates show strong overall gene synteny and share a highly conserved plasmid backbone (100.8 kb, >80% nucleotide sequence identity). This IncA/C core plasmid carries 100 protein-encoding sequences and is largely identical to the plasmid backbone identified on pIP1202 from Yersinia pestis IP275, pYR1 from Y. ruckeri YR71, and pSN254 from Salmonella enterica Newport SL254. This backbone is common among MDR Salmonella isolates from food sources (45). The most prominent IncA/C backbone features are the IncA/C origin of replication and a type IV secretion-like conjugative plasmid transfer system consisting of 16 genes located in three gene clusters at two distinct loci of the plasmid backbone (Fig. 1). Comparison of different pRA1 transfer proteins with the nonredundant protein database at NCBI (blast.ncbi.nlm.nih.gov/Blast.cgi) identified a number of integrating conjugative elements (ICEs) as the closest relatives of IncA/C plasmids (see Table S1 in the supplemental material). Consequently, the nomenclature established for one of these ICEs, SXT from Vibrio cholerae, which was based on experimental characterizations (3), was used throughout the annotation of pRA1. In contrast to all other sequenced IncA/C plasmids and ICEs, pRA1 lacks ssb, the gene encoding the single-stranded DNA binding protein.
FIG. 1.
Circular representation of pRA1 and pRAx compared to previously sequenced IncA/C plasmids. Circles from inside to outside: 1, pRAx (A. hydrophila); 2, pRA1 (A. hydrophila); 3, pYR1 (Y. ruckeri); 4, pIP1202 (Y. pestis); 5, pP99-018 (P. damselae subsp. piscicida); 6, pP91278 (P. damselae subsp. piscicida); 7, pSN254 (S. enterica serovar Newport). Genes were color coded, depending on functional annotations, as follows: transposition/recombination, gold; plasmid maintenance, blue; conjugative plasmid transfer, green; antimicrobial and heavy metal resistance, red; other functions, black; and hypothetical proteins, gray. Sequence fragments present in pRA1 but absent from pRAx are shaded in green. Major differences between pRA1 and pYR1, pIP1202, pP99-018, pP91278, and pSN254 such as additional or different sequence fragments are shown with their specific locations and shaded in blue. Locations of minor differences between pRA1 and all other plasmids (except pRAx) or between pRA1 and any of the other plasmids (except pRAx) are indicated by black and gray arrows, respectively.
TABLE 1.
General features of completely sequenced IncA/C plasmids
| Parameter | Characteristic of plasmid:
|
||||||
|---|---|---|---|---|---|---|---|
|
A. hydrophila
|
Y. ruckeri pYR1 | Y. pestis pIP1202 |
P. damselae
|
S. enterica pSN254 | |||
| pRAx | pRA1 | pP91278 | pP99-018 | ||||
| Species | |||||||
| Subclassification | Biovar Orientalis | Subsp. piscicida | Subsp. Enterica serovar Newport | ||||
| Strain | NAc | NA | YR71 | IP275 (17/95) | USA91278 | PT99-018 | SL254 |
| Isolation | NA | NA | United States | Madagascar | United States | Japan | United States |
| Yr | NA | NA | 1996 | 1995 | 1991 | 1999 | 2000 |
| Reference | 2 | 45 | 45 | 21 | 21 | 45 | |
| Plasmid | |||||||
| Estimated copy no. | NA | NA | 1 | 1 | NA | NA | 1 |
| Size (bp) | 49,755 | 143,963 | 158,038 | 182,913 | 131,520 | 150,157 | 176,473 |
| G+C content (%) | 52.0 | 50.6 | 50.9 | 52.8 | 51.7 | 51.4 | 52.8 |
| Protein-coding genes | 66 | 158 | 185 | 212 | 161 | 187 | 190 |
| Coding sequence (%) | 83 | 85 | 89 | 87 | 89 | 89 | 87 |
| No. (%) of genes on positive stranda | 48 (73) | 131 (83) | 156 (84) | 170 (80) | 137 (85) | 156 (83) | 146 (77) |
| No. (%) of hypothetical protein genes | 45 (68) | 102 (65) | 109 (59) | 105 (50) | 98 (61) | 105 (56) | 99 (52) |
| No. of transfer genes | 0 | 16 | 16 | 16 (+3) | 16 | 16 (+3) | 16 |
| No. of SNPsb | NA | 0 | 5,702 | 5,776 | 5,778 | 5,776 | 5,956 |
| Resistance genes | |||||||
| No. of genes | 5 | 5 | 6 | 18 | 4 | 5 | 21 |
| Unspecific | hipAB | hipAB | |||||
| β-Lactams | blaSHV | blaCMY-21, blaCMY-22 | |||||
| Aminoglycosides | strAB | strAB, aadA, aphA | aphA | strAB, aacC, aadA | |||
| Tetracyclines | tetRAclassD | tetRAclassD | tetRAclassB | tetRAclassD | tetRAclassD | tetRAclassD | tetRAclassA |
| Chloramphenicols | cat | cat | floR | ||||
| Sulfonamides | sul2 | sul2 | sul2 | sul1, sul2 | sul2 | sul2 | sul1, sul2 |
| Trimethoprim | dhfr | dhfr | |||||
| Quaternary ammonium compounds | qacEΔ1 | qacEΔ1, sugE1, sugE2 | |||||
| Mercury ions | merRTPCADE | merRTPABDE | |||||
As defined by repA.
Based on comparison to the pRA1 gene set of 100 genes from the IncA/C plasmid backbone.
NA, not applicable.
pRA1 carries three potential resistance genes or gene clusters (Table 1). Sulfonamide resistance is conferred by dihydropteroate synthase encoded by a sul2 gene next to a truncated ISVsa3 element. This conserved resistance gene array is found at different locations on all sequenced IncA/C plasmids (Fig. 1). Adjacent to the sulfonamide resistance gene array and encompassed within two IS26 elements, pRA1 carries a tetracycline resistance gene cluster (tetRA) belonging to class D. Different classes of tetRA gene clusters are found on all sequenced IncA/C plasmids. Located in the same plasmid region as the sulfonamide and tetracycline resistance determinants, pRA1 also contains a hipAB-related gene cluster next to a phage integrase, both of which are absent from all other sequenced IncA/C plasmids. hipAB is a toxin-antitoxin (TA) module and was shown to be responsible for the generation of persister subpopulations in Escherichia coli, which show increased survival rates under antimicrobial selection (10, 32). The IncA/C-related plasmid pAsa4 from the fish pathogen Aeromonas salmonicida subsp. salmonicida (Fig. 2) (38) carries the same hipAB-integrase gene array as pRA1. Although both a tetRA gene cluster (class E) and a sul2 resistance gene are also present on pAsa4, the strongest sequence conservation between pRA1 and pAsa4 is seen at the hipAB-like gene cluster locus (80% nucleotide identity). A third, however, less-related, plasmid-encoded hipAB-like gene cluster is found on the IncHI MDR plasmid pK29 from Klebsiella pneumoniae (9).
FIG. 2.
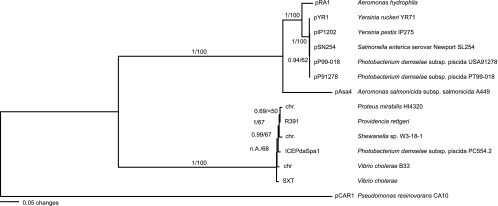
Phylogenetic relationships of conjugative transfer systems found on IncA/C plasmids and integrating conjugative elements, based on maximum likelihood and Bayesian methods. The tree was created based on the alignment of concatenated protein sequences of TraD, TraB, and TraF. Bootstrap values and posterior probabilities are indicated on branches (see Materials and Methods for details).
Comparative analysis of historical and current IncA/C plasmids.
In addition to pRA1, five recent IncA/C plasmid isolates have been sequenced, all of which were characterized based on MDR phenotypes (Table 1). pIP1202, pSN254, and pYR1 were isolated from the human pathogens Y. pestis and S. enterica Newport and from the fish pathogen Y. ruckeri, respectively (45). pP99-018 and pP91278 originated from U.S. and Japanese isolates of the fish pathogen Photobacterium damselae subsp. piscicida (formerly Pasteurella piscicida) (21). Sequence diversity within the shared backbone sequences from all sequenced IncA/C plasmids is low. Using a set of 100 shared backbone genes, the number of SNPs ranges from zero (pIP1202 and pP99-018 comparison) to 5,956 (pRA1 and pSN254 comparison). repAIncA/C genes from pIP1202 (Y. pestis), pSN254 (S. enterica serovar Newport), pP91278, pP99-018 (P. damselae), and pMRV150 (V. cholerae) are 100% identical at the nucleotide sequence level. All antimicrobial resistance determinants are carried within specific, but partially overlapping, mosaic regions that are integrated at few sites within the conserved plasmid backbone. Sequence composition and codon usage deviate in these regions from the conserved plasmid backbone and are indicative of horizontal gene transfer (44). Most resistance determinants are located within resistance gene arrays, composed of resistance genes or gene clusters and mobile genetic elements such as IS elements, transposons or integrons. Several conserved resistance gene arrays, e.g., those coding for sulfonamide (sul2 plus ISVsa3-like IS element) or mercury resistance (Tn21), are found on different plasmids, albeit they are integrated at different sites in the conserved IncA/C plasmid backbone.
An experimental conjugative plasmid transfer system was set up to confirm the functional predictions inferred from the plasmid annotations and the sequence analyses and to determine the transfer efficiency of pRA1 and other IncA/C plasmids to the plasmid-free E. coli strain DH10B (see Table S2 in the supplemental material). In addition to pRA1 host Aeromonas hydrophila, four other IncA/C plasmid-carrying strains were tested: Salmonella enterica serovar Newport SL254(pSN254), Salmonella enterica serovar Heidelberg SL418(pN418), Yersinia ruckeri YR71 (pYR1), and E. coli D7-3 (pRAx). Transfer of the IncF plasmid pCVM29188_146 (GenBank accession no. CP001122) from Salmonella enterica serovar Kentucky strain CVM29188 was used for the comparison of transfer rates between IncA/C and IncF plasmids. Of these plasmids, only pRA1, pYR1, pN418, and pCVM29188_146 were transferable to the E. coli recipient strain. pYR1 showed the highest transfer rate of 5.0 × 10−2 transconjugants (tc) per CFU of donor, followed by pRA1 (9.2 × 10−3 tc/CFU donor) and pN418 (2.9 × 10−3 tc/CFU donor). All conjugative IncA/C plasmids transferred more efficiently than the S. enterica serovar Kentucky IncF plasmid pCVM29188_146 (3.0 × 10−4 tc/CFU donor). To identify conjugative transfer elements related to those found on IncA/C plasmids, three highly conserved transfer proteins (TraBDF) were compared with the nonredundant protein database at NCBI (see Table S1 in the supplemental material). Protein sequences from all IncA/C plasmids and the resulting best matches at NCBI were concatenated, aligned, and used in phylogenetic analysis (Fig. 2). Apart from a set of proteins from the distantly related Pseudomonas resinovorans plasmid pCAR1 that were used as an outgroup, all related transfer proteins were chromosomally encoded. Among these were several protein sets from ICEs such as SXT from V. cholerae (3), R391 from Providencia rettgeri (4), and ICEPdaSpaI from P. damselae (33). Phylogenetic tree predictions based on maximum likelihood and Bayesian inference methods show similar tree topologies and identical branching patterns and suggest that the group of chromosomally encoded ICEs shares a recent common ancestor with the IncA/C plasmids (Fig. 2).
Analysis of the cryptic IncA/C plasmid pRAx.
pRAx (49,763 bp) is about one-third of the size of pRA1. A 2,360-nucleotide (nt)-sequence region from pRAx carrying a ParA-like plasmid partitioning resolvase and two genes annotated as hypothetical proteins is the only sequence fragment that is absent from pRA1, but it can be found on pIP1202, pP99-018, pP91278, and pSN254. Conserved sequence regions between pRAx and pRA1 are distributed over six syntenic blocks that vary in size between 400 and 17,500 bp and share 100% nucleotide sequence identity (see Fig. S1 in the supplemental material). All syntenic sequence blocks that are shared between pRAx and pRA1 overlap by either 2 or 4 nt on pRAx: i.e., the last 2 or 4 nt of one conserved sequence fragment are identical to the first 2 nt of the next conserved sequence fragment (see Fig. S1 in the supplemental material). In contrast, no such sequence overlap is seen between the two syntenic blocks that carry the single sequence fragment from pRAx, which is absent in pRA1. pRAx contains 66 genes, 48 of which are encoded on the same strand as the IncA/C-specific repA. The IncA/C plasmid core that is shared between pRAx and all other sequenced plasmids consists of 25.0 kb (23 genes) and is involved in plasmid replication (origin and terminus of replication), maintenance (plasmid partitioning), and restriction/modification (DNA methylase). The corresponding gene set includes repA (replication initiation protein A), tus (terminus site-binding protein), parAB (plasmid partitioning proteins), and a cytosine-specific DNA methylase gene. pRAx, on the other hand, lacks 67 genes found on all other IncA/C plasmids, encoding the complete conjugative transfer apparatus, topoisomerase III, a primase/helicase, and a second cytosine-specific DNA methylase.
DISCUSSION
pRA1 was isolated in 1971 as the first member of the IncA/C group of MDR plasmids (2), members of which are now commonly identified among agricultural and clinical isolates in the United States and elsewhere. This group of plasmids is regarded as a considerable public health threat due to its ability to spread across taxonomic borders (8, 20, 29, 34, 44, 45). In this study, we present the complete plasmid sequence of pRA1 and provide new insights into the evolution of MDR IncA/C plasmids that allow for a better understanding of the underlying principles of plasmid propagation.
Previously, it was observed that IncA/C plasmids share a common structural composition that consists of a highly conserved plasmid backbone into which specific horizontally acquired resistance fragments were integrated at a few conserved sites (45). This plasmid architecture is also apparent in pRA1 and other IncA/C plasmids that have been sequenced recently (Fig. 1). It is suggestive of an evolutionary model in which each IncA/C plasmid diverged from a common ancestor through a specific process of stepwise integration events of horizontally acquired resistance gene arrays.
pRA1 shows a reduced antimicrobial resistance spectrum limited to sulfonamides (sul2) and tetracyclines (tetRA), as compared with all other sequenced IncA/C plasmids (Table 1). This spectrum possibly reflects an earlier stage of MDR plasmid evolution, at a time when selective pressure due to antimicrobial use was more limited. At the same time, a bias in the selection of the sequenced IncA/C plasmids cannot be ruled out, since all plasmid-carrying strains were isolated based on their antimicrobial resistance phenotypes. The Y. pestis MDR plasmid pIP1202, however, was isolated from a patient with bubonic plague (16). Since antimicrobial therapy in this case was very short and antimicrobial resistance phenotypes have otherwise very rarely been found in Y. pestis (1, 15), resistance determinants on pIP1202 were probably acquired prior to the infection of the human host with Y. pestis IP275. Interestingly, sulfonamides and tetracyclines were among the first antimicrobial compounds used in human and animal medicine (12, 13). As tetRA gene clusters on IncA/C plasmids belong to different classes, it is hypothesized that these clusters were most likely acquired through independent horizontal gene transfer events. The sul2-dependent sulfonamide resistance phenotype, on the other hand, is likely to represent the first antimicrobial resistance phenotype to have been acquired by a putative IncA/C precursor. Sulfonamides, in contrast to tetracyclines, are synthetic antimicrobials and were introduced as the first commercially available antimicrobial agent in the 1930s (Prontosil Red; Bayer Laboratories). It is conceivable that sulfonamide resistance, in contrast to tetracycline resistance, which was already present in the environment, had to evolve de novo and could therefore have provided a stronger selective benefit in the early evolution of the IncA/C plasmid family. Of note, sulfonamides are water soluble, often detected in sewage and surface water (17, 22), and the pRA1 host species A. hydrophila is a waterborne pathogen found in brackish, fresh, estuarine, marine, chlorinated, and unchlorinated water supplies worldwide (19). The combination of a waterborne host with antimicrobial exposure in the aquatic environment could have readily facilitated the evolution of the pRA1 MDR phenotype. Moreover, its ubiquity would make A. hydrophila a likely candidate for IncA/C plasmid transfer between and among different environmental and clinical settings.
The presence of a hipAB-related gene cluster on pRA1 and on the IncA/C-like MDR plasmid pAsa4 from A. salmonicida, both of which are likely to have evolved from a common ancestor (Fig. 2), raises interesting questions about the potential evolutionary relationship between toxin-antitoxin modules and antimicrobial resistance. In Escherichia coli, a chromosomally encoded hipAB gene cluster has been associated with the formation of persister subpopulations, which are able to provide a general unspecific antimicrobial resistance mechanism (23). It is possible that plasmid-mediated persistence might also have served as a broad-spectrum antimicrobial resistance mechanism in the early evolution of the IncA/C group of plasmids.
A key function for the horizontal spread of IncA/C plasmids lies in their conjugative self-transferability. Phylogenetic comparisons, based on conjugative transfer operons, predict that IncA/C plasmids are closely related to a group of ICEs (Fig. 2). During vegetative growth, ICEs stably integrate into a host chromosome but excise themselves prior to conjugation, integrating into a new host chromosome after conjugation (5). ICEs were shown to build cointegrates with plasmids, in this case mobilizing entire, otherwise nontransferable, plasmids through conjugation (33). ICEs, like IncA/C plasmids, are widespread among gram-negative enterobacteria (5), and members of both groups have been identified in P. damselae subsp. piscicida and V. cholerae (Fig. 2). While certainly more comparative sequence analysis will be necessary to develop sound hypotheses in the future, two possible models could explain the evolution of conjugative IncA/C plasmids and ICEs: (i) IncA/C plasmids became transferable through the stable integration of an ICE precursor into the IncA/C plasmid backbone, or (ii) ICEs were derived from conjugative plasmids through excision of the conjugative transfer operon.
We tested and compared four of the sequenced IncA/C plasmids for self-transferability to E. coli; successful transfer of pIP1202 from Y. pestis to E. coli had been shown previously (16). pRA1, pYR1, and pN418 (which has not been sequenced yet) were transferable at comparable rates into E. coli and other enteric bacteria, whereas pSN254 and pRAx could not be transferred. Compared to the F-type S. enterica serovar Kentucky plasmid pCVM29188_146, the transferable IncA/C plasmids showed an at least 10-fold increase in transfer efficiency. However, previous transfer experiments with IncA/C plasmids from agricultural sources have shown that transfer rates for different plasmids can vary by as much as 104-fold (45). An ssb gene is found on all sequenced IncA/C plasmids and ICEs, except for pRA1, suggesting that the single-stranded DNA binding protein is not necessary for conjugative transfer. pSN254 contains the complete set of conjugative transfer genes identified on other transferable plasmids. The only larger fragment, which is absent from pSN254 but present on all transferable plasmids, is a 5.8-kb region upstream of the parAB gene cluster. A screening of several transferable and nontransferable IncA/C plasmids for this fragment, using three specific PCR primer pairs, showed no consistent correlation between the presence of this fragment and plasmid self-transferability (data not shown). Welch et al. demonstrated that among the three most abundant agricultural Salmonella isolates harboring IncA/C, only S. enterica serovar Newport strains failed invariably in plasmid transfer (45). The nontransferability phenotype might therefore be due to chromosomal factors rather than plasmid-encoded features or depend on the presence of additional unknown helper plasmids.
Both pIP1202 and pP99-018 carry an additional transfer-related gene cluster within one of their resistance fragments, which is absent from all other sequenced IncA/C plasmids and ICEs. This entire cluster is most similar to the VirB mating pair formation locus identified on the Salmonella IncN plasmid pKM101 (35). It consists of a truncated virB8-like gene (similar to traEpKM101) and virB9-, virB10-, and virB11-like genes (corresponding to traO, traF, and traG from pKM101, respectively), as well as endonuclease (nuc) and conjugal transfer inhibition protein (fipA) genes (40). Truncated genes and IS elements next to the VirB fragment suggest that it was acquired via horizontal gene transfer, possibly from another plasmid. The TraG coupling factor, which is encoded in the VirB fragment, has been shown to link mating bridge and plasmid relaxosome during conjugation (6, 18), an effect that is inhibited by FipA (40). Whether VirB fragments on pIP1202 and pP99-018 allow these plasmids to hijack external conjugation machineries encoded on other conjugative plasmids warrants further investigation.
The complete plasmid sequence of the cryptic pRA1-derivate pRAx provides new insights into the minimum requirements for IncA/C plasmid maintenance. Using a PCR-based screening assay for 12 loci (in addition to repA) from the conserved IncA/C plasmid backbone, shared between pIP1202, pYR1, and pSN254, Welch et al. showed that of 70 IncA/C plasmids from agricultural sources, each tested positive for at least four loci (one isolate) and on average nine loci (45). pRAx, however, contains only 1 out of these 12 loci, and pRA1 contains 5. These screening results demonstrate that pRAx is likely to contain a reduced plasmid backbone compared to any of the recent IncA/C plasmid isolates. Little is known about the laboratory history of the pRAx-carrying E. coli strain D7-3 between the acquisition of pRA1 (30) and the use for plasmid DNA isolation in this study. However, a massive reduction in plasmid size and gene content is apparent from the comparative analysis of pRAx and pRA1. Only 25.0 kb is conserved between pRAx and all other sequenced IncA/C plasmids (Fig. 1), reducing the IncA/C plasmid core sequence to four segments, responsible for basic functions in plasmid replication, maintenance, and inheritance. The fact that pRAx, in spite of the substantial sequence loss, did not a acquire a single point mutation compared to pRA1 indicates the possibility that plasmid evolution within the IncA/C group progresses faster through recombination than through the accumulation of SNPs. This is also supported by the observation that pIP1202, pP99-018, and pP91278 have almost identical plasmid backbones (only one and two SNPs difference, respectively [see below]) but very different resistance islands (Fig. 1). If plasmid variations arose due to recombination events during repeated growth of IncA/C plasmid-carrying strains in culture, a procedure which was used to prepare plasmid DNA of pRA1 and pRAx for sequencing (see Materials and Methods), the presence of the 2.3-kb fragment on pRAx and all recently isolated IncA/C plasmids, except for pYR1, would indicate that the sequenced variant represents an incomplete derivate of pRA1. While the events that lead to the plasmid reduction of pRAx are unknown, selective pressures alone are unlikely to account for the observed degree of sequence conservation between pRAx and pRA1 since conserved fragments include several truncated IS elements, which confer no apparent selective benefits. Recently, an IncA/C-type origin of replication has also been found on the small metabolic plasmid pBPs2 (estimated size, 12 to 15 kb) isolated from the obligate intracellular aphid symbiont Buchnera aphidicola (43). This might indicate that components involved in core plasmid functions are subject to recombination even between unrelated plasmids.
The isolation of Y. pestis strain IP275 with resistance to most antimicrobial drugs recommended for acute or preventive treatment of plague has caused considerable concern in the public health community, not the least due to potential biodefense concerns (1, 15). In this context, it is revealing that the 100-kb plasmid backbones of pIP1202, which was isolated in Madagascar in 1995, and pP99-018, which was isolated in Japan in 1999, lack any SNPs and that the core plasmids of pIP1202 and pP91278, isolated in the United States in 1991, differ by only two SNPs. Moreover, pIP1202 contains 98% of the sequence of pP99-018, including a complete mosaic resistance fragment (Fig. 1). Not only does this suggest that these three plasmids diverged very recently from a common ancestor, it also could indicate that the origin of pIP1202 is in the aquatic environment. Further comparative sequence analysis of different IncA/C plasmids will be necessary to confirm this hypothesis and deepen our understanding of IncA/C plasmid evolution.
Supplementary Material
Acknowledgments
We thank Catherine Llanes and Stuart Levy for graciously providing plasmid-carrying strains for pRA1 and pRAx, respectively.
The sequencing of pRAx was supported with federal funds from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, Department of Health and Human Services, under NIAID contract N01-AI-30071.
Footnotes
Published ahead of print on 29 May 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anisimov, A. P., and K. K. Amoako. 2006. Treatment of plague: promising alternatives to antibiotics. J. Med. Microbiol. 551461-1475. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, T., S. Egusa, Y. Ogata, and T. Watanabe. 1971. Detection of resistance factors in fish pathogen Aeromonas liquefaciens. J. Gen. Microbiol. 65343-349. [DOI] [PubMed] [Google Scholar]
- 3.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 1844259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 1845158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155376-386. [DOI] [PubMed] [Google Scholar]
- 6.Cabézon, E., E. Lanka, and F. de la Cruz. 1994. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J. Bacteriol. 1764455-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63219-228. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli, A., V. Miriagou, A. Bertini, A. Loli, C. Colinon, L. Villa, J. M. Whichard, and G. M. Rossolini. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg. Infect. Dis. 121145-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y.-T., T.-L. Lauderdale, T.-L. Liao, Y.-R. Shiau, H.-Y. Shu, K.-M. Wu, J.-J. Yan, I.-J. Su, and S.-F. Tsai. 2007. Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 β-lactamases in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 513004-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Correia, F. F., A. D'Onofrio, T. Rejtar, L. Li, B. L. Karger, K. Makarova, E. V. Koonin, and K. Lewis. 2006. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 1888360-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delcher, A. L., S. L. Salzberg, and A. M. Phillippy. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinformatics Unit 10.3 doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed]
- 12.Domagk, G. J. 1935. Ein Beitrag zur Chemotherapie der bakteriellen Infektionen. Dtsch. Med. Wochenschr. 61250-253. [Google Scholar]
- 13.Duggar, B. M. 1948. Aureomycin: a product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 51177-181. [DOI] [PubMed] [Google Scholar]
- 14.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 321792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galimand, M., E. Carniel, and P. Courvalin. 2006. Resistance of Yersinia pestis to antimicrobial agents. Antimicrob. Agents Chemother. 503233-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galimand, M., A. Guiyoule, G. Gerbaud, B. Rasoamanana, S. Chanteau, E. Carniel, and P. Courvalin. 1997. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N. Engl. J. Med. 337677-680. [DOI] [PubMed] [Google Scholar]
- 17.Gobel, A., C. S. McArdell, A. Joss, H. Siegrist, and W. Giger. 2007. Fate of sulfonamides, macrolides, and trimethoprim in different wastewater treatment technologies. Sci. Total Environ. 372361-371. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton, C. M., H. Lee, P.-L. Li, D. M. Cook, K. R. Piper, S. B. von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 1821541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazen, T. C., C. B. Fliermans, R. P. Hirsch, and G. W. Esch. 1978. Prevalence and distribution of Aeromonas hydrophila in the United States. Appl. Environ. Microbiol. 36731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, H. Y., K. Y. Kim, J. Kim, J. C. Lee, Y. C. Lee, D. T. Cho, and S. Y. Seol. 2008. Distribution of conjugative-plasmid-mediated 16S rRNA methylase genes among amikacin-resistant Enterobacteriaceae isolates collected in 1995 to 1998 and 2001 to 2006 at a university hospital in South Korea and identification of conjugative plasmids mediating dissemination of 16S rRNA methylase. J. Clin. Microbiol. 46700-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, M.-J., I. Hirono, K. Kurokawa, T. Maki, J. Hawke, H. Kondo, M. D. Santos, and T. Aoki. 2008. Complete DNA sequence and analysis of the transferable multiple-drug resistance plasmids (R plasmids) from Photobacterium damselae subsp. piscicida isolates collected in Japan and the United States. Antimicrob. Agents Chemother. 52606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolpin, D. W., E. T. Furlong, M. T. Meyer, E. M. Thurman, S. D. Zaugg, L. B. Barber, and H. T. Buxton. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999-2000: a national reconnaissance. Environ. Sci. Technol. 361202-1211. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322107-131. [DOI] [PubMed] [Google Scholar]
- 24.Lindsey, R. L., P. J. Fedorka-Cray, J. G. Frye, and R. J. Meinersmann. 2009. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 751908-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llanes, C., P. Gabant, M. Couturier, L. Bayer, and P. Plesiat. 1996. Molecular analysis of the replication elements of the broad-host-range RepA/C replicon. Plasmid 3626-35. [DOI] [PubMed] [Google Scholar]
- 26.Llanes, C., P. Gabant, M. Couturier, and Y. Michel-Briand. 1994. Cloning and characterization of the Inc A/C plasmid RA1 replicon. J. Bacteriol. 1763403-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathew, A. G., R. Cissell, and S. Liamthong. 2007. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog. Dis. 4115-133. [DOI] [PubMed] [Google Scholar]
- 28.McDermott, P. F., S. Zhao, D. D. Wagner, S. Simjee, R. D. Walker, and D. G. White. 2002. The food safety perspective of antibiotic resistance. Anim. Biotechnol. 1371-84. [DOI] [PubMed] [Google Scholar]
- 29.McIntosh, D., M. Cunningham, B. Ji, F. A. Fekete, E. M. Parry, S. E. Clark, Z. B. Zalinger, I. C. Gilg, G. R. Danner, K. A. Johnson, M. Beattie, and R. Ritchie. 2008. Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J. Antimicrob. Chemother. 611221-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurry, L., R. E. Petrucci, Jr., and S. B. Levy. 1980. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA 773974-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez, B., C. Tachibana, and S. B. Levy. 1980. Heterogeneity of tetracycline resistance determinants. Plasmid 399-108. [DOI] [PubMed] [Google Scholar]
- 32.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osorio, C. R., J. Marrero, R. A. F. Wozniak, M. L. Lemos, V. Burrus, and M. K. Waldor. 2008. Genomic and functional analysis of ICEPdaSpa1, a fish-pathogen-derived SXT-related integrating conjugative element that can mobilize a virulence plasmid. J. Bacteriol. 1903353-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan, J.-C., R. Ye, H.-Q. Wang, H.-Q. Xiang, W. Zhang, X.-F. Yu, D.-M. Meng, and Z.-S. He. 2008. Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob. Agents Chemother. 523829-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pohlman, R. F., H. D. Genetti, and S. C. Winans. 1994. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol. Microbiol. 14655-668. [DOI] [PubMed] [Google Scholar]
- 36.Rasko, D. A., G. S. Myers, and J. Ravel. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 42381-86. [DOI] [PubMed] [Google Scholar]
- 38.Reith, M. E., R. K. Singh, B. Curtis, J. M. Boyd, A. Bouevitch, J. Kimball, J. Munholland, C. Murphy, D. Sarty, J. Williams, J. H. Nash, S. C. Johnson, and L. L. Brown. 2008. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 191572-1574. [DOI] [PubMed] [Google Scholar]
- 40.Santini, J. M., and V. A. Stanisich. 1998. Both the fipA gene of pKM101 and the pifC gene of F inhibit conjugal transfer of RP1 by an effect on traG. J. Bacteriol. 1804093-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4.0b10 ed. Sinauer Associates, Sunderland, MA.
- 42.Tenover, F. C. 24 June 2008, posting date. CDC's role in monitoring and preventing antimicrobial resistance. Testimony before the Health, Education, Labor and Pensions Committee, United States Senate. http://help.senate.gov/Hearings/2008_06_24/Tenover.pdf.
- 43.Van Ham, R. C. H. J., D. Martinez-Torres, A. Moya, and A. Latorre. 1999. Plasmid-encoded anthranilate synthase (TrpEG) in Buchnera aphidicola from aphids of the family Pemphigidae. Appl. Environ. Microbiol. 65117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch, T. J., J. Evenhuis, D. G. White, P. F. McDermott, H. Harbottle, R. A. Miller, M. Griffin, and D. Wise. 2009. IncA/C plasmid-mediated florfenicol resistance in the catfish pathogen Edwardsiella ictaluri. Antimicrob. Agents Chemother. 53845-846. (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welch, T. J., W. F. Fricke, P. F. McDermott, D. G. White, M. L. Rosso, D. A. Rasko, M. K. Mammel, M. Eppinger, M. J. Rosovitz, D. Wagner, L. Rahalison, J. E. Leclerc, J. M. Hinshaw, L. E. Lindler, T. A. Cebula, E. Carniel, and J. Ravel. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE 2e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams, L. E., C. Detter, K. Barry, A. Lapidus, and A. O. Summers. 2006. Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Appl. Environ. Microbiol. 724899-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.