Abstract
Pseudomonas reinekei MT1 has previously been reported to degrade 4- and 5-chlorosalicylate by a pathway with 4-chlorocatechol, 3-chloromuconate, 4-chloromuconolactone, and maleylacetate as intermediates, and a gene cluster channeling various salicylates into an intradiol cleavage route has been reported. We now report that during growth on 5-chlorosalicylate, besides a novel (chloro)catechol 1,2-dioxygenase, C12OccaA, a novel (chloro)muconate cycloisomerase, MCIccaB, which showed features not yet reported, was induced. This cycloisomerase, which was practically inactive with muconate, evolved for the turnover of 3-substituted muconates and transforms 3-chloromuconate into equal amounts of cis-dienelactone and protoanemonin, suggesting that it is a functional intermediate between chloromuconate cycloisomerases and muconate cycloisomerases. The corresponding genes, ccaA (C12OccaA) and ccaB (MCIccaB), were located in a 5.1-kb genomic region clustered with genes encoding trans-dienelactone hydrolase (ccaC) and maleylacetate reductase (ccaD) and a putative regulatory gene, ccaR, homologous to regulators of the IclR-type family. Thus, this region includes genes sufficient to enable MT1 to transform 4-chlorocatechol to 3-oxoadipate. Phylogenetic analysis showed that C12OccaA and MCIccaB are only distantly related to previously described catechol 1,2-dioxygenases and muconate cycloisomerases. Kinetic analysis indicated that MCIccaB and the previously identified C12OsalD, rather than C12OccaA, are crucial for 5-chlorosalicylate degradation. Thus, MT1 uses enzymes encoded by a completely novel gene cluster for degradation of chlorosalicylates, which, together with a gene cluster encoding enzymes for channeling salicylates into the ortho-cleavage pathway, form an effective pathway for 4- and 5-chlorosalicylate mineralization.
The aerobic degradation of chloroaromatic compounds usually proceeds via chlorocatechols as central intermediates (20, 47), which in most of the cases reported thus far, are further degraded by enzymes of the chlorocatechol pathway (44). This pathway involves ortho-cleavage by a chlorocatechol 1,2-dioxygenase with high activity for chlorocatechols (12), a chloromuconate cycloisomerase with high activity for chloromuconates (54), a dienelactone hydrolase active with both cis- and trans-dienelactone (4-carboxymethylenebut-2-en-4-olide) (54), and a maleylacetate reductase (MAR) (28).
However, it has become evident in recent years that microorganisms have evolved various alternative strategies to mineralize chlorocatechols. Pseudomonas putida GJ31 was found to degrade chlorobenzene rapidly via 3-chlorocatechol using a catechol meta-cleavage pathway (33). Two alternative pathways for 3- and 4-chlorocatechol degradation that involve reactions known from the chlorocatechol, as well as the 3-oxoadipate, pathway have recently been observed in Rhodococcus opacus 1CP (35) and Pseudomonas reinekei MT1 (39). In R. opacus 1CP, 3-chloro- and 2,4-dichloro-cis,cis-muconate (the ring cleavage products of 4-chlorocatechol and 3,5-dichlorocatechol, respectively) are converted to the respective cis-dienelactones (35, 58), similar to the reaction described for proteobacterial chloromuconate cycloisomerases (54). However, proteobacterial chloromuconate cycloisomerase can dehalogenate 2-chloromuconate (the ring cleavage product of 3-chlorocatechol) and transform this compound via 5-chloromuconolactone into trans-dienelactone (54, 65), whereas none of the described chloromuconate cycloisomerases of R. opacus 1CP can catalyze such a dehalogenation, and 5-chloromuconolactone is the product of the cycloisomerization reaction (35, 58). Dehalogenation is achieved by an enzyme with high sequence similarity to muconolactone isomerases (35), which in proteobacteria have been shown to be capable of dehalogenating 5-chloromuconolactone to cis-dienelactone (46).
In P. reinekei MT1, a trans-dienelactone hydrolase (trans-DLH) was identified as the key enzyme involved in the degradation of 4- and 5-chlorosalicylate via 4-chlorocatechol as an intermediate (39). In contrast to all previously described dienelactone hydrolases involved in chlorocatechol degradation, which belong to the α/β hydrolase fold enzymes with a catalytic triad consisting of Cys, His, and Asp (10), trans-DLH was shown to be a zinc-dependent hydrolase (8). The function of this enzyme in the 4-chlorocatechol metabolic pathway was to interact with the muconate cycloisomerase (MCI)-mediated transformation of 3-chloromuconate into protoanemonin. By acting on the reaction intermediate 4-chloromuconolactone, trans-DLH prevents the formation of protoanemonin by catalyzing its hydrolysis to maleylacetate (39). Maleylacetate, in turn, is reduced by MAR to 3-oxoadipate.
A more detailed genetic and biochemical analysis of the degradation of differently substituted salicylates (7) had shown the presence of two catabolic gene clusters in MT1. An archetype catRBCA gene cluster was shown to be involved in salicylate degradation. The second gene cluster (sal) had a novel gene arrangement, with salA, encoding a salicylate 1-hydroxylase, clustered with the salCD genes, encoding MCI and catechol 1,2-dioxygenase (C12O), respectively. As these genes were expressed during growth on differently substituted salicylates, it was proposed that the function of the sal gene cluster is to channel both chlorosubstituted and methylsubstituted salicylates into a catechol ortho-cleavage pathway, followed by dismantling of the formed substituted muconolactones through specific pathways. However, previous analyses had indicated the presence of an additional and thus third (chloro)muconate cycloisomerase in MT1 during growth on chlorosalicylate, which is distinct from both previously described MCIs encoded by the cat cluster (MCIcatB) and the sal cluster (MCIsalC), as it transforms 3-chloromuconate into approximately equal amounts of cis-dienelactone and protoanemonin (39). In the present report, this cycloisomerase is biochemically and genetically described and shown to be located in a third gene cluster involved in the degradation of 5-chlorosalicylate by strain MT1. This cluster comprises genes encoding a third C12O, trans-DLH (8), and a MAR. Evidently, P. reinekei MT1 is the first microorganism in which such a complex net of genes involved in chlorocatechol degradation has been described.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
P. reinekei MT1 was grown and cell extracts were prepared as previously described (39).
Enzyme assays.
C12O, MCI, trans-DLH, and MAR activities were determined spectrophotometrically as previously described (27, 39, 54). The activity of MCIccaB with 3-chloromuconate was determined by high-performance liquid chromatography (HPLC) (39) following substrate depletion and product formation. To more sensitively follow the activity of MCIccaB with muconate and 2-chloromuconate, the transformation of these substrates (100 μM) was also followed by HPLC using up to 10 U/ml (measured with 100 μM 3-chloromuconate) of purified MCIccaB. Specific activities are expressed as μmol of substrate converted or product formed per minute per gram of protein at 25°C. Protein concentrations were determined by the Bradford procedure using the Bio-Rad protein assay with bovine serum albumin as a protein standard (5).
Analysis of kinetic data.
The Vmax, kcat, and apparent Km values of C12OccaA with catechol, 3-methylcatechol, 4-methylcatechol, and 4-chlorocatechol were determined using 1 to 100 μM of substrate in air-saturated buffer, and the kinetic data were calculated from the initial velocities using the Michaelis-Menten equation by nonlinear regression (KaleidaGraph; Synergy Software). As very low Km values were indicated by this method, kinetic data were finally determined from progress curves obtained from reactions with initial substrate concentrations of 10 μM, as previously described (7). Vmax, kcat, and apparent Km values of MCIccaB with 2-methylmuconate, and 3-methylmuconate were determined using 2 to 100 μM of substrate. Transformation of 3-chloromuconate was determined by HPLC analysis at substrate concentrations of 50 μM to 500 μM. Samples were taken during the reaction time, and the formation of protoanemonin and cis-dienelactone was directly quantified by HPLC analysis. At least two independent experiments were performed for each value. Km and Vmax values were calculated by nonlinear regression to the Michaelis-Menten equation, using KaleidaGraph (Synergy Software). Turnover numbers (kcat values) were calculated assuming subunit molecular masses of 29,424 (C12OccaA) and 39,764 (MCIccaB) Da, respectively.
Enzyme purification.
C12OccaA and MCIccaB were purified using a Fast Protein Liquid Chromatography system (Amersham Biosciences). Cells were harvested during late exponential growth with 5-chlorosalicylate or 4-methylsalicylate. Cell disruption and all protein elutions were performed in 50 mM Tris-HCl, pH 7.5, 2 mM MnCl2.
For analyzing the presence and abundances of different C12Os and MCIs under different growth conditions, either cell extracts (usually containing 35 mg of protein per ml) were applied directly to a MonoQ HR5/5 (Amersham Pharmacia Biotech) and proteins were eluted by a linear gradient of 0 to 0.5 M NaCl over 25 ml with a flow of 0.5 ml/min, or the cell extract was mixed with 4 M (NH4)2SO4 to give a final concentration of 1 M (NH4)2SO4 and applied to a Source 15PHE PE 4.6/100 (hydrophobic interaction) column (Amersham Pharmacia Biotech). Proteins were eluted from the Source column by a linear gradient of (NH4)2SO4 (1 M to 0 M) over 25 ml with a flow of 0.5 ml/min. Fraction volumes were 0.5 ml. Hydrophobic interaction chromatography (HIC) separated C12OccaA (0.52 ± 0.02 M), C12OsalD (0.45 ± 0.04 M), C12OcatA (0.16 ± 0.04 M), MCIccaB (0.25 ± 0.04 M), MCIsalC (0.06 ± 0.06 M), and MCIcatB (0.12 ± 0.06 M), thus excluding interference between their activities. During anion-exchange chromatography, C12OccaA eluted at 0.23 ± 0.01 M NaCl, whereas MCIccaB eluted at 0.37 ± 0.02 M NaCl. Under these conditions, C12OcatA and C12OsalD had been shown to coelute at 0.28 ± 0.02 M NaCl, whereas MCIcatB and MCIsalC coeluted at 0.24 ± 0.02 M NaCl (7).
For purification of C12OccaA, 35 mg of protein from 5-chlorosalicylate-grown cells was applied to the MonoQ HR 5/5 (Amersham Pharmacia Biotech), and proteins were eluted as described above. Fractions containing C12OccaA activity were combined, supplemented with 4 M (NH4)2SO4 to give a final concentration of 1 M (NH4)2SO4, and loaded on a Source 15PHE PE 4.6/100 (hydrophobic interaction) column (Amersham Pharmacia Biotech) as described above.
For purification of MCIccaB, up to 400 mg of protein from 5-chlorosalicylate-grown cells was applied to a MonoQ HR 10/10 (Amersham Pharmacia Biotech). A stepwise gradient of 0 to 60 mM NaCl over 40 ml, 60 to 380 mM NaCl over 120 ml, and 380 to 2,000 mM NaCl over 40 ml was applied. The flow rate was 0.3 ml/min. The eluate was collected in fractions of 5 ml. All fractions eluting at NaCl concentrations of 90 to 330 mM were pooled and concentrated to a final volume of 4.25 ml using ultrafiltration by Centriprep YM-50 (Millipore) according to the protocol of the manufacturer. The protein solution was supplemented with 4 M (NH4)2SO4 to give a final concentration of 0.8 M (NH4)2SO4 and centrifuged directly before application of the soluble proteins to the Source column. Aliquots comprising 40 mg of protein were separated as described above. Fractions containing MCIccaB were combined and concentrated by a Centricon YM-50 (Millipore). Further purification was achieved by gel filtration using a Superose 12 HR10/10 column (Amersham Pharmacia Biotech). Proteins were eluted with 50 mM Tris-HCl, 2 mM MnCl2, pH 7.5, over 15 ml (flow rate, 0.2 ml/min; fraction volume, 0.5 ml). The fractions containing high MCIccaB activity (eluting at 10.5 to 11.5 ml) were applied to a MonoQ HR5/5 (anionic-exchange) column (Amersham Pharmacia Biotech), and the proteins were eluted by a linear gradient of 0 to 0.4 M NaCl over 25 ml with a flow of 0.2 ml/min. Homogeneity was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). trans-DLH was purified as previously described (8).
Transformation of 3-chloromuconate by enzyme mixtures.
Product formation from 3-chloromuconate by purified MCIccaB and in the presence of purified trans-DLH was analyzed by HPLC in assays performed at room temperature in 150 μl Tris-HCl (50 mM), 2 mM MnCl2, pH 7.5, with 120 μM 3-chloromuconate as a substrate. MCIccaB was added to give an activity of 53 mU/ml (determined by the transformation of 100 μM 3-chloro-cis,cis-muconate), corresponding to 8.8 nM MCIccaB, whereas trans-DLH was applied in amounts ranging from 1.32 to 1,320 mU/ml (determined by the transformation of 50 μM trans-dienelactone), corresponding to 0.88 to 88 nM trans-DLH.
Determination of molecular mass.
The molecular mass of MCIccaB was determined by gel filtration using a Superose 12 column as described above. The column was calibrated for molecular mass determinations using ovalbumin (43 kDa), aldolase (158 kDa), catalase (232 kDa), and ferritin (440 kDa) from Bio-Rad.
Electrophoretic methods.
SDS-PAGE was performed on a Bio-Rad Miniprotein II as previously described (32), with acrylamide concentrations of 5 and 10% (wt/vol) used for the concentrating and separating gels, respectively. The proteins were stained with Coomassie brilliant blue (Serva). A PageRuler Protein Ladder (Fermentas) was used as a marker.
Amino acid sequencing.
N-terminal amino acid sequences were determined as described previously (26).
Identification of the gene encoding MCIccaB of strain MT1.
Part of the gene encoding MCIccaB was amplified by PCR using the degenerate primers NT1B (WSNCARGGNTTYGTNATCGG) and NTREV2A (AANWSCATNCKDATNGGCTG), which were designed based on the determined N-terminal protein sequence (underlined) SQGFVIGRVLAQRLDIPFSQPIRMSFGTLD. Touchdown PCR consisted of an initial denaturation (94°C for 4 min), followed by 10 cycles of denaturation (94°C for 45 s), annealing (60°C for 30 s − 1°C per cycle), and elongation (72°C for 30 s), followed by 25 cycles with an annealing temperature of 50°C for 45 s and a final elongation step (72°C for 7 min). A 72-bp fragment was obtained, cloned into the pGEM-T Easy vector (Promega), and transformed into Escherichia coli XL10-Gold (Stratagene), and inserts of the clones generated were then sequenced. The deduced amino acid sequence matched that of the N-terminal amino acid sequence.
An extended part of the gene encoding MCIccaB was amplified by PCR using the primers MCIB1 (GCAACGGCTGGATATACCTT) and MCIBR2 (GTRTCGCCRCTSGCSARCGTCC), which were designed based on the DNA sequence generated above and a protein sequence, WTLASGDT, identified by protein sequence alignment to be conserved in proteobacterial muconate and chloromuconate cycloisomerases. The touchdown PCR conditions included 10 cycles as described above, followed by 25 cycles at an annealing temperature of 55°C. An approximately 400-bp fragment was obtained, cloned, and sequenced as mentioned above. The DNA sequence matched the sequence deduced from the N-terminal sequence, clearly confirming that the cloned PCR product corresponded to part of the gene encoding MCIccaB.
DNA isolation, fosmid library construction, and identification of the cca gene cluster.
Preparation of the fosmid library in pCC1FOS, which comprised a total of 282 individual clones, was previously described (7). The fosmid library was screened by PCR using primers specific for ccaB (MCIB1 [GCAACGGCTGGATATACCTT] and inMCIB [AGCAGAAACACCCAACTGCT], with an annealing temperature of 59°C). Fosmid clones harboring the expected 340-bp ccaB gene fragment were subsequently checked by PCR for the presence of the ccaC gene, encoding trans-DLH (TransFOR [AATCCCTGCCGACATACAAG] and TransREV [CGTCAGCATGAAGGTGTAGC]). From the three fosmids carrying both the ccaB and the ccaC gene fragments, one fosmid was chosen and purified with the FosmidMAX DNA purification kit (Epicentre), and the complete cca gene cluster was obtained by direct sequencing (Seqlab, Göttingen, Germany) from the purified fosmid with a sixfold coverage of the insert.
DNA sequencing and sequence analysis.
PCR products were purified with the QIAquick PCR Purification Kit (Qiagen) and sequenced using the ABI Prism BigDye Terminator v1.1 Ready Reaction Cycle Sequencing Kit (Applied Biosystems) and a DNA capillary sequencer, the 3130xl Genetic Analyzer (Applied Biosystems). Raw sequence data from both strands were assembled with Sequencher software version 4.0.5 (Gene Codes Corporation). DNA and protein similarity searches were performed using the BLASTN and BLASTP programs from the NCBI website. Translated protein sequences were aligned with CLUSTALX 1.83 using default values (61). The evolutionary history was inferred with MEGA4 (59) using the neighbor-joining algorithm with p-distance correction and pairwise deletion of gaps and missing data. A total of 100 bootstrap replications were performed to test for branch robustness.
Gene expression studies.
Harvest of P. reinekei MT1 cells and RNA extraction were done as previously described (7). Reverse transcription (RT) and quantitative real-time PCR were performed using a QuantiTect SYBR green RT-PCR kit (Qiagen) for one-step RT-PCR in a Rotor-Gene 2000 real-time PCR machine (Corbett Research). Transcripts of ccaA, ccaB, ccaC, and ccaD were quantified with the following primer pairs: CcaA-F (GGGCGCTTTCACACCAATGACC) and CcaA-R (GCAGGTGAGCGGGTCGGAAGTA), CcaB-F (GCAGTTGAGGCGGCGGTTGTTA) and CcaB-R (GCTTGCCAACCAGGTCGAATGC), CcaC-F (TGACACGTCCAAATCCCTGCCG) and CcaC-R (GCAAGCGTGCGGCGTTATCAAT), and CcaD-F (GATGGCGTTGTCGGTCTTGG) and CcaD-R (TGACGGTTTCAGGGCGGATA). A housekeeping reference gene (ribosomal rpsL) was selected to normalize the results obtained (9, 13). Real-time PCRs were carried out and relative expression ratios were determined as previously described (7).
Mathematical calculations.
Numerical calculations were performed with a kinetic model built in SIMULINK v6.4.1 under the MATLAB v7.2.0.232 environment (The MathWorks, Inc., Natick, MA) based on Michaelis-Menten kinetics using the kinetic constants experimentally determined here or previously (7) and assuming a constant concentration of enzyme and zero-order kinetics for oxygen and NADH.
Analytical methods.
HPLC was performed as previously described (7).
Chemicals.
3-Chlorocatechol, 4-chlorocatechol, 3-methylcatechol, and 4-methylcatechol were obtained from Helix Biotech. 2-Methylmuconate, 3-methylmuconate, and 3-chloro-cis,cis-muconate were freshly prepared from 3-methylcatechol, 4-methylcatechol, and 4-chlorocatechol, respectively, in 50 mM Tris-HCl, pH 7.5, 2 mM MnCl2 using chlorocatechol 1,2-dioxygenase TetC of Pseudomonas chlororaphis RW71 (45) or partially purified C12OsalC free of muconate cycloisomerizing activity. cis-Dienelactone was kindly provided by Walter Reineke (Bergische Universität-Gesamthochschule, Wuppertal, Germany) and Stefan Kaschabeck (TU Bergakademie, Freiberg, Germany). Protoanemonin, 2-chloro-cis,cis-muconate, and trans-dienelactone were prepared as previously described (4, 48).
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was deposited in the DDBJ/EMBL/GenBank databases under accession number EF159980.
RESULTS
Characterization of a cycloisomerase transforming 3-chloromuconate into both cis-dienelactone and protoanemonin.
Two MCIs, both transforming 3-chloromuconate into protoanemonin, with minor quantities of cis-dienelactone, had previously been characterized from P. reinekei MT1, and the encoding genes had been localized (7). However, during growth on 5-chlorosalicylate, the presence of a distinct enzyme capable of transforming 3-chloromuconate was evident. This enzyme, termed MCIccaB, eluted at 0.25 ± 0.04 M during HIC, and as previously indicated (39), approximately equal amounts of protoanemonin (50% ± 3%) and cis-dienelactone (47% ± 5%) were formed when proteins of such fractions were supplemented with 3-chloromuconate. As the formation of such a product mixture by any muconate or chloromuconate cycloisomerase had not been previously observed, the enzyme was purified to homogeneity. The native molecular mass of MCIccaB was estimated by gel filtration to be 350 ± 20 kDa, and a single band of 43 ± 3 kDa was observed on SDS gels. Thus, MCIccaB, like MCI of P. putida PRS2000 (22) or chloromuconate cycloisomerase from Cupriavidus necator JMP 134 (23), may be a homo-octamer. N-terminal amino acid analysis (SQGFVIGRVLAQRLDIPFSQPIRMSFGTLD) revealed no significant similarity when these sequences were compared to the sequences of other cycloisomerases available in databases.
Of the substrates tested, only 3-chloromuconate and 3-methylmuconate were transformed with high activity by this enzyme. The highest turnover rate, 10-fold higher than with 3-methylmuconate, was observed with 3-chloromuconate (Table 1). However, the specificity constants of 3-chloromuconate and 3-methylmuconate were almost equal, due to the significantly higher Km value with 3-chloromuconate. Activity of the enzyme with muconate was negligible, and at a substrate concentration of 0.1 mM substrate, the activity was only 0.4% of that with 3-chloromuconate. Thus, from the substrate utilization profile, MCIccaB is clearly different from previously reported MCIs, which are characterized by their high activity with muconate (53, 54). It also differed from MCIsalC of MT1, which has previously been characterized as being adapted for the turnover of 3-methylmuconate (7) but retained a significant activity with muconate. MCIccaB was practically inactive with 2-chloromuconate, which is transformed at high rates by most proteobacterial chloromuconate cycloisomerases described thus far (31, 63, 64).
TABLE 1.
Substrate specificities of C12OccaA and MCIccaB from P. reinekei MT1a
| Enzyme | Substrate | Activity with 0.1 mM substrate (U/mg) | Km (μM) | kcat (s−1) | kcat/Km (s−1 μM−1) |
|---|---|---|---|---|---|
| C12OccaA | Catechol | 2.5 ± 0.1 | 2.0 ± 0.4 | 1.2 ± 0.05 | 0.6 |
| 4-Chlorocatechol | 0.24 ± 0.02 | 0.6 ± 0.1 | 0.12 ± 0.01 | 0.2 | |
| 3-Chlorocatechol | 0.08 ± 0.01 | ND | ND | ND | |
| 4-Methylcatechol | 24.0 ± 1.2 | 0.6 ± 0.1 | 11.5 ± 0.6 | 19.2 | |
| 3-Methylcatechol | 12.4 ± 0.3 | 21.5 ± 2.5 | 6.0 ± 0.15 | 0.3 | |
| MCIccaB | Muconate | 0.55 ± 0.1 | ND | ND | ND |
| 3-Chloromuconate | 140 ± 10 | 105 ± 15 | 111 ± 8 | 1.1 | |
| 2-Chloromuconate | <0.003 | ND | ND | ND | |
| 3-Methylmuconate | 26 ± 2.2 | 10.6 ± 1.2 | 11 ± 0.5 | 1.0 | |
| 2-Methylmuconate | 0.95 ± 0.1 | 40 ± 8 | 0.5 ± 0.05 | 0.01 |
The kinetic constants were determined as described in Materials and Methods. Standard deviations were calculated with the KaleidaGraph program. ND, not determined.
The fact that purified MCIccaB transformed 3-chloromuconate stoichiometrically into equal amounts of protoanemonin and cis-dienelactone contrasts with all previously described cycloisomerases, which form either protoanemonin (MCIs) or cis-dienelactone (chloromuconate cycloisomerases) as the predominant product (4, 39, 53, 54, 58). Following 3-chloromuconate transformation over time showed that both products were formed at a constant ratio, indicating that the reaction mechanism was independent of the substrate concentration.
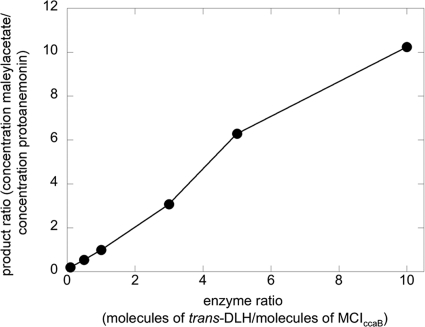
It has previously been shown that trans-DLH of strain MT1 interferes with the cycloisomerization of 3-chloromuconate catalyzed by MCIsalC (39), an enzyme encoded by the sal cluster and induced during growth on chlorosalicylates (7), and it was suggested that trans-DLH acts on intermediate 4-chloromuconolactone to form maleylacetate, thereby preventing protoanemonin formation. To validate the notion that trans-DLH can similarly interact with MCIccaB, 3-chloromuconate (0.12 mM) was transformed by enzyme mixtures comprising MCIccaB (8.8 nM) and various amounts of trans-DLH (0.88 to 88 nM). As previously observed for MCIsalC (39), the simultaneous presence of trans-DLH decreased the amount of protoanemonin formed (Fig. 1) but did not influence the extent of cis-dienelactone formation, which was always 47% ± 5% of the substrate transformed.
FIG. 1.
Ratio of maleylacetate and protoanemonin formed from 3-chloromuconate by mixtures of MCIccaB (8.8 nM) with various amounts of trans-DLH (0 to 88 nM) of P. reinekei MT1. The reaction mixtures contained 50 mM Tris-HCl, 2 mM MnCl2, pH 7.5, and 120 μM 3-chloromuconate. Substrate and product concentrations were analyzed by HPLC.
Characterization of a C12O specifically induced during growth on 5-chlorosalicylate.
As an MCI that was not encoded by the previously described cat or sal gene cluster was induced during growth on chlorosalicylate (7), we assessed whether a distinct C12O was also induced under such conditions. In fact, C12O activity was observed in protein fractions of cell extracts, eluting at 0.23 ± 0.01 M NaCl during anionic-exchange chromatography, in addition to previously described C12OsalC, eluting at 0.29 ± 0.01 M NaCl. HIC confirmed the presence of a previously uncharacterized catechol dioxygenase, termed C12OccaA, eluting at 0.52 ± 0.02 M (NH4)2SO4 in 5-chlorosalicylate-grown cells.
C12OccaA was purified to 95% purity by a two-step procedure (see Materials and Methods). A prominent band of 30 ± 2 kDa observed after SDS-PAGE was subjected to N-terminal sequencing. The determined N terminus (AVSRLAELVTALESD) showed no significant similarity to any proteins in public databases. It thus seems that C12OccaA is only distantly related to previously characterized C12Os.
Kinetic data were measured directly in fractions comprising C12OccaA with a purity of at least 95% of the total protein. Thus, it can be calculated that maximum turnover rates with catechol of 2,375 U/g of protein correspond to activities of 2,500 ± 100 U/g C12OccaA and, based on a subunit molecular mass of 29.424 kDa (as supposed for the predicted amino acid sequence of C12OccaA [see below]), to a kcat value for catechol of 1.2 ± 0.05 s−1 (Table 1). This was approximately 1 order of magnitude lower than those previously reported for C12OcatA and C12OsalD and for other previously analyzed proteobacterial C12Os (6, 37, 49, 51). A high turnover rate was observed only for 4-methylcatechol, and a comparison of specificity constants (kcat/Km) showed 4-methylcatechol to be the highly preferred substrate (Table 1). A similar substrate profile has so far been observed only for C12OsalD, and it contrasts with that reported for either catechol or chlorocatechol 1,2-dioxygenases (3, 6, 11, 45). However, the degree of specificity of C12OccaA was even more remarkable than that of C12OsalD, as specificity constants for 4-methylcatechol compared to those for catechol, 4-chlorocatechol, and 3-methylcatechol differed by factors of 30 to 100. Surprisingly, activity of C12OccaA against 4-chlorocatechol was rather poor and was similar to those of previously described C12Os (11, 30, 38, 51).
Characterization of the cca gene cluster.
To localize genes encoding C12OccaA and MCIccaB, degenerate primers based on the N-terminal sequence were used for the amplification from genomic DNA of a 72-bp DNA segment encoding part of MCIccaB. This allowed the design of a specific primer that, together with a degenerate primer based on a conserved sequence motif identified in both proteobacterial muconate and chloromuconate cycloisomerases, resulted in the amplification of an ∼400-bp DNA fragment. PCR-based screening of a fosmid library of the genome of strain MT1 using primers specific for the gene encoding MCIccaB and that encoding trans-DLH (8) showed that both genes were carried on the same fosmid, which contained an approximately 37.6-kb DNA fragment from MT1.
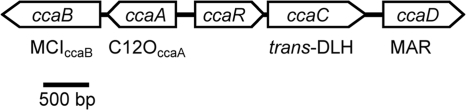
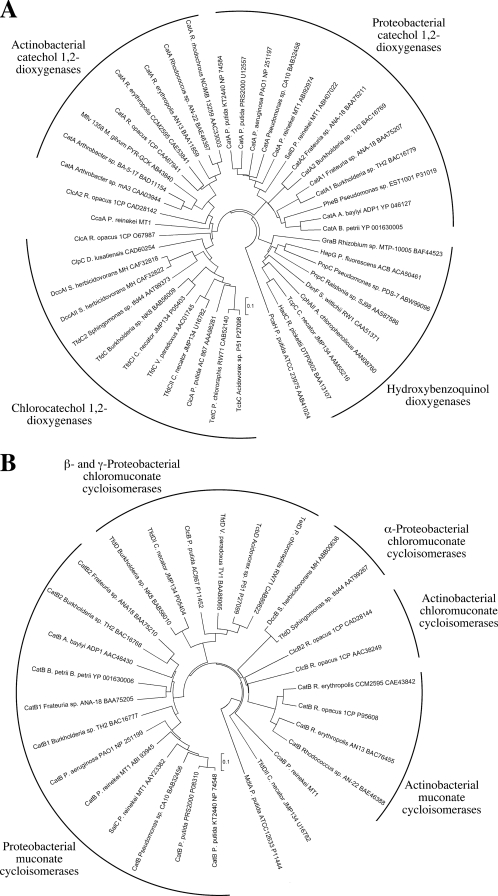
Sequencing of the insert revealed an approximately 5,100-bp region with five open reading frames (ORFs) (Fig. 2) probably involved in the degradation of aromatic compounds by strain MT1. One ORF, designated ccaB, contained the above-identified 400-bp fragment encoding part of MCIccaB and can thus be supposed to encode MCIccaB. The ccaB gene product showed only moderate identity to proteobacterial MCIs (35% to 42%), proteobacterial chloromuconate cycloisomerases (33% to 40%), or muconate and chloromuconate cycloisomerases (35% to 37%) identified in gram-positive microorganisms, which in a phylogenetic analysis form separate branches with low sequence identity to one another (Fig. 3). This indicated that MCIccaB of strain MT1 forms a new branch, illustrating a distinct evolutionary history. Upstream of ccaB, ccaA encoded an enzyme with a deduced N-terminal sequence identical to that of the above-characterized C12OccaA protein. As observed for MCIccaB, in a phylogenetic analysis, C12OccaA does not cluster with any of the previously described separate branches observed in intradiol dioxygenases (Fig. 3) and showed only moderate identities with proteobacterial C12Os (30% to 38%), proteobacterial chlorocatechol 1,2-dioxygenases (32% to 37%), or catechol and chlorocatechol 1,2-dioxygenases (31% to 43%) from gram-positive microorganisms. Lower sequence identity (27% to 33%) was observed with members of the hydroxyquinol branch of intradiol dioxygenases (1, 17). The predicted amino acid sequence of the ORF transcribed divergently toward ccaA and designated ccaR showed up to 47% sequence identity with identified and putative transcriptional regulators of the IclR family, specifically with those of the PobR subfamily of IclR-type regulators, comprising, among others, proteins involved in the transcriptional regulation of protocatechuate or 4-hydroxybenzoate degradative genes (62). The highest sequence identity was observed with a putative IclR regulator of Corynebacterium efficiensYS-314 (accession number BAC19104); however, only slightly lower sequence identity was observed with regulators with identified functions (40% sequence identity with pcaR of P. putida PRS2000, involved in regulation of protocatechuate degradation [50], and 39% sequence identity with pcaR of P. putida WCS358 [2]).
FIG. 2.
Gene organization of a 5,129-bp region from P. reinekei MT1 containing the cca gene cluster. The arrows indicate gene orientations: ccaA, C12O gene; ccaB, MCI gene; ccaC, trans-DLH gene; ccaD, putative MAR gene; and ccaR, putative transcriptional regulator gene. The encoded enzymes are given below the gene clusters.
FIG. 3.
Dendrograms showing the relatedness of intradiol dioxygenases (A) and MCIs (B). The evolutionary history was inferred with MEGA4 (59) using the neighbor-joining algorithm with p-distance correction and pairwise deletion of gaps and missing data. A total of 100 bootstrap replications were performed to test for branch robustness. The scale bars indicate amino acid differences per site.
Downstream of ccaR, the previously described gene encoding trans-DLH (8) and designated ccaC could be localized. The deduced product of the downstream ccaD gene showed the highest sequence homology with MARs, with the highest identity (59%) being observed with MAR TfdF2 of the 2,4-dichlorophenoxyacetic acid-degrading Sphingomonas sp. strain TFD44 (60).
RT-PCR analysis of the cca cluster.
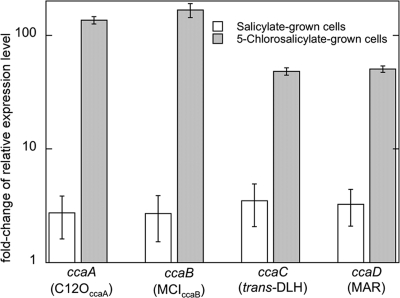
The accumulation of transcripts of ccaA, ccaB, ccaC, and ccaD was measured during growth on 5-chlorosalicylate, salicylate, and acetate (noninducing negative control). When the relative expression levels between the target and the reference gene (rpsL) were compared to those under noninducing conditions (at a ratio of 1), significantly higher levels of ccaA, ccaB, ccaC, and ccaD transcripts were observed only in 5-chlorosalicylate-grown cells (50- to 150-fold) and not in salicylate-grown cells (Fig. 4).
FIG. 4.
Relative expression levels of catabolic genes in salicylate- and 5-chlorosalicylate-grown cells of P. reinekei MT1 as determined by quantitative RT-PCR. The values represent n-fold change (mean of triplicate samples) in the ratio of gene expression between the target gene and the reference gene (rpsL) compared to expression under noninducing conditions (for acetate-grown cells, this ratio was set at 1). The error bars indicate standard deviations.
Induction of C12OccaA and MCIccaB during growth on 5-chlorosalicylate and 4-methylsalicylate.
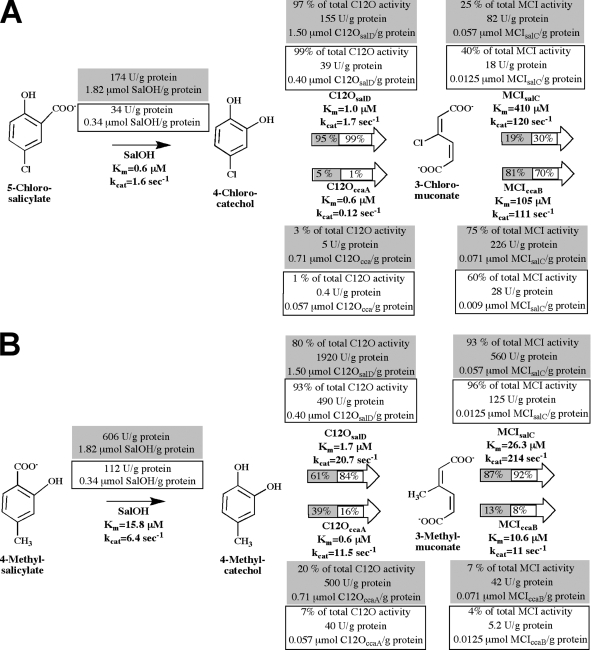
As two of the three C12O-encoding catabolic gene clusters of strain MT1 (the sal gene cluster and the cca gene cluster) were expressed during growth on 5-chlorosalicylate, the importance of the encoded C12Os and MCIs was assessed after growth on 5-chlorosalicylate and 4-methylsalicylate. Cell extracts were separated by anionic-exchange chromatography, fractions were monitored for transformation of 4-methylcatechol and 3-methylmuconate, and the activities were quantified. Both C12O and muconate cycloisomerizing activities could be nearly quantitatively recovered (recovery was >90% for C12O activity against 4-methylcatechol and 85 to 95% for MCI activity against 3-methylmuconate).
Fractions of cell extracts of 5-chlorosalicylate-grown cells eluting at 0.23 ± 0.01 M NaCl and thus containing C12OccaA accounted for only 20% ± 5% of the total activity against 0.1 mM 4-methylcatechol, whereas fractions eluting at 0.28 ± 0.02 M NaCl and corresponding to C12OsalD accounted for 80% ± 5% of the total activity against 0.1 mM 4-methylcatechol (Fig. 5). Analysis of cell extracts from 4-methylsalicylate-grown cells showed that only 7% ± 2% of the total activity against 4-methylcatechol was due to C12OccaA. Similar results were obtained when activities against 0.1 mM 3-methylmuconate were analyzed, with only 7% ± 2% (cell extracts of 5-chlorosalicylate-grown cells) and 4% ± 1% (cell extracts of 4-methylsalicylate-grown cells) of the total activity due to MCIccaB. This indicated that C12OccaA and MCIccaB were of only minor importance during the degradation of 4-methylsalicylate. In contrast, a calculation of the respective activities against 0.1 mM 3-chloromuconate indicated that 75% ± 5% of the total activity in extracts of 5-chlorosalicylate-grown cells was due to induction of MCIccaB, whereas C12OccaA seemed to be of minor importance for 4-chlorocatechol turnover (approximately 1% of the total recovered activity against 0.1 mM 4-chlorocatechol). Calculation of the metabolic flux of 0.1 mM 5-chlorosalicylate or 4-methylsalicylate in cells pregrown in each, based on the kinetic parameters obtained in this study or obtained previously (7) (Fig. 5), supported the notion that 5-chlorosalicylate degradation is driven predominantly by C12OsalD and MCIccaB (95% and 81% of the overall flux in 5-chlorosalicylate-grown cells, respectively) and that C120ccaB is of minor importance. C12OsalD and MCIsalC were of major importance for 4-methylsalicylate degradation (84% and 92% of the overall flux in 4-methylsalicylate-grown cells). It should be noted, however, that the kinetic parameters used for these calculations reflect their activities in the enzymatic test and not necessarily their activities in situ.
FIG. 5.
Metabolism of 5-chlorosalicylate (A) or 4-methylsalicylate (B) by P. reinekei MT1. The kinetic constants of SalOH, C12OsalD, C12OccaA, MCIsalC, and MCIccaB are indicated. The specific activity (U/g protein) was determined in cell extracts, and the contribution of each of the (chloro)catechol 1,2-dioxygenases or (chloro)muconate cycloisomerases to the total activity against 0.1 mM 4-chlorocatechol or 0.1 mM 3-chloromuconate (A) or against 0.1 mM 4-methylcatechol or 0.1 mM 3-methylmuconate (B) in 5-chlorosalicylate-grown (gray) or 4-methylsalicylate-grown (boxed) cells was calculated after enzyme partial purification (given in percent and U/g protein). The enzyme concentrations (μmol/g protein) in the cell extracts were calculated based on the kinetic parameters of the enzyme of interest. The contributions of isoenzymes to the total metabolic flux of 0.1 mM 5-chlorosalicylate or 4-methylsalicylate by 5-chlorosalicylate-grown (gray) or 4-methylsalicylate-grown (boxed) cells were calculated by MATLAB and are given in percentages in the arrows.
DISCUSSION
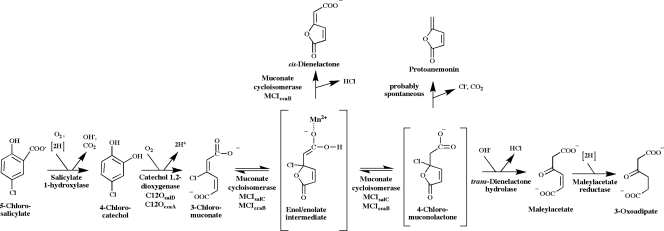
Here, we report the identification of a set of five genes that are located in a 5.1-kb region of the genome of P. reinekei MT1 and that encode enzymes involved in the degradation of 5-chlorosalicylate via 4-chlorocatechol (Fig. 6).
FIG. 6.
Degradation of 5-chlorosalicylate by P. reinekei MT1. Designations of gene products are given below the reaction steps.
In addition to the ccaC gene, encoding trans-DLH (8), this gene cluster comprised genes encoding functional C12OccaA and MCIccaB proteins that were induced when the strain was grown on 5-chlorosalicylate (but also on 4-methylsalicylate). The presence of three distinct sets of (chloro)catechol 1,2-dioxygenases and (chloro)muconate cycloisomerases raises the question of their functions for the degradation of differently substituted salicylates in strain MT1. On one hand, the induction of C12OccaA and MCIccaB during growth on chlorosalicylate indicates their involvement in the degradation of chloroaromatics. On the other hand, C12OccaA was found to be only poorly active against 4-chlorocatechol, the central intermediate of chlorosalicylate degradation by MT1, and in its kinetic properties against catechol and 4-chlorocatechol, this enzyme resembles proteobacterial C12Os (6, 37, 49, 51). In contrast, C12OsalD, being coinduced during growth on chlorosalicylate, was reported to exhibit increased 4-chlorocatechol turnover rates compared with other proteobacterial C12Os (7). In fact, calculation of the relative activities against 4-chlorocatechol in cell extracts and of the metabolic flux indicated that C12OsalD, rather than C12OccaA, drives 4-chlorocatechol metabolism but indicated some importance of C12OccaA for 4-methylcatechol metabolism.
The turnover of intermediate 4-chlorocatechol has been reported to be a pathway bottleneck for the growth of strain MT1 on chlorosalicylates (42), and at higher chlorosalicylate loads, 4-chlorocatechol was shown to accumulate. As chlorinated catechols are highly toxic to eukaryotic and bacterial cells (55), the concomitant accumulation of 4-chlorocatechol results in cell death and termination of degradative performance (43). The induction of two C12Os may result in a more robust degradative phenotype, avoiding to a significant extent the accumulation of 4-chlorocatechol. Accordingly, Perez-Pantoja et al. (43) showed that an efficient turnover of chlorocatechols is essential for the growth of C. necator JMP134 on 3-chlorobenzoate and that multiple copies of a chlorocatechol 1,2-dioxygenase gene are necessary to efficiently deplete chlorocatechols produced during 3-chlorobenzoate turnover by this strain. Taking into account the low turnover rate of both C12OsalD and C12OccaA for 4-chlorocatechol, it can be reasoned that their combined actions are necessary for efficient degradation.
P. reinekei MT1 was originally isolated from a four-member 4-chlorosalicylate-degrading bacterial community in which two other community members, namely, Achromobacter spanius MT3 and Pseudomonas veronii MT4 (41), were supposed to support degradation by depleting toxic metabolites, 4-chlorocatechol and protoanemonin, formed by MT1 during chlorosalicylate metabolism (42). Thus, it seems that MT1 is specifically adapted to degrade chlorosalicylates in concert with those strains due to rather ineffective chlorocatechol-transforming enzymes that are not suited for highly effective mineralization of chlorosalicylates in pure culture (41).
As for ring cleavage activities, two muconate-cycloisomerizing activities were also induced during growth of MT1 on chlorosalicylates. The major difference between these enzymes is the fact that MCIsalC predominantly catalyzes the formation of protoanemonin, a reaction that trans-DLH can interfere with to produce maleylacetate whereas MCIccaB catalyzes the transformation to approximately equal amounts of protoanemonin and cis-dienelactone. As trans-DLH cannot interfere with cis-dienelactone formation, MCIccaB can ensure a rapid metabolism of intermediate 3-chloromuconate but increases the formation of the cis-dienelactone dead-end intermediate. The presence of two MCIs assisting in the metabolism of chlorosalicylates may equip MT1 with a certain level of metabolic flexibility. Evidently, strain MT1 mineralizes 5-chlorosalicylate through a complex metabolic interplay between enzymes encoded by the cca and sal gene clusters.
Specific inactivation of genes of the sal and cca gene clusters will in future clarify their importance for the degradation of chlorosalicylates by strain MT1 and the effects exerted when mutant MT1 strains have to interact with the above-described community members.
Two other catabolic enzymes are encoded in the cca gene cluster. The ccaC gene product (trans-DLH) has recently been described as a zinc-dependent hydrolase (8) that interacts with the cycloisomerization of 3-chloromuconate by hydrolyzing the intermediate 4-chloromuconolactone to maleylacetate (Fig. 6). The ccaD gene obviously encodes a MAR. Genes encoding MARs have initially been observed in chlorocatechol gene operons (28, 36, 56, 57), where the encoding enzymes catalyze a crucial degradation step channeling the substrate into the 3-oxoadipate pathway (47). MARs are also involved in the degradation of chloroaromatics via hydroxybenzoquinols, such as in the degradation of 2,4,5-trichlorophenoxyacetate (25) or 2,4,6-trichlorophenol (34); in the degradation of sulfoaromatics (16, 21); and in the degradation of natural aromatics, such as resorcinol (9, 24).
The cca gene cluster of MT1 not only presents a novel gene arrangement, but specifically comprises enzymes only distantly related (C12OccaA and MCIccaB) or completely unrelated (trans-DLH) to enzymes previously described as involved in catechol or chlorocatechol metabolism. Also unexpected was the observation of a gene encoding an IclR-type regulator transcribed divergently compared to the ccaA and ccaB genes, as catechol and chlorocatechol catabolic gene clusters are commonly under the control of a LysR-type regulator (62). Protocatechuate catabolic gene clusters, in contrast, are usually regulated by IclR-type regulators, such as PcaR of P. putida (50), PcaU of Acinetobacter sp. strain ADP1 (18), PcaR of R. opacus 1CP (14), and PcaQ of Agrobacterium tumefaciens (40). A gene organization similar to that in MT1 has so far been described only by Eulberg and Schlömann (15) for the catABC gene cluster from R. opacus 1CP, where a gene encoding an IclR-type regulator is transcribed divergently to a gene encoding C12O. However, in contrast to the observation by Eulberg and Schlömann, who argued that after the divergence of the cat genes found in Rhodococcus from other catechol genes the original LysR-type regulator gene was replaced by one belonging to the PobR subfamily of IclR-type regulators, no indications of the evolutionary events leading to the development of the MT1 cca cluster can be given at this time, as both C12Occa and MCIccaB seem to represent a new lineage in the phylogeny of intradiol dioxygenases.
It is astonishing that despite the tremendous efforts in sequencing isolates and in isolating new organisms with new catabolic properties, these new lineages have not yet been observed. One of the possible reasons may be the restricted substrate specificity for metabolism of specifically p-substituted catechols and m-substituted muconates. Specifically, the catabolic properties of MCIccaB deserve special attention, as it showed metabolic properties not yet reported for any cycloisomerase, producing both cis-dienelactone (as do chloromuconate cycloisomerases) and protoanemonin (as do MCIs) (4, 39, 53, 54, 58). Studies of the mechanism of MCI have suggested that the reaction proceeds via an enol/enolate to which a proton is added to form muconolactone (19), as depicted in Fig. 6. Similarly, the formation of protoanemonin from 3-chloro-cis,cis-muconate involves a protonation reaction, whereas in the reaction of chloromuconate cycloisomerases with 3-chloromuconate, the corresponding enol/enolate intermediate is not protonated but rather loses the negative charge by chloride abstraction (29). Replacement of Lys169 of P. putida PRS2000 MCI, which is known to provide the proton for the protonation reaction (19, 52), by alanine resulted in mutants that were not able to form protoanemonin but rather formed cis-dienelactone (29). However, as a protonating lysine residue is also conserved in chloromuconate cycloisomerases, as it is in MCIccaB, it was proposed that during the divergence of chloromuconate cycloisomerases from MCIs the rate of chloride elimination from the enol/enolate intermediate was enhanced, even though residues that could accelerate chloride elimination could not yet be identified in chloromuconate cycloisomerases (29). MCIccaB appears from the mechanistic and genetic points of view to be an evolutionary intermediate between chloromuconate cycloisomerases and MCIs, in which the rate of dechlorination was enhanced compared to those of MCIs (as was evident from the formation of cis-dienelactone) but significant rates of proton addition were also observed (as was evident from the formation of protoanemonin). Thus, a detailed analysis of the substrate binding pocket of MCIccaB could reveal important information about residues crucial for dehalogenation.
Acknowledgments
The work was supported by the DFG-European Graduate College 653.
We thank Rita Getzlaff (HZI) for N-terminal protein amino acid sequencing. We gratefully acknowledge Iris Plumeier and Agnes Waliczek for their excellent technical support and Melissa Wos-Oxley for critical reading of the manuscript.
Footnotes
Published ahead of print on 22 May 2009.
REFERENCES
- 1.Armengaud, J., K. N. Timmis, and R. M. Wittich. 1999. A functional 4- hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J. Bacteriol. 1813452-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertani, I., M. Kojic, and V. Venturi. 2001. Regulation of the p-hydroxybenzoic acid hydroxylase gene (pobA) in plant-growth-promoting Pseudomonas putida WCS358. Microbiology 1471611-1620. [DOI] [PubMed] [Google Scholar]
- 3.Bhat, M. A., T. Ishida, K. Horiike, C. S. Vaidyanathan, and M. Nozaki. 1993. Purification of 3,5-dichlorocatechol 1,2-dioxygenase, a nonheme iron dioxygenase and a key enzyme in the biodegradation of a herbicide, 2,4-dichlorophenoxyacetic acid (2,4-D), from Pseudomonas cepacia CSV90. Arch. Biochem. Biophys. 300738-746. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, R., R.-M. Wittich, M. Mallavarapu, K. N. Timmis, and D. H. Pieper. 1995. From xenobiotic to antibiotic. Formation of protoanemonin from 4-chlorocatechol by enzymes of the 3-oxoadipate pathway. J. Biol. Chem. 27029229-29235. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Briganti, F., E. Pessione, C. Giunta, and A. Scozzafava. 1997. Purification, biochemical properties and substrate specificity of a catechol 1,2-dioxygenase from a phenol degrading Acinetobacter radioresistens. FEBS Lett. 41661-64. [DOI] [PubMed] [Google Scholar]
- 7.Cámara, B., P. Bielecki, F. Kaminski, V. M. dos Santos, I. Plumeier, P. Nikodem, and D. H. Pieper. 2007. A gene cluster involved in degradation of substituted salicylates via ortho cleavage in Pseudomonas sp. strain MT1 encodes enzymes specifically adapted for transformation of 4-methylcatechol and 3-methylmuconate. J. Bacteriol. 1891664-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cámara, B., M. Marín, M. Schlömann, H. J. Hecht, H. Junca, and D. H. Pieper. 2008. trans-Dienelactone hydrolase from Pseudomonas reinekei MT1, a novel zinc-dependent hydrolase. Biochem. Biophys. Res. Commun. 376423-428. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, P. J., and D. W. Ribbons. 1976. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J. Bacteriol. 125985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheah, E., C. Austin, G. W. Ashley, and D. Ollis. 1993. Substrate-induced activation of dienelactone hydrolase: an enzyme with a naturally occuring Cys-His-Asp triad. Protein Eng. 6575-583. [DOI] [PubMed] [Google Scholar]
- 11.Dorn, E., and H.-J. Knackmuss. 1978. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem. J. 17485-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorn, E., and H.-J. Knackmuss. 1978. Chemical structure and biodegradability of halogenated aromatic compounds. Two catechol 1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad. Biochem. J. 17473-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumas, J., C. van Delden, K. Perron, and T. Koehler. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 254217-225. [DOI] [PubMed] [Google Scholar]
- 14.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlömann. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 1801072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulberg, D., and M. Schlömann. 1998. The putative regulator of catechol catabolism in Rhodococcus opacus 1CP—an IclR-type, not a LysR-type transcriptional regulator. Antonie van Leeuwenhoek 7471-82. [DOI] [PubMed] [Google Scholar]
- 16.Feigel, B. J., and H.-J. Knackmuss. 1993. Syntrophic interactions during degradation of 4-aminobenzenesulfonic acid by a two species bacterial culture. Arch. Microbiol. 159124-130. [DOI] [PubMed] [Google Scholar]
- 17.Ferraroni, M., J. Seifert, V. M. Travkin, M. Thiel, S. Kaschabek, A. Scozzafava, L. Golovleva, M. Schlömann, and F. Briganti. 2005. Crystal structure of the hydroxyquinol 1,2-dioxygenase from Nocardioides simplex 3E, a key enzyme involved in polychlorinated aromatics biodegradation. J. Biol. Chem. 28021144-21154. [DOI] [PubMed] [Google Scholar]
- 18.Gerischer, U., A. Segura, and L. N. Ornston. 1998. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J. Bacteriol. 1801512-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlt, J. A., and P. G. Gassmann. 1992. Understanding enzyme-catalyzed proton abstraction from carbon acids: details of stepwise mechanisms for β-elimination reactions. J. Am. Chem. Soc. 1145928-5934. [Google Scholar]
- 20.Häggblom, M. M. 1992. Microbial breakdown of halogenated aromatics pesticides and related compounds. FEMS Microbiol. Rev. 10329-72. [DOI] [PubMed] [Google Scholar]
- 21.Halak, S., T. Basta, S. Burger, M. Contzen, and A. Stolz. 2006. Characterization of the genes encoding the 3-carboxy-cis,cis-muconate-lactonizing enzymes from the 4-sulfocatechol degradative pathways of Hydrogenophaga intermedia S1 and Agrobacterium radiobacter S2. Microbiology 1523207-3216. [DOI] [PubMed] [Google Scholar]
- 22.Helin, S., P. C. Kahn, B. L. Guha, D. G. Mallows, and A. Goldman. 1995. The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 Å resolution. J. Mol. Biol. 254918-941. [DOI] [PubMed] [Google Scholar]
- 23.Hoier, H., M. Schlömann, A. Hammer, J. P. Glusker, H. L. Carrell, A. Goldman, J. J. Stezowski, and U. Heinemann. 1994. Crystal structure of cloromuconate cycloisomerase from Alcaligenes eutrophus JMP134 (pJP4) at 3 Å resolution. Acta Crystallogr. 5075-84. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Y., K. X. Zhao, X. H. Shen, M. T. Chaudhry, C. Y. Jiang, and S. J. Liu. 2006. Genetic characterization of the resorcinol catabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 727238-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hübner, A., C. E. Danganan, L. Y. Xun, A. M. Chakrabarty, and W. Hendrickson. 1998. Genes for 2,4,5-trichlorophenoxyacetic acid metabolism in Burkholderia cepacia AC1100: characterization of the tftC and tftD genes and locations of the tft operons on multiple replicons. Appl. Environ. Microbiol. 642086-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Junca, H., and D. H. Pieper. 2004. Functional gene diversity analysis in BTEX contaminated soils by means of PCR-SSCP DNA fingerprinting: comparative diversity assessment against bacterial isolates and PCR-DNA clone libraries. Environ. Microbiol. 695-110. [DOI] [PubMed] [Google Scholar]
- 27.Kaschabek, S. R., and W. Reineke. 1993. Degradation of chloroaromatics: purification and characterization of maleylacetate reductase from Pseudomonas sp. strain B13. J. Bacteriol. 1756075-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaschabek, S. R., and W. Reineke. 1992. Maleylacetate reductase of Pseudomonas sp. strain B13: dechlorination of chloromaleylacetates, metabolites in the degradation of chloroaromatic compounds. Arch. Microbiol. 158412-417. [DOI] [PubMed] [Google Scholar]
- 29.Kaulmann, U., S. R. Kaschabek, and M. Schlömann. 2001. Mechanism of chloride elimination from 3-chloro- and 2,4-dichloro-cis,cis-muconate: new insight obtained from analysis of muconate cycloisomerase variant CatB-K169A. J. Bacteriol. 1834551-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, S. I., S. H. Leem, J. S. Choi, Y. H. Chung, S. Kim, Y. M. Park, Y. K. Park, Y. N. Lee, and K. S. Ha. 1997. Cloning and characterization of two catA genes in Acinetobacter lwoffii K24. J. Bacteriol. 1795226-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhm, A. E., M. Schlömann, H.-J. Knackmuss, and D. H. Pieper. 1990. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP134. Biochem. J. 266877-883. [PMC free article] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 33.Mars, A. E., T. Kasberg, S. R. Kaschabek, M. H. van Agteren, D. B. Janssen, and W. Reineke. 1997. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J. Bacteriol. 1794530-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matus, V., M. A. Sánchez, M. Martínez, and B. González. 2003. Efficient degradation of 2,4,6-trichlorophenol requires a set of catabolic genes related to tcp genes from Ralstonia eutropha JMP134 (pJP4). Appl. Environ. Microbiol. 697108-7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moiseeva, O. V., I. P. Solyanikova, S. R. Kaschabek, J. Groning, M. Thiel, L. A. Golovleva, and M. Schlömann. 2002. A new modified ortho cleavage pathway of 3-chlorocatechol degradation by Rhodococcus opacus 1CP: genetic and biochemical evidence. J. Bacteriol. 1845282-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller, D., M. Schlömann, and W. Reineke. 1996. Maleylacetate reductases in chloroaromatic-degrading bacteria using the modified ortho pathway: comparison of catalytic properties. J. Bacteriol. 178298-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakai, C., K. Horiike, S. Kuramitsu, H. Kagamiyama, and M. Nozaki. 1990. Three isoenzymes of catechol 1,2-dioxygenase (Pyrocatechase), αα, αβ, and ββ, from Pseudomonas arvilla C-1. J. Biol. Chem. 265660-665. [PubMed] [Google Scholar]
- 38.Nakai, C., T. Nakazawa, and M. Nozaki. 1988. Purification and properties of catechol 1,2-dioxygenase (pyrocatechase) from Pseudomonas putida mt-2 in comparison with that from Pseudomonas arvilla C-1. Arch. Biochem. Biophys. 267701-713. [DOI] [PubMed] [Google Scholar]
- 39.Nikodem, P., V. Hecht, M. Schlömann, and D. H. Pieper. 2003. New bacterial pathway for 4- and 5-chlorosalicylate degradation via 4-chlorocatechol and maleylacetate in Pseudomonas sp. strain MT1. J. Bacteriol. 1856790-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parke, D. 1995. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J. Bacteriol. 1773808-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pawelczyk, S., W. R. Abraham, H. Harms, and S. Müller. 2008. Community-based degradation of 4-chlorosalicylate tracked on the single cell level. J. Microbiol. Methods 75117-126. [DOI] [PubMed] [Google Scholar]
- 42.Pelz, O., M. Tesar, R. M. Wittich, E. R. B. Moore, K. N. Timmis, and W. R. Abraham. 1999. Towards elucidation of microbial community metabolic pathways: unravelling the network of carbon sharing in a pollutant-degrading bacterial consortium by immunocapture and isotopic ratio mass spectrometry. Environ. Microbiol. 1167-174. [DOI] [PubMed] [Google Scholar]
- 43.Perez-Pantoja, D., T. Ledger, D. H. Pieper, and B. Gonzalez. 2003. Efficient turnover of chlorocatechols is essential for growth of Ralstonia eutropha JMP134(pJP4) in 3-chlorobenzoic acid. J. Bacteriol. 1851534-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pieper, D. H. 2005. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 67170-191. [DOI] [PubMed] [Google Scholar]
- 45.Potrawfke, T., J. Armengaud, and R. M. Wittich. 2001. Chlorocatechols at positions 4 and 5 are substrates of the broad-spectrum chlorocatechol 1,2-dioxygenase Pseudomonas chlororaphis RW71. J. Bacteriol. 183997-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prucha, M., V. Wray, and D. H. Pieper. 1996. Metabolism of 5-chlorosubstituted muconolactones. Eur. J. Biochem. 237357-366. [DOI] [PubMed] [Google Scholar]
- 47.Reineke, W., and H.-J. Knackmuss. 1988. Microbial degradation of haloaromatics. Annu. Rev. Microbiol. 42263-287. [DOI] [PubMed] [Google Scholar]
- 48.Reineke, W., and H.-J. Knackmuss. 1984. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl. Environ. Microbiol. 47395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ridder, L., F. Briganti, M. Boersma, S. Boeren, E. Vis, A. Scozzafava, C. Veeger, and I. Rietjens. 1998. Quantitative structure/activity relationship for the rate of conversion of C4-substituted catechols by catechol-1,2-dioxygenase from Pseudomonas putida (arvilla) C1. Eur. J. Biochem. 25792-100. [DOI] [PubMed] [Google Scholar]
- 50.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J. Bacteriol. 1765771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sauret-Ignazi, G., J. Gagnon, C. Beguin, M. Barrelle, Y. Markowicz, J. Pelmont, and A. Toussaint. 1996. Characterisation of a chromosomally encoded catechol 1,2-dioxygenase (EC 1.13.11.1) from Alcaligenes eutrophus CH34. Arch. Microbiol. 16642-50. [DOI] [PubMed] [Google Scholar]
- 52.Schell, U., S. Helin, T. Kajander, M. Schlömann, and A. Goldman. 1999. Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates. Proteins 34125-136. [PubMed] [Google Scholar]
- 53.Schlömann, M. 1994. Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation 5301-321. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt, E., and H.-J. Knackmuss. 1980. Chemical structure and biodegradability of halogenated aromatic compounds. Conversion of chlorinated muconic acids into maleoylacetic acid. Biochem. J. 192339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schweigert, N., A. J. B. Zehnder, and R. I. L. Eggen. 2001. Chemical properties of catechols and their molecular modes of toxic action in cells, from microorganisms to mammals. Environ. Microbiol. 381-91. [DOI] [PubMed] [Google Scholar]
- 56.Seibert, V., E. M. Kourbatova, L. A. Golovleva, and M. Schlomann. 1998. Characterization of the maleylacetate reductase MacA of Rhodococcus opacus 1CP and evidence for the presence of an isofunctional enzyme. J. Bacteriol. 1803503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seibert, V., K. Stadler-Fritzsche, and M. Schlömann. 1993. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 1756745-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Solyanikova, I. P., O. V. Malteva, M. D. Vollmer, L. A. Golovleva, and M. Schlömann. 1995. Characterization of muconate and chloromuconate cycloisomerase from Rhodococcus erythropolis 1CP: indications for functionally convergent evolution among bacterial cycloisomerases. J. Bacteriol. 1772821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetic analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 60.Thiel, M., S. Kaschabek, J. Gröning, M. Mau, and M. Schlömann. 2005. Two unusual chlorocatechol catabolic gene clusters in Sphingomonas sp. TFD44. Arch. Microbiol. 18380-94. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tropel, D., and J. R. van der Meer. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68474-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vollmer, M. D., H. Hoier, H. J. Hecht, U. Schell, J. Groning, A. Goldman, and M. Schlömann. 1998. Substrate specificity of and product formation by muconate cycloisomerases: an analysis of wild-type enzymes and engineered variants. Appl. Environ. Microbiol. 643290-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vollmer, M. D., U. Schell, V. Seibert, S. Lakner, and M. Schlömann. 1999. Substrate specificities of the chloromuconate cycloisomerases from Pseudomonas sp. B13, Ralstonia eutropha JMP134 and Pseudomonas sp. P51. Appl. Microbiol. Biotechnol. 51598-605. [DOI] [PubMed] [Google Scholar]
- 65.Vollmer, M. K., and M. Schlömann. 1995. Conversion of 2-chloro-cis,cis-muconate and its matabolites 2-chloro- and 5-chloromuconolactone by chloromuconate cycloisomerases of pJP4 and pAC27. J. Bacteriol. 1772938-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]