Abstract
The NKG2D receptor is one of the most potent activating natural killer cell receptors involved in antiviral responses. The mouse NKG2D ligands MULT-1, RAE-1, and H60 are regulated by murine cytomegalovirus (MCMV) proteins m145, m152, and m155, respectively. In addition, the m138 protein interferes with the expression of both MULT-1 and H60. We show here that one of five RAE-1 isoforms, RAE-1δ, is resistant to downregulation by MCMV and that this escape has functional importance in vivo. Although m152 retained newly synthesized RAE-1δ and RAE-1γ in the endoplasmic reticulum, no viral regulator was able to affect the mature RAE-1δ form which remains expressed on the surfaces of infected cells. This differential susceptibility to downregulation by MCMV is not a consequence of faster maturation of RAE-1δ compared to RAE-1γ but rather an intrinsic property of the mature surface-resident protein. This difference can be attributed to the absence of a PLWY motif from RAE-1δ. Altogether, these findings provide evidence for a novel mechanism of host escape from viral immunoevasion of NKG2D-dependent control.
Cytomegaloviruses (CMVs) are ubiquitous pathogens causing morbidity in immune suppressed and immunodeficient hosts (34). Since CMVs are strictly species-specific viruses, the infection of mice with murine CMV (MCMV) represents a widely used model for studying CMV infection and disease (22, 40).
Natural killer (NK) cells play a crucial role in the control of many viruses and are among the first cells to sense proinflammatory cytokines, as well as the perturbations in the expression of major histocompatibility complex (MHC) class I molecules and other surface molecules induced by viral infection (13). Both human CMV (HCMV) and MCMV have evolved strategies to compromise innate immunity-mediated by NK cells (20, 49).
Although proinflammatory cytokines released during the early stage of MCMV infection induce NK cell activation, this is usually not sufficient for virus control (11). Namely, most mouse strains fail to mount an effector phase of NK cell response against infected cells (42), in spite of the fact that MCMV infection causes the downmodulation of MHC I molecules (17), which should activate NK cells via a “missing-self” mechanism (28). The lack of NK cell activation by MCMV is even more puzzling considering that NK cells possess activating receptors that recognize cellular ligands induced by infection. Among these is the activating receptor NKG2D, a C-type lectinlike receptor encoded by a single gene in humans and rodents (39). Engagement of NKG2D transduces a strong activating signal to promote NK cell stimulation. NKG2D also serves as a costimulatory receptor on CD8+ T cells (2). Several NKG2D ligands have been described in mice: MULT-1, H60a, H60b, H60c, and RAE-1α, -1β, -1γ, -1δ, and -1ɛ isoforms (4-6, 10, 14, 32, 35, 44). What prevents the activation of NK cells via the NKG2D receptor during MCMV infection? We and others have characterized four MCMV proteins involved in the downmodulation of NKG2D ligands (15, 23, 24, 26, 29, 30). Furthermore, the deletion of any of the four MCMV inhibitors of NKG2D ligands rendered virus mutants susceptible to NK cell control in vivo. The MCMV immunoevasin of NKG2D described first was the glycoprotein gp40, encoded by the gene m152 (23). Note that m152 also compromises the CD8+ T-cell response by downregulation of MHC class I molecules (25, 54). Later, it was noticed that m152 also affects the expression of RAE-1 proteins (29). It is important to point out that mouse strains express different RAE-1 isoforms. Some strains, such as BALB/c, express RAE-1α, -1β, and -1γ, while others, such as C57BL/6, express RAE-1δ and -1ɛ (29). All five RAE-1 isoforms are glycosylphosphatidylinositol (GPI)-linked proteins and contain MHC class I-like α1 and α2 domains (6, 10, 14, 35).
Based on our initial observation that there is NKG2D-dependent control of wild type (WT) MCMV in certain mouse strains, we postulated NKG2D ligands that resist virus mediated downmodulation. We show here that the RAE-1 proteins differ in their susceptibility to downregulation by MCMV. In contrast to RAE-1γ, representing the sensitive isoform, surface-resident RAE-1δ remains present on MCMV-infected cells. The differential downmodulation of RAE-1 isoforms during MCMV infection is caused by differences in the stability of the mature RAE-1 molecules associated with a sequence motif absent in RAE-1δ.
MATERIALS AND METHODS
Mice and blockade of NKG2D.
BALB/c (H-2d), C3H/J (H-2k), CBA/J (H-2k), C57BL/6 (H-2b), and DBA/2 (H-2d) mice used in the present study were housed under spefic-pathogen-free conditions at the Central Animal Facility, Faculty of Medicine, University of Rijeka. Mice were injected intravenously (i.v.) with 3 × 105 PFU of tissue culture-grown WT or Δm157 MCMV (3). Salivary gland-derived (SGV) MCMV was injected intraperitoneally (i.p.) at a dose of 5 × 104 PFU. Virus titers in tissues were analyzed at indicated days postinfection (p.i.) as described previously (21). All of the protocols used for breeding of mice and different kinds of treatment were approved by the Ethics Committee of the University of Rijeka and were performed in accordance with Croatian Law for the Protection of Laboratory Animals, which has been harmonized with the existing EU legislation (EC Directive 86/609/EEC). Handling of animals, identification, surgical procedures, and anesthesia were performed using the recommendation described in the Guide for the Care and Use of Laboratory Animals (National Academies Press) and the Act on the Welfare of Animals (NN 19/1999).
The blocking of NKG2D in vivo was performed by i.p. injection of anti-mouse NKG2D monoclonal antibody (MAb; clone C7 [kindly provided by W. M. Yokoyama]) (18) 1 day before infection. CD4 and CD8 T cells were depleted by i.p. injection of the rat MAbs YTS191.1 and YTS169.4, respectively (8). The statistical significance of the difference between experimental groups was determined by the Mann-Whitney exact-rank test.
Cells and transfection conditions.
SVEC4-10 (ATCC CRL-2181) and NIH 3T3 (ATCC CRL-1658) cells were cultivated in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). Mouse embryonic fibroblasts (MEFs) prepared from C3H/J mice were cultivated in DMEM supplemented with 3% FCS (21). FLAG-tagged RAE-1γ was cloned into the SalI restriction site of pB45-Neo plasmid (kindly provided by E. R. Podack, University of Miami School of Medicine, Miami, FL) (37) to generate pB45-Neo-RAE-1γ (described in reference 1). The FLAG tag was introduced into the RAE-1δ gene (GenBank accession no. AAF97631) and the RAE-1δ gene lacking the PLWY by gene synthesis (Ezbiolab). Both synthetic open reading frames were recloned into the SalI site of the pB45-Neo plasmid, generating pB45-Neo-RAE-1δ and pB45-Neo-RAE-1δ-PLWY. These plasmids carrying FLAG-tagged RAE-1γ, RAE-1δ, and RAE-1δ-PLWY constructs were then transfected into NIH 3T3 fibroblasts using SuperFect transfection reagent (Qiagen) according to the manufacturer's instructions, and the cell lines NIH-RAE-1γ, NIH-RAE-1δ, and NIH-RAE-1δ-PLWY expressing the respective constructs were selected in DMEM supplemented with 10% FCS and 750 μg of G418 (Sigma)/ml.
Viruses.
A bacterial artificial chromosome (BAC)-derived MCMV, MW97.01, has previously been shown to be biologically equivalent to the MCMV Smith strain (ATCC VR-1399) and is referred to here as WT MCMV (47). MCMV-gfp recombinant virus, a derivative of the MCMV strain MW97.01 is here referred to as WT-gfp MCMV (33). The recombinant strains lacking the m152 or m157 gene (MCMV-Δm152 and MCMV-Δm157, respectively) were propagated as described previously (3, 46). Tissue culture-grown virus preparations were used for in vivo and in vitro experiments (21). SGV-MCMV was used as a third passage and prepared as described elsewhere (21).
Flow cytometry.
SVEC4-10, NIH 3T3, or RAE-1-transfected cells or C3H/J MEFs were infected with WT MCMV or mutant viruses. Cells were harvested after 2 to 12 h, washed in fluorescence-activated cell sorting medium, and then stained with phycoerythrin (PE)-labeled NKG2D-tetramer (23), biotinylated-mouse anti-FLAG M2 (Sigma) MAb, rat anti-MULT-1 MAb (24), rat anti-RAE-1αβγ (R&D Systems), rat anti-RAE-1αβδɛ (R&D Systems), rat anti-RAE-1ɛ MAb (R&D Systems), or mouse anti-RAE-1δ MAb (immunoglobulin G1 [IgG1], clone RD33). The latter was generated by immunizing mice with baculovirus-expressed RAE-1δ (4), followed by standard hybridoma generation using the Ag8x63 fusion partner. The binding of primary MAbs was visualized by streptavidin-PE, PE-labeled goat anti-rat IgG (Caltag), fluorescein isothiocyanate-labeled goat anti-rat IgG F(ab)2 (Santa Cruz Biotechnology), or biotinylated goat anti-mouse IgG following fluorescein isothiocyanate- or PE-labeled streptavidin (BD Pharmingen).
Confocal microscopic analysis.
RAE-1-transfected NIH 3T3 cells were infected for 12 h with 4 PFU per cell of WT MCMV or Δm152 MCMV or left uninfected. The cells were washed in phosphate-buffered saline (PBS) and fixed and permeabilized using ice-cold methanol or fixed in 2.5% paraformaldehyde and permeabilized using 0.1% Triton X-100. Unspecific binding was blocked in 0.2% fish skin gelatin. The MAbs used are described above in the flow cytometry section. Samples were analyzed with Olympus FV300 confocal laser scanning microscope.
Immunoprecipitation and Western blotting.
The cells were mock treated or infected, as indicated in the specific experiments. The proteins were extracted by using NP-40 lysis buffer (150 mM NaCl, 10 mM Na3PO4 [pH 7.2], 1 mM phenylmethylsulfonyl fluoride, 2 mM EDTA [pH 8], 1% NP-40) for 20 min on ice. To analyze the RAE-1 maturation by Western blotting the cell lysates (100 to 150 μg of proteins) were incubated with 5 mU of EndoH (Roche) at 37°C overnight. If it was required to inhibit glycosylation, the cells were treated before the lysis with tunicamycin (2 μg/ml) for the indicated period of time. For immunoprecipitation, the lysates were precleared with protein G-Sepharose (Amersham), followed by the incubation with anti-FLAG M2 Sepharose (Sigma) for 60 min. The Sepharose beads were then washed and eluted (5 min, 95°C) with nonreducing loading buffer (200 mM Tris [pH 8.8], 1 M sucrose, 1% sodium dodecyl sulfate [SDS], 5 mM EDTA [pH 8.0], 0.02% bromophenol blue) and separated by SDS-polyacrylamide gel electrophoresis (PAGE) using an 11.5% gel. The separated proteins were transferred to Hybond-C membrane (Amersham), and the specific signals were obtained by using rat anti-RAE-1αβδɛ MAbs (R&D Systems) and polyclonal goat anti-RAE-1γ (R&D Systems), followed by peroxidase-labeled goat anti-rat F(ab)2 antibodies (Amersham) and rabbit anti-goat IgG (Wako), respectively. Detection was performed with BM chemiluminescence Western blot kit (Roche).
For cell surface biotinylation, RAE-1 transfectants were grown in 12-well plates, washed twice with PBS before biotinylation buffer (10 mM Na2B4O7·10H2O, 5 mM NaCl, 2 mM CaCl2, 1 mM MgCl2 [pH 8.8]) containing 0.05 mg of Sulfo-NHS-SS-Biotin (Pierce)/ml was added. The biotinylation reaction was performed on ice and stopped after 20 min with the cold stop solution (10 mM glycine in PBS) for 10 min on ice. After three washes with PBS, medium supplemented with 10% fetal bovine serum was added, and the cells were incubated for different periods of time. Cells were lysed by using NP-40 lysis buffer for 20 min on ice. The immunoprecipitation was performed with anti-FLAG M2 Sepharose (Sigma), followed by immunoblotting by SA-POD (Roche). Detection was performed with a BM chemiluminescence Western blot kit (Roche).
For metabolic labeling, cells were labeled either with 300 μCi of [35S]methionine (Amersham)/ml for 2 h or with 500 μCi of [35S]methionine/ml for 1 h at 37°C, washed, and chased at 37°C for the indicated periods of time in DMEM supplemented with 10% FCS. The cells were then washed in PBS and lysed in NP-40 lysis buffer. The cell lysates were precleared with protein G-Sepharose (Amersham), incubated for 1.5 h at 4°C either with 3 μg of anti-RAE-1αβδɛ MAbs (R&D Systems), goat anti-RAE-1γ antibodies (R&D Systems), or anti-RAE-1γ MAbs (eBioscience), followed by 1.5 h of incubation with 60 μl of protein G-Sepharose (Amersham). Samples were eluted (5 min, 95°C) with the reducing loading buffer (200 mM Tris [pH 8.8], 1 M sucrose, 1% SDS, 5 mM EDTA [pH 8.0], 0.02% bromophenol blue, 0.05 M dithiothreitol). If EndoH treatment was performed, the beads were eluted in 100 mM citrate buffer (pH 5.6) containing 10 mM β-mercaptoethanol (Sigma) and 2 mM phenylmethylsulfonyl fluoride (5 min, 95°C) and then treated with 10 mU of EndoH for 12 h or left untreated. Samples were boiled on 95°C and separated by SDS-11.5% PAGE. Dried gels were analyzed by autoradiography by exposition of BioMaxMR films (Kodak) at −80°C for 2 to 3 days.
RESULTS
NKG2D-dependent control of MCMV in vivo.
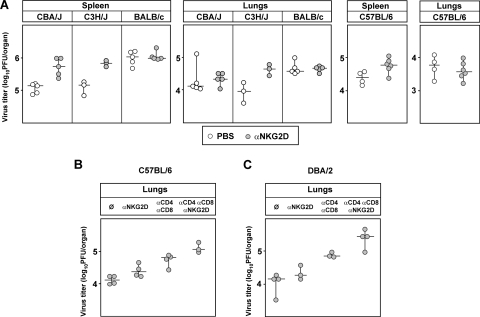
We and others reported that downmodulation of NKG2D ligands prevents the NK cell response in BALB/c mice to WT MCMV infection (23, 29). This is in accordance with in vitro studies showing the complete absence of NKG2D ligands on BALB/c-derived fibroblasts infected with WT MCMV using NKG2D tetramers. In an attempt to explain the resistance of different mouse strains to early MCMV infection, we noticed that in some mouse strains the blocking of NKG2D activity by in vivo administration of anti-NKG2D MAbs resulted in a modest but consistent increase of virus titers in spleen compared to BALB/c mice, where no effect could be detected (Fig. 1A). This effect of NKG2D blocking was less pronounced in lungs, as well as in liver (data not shown). Note that CBA/J, C3H/J, and C57BL/6 mice express RAE-1δ and -1ɛ in contrast to BALB/c mice, which express RAE-1α, -1β, and -1γ (see supplemental Fig. S1 [http://www.medri.hr/∼jstipan/jonjic/]). To test the putative role of NKG2D at later stages of infection, C57BL/6 mice were treated with blocking anti-NKG2D MAbs either alone or in combination with anti-CD4 and anti-CD8 MAbs to deplete T cells. As shown in Fig. 1B, even at 7 days p.i. anti-NKG2D treatment had only modest effects on virus titers in lungs. However, when anti-NKG2D was combined with T-cell depletion, an increase of virus titer was observed exceeding the titers observed by depletion of T cells alone. Thus, the results indicated that the role of NKG2D in NK cell-dependent virus control is detectable in the absence of T cells. The effect of NKG2D blocking was not restricted to control of low-virulent tissue culture-grown MCMV since similar results were obtained in DBA/2 mice 11 days p.i. with the more virulent SGV MCMV (Fig. 1C). These findings prompted us to investigate whether the NKG2D-dependent antiviral activity in WT MCMV-infected mice is a consequence of differential susceptibility of NKG2D ligands to viral regulation.
FIG. 1.
NKG2D-dependent virus control in vivo. (A) CBA/J, C3H/J, and BALB/c mice were injected i.v. with 3 × 105 PFU of WT MCMV (MW97.01). C57BL/6 mice received the same dose of Δm157 MCMV. Mice were injected i.p. with PBS or with blocking anti-NKG2D MAbs. Virus titers were determined 3 days p.i. There were significant differences in virus titers in spleen between the groups of untreated mice and groups treated with anti-NKG2D MAbs: for CBA/J mice (P = 0.008) and for C57BL/6 mice (P = 0.038). Because of small number of animals per group (n = 3) for C3H/J mice, although the virus titer differences are indicative, there were no statistically significant differences. (B) C57BL/6 mice, injected i.p. with either PBS or blocking NKG2D MAbs either alone or in combination with cytolytic anti-CD4 and anti-CD8 MAbs, were injected i.v. with 2 × 105 PFU of Δm157 virus. Virus titers were determined 7 days p.i. There were significant differences between the untreated group and the T-cell-depleted group (P = 0.029), as well as between the untreated group and the group treated with anti-NKG2D, anti-CD4, and anti-CD8 MAbs (P = 0.049). (C) DBA/2 mice were injected i.p. with PBS or blocking NKG2D MAbs or, in addition, with cytolytic anti-CD4 and anti-CD8 MAbs. The mice were also injected i.p. with 5 × 104 PFU of SGV-WT MCMV. Virus titers were determined 11 days p.i. There were significant differences between the untreated group and the T-cell-depleted group (P = 0.049), as well as the untreated group and the group treated with anti-NKG2D, anti-CD4, and anti-CD8 MAbs (P = 0.029). Titers for individual mice (circles) and median values (horizontal bars) are shown.
RAE-1δ is exempt from the downregulation by MCMV.
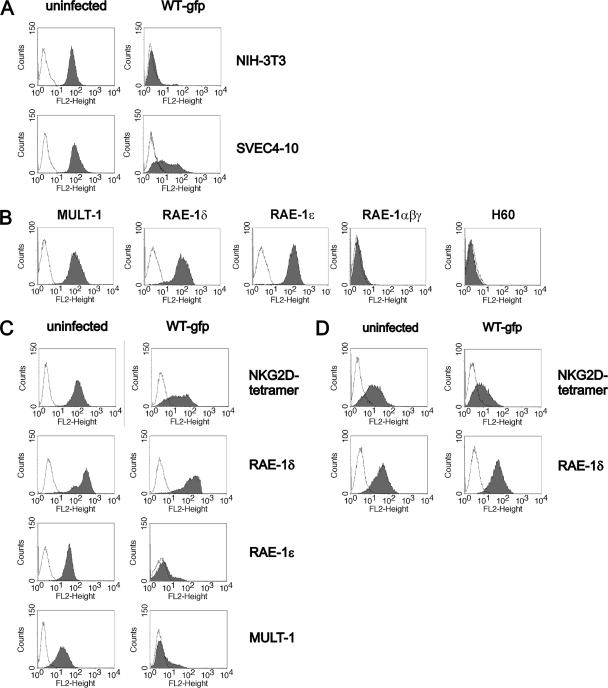
We analyzed the expression of NKG2D ligands on the cell lines derived from individual mouse strains. In contrast to WT MCMV-infected NIH 3T3 cells, in which infection abolishes the expression of NKG2D ligands, SVEC4-10 cells (C3H/J derived) showed only partial reduction of NKG2D binding (Fig. 2A). Specific MAbs demonstrated MULT-1, RAE-1δ, and RAE-1ɛ on the plasma membrane of SVEC4-10 cells, whereas H60 and RAE-1α, -1β, and -1γ were absent (Fig. 2B). In addition, WT MCMV infection resulted in complete downmodulation of surface MULT-1 and RAE-1ɛ, whereas the expression of RAE-1δ was preserved (Fig. 2C). Similar results were obtained with primary MEFs derived from C3H/J (Fig. 2D), CBA/J, and C57BL/6 mice, as well as with C57BL/6-derived cell lines TpnT and IC-21 (data not shown). Altogether, the results showed that RAE-1δ escapes MCMV-mediated downregulation.
FIG. 2.
Different susceptibilities of NKG2D ligands to MCMV. (A) NIH 3T3 and SVEC4-10 cells were infected for 12 h with one PFU of WT-gfp MCMV/cell or left uninfected. Cells were stained with PE-labeled NKG2D tetramer (filled histograms). AV-PE was used as a control (open histograms). (B) Expression of NKG2D ligands on SVEC4-10 cells was tested by specific MAbs: rat anti-MULT-1, mouse anti-RAE-1δ, rat anti-RAE-1ɛ, rat anti-RAE-1αβγ, and rat anti-H60, followed by biotinylated-goat anti-rat IgG or biotinylated-goat anti-mouse IgG and PE-labeled streptavidin (filled histograms). Isotype-matched rat IgG2a and mouse IgG1 MAbs were used as a negative control (open histograms). (C) SVEC4-10 and (D) C3H/J derived MEFs were infected for 12 h with one PFU of WT-gfp MCMV/cell or left uninfected. Cells were stained with PE-labeled NKG2D tetramer or with MAbs to RAE-1δ, RAE-1ɛ, and MULT-1 (filled histograms). Isotype-matched irrelevant MAbs and AV-PE were used as controls for staining with specific antibodies and tetramer, respectively (open histograms).
m152 abolishes the surface expression of RAE-1γ but does not prevent the expression of RAE-1δ.
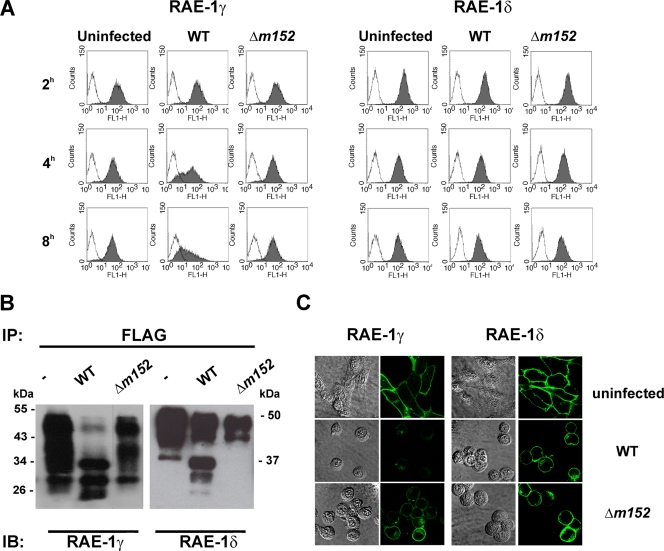
Downmodulation of RAE-1 molecules is mediated by MCMV m152 (23, 29). Conventional mouse strains express either RAE-1α, -1β, and -1γ or RAE-1δ and -1ɛ (29). To avoid the possible impact of MHC class I haplotypes on the susceptibility of RAE-1 isoforms to regulation by MCMV, we generated NIH 3T3 cells stably transfected with RAE-1γ and RAE-1δ. The cell lines were infected with either WT MCMV, the mutant lacking the m152 gene (Δm152), or left uninfected, and the surface expression of RAE-1 was analyzed at different time points p.i. In accordance with published data (29), MCMV infection led to downmodulation of RAE-1γ, which was visible already 4 h p.i. (Fig. 3A, left). The effect was m152 dependent, since the level of RAE-1γ expression in Δm152 mutant-infected cells was comparable to that of not infected cells. In contrast, the expression of RAE-1δ on infected cells was only marginally affected (Fig. 3A, right). Similar to WT MCMV, the m152 revertant virus strain (19) showed differential effect on RAE-1γ and RAE-1δ (see supplemental Fig. S2A [http://www.medri.hr/∼jstipan/jonjic/]).
FIG. 3.
m152 affects the maturation of RAE-1 proteins. RAE-1γ- and RAE-1δ-transfected NIH 3T3 cells were infected with four PFU of WT MCMV or Δm152 MCMV/cell or left uninfected. Cells were analyzed with specific anti-RAE-1 MAbs by flow cytometry (A), by immunoblotting (IB) after immunoprecipitation (IP) with anti-FLAG M2 Sepharose (B), and by confocal microscopy (C).
m152 differentially affects the maturation pattern of RAE-1δ and RAE-1γ.
To gain insight into the mechanism of RAE-1 isoform control by MCMV, we performed immunoprecipitation and confocal analysis on the RAE-1γ and RAE-1δ transfectants. Uninfected, WT MCMV-infected, or Δm152 mutant-infected cells were analyzed 12 h p.i. In contrast to RAE-1δ precipitated from control cells or cells infected with Δm152 mutant, additional RAE-1δ forms of lower molecular size were immunoprecipitated from the lysate of WT MCMV-infected cells (Fig. 3B). The same results were obtained on transfectans expressing RAE-1γ and RAE-1δ after infection with BAC-derived m152 revertant virus (see supplemental Fig. S2B posted at http://www.medri.hr/∼jstipan/jonjic/). The effect of m152 on RAE-1γ was also revealed by accumulation of bands of lower molecular weight representing the immature protein. However, in contrast to RAE-1δ, the loss of mature RAE-1γ protein was almost complete in lysates of WT MCMV-infected cells and RAE-1γ was also not detectable on the surface of WT MCMV-infected cells by confocal microscopy (Fig. 3C). The infection with Δm152 MCMV resulted in the preservation of RAE-1γ display.
This accumulation of lower-molecular-weight forms of RAE-1δ and RAE-1γ resembles the m152 effect on MHC class I molecules, which are retained in the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) (54). However, the RAE-1δ expression on the surface of infected cells is in clear contrast to RAE-1γ and MHC class I downregulation. One explanation for this could be that newly synthesized RAE-1δ can bypass the m152 block; alternatively, the mature cell membrane form of RAE-1δ might be more stable than RAE-1γ, or RAE-1δ might mature and transverse to the cell surface at a faster rate, thus being less sensitive to control by an MCMV early gene such as m152.
The PLWY motif determines resistance of RAE-1δ to MCMV.
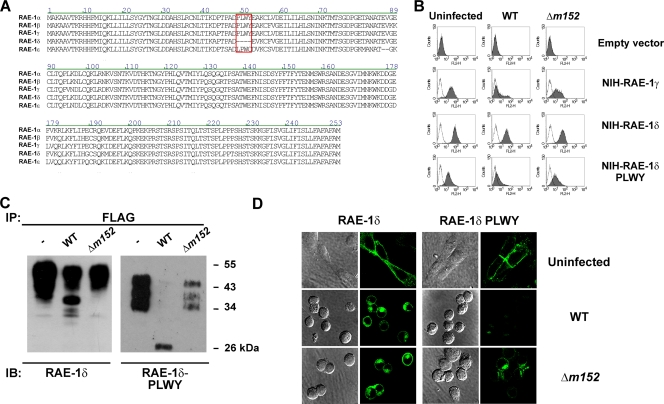
In an attempt to explain the differential susceptibility of RAE-1 to MCMV, we aligned the sequence of RAE-1 isoforms (Vector NTI AlignX; NCBI Pub-Med database search) (Fig. 4A). As described previously (6), the sequences are highly conserved. However, among other differences, the amino acid sequence PLWY (amino acids 49 to 52) present in RAE-1α, -1β, and -1γ was absent in RAE-1δ, whereas a LPWC sequence in RAE-1ɛ was found at the same position. Therefore, transfectants of RAE-1δ with an insertion of the PLWY were generated (RAE-1δ-PLWY) and tested for the effect of WT MCMV and Δm152 MCMV (Fig. 4B). The insertion of the PLWY motif into the RAE-1δ sequence indicated RAE-1δ susceptibility to MCMV regulation. These findings were corroborated by IP (Fig. 4C) and confocal studies (Fig. 4D). Clearly, and in contrast to WT RAE-1δ showing both immature and mature forms in lysates of cells infected with WT MCMV, only the immature form of the RAE-1δ-PLWY could be precipitated from WT MCMV-infected cells. Two models of PLWY regulation of RAE-1 isoforms are possible. First, PLWY itself could serve as a site for the interaction of RAE-1γ with m152 and, second, the PLWY sequence might affects the maturation of the RAE-1 molecules. The lower molecular weight of the mature RAE-1δ-PLWY suggests an effect of the motif on RAE-1 glycosylation (Fig. 4C).
FIG. 4.
The PLWY motif of RAE-1 protein modulates sensitivity to MCMV. (A) Comparison of the amino acid sequences of RAE-1 isoforms using the Vector NT AlignX program (NCBI Pub-Med database search). The PLWY motif (amino acids 49 to 52) is marked. (B) RAE-1γ-, RAE-1δ-, and RAE-1δ-PLWY-transfected NIH 3T3 cells were infected for 12 h with the indicated viruses and analyzed for the expression of RAE-1 isoforms using the anti-FLAG M2 MAb. (C and D) RAE-1δ- and RAE-1δ-PLWY-transfected NIH 3T3 cells were infected for 12 h with four PFU of indicated viruses/cell and analyzed either by immunoblotting (IB) using the anti-RAE-1 MAbs after the immunoprecipitation (IP) with anti-FLAG-M2 Sepharose (C) or by confocal microscopy (D).
m152 does not affect the mature form of RAE-1.
Due to the presence of m152 in MCMV-infected cells, the MHC class I proteins mature only to the stage of EndoH-sensitive glycosylation, indicating that the protein does not exit the ER (54). We tested whether the RAE-1δ forms of lower molecular weight represent proteins retained by the m152 in a compartment with ERGIC/cis-Golgi properties (Fig. 5A). The EndoH treatment of lysates from WT MCMV-infected cells showed that the lower molecular band of RAE-1δ is EndoH sensitive. However, we noticed a shift in mobility after EndoH treatment even in case of mature forms of RAE-1δ and RAE-1γ, suggesting that some of the N-linked carbohydrate chains are still EndoH sensitive. Similar findings were observed for some other cellular (38, 52) and viral (31) glycoproteins.
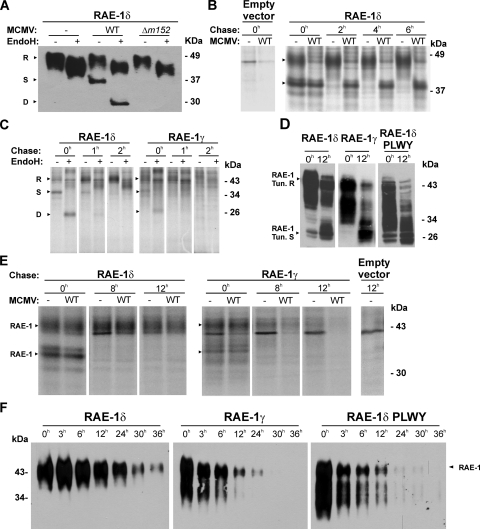
FIG. 5.
Differential stability of RAE-1δ and RAE-1γ mature forms. (A) RAE-1δ-transfected NIH 3T3 cells were infected with four PFU of WT MCMV or Δm152 MCMV/cell or left uninfected. RAE-1δ was immunoblotted from EndoH-treated or untreated lysates with anti-RAE-1δ MAbs. (B) RAE-1δ- or empty vector-transfected NIH 3T3 cells were infected with four PFU of WT MCMV or left uninfected. Cells were metabolically labeled with 300 μCi of [35S]methionine/ml 6 h to 8 h after infection and chased for the indicated periods of time, and immunoprecipitation was performed using anti-RAE-1δ MAbs, followed by protein G-Sepharose. (C) RAE-1δ- or RAE-1γ-transfected NIH 3T3 cells were labeled with 500 μCi of [35S]methionine/ml for 1 h and chased for the indicated periods of time. After immunoprecipitation with either anti-RAE-1δ or anti-RAE-1γ MAbs followed by protein G-Sepharose, eluted proteins were treated with EndoH or left untreated. (D) RAE-1γ-, RAE-1δ-, and RAE-1δ-PLWY-transfected NIH 3T3 cells were untreated or treated for 12 h with tunicamycin (2 μg/ml). Cell lysates were immunoblotted using specific anti-RAE-1 antibodies. (E) RAE-1δ-, RAE-1γ-, or empty vector-transfected NIH 3T3 cells were metabolically labeled with 300 μCi of [35S]methionine/ml for 2 h before infection with four PFU of WT MCMV. Cells were chased for indicated periods of time and immunoprecipitation was performed with either anti-RAE-1δ MAbs or anti-RAE-1γ antibodies followed by protein G-Sepharose. (F) RAE-1δ-, RAE-1γ-, and RAE-1δ-PLWY-transfected NIH 3T3 cells were surface biotinylated. After the indicated periods of time, RAE-1 molecules were immunoprecipitated using anti-FLAG M2-Sepharose, followed by immunoblotting with SA-POD. Arrows indicate different maturation forms of RAE-1 proteins: R, resistant to EndoH; S, sensitive to EndoH; D, digested with EndoH. RAE-1 Tun.R indicates the matured form of RAE-1 which is resistant to tunicamycin, whereas RAE-1 Tun.S indicates the deglycosylated form of RAE-1 proteins by tunicamycin.
The results presented above demonstrated that m152 can retain immature RAE-1δ in the ER despite simultaneous surface expression of this ligand observed on the WT MCMV-infected cells. We therefore tested whether this discrepancy could be explained by the incomplete block of newly synthesized RAE-1δ. Τo that aim, the cells were labeled with [35S]methionine between 6 to 8 h p.i. and chased for different periods of time (Fig. 5B). From uninfected cells at zero time of chase both RAE-1δ forms, the lower- and higher-molecular-weight bands, corresponding to immature and mature forms, were immunoprecipitated. As early as after 2 h of chase, a complete maturation of RAE-1δ was observed. However, in WT MCMV-infected cells the protein remained immature throughout the chase period (Fig. 5B), similar to the effect of m152 on MHC class I molecules (54).
Next, we tested RAE-1δ maturation in comparison to RAE-1γ maturation. To that aim, RAE-1δ and RAE-1γ transfectants were pulsed with [35S]methionine for 1 h and chased. As shown in the Fig. 5C, the rate of maturation of RAE-1δ was comparable to that of RAE-1γ. As early as after 1 h of chase, RAE-1γ reached the fully mature form, whereas a small proportion of RAE-1δ remained EndoH sensitive. These results strongly argue against different maturation rates as a reason for resistance to MCMV infection.
We then tested whether surface expression of RAE-1δ in WT MCMV-infected cells is a consequence of the inability of the virus to affect the RAE-1δ that had matured already prior to infection and m152 expression. To that aim, RAE-1δ or RAE-1γ transfectants were pulsed with [35S]methionine for 2 h prior to infection and chased for up to 12 h. As shown in Fig. 5E, RAE-1δ reached the mature form and remained in this form stably expressed also in WT MCMV-infected cells. The results document a differential susceptibility of immature and mature forms of RAE-1δ to MCMV. In addition, there were differences between the half-lives of mature RAE-1δ and RAE-1γ proteins (Fig. 5E). Prior to chase, both immature and mature RAE-1γ forms were present, similar to RAE-1δ, whereas during chase mature RAE-1γ protein were also lost from the lysates of uninfected and MCMV-infected cells. However, the disappearance of the mature RAE-1γ protein was faster during infection conditions.
The PLWY motif determines the stability of RAE-1 proteins.
The lack of a PLWY motif determined the resistance of surface RAE-1δ to MCMV (Fig. 4), and its role in the RAE-1 glycosylation pattern has been indicated. Since RAE-1δ surface expression in WT MCMV-infected cells cannot be explained by escape of newly synthesized RAE-1δ from the m152-mediated export block (Fig. 5B), we tested whether the PLWY motif defines the stability of RAE-1 isoforms. Therefore, RAE-1γ, RAE-1δ, and RAE-1δ-PLWY transfectants were treated with the glycosylation inhibitor, tunicamycin, and surface biotinylation was carried out. RAE-1 proteins were immunoblotted from the lysates of the untreated cells or cells incubated with tunicamycin (Fig. 5D). The mature form of RAE-1γ was visualized in untreated cells, whereas, after 12 h of tunicamycin treatment, mainly the immature, unglycosylated form of protein was detected. A similar effect on RAE-1γ was observed upon the cycloheximide treatment (data not shown). In contrast, a significant fraction of mature RAE-1δ was still present after 12 h of tunicamycin treatment. The differential stability of the mature RAE-1δ and RAE-1γ forms was affected by the presence of PLWY, since the mature form of RAE-1δ-PLWY showed the pattern of RAE-1γ. The same differences between RAE-1δ, RAE-1γ, and RAE-1δ-PLWY were seen when the analysis addressed mainly the stability of surface resident molecules by biotinylation (Fig. 5F). The reduction of the RAE-1δ signal during a 12 h period after surface labeling is small compared to the short half-life of RAE-1γ and RAE-1δ-PLWY. Altogether, the differential susceptibility of surface RAE-1γ and RAE-1δ to downregulation during MCMV infection is determined by the PLWY motif which affects the intrinsic stability of the mature protein forms.
DISCUSSION
NKG2D is a dominant NK cell activating receptor present on almost all human and mouse NK cells. HCMV and MCMV encode a set of immunevasins which avoid activation of this receptor through downmodulation of NKG2D ligands (20). We report here the inability of MCMV to downmodulate RAE-1δ from the surfaces of infected cells. Our results also showed the functional significance of RAE-1δ escape from MCMV regulation. Although m152 is able to affect all RAE-1 isoforms, it affects only their immature forms. We show that RAE-1γ and RAE-1δ isoforms possess intrinsic differences with respect to the stability of their mature forms. The differential stability of RAE-1 proteins and their susceptibility to MCMV is associated with a PLWY motif which affects RAE-1 glycosylation and leads to the shorter half-life of the mature RAE-1 that possesses this motif.
After cotransfection of 293T cells with m152 and RAE-1 cDNAs (α, β, γ, δ, or ɛ), all RAE-1 proteins were found to be susceptible to the impact of m152 (29). We have shown that the molecular mechanism of the m152-mediated effect is the retention of RAE-1 molecules in the ER/cis-Golgi compartment. However, in contrast to the model used by Lodoen et al. in which the m152 effect was tested in isolation, we used here cell lines that constitutively or stably express RAE-1δ. In the present study, we found that MCMV is unable to downregulate surface-resident RAE-1δ. It is of note that RAE-1 proteins are rapidly induced (6, 16) also upon MCMV infection (29, 44). We demonstrate that newly synthesized RAE-1 molecules mature in less than 2 h. It is to be expected that after MCMV infection significant amounts of RAE-1 proteins are displayed on the plasma membrane well before the function of m152 can take place.
The m152 was first described by its ability to cause the retention of MHC class I in the ER/cis-Golgi compartment (54). The cytoplasmic tail and the transmembrane region of m152 are not relevant for the MHC class I retention by m152 (53). Since RAE-1 proteins are GPI anchored (6, 10), the logical presumption was that the luminal domain of m152 is also involved in its effect on RAE-1. The m152 and MHC class I molecules do not coprecipitate (53), and we were also unable to find complexes of the m152 with RAE-1. Although m152 is able to affect all mouse MHC class I haplotypes (54), there are allele-specific differences in MHC class I downregulation (46). Although all RAE-1 proteins are targets for m152, the RAE-1β isoform shows the highest degree of susceptibility (29). When we first noticed RAE-1δ resistance to MCMV, we predicted that m152 might have lower affinity for this isoform and, in that way, a substantial portion of RAE-1δ could escape m152-mediated blocking. However, immunoprecipitation-Western blot analysis showed nascent RAE-1δ susceptibility. The m152-retained RAE-1δ molecules were not degraded, which resembles the m152 effect on MHC class I molecules (53).
The differential stability of the mature RAE-1γ and -1δ was lost after the insertion of PLWY motif in the RAE-1δ sequence. The PLWY motif did not affect RAE-1δ susceptibility to m152 but affected the stability of the ligand. Insertion of PLWY in RAE-1δ resulted not only in the altered sensitivity to MCMV but also in a significantly lower molecular weight of precipitated proteins resembling the pattern of RAE-1γ. One could speculate that RAE-1δ is differently glycosylated due to the structural effect of the missing PLWY. The crystal structure of murine RAE-1β (27) can serve as a template to determine the PLWY-containing loop and its relationship to the N-linked carbohydrate addition sites. As shown elsewhere (see supplemental Fig. S3 [http://www.medri.hr/∼jstipan/jonjic/]), the PLWY loop lies close to the N70 glycosylation site, while the surface representation shows that both are in a prominent solvent-exposed region. The lack of PLWY, as observed in RAE-1δ, would eliminate the bulky PLWY exposed loop and possibly make N70 more accessible as well. In other words, PLWY might limit the accessibility of N70 to the glycosylation machinery needed for the full maturation of RAE-1 molecules.
Certain human ligands of NKG2D receptor also show differential resistance to viral inhibitors. HCMV UL16 arrests ULBP-1, ULBP-2, and MICB proteins in the ER, but not the ULBP-3, ULBP-4, and MICA family members (9, 12, 41, 48, 51). Sequence differences in the MICA and MICB α2 domains precluded UL16 from binding to MICA and, in addition, the replacement of the MICA α2 domain with the MICB α2 domain resulted in the UL16-sensitive recombinant protein (43). However, another HCMV-encoded viral inhibitor, UL142, was later characterized as a specific downregulator of MICA (7, 50). Interestingly, UL142 is not able to affect surface expression of all MICA alleles; the truncated form of MICA, which is the most common, is completely resistant to the HCMV regulation (55). The K5 protein of the Kaposi sarcoma-associated herpesvirus downregulates the NKG2D ligands MICA and MICB, as well as the AICL ligand for the NKp80 receptor (45). As in the case of UL142, the truncated MICA form lacking the cytoplasmic tail is resistant to K5.
Variations in RAE-1 isoforms are probably consequences of selective pressure of the virus during the evolution. The unique absence of PLWY motif in RAE-1δ may represent a host escape from MCMV. Another counterpart of RAE-1, RAE-1ɛ, possesses a LPWC motif instead of PLWY. Recently, we could show that deletion of m152 only is not sufficient to prevent downmodulation of RAE-1ɛ. MCMV has other means to complete RAE-1ɛ downmodulation, namely, the viral protein m138/fcr1 (1). RAE-1ɛ possesses the highest affinity for the NKG2D receptor of all RAE-1 isoforms. Notably, RAE-1δ has the lowest affinity (5, 36). This might explain a more intensive viral effort to regulate RAE-1ɛ than RAE-1δ surface expression. Still, the phenomenon of RAE-1δ resistance in vivo points to a threat to the virus since the increased resistance of some mouse strains to acute MCMV infection can be attributed to NK/NKG2D-mediated control. Thus, the results presented here emphasize a continuous evolutional struggle between viruses and their hosts. NKG2D, as a dominant activation receptor important not only for the innate but also for the adaptive immune response, is an obvious target.
Acknowledgments
We thank David H. Margulies and Li Zhi for help and critical reading of the manuscript, Mark Lötzerich and Frederic Lemmnitzer for valuable information, Hermine Muehlbach and Christian Mohr for help with transfectants and Edvard Razic for technical help.
This study was supported by Croatian Ministry of Science grants 0621261-1263 (S.J.) and 0621261-1268 (A.K.). A.K. is supported by the Howard Hughes Medical Institute International Research Scholars grant. U.H.K. and Z.R. are supported by the Deutsche Forschungsgemeinschaft through SFB 455. L.N.C. was supported by NIAID K08 AI057361.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Arapovic, J., T. Lenac Rovis, A. B. Reddy, A. Krmpotic, and S. Jonjic. 16 March 2009. Promiscuity of MCMV immunoevasin of NKG2D: m138/fcr-1 down-modulates RAE-1varepsilon in addition to MULT-1 and H60. Mol. Immunol. doi: 10.1016/j.molimm.2009.02.010. [DOI] [PubMed]
- 2.Bauer, S., V. Groh, J. Wu, A. Steinle, J. H. Phillips, L. L. Lanier, and T. Spies. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285727-729. [DOI] [PubMed] [Google Scholar]
- 3.Bubic, I., M. Wagner, A. Krmpotic, T. Saulig, S. Kim, W. M. Yokoyama, S. Jonjic, and U. H. Koszinowski. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 787536-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carayannopoulos, L. N., O. V. Naidenko, D. H. Fremont, and W. M. Yokoyama. 2002. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 1694079-4083. [DOI] [PubMed] [Google Scholar]
- 5.Carayannopoulos, L. N., O. V. Naidenko, J. Kinder, E. L. Ho, D. H. Fremont, and W. M. Yokoyama. 2002. Ligands for murine NKG2D display heterogeneous binding behavior. Eur. J. Immunol. 32597-605. [DOI] [PubMed] [Google Scholar]
- 6.Cerwenka, A., A. B. Bakker, T. McClanahan, J. Wagner, J. Wu, J. H. Phillips, and L. L. Lanier. 2000. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity 12721-727. [DOI] [PubMed] [Google Scholar]
- 7.Chalupny, N. J., A. Rein-Weston, S. Dosch, and D. Cosman. 2006. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem. Biophys. Res. Commun. 346175-181. [DOI] [PubMed] [Google Scholar]
- 8.Cobbold, S. P., A. Jayasuriya, A. Nash, T. D. Prospero, and H. Waldmann. 1984. Therapy with monoclonal antibodies by elimination of T-cell subsets in vivo. Nature 312548-551. [DOI] [PubMed] [Google Scholar]
- 9.Cosman, D., J. Mullberg, C. L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N. J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14123-133. [DOI] [PubMed] [Google Scholar]
- 10.Diefenbach, A., A. M. Jamieson, S. D. Liu, N. Shastri, and D. H. Raulet. 2000. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 1119-126. [DOI] [PubMed] [Google Scholar]
- 11.Dokun, A. O., S. Kim, H. R. Smith, H. S. Kang, D. T. Chu, and W. M. Yokoyama. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2951-956. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, C., N. J. Chalupny, C. L. Sutherland, S. Dosch, P. V. Sivakumar, D. C. Johnson, and D. Cosman. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J. Exp. Med. 1971427-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French, A. R., and W. M. Yokoyama. 2003. Natural killer cells and viral infections. Curr. Opin. Immunol. 1545-51. [DOI] [PubMed] [Google Scholar]
- 14.Girardi, M., D. E. Oppenheim, C. R. Steele, J. M. Lewis, E. Glusac, R. Filler, P. Hobby, B. Sutton, R. E. Tigelaar, and A. C. Hayday. 2001. Regulation of cutaneous malignancy by gammadelta T cells. Science 294605-609. [DOI] [PubMed] [Google Scholar]
- 15.Hasan, M., A. Krmpotic, Z. Ruzsics, I. Bubic, T. Lenac, A. Halenius, A. Loewendorf, M. Messerle, H. Hengel, S. Jonjic, and U. H. Koszinowski. 2005. Selective down-regulation of the NKG2D ligand H60 by mouse cytomegalovirus m155 glycoprotein. J. Virol. 792920-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayakawa, Y., J. M. Kelly, J. A. Westwood, P. K. Darcy, A. Diefenbach, D. Raulet, and M. J. Smyth. 2002. Cutting edge: tumor rejection mediated by NKG2D receptor-ligand interaction is dependent upon perforin. J. Immunol. 1695377-5381. [DOI] [PubMed] [Google Scholar]
- 17.Hengel, H., U. Reusch, A. Gutermann, H. Ziegler, S. Jonjic, P. Lucin, and U. H. Koszinowski. 1999. Cytomegaloviral control of MHC class I function in the mouse. Immunol. Rev. 168167-176. [DOI] [PubMed] [Google Scholar]
- 18.Ho, E. L., L. N. Carayannopoulos, J. Poursine-Laurent, J. Kinder, B. Plougastel, H. R. Smith, and W. M. Yokoyama. 2002. Costimulation of multiple NK cell activation receptors by NKG2D. J. Immunol. 1693667-3675. [DOI] [PubMed] [Google Scholar]
- 19.Holtappels, R., J. Podlech, M. F. Pahl-Seibert, M. Julch, D. Thomas, C. O. Simon, M. Wagner, and M. J. Reddehase. 2004. Cytomegalovirus misleads its host by priming of CD8 T cells specific for an epitope not presented in infected tissues. J. Exp. Med. 199131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonjic, S., M. Babic, B. Polic, and A. Krmpotic. 2008. Immune evasion of natural killer cells by viruses. Curr. Opin. Immunol. 2030-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonjic, S., A. Krmpotic, J. Arapovic, and U. H. Koszinowski. 2008. Dissection of the antiviral NK cell response by MCMV mutants. Methods Mol. Biol. 415127-149. [DOI] [PubMed] [Google Scholar]
- 22.Krmpotic, A., I. Bubic, B. Polic, P. Lucin, and S. Jonjic. 2003. Pathogenesis of murine cytomegalovirus infection. Microbes Infect. 51263-1277. [DOI] [PubMed] [Google Scholar]
- 23.Krmpotic, A., D. H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A. A. Scalzo, U. H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3529-535. [DOI] [PubMed] [Google Scholar]
- 24.Krmpotic, A., M. Hasan, A. Loewendorf, T. Saulig, A. Halenius, T. Lenac, B. Polic, I. Bubic, A. Kriegeskorte, E. Pernjak-Pugel, M. Messerle, H. Hengel, D. H. Busch, U. H. Koszinowski, and S. Jonjic. 2005. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J. Exp. Med. 201211-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krmpotic, A., M. Messerle, I. Crnkovic-Mertens, B. Polic, S. Jonjic, and U. H. Koszinowski. 1999. The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T-cell control in vivo. J. Exp. Med. 1901285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenac, T., M. Budt, J. Arapovic, M. Hasan, A. Zimmermann, H. Simic, A. Krmpotic, M. Messerle, Z. Ruzsics, U. H. Koszinowski, H. Hengel, and S. Jonjic. 2006. The herpesviral Fc receptor fcr-1 down-regulates the NKG2D ligands MULT-1 and H60. J. Exp. Med. 2031843-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, P., G. McDermott, and R. K. Strong. 2002. Crystal structures of RAE-1β and its complex with the activating immunoreceptor NKG2D. Immunity 1677-86. [DOI] [PubMed] [Google Scholar]
- 28.Ljunggren, H. G., and K. Karre. 1990. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 11237-244. [DOI] [PubMed] [Google Scholar]
- 29.Lodoen, M., K. Ogasawara, J. A. Hamerman, H. Arase, J. P. Houchins, E. S. Mocarski, and L. L. Lanier. 2003. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J. Exp. Med. 1971245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lodoen, M. B., G. Abenes, S. Umamoto, J. P. Houchins, F. Liu, and L. L. Lanier. 2004. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J. Exp. Med. 2001075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, X., D. G. Kavanagh, and A. B. Hill. 2006. Cellular and molecular requirements for association of the murine cytomegalovirus protein m4/gp34 with major histocompatibility complex class I molecules. J. Virol. 806048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malarkannan, S., P. P. Shih, P. A. Eden, T. Horng, A. R. Zuberi, G. Christianson, D. Roopenian, and N. Shastri. 1998. The molecular and functional characterization of a dominant minor H antigen, H60. J. Immunol. 1613501-3509. [PubMed] [Google Scholar]
- 33.Mathys, S., T. Schroeder, J. Ellwart, U. H. Koszinowski, M. Messerle, and U. Just. 2003. Dendritic cells under influence of mouse cytomegalovirus have a physiologic dual role: to initiate and to restrict T-cell activation. J. Infect. Dis. 187988-999. [DOI] [PubMed] [Google Scholar]
- 34.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 35.Nomura, M., Z. Zou, T. Joh, Y. Takihara, Y. Matsuda, and K. Shimada. 1996. Genomic structures and characterization of Rae1 family members encoding GPI-anchored cell surface proteins and expressed predominantly in embryonic mouse brain. J. Biochem. 120987-995. [DOI] [PubMed] [Google Scholar]
- 36.O'Callaghan, C. A., A. Cerwenka, B. E. Willcox, L. L. Lanier, and P. J. Bjorkman. 2001. Molecular competition for NKG2D: H60 and RAE1 compete unequally for NKG2D with dominance of H60. Immunity 15201-211. [DOI] [PubMed] [Google Scholar]
- 37.Ohe, Y., D. Zhao, N. Saijo, and E. R. Podack. 1995. Construction of a novel bovine papillomavirus vector without detectable transforming activity suitable for gene transfer. Hum. Gene Ther. 6325-333. [DOI] [PubMed] [Google Scholar]
- 38.Omary, M. B., and I. S. Trowbridge. 1981. Biosynthesis of the human transferrin receptor in cultured cells. J. Biol. Chem. 25612888-12892. [PubMed] [Google Scholar]
- 39.Raulet, D. H. 2003. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 3781-790. [DOI] [PubMed] [Google Scholar]
- 40.Reddehase, M. J., J. Podlech, and N. K. Grzimek. 2002. Mouse models of cytomegalovirus latency: overview. J. Clin. Virol. 25(Suppl. 2)S23-S36. [DOI] [PubMed] [Google Scholar]
- 41.Rolle, A., M. Mousavi-Jazi, M. Eriksson, J. Odeberg, C. Soderberg-Naucler, D. Cosman, K. Karre, and C. Cerboni. 2003. Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein. J. Immunol. 171902-908. [DOI] [PubMed] [Google Scholar]
- 42.Scalzo, A. A., A. J. Corbett, W. D. Rawlinson, G. M. Scott, and M. A. Degli-Esposti. 2007. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol. Cell Biol. 8546-54. [DOI] [PubMed] [Google Scholar]
- 43.Spreu, J., T. Stehle, and A. Steinle. 2006. Human cytomegalovirus-encoded UL16 discriminates MIC molecules by their alpha2 domains. J. Immunol. 1773143-3149. [DOI] [PubMed] [Google Scholar]
- 44.Takada, A., S. Yoshida, M. Kajikawa, Y. Miyatake, U. Tomaru, M. Sakai, H. Chiba, K. Maenaka, D. Kohda, K. Fugo, and M. Kasahara. 2008. Two novel NKG2D ligands of the mouse H60 family with differential expression patterns and binding affinities to NKG2D. J. Immunol. 1801678-1685. [DOI] [PubMed] [Google Scholar]
- 45.Thomas, M., J. M. Boname, S. Field, S. Nejentsev, M. Salio, V. Cerundolo, M. Wills, and P. J. Lehner. 2008. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 1051656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 737056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welte, S. A., C. Sinzger, S. Z. Lutz, H. Singh-Jasuja, K. L. Sampaio, U. Eknigk, H. G. Rammensee, and A. Steinle. 2003. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur. J. Immunol. 33194-203. [DOI] [PubMed] [Google Scholar]
- 49.Wilkinson, G. W., P. Tomasec, R. J. Stanton, M. Armstrong, V. Prod'homme, R. Aicheler, B. P. McSharry, C. R. Rickards, D. Cochrane, S. Llewellyn-Lacey, E. C. Wang, C. A. Griffin, and A. J. Davison. 2008. Modulation of natural killer cells by human cytomegalovirus. J. Clin. Virol. 41206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wills, M. R., O. Ashiru, M. B. Reeves, G. Okecha, J. Trowsdale, P. Tomasec, G. W. Wilkinson, J. Sinclair, and J. G. Sissons. 2005. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J. Immunol. 1757457-7465. [DOI] [PubMed] [Google Scholar]
- 51.Wu, J., N. J. Chalupny, T. J. Manley, S. R. Riddell, D. Cosman, and T. Spies. 2003. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J. Immunol. 1704196-4200. [DOI] [PubMed] [Google Scholar]
- 52.Yuan, W., A. Dasgupta, and P. Cresswell. 2006. Herpes simplex virus evades natural killer T-cell recognition by suppressing CD1d recycling. Nat. Immunol. 7835-842. [DOI] [PubMed] [Google Scholar]
- 53.Ziegler, H., W. Muranyi, H. G. Burgert, E. Kremmer, and U. H. Koszinowski. 2000. The luminal part of the murine cytomegalovirus glycoprotein gp40 catalyzes the retention of MHC class I molecules. EMBO J. 19870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziegler, H., R. Thäle, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U. H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 657-66. [DOI] [PubMed] [Google Scholar]
- 55.Zou, Y., W. Bresnahan, R. T. Taylor, and P. Stastny. 2005. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J. Immunol. 1743098-3104. [DOI] [PubMed] [Google Scholar]