Abstract
The positive-strand RNA genome of Japanese encephalitis virus (JEV) terminates in a highly conserved 3′-noncoding region (3′NCR) of six domains (V, X, I, II-1, II-2, and III in the 5′-to-3′ direction). By manipulating the JEV genomic RNA, we have identified important roles for RNA elements present within the 574-nucleotide 3′NCR in viral replication. The two 3′-proximal domains (II-2 and III) were sufficient for RNA replication and virus production, whereas the remaining four (V, X, I, and II-1) were dispensable for RNA replication competence but required for maximal replication efficiency. Surprisingly, a lethal mutant lacking all of the 3′NCR except domain III regained viability through pseudoreversion by duplicating an 83-nucleotide sequence from the 3′-terminal region of the viral open reading frame. Also, two viable mutants displayed severe genetic instability; these two mutants rapidly developed 12 point mutations in domain II-2 in the mutant lacking domains V, X, I, and II-1 and showed the duplication of seven upstream sequences of various sizes at the junction between domains II-1 and II-2 in the mutant lacking domains V, X, and I. In all cases, the introduction of these spontaneous mutations led to an increase in RNA production that paralleled the level of protein accumulation and virus yield. Interestingly, the mutant lacking domains V, X, I, and II-1 was able to replicate in hamster BHK-21 and human neuroblastoma SH-SY5Y cells but not in mosquito C6/36 cells, indicating a cell type-specific restriction of its viral replication. Thus, our findings provide the basis for a detailed map of the 3′ cis-acting elements in JEV genomic RNA, which play an essential role in viral replication. They also provide experimental evidence for the function of 3′ direct repeat sequences and suggest possible mechanisms for the emergence of these sequences in the 3′NCR of JEV and perhaps in other flaviviruses.
Japanese encephalitis virus (JEV), a mosquito-borne flavivirus of the family Flaviviridae, is serologically related to several significant human pathogens, including West Nile virus (WNV), Kunjin virus (KUNV), St. Louis encephalitis virus, and Murray Valley encephalitis virus. It is also phylogenetically close to other clinically important human pathogens, including yellow fever virus (YFV) and dengue virus (DENV) (11, 67). JEV is the leading cause of viral encephalitis in Southeast Asia, including China, Japan, Korea, the Philippines, Thailand, and India, and it has begun to expand throughout the Indonesian archipelago and as far as Australia (21, 43). Despite the fact that JEV is generally asymptomatic, ∼50,000 cases are reported annually, and the disease has a mortality rate of ∼25%, mainly in children and young adults (29, 63). Thus, the geographic expansion and clinical importance of JEV infection have drawn increasing attention from the international public health community (44, 71).
Like other flaviviruses, JEV is a spherical enveloped virus (∼50 nm diameter) with a single-stranded positive-sense RNA genome that contains a 5′ cap structure but lacks a 3′ polyadenylated tail. Its genomic RNA of ∼11,000 nucleotides (nt) consists of a single long open reading frame (ORF) with two noncoding regions (NCRs) at the 5′ and 3′ ends (41, 84). The ORF is translated into an ∼3,400-amino acid polyprotein precursor, which is co- or posttranslationally cleaved by a cellular protease(s) or a viral protease complex into 10 mature proteins: (i) three structural proteins, the capsid (C), premembrane (prM; which is further processed into pr and M), and envelope (E) proteins; and (ii) seven nonstructural proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5, as arranged in the genome (13, 41, 84). The nonstructural proteins, together with cellular factors, form a viral replicase complex that directs the replication of the genomic RNA in the cytoplasm of the host cell in association with perinuclear membranes (40, 74). For the synthesis of the genomic RNA to take place, this replicase complex must specifically recognize viral cis-acting RNA elements, defined by primary sequences or secondary/tertiary structures. These RNA elements are found in various locations within the genome but most frequently are located in the 5′- and 3′NCRs (23, 47). The identification and characterization of these cis-acting RNA elements is critical for understanding the complete cycle of JEV genome replication.
The availability of the complete nucleotide sequence of YFV genomic RNA (57) has led to the identification of three major conserved elements in the 5′- and 3′-terminal regions of the genomic RNA that contain the short primary sequences and secondary structures required for flavivirus RNA replication. (i) Both ends of the genomic RNA terminate with the conserved dinucleotides 5′-AG and CU-3′ (9, 10, 32, 45, 57, 72, 73) in all flaviviruses except an insect cell fusing agent virus (12). Mutations substituting another nucleotide for one of these four nucleotides in KUNV or WNV replicon RNA are known to abolish or compromise RNA replication (35, 69). (ii) A 3′ stem-loop structure (3′SL) has been recognized in all flaviviruses within the ∼90-nt 3′-terminal region of the genomic RNA (9, 45, 57). The structural and functional importance of this 3′SL in RNA replication has been demonstrated in several flaviviruses (9, 18, 49, 50, 61, 70, 82, 86). (iii) The presence of short 5′ and 3′ cyclization sequences (5′CYC and 3′CYC, respectively) in all mosquito-borne flaviviruses suggests that flavivirus genomes can cyclize via 5′-3′ long-range base-pairing interaction, since the 3′CYC upstream of the 3′SL is complementary to the 5′CYC in the 5′ coding region of the C protein (30). The role of these CYC motifs in RNA replication has been well characterized via cell-based assays in many mosquito-borne flaviviruses, including KUNV (34), WNV (42), YFV (8, 14), and DENV (2, 22, 49), and in cell-free systems in the case of WNV (51) and DENV (1, 3, 79, 80). Other RNA elements that have recently been shown to be important for RNA replication in DENV and WNV include an additional pair of complementary sequences (designated 5′- and 3′UARs) that participate in genome cyclization (3, 4, 17, 87) and a 5′ stem-loop structure (designated 5′SLA) present within the 5′NCR that promotes RNA synthesis in association with the 3′NCR (22).
In all flaviviruses, the 3′NCR of the genomic RNA is relatively long (∼400 to ∼800 nt), with an array of conserved primary sequences and secondary structures. Although significant progress has been made in identifying cis-acting elements within the 3′NCRs that are essential for RNA replication, most of these elements (i.e., the 3′CYC, 3′SL, and CU-3′) are limited to the ∼100-nt 3′-terminal region that is highly conserved in these viruses (see recent reviews in references 23 and 47). However, the functional importance of the remaining 5′-proximal region of the 3′NCR, which differs in sequence between the various serological groups, is poorly understood. In particular, comparative sequence analyses and genetic algorithm-based computer modeling have suggested that in addition to the well-studied ∼100-nt 3′-proximal region, the remaining ∼474-nt 5′-proximal region of the 574-nt JEV 3′NCR also contains several RNA elements that may play critical roles in the viral life cycle (52, 55, 56, 68). To date, however, experimental evidence for the functional importance of these potential RNA elements in JEV genomic RNA replication is lacking.
In the present study, we have identified and characterized the 3′ cis-acting RNA elements within the JEV 3′NCR and shown that they play an essential and/or regulatory role in genomic RNA replication. In particular, we have constructed and functionally characterized genome-length JEV mutant cDNAs with a series of 5′-to-3′ or 3′-to-5′ progressive deletions within the 3′NCR. In addition to identifying particular mutations within this region that affect either the competence or efficiency of genomic RNA replication, we found that the serial passaging of these mutants in susceptible BHK-21 cells produced a large number of pseudorevertants bearing a wide variety of spontaneous point mutations and sequence duplications, some of which were capable of restoring the replication competence of the defective mutants or enhancing replication efficiency. In addition, we assessed the replication of these mutants in three different cell types (BHK-21, SH-SY5Y, and C6/36 cells). Collectively, these data offer new insights into the functional importance of 3′ cis-acting RNA elements that regulate the cell type-dependent replication of JEV and perhaps other closely related mosquito-borne flaviviruses. Our findings also provide experimental evidence for the emergence of functional 3′ direct repeat sequences that are duplicated from the coding region and 3′NCR of JEV genomic RNA.
MATERIALS AND METHODS
Cells.
The three cell lines used in this study, baby hamster kidney (BHK-21), human neuroblastoma (SH-SY5Y), and mosquito Aedes albopictus (C6/36), were cultivated as described previously (36). All reagents used for cell culture were purchased from Invitrogen-Gibco, Carlsbad, CA.
Antibodies.
A mouse polyclonal JEV-specific antiserum was purchased from the American Type Culture Collection (36). A rabbit polyclonal antiserum specific for JEV NS1 (166 amino acids, corresponding to nt 2,478 to 2,975) was produced by immunizing New Zealand White rabbits with a glutathione S-transferase-tagged recombinant protein consisting of the defined coding region of NS1 fused to the C terminus of glutathione S-transferase (64). Nucleotide positions refer to the complete genome sequence of the JEV CNU/LP2 strain (GenBank accession no. AY585243). A rabbit polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antiserum was obtained from LabFrontier, Seoul, South Korea (36). Horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG) and alkaline phosphatase (AP)-conjugated goat anti-mouse and anti-rabbit IgGs were purchased from Jackson ImmunoResearch Laboratories, West Grove, PA.
Mutagenesis of the full-length infectious JEV cDNA.
Standard recombinant DNA techniques were used to construct the recombinant molecular clones for JEV (59). All JEV 3′NCR mutations were introduced into the full-length infectious JEV cDNA molecular clone, pBACSP6/JVFLx/XbaI (83), designated the wild type (WT) in this report; the complete genome sequence of WT-derived JEV (CNU/LP2x) has been deposited under GenBank accession no. GQ199609. Mutations were generated by PCR-based mutagenesis, and all PCR-derived DNA fragments were sequenced to confirm the presence of the intended mutations and the absence of undesired mutations. The recombinant plasmids were purified by standard CsCl/ethidium bromide density centrifugation; the integrity of all plasmids was checked by extensive restriction endonuclease mapping and sequencing. The oligonucleotides used for cDNA synthesis, PCR amplification, and mutagenesis in this study are summarized in Table 1.
TABLE 1.
Oligonucleotides used for cDNA synthesis, PCR amplification, and mutagenesis
| Oligonucleotide | Sequencea (5′ to 3′) | Polarity |
|---|---|---|
| JE3ncrF | AAGCATATGCACAGATGTGGCTACTCC | Sense |
| JE3ncrR | agtcggtccgcggccgctctagAGATCCTGTG | Antisense |
| JEΔ133F | AGGGTCATCTAGCAAATCTGACAACAGAAAGT | Sense |
| JEΔ133R | TTGTCAGATTTGCTAGATGACCCTGTCTTCCT | Antisense |
| JEΔ211F | AGGGTCATCTAGCAACGTCAGGCCACGAATTT | Sense |
| JEΔ211R | GGCCTGACGTTGCTAGATGACCCTGTCTTCCT | Antisense |
| JEΔ284F | AGGGTCATCTAGCAAAGCCGTTGAGGCCCCCA | Sense |
| JEΔ284R | TCAACGGCTTTGCTAGATGACCCTGTCTTCCT | Antisense |
| JEΔ381F | AGGGTCATCTAGCATGCGGCCCAAGCCCCCTC | Sense |
| JEΔ381R | TTGGGCCGCATGCTAGATGACCCTGTCTTCCT | Antisense |
| JEΔ450F | AGGGTCATCTAGTTGCATCAAACAGCATATTG | Sense |
| JEΔ450R | TGTTTGATGCAACTAGATGACCCTGTCTTCCT | Antisense |
| JEΔ473F | AGGGTCATCTAGCCTGGGAATAGACTGGGAGA | Sense |
| JEΔ473R | TCTATTCCCAGGCTAGATGACCCTGTCTTCCT | Antisense |
| JEΔ574R | tccgcggccgctctagAGGTGTCAATATGCTGTTT | Antisense |
| JEcircleF | gatgagctcCACTCAGGAGATACGAAGAC | Sense |
| OligoT | ccagtgttgtggcctgcagggcgaatt | Sense |
| OligoTR | gatgaattcgccctgcaggccacaaca | Antisense |
JEV-specific sequences are shown in uppercase letters, and sequences unrelated to JEV are indicated in lowercase letters. Restriction endonuclease recognition sites used for cDNA cloning are underlined.
(i) Construction of JEV 3′NCR mutant cDNAs.
We constructed a total of seven JEV 3′NCR mutant cDNA clones, each carrying a deletion of a different size within the 3′NCR of the full-length WT JEV cDNA. The first six deletion mutants (MUTΔ1-133, MUTΔ1-211, MUTΔ1-284, MUTΔ1-381, MUTΔ1-450, and MUTΔ1-473) were generated by overlapping extension PCR with the WT as the template. Initially, two primary overlapping fragments were amplified by a first round of PCR with two pairs of primers: (i) JE3ncrF+JEΔ133R and JEΔ133F+JE3ncrR for MUTΔ1-133, (ii) JE3ncrF+JEΔ211R and JEΔ211F+JE3ncrR for MUTΔ1-211, (iii) JE3ncrF+JEΔ284R and JEΔ284F+JE3ncrR for MUTΔ1-284, (iv) JE3ncrF+JEΔ381R and JEΔ381F+JE3ncrR for MUTΔ1-381, (v) JE3ncrF+JEΔ450R and JEΔ450F+JE3ncrR for MUTΔ1-450, and (vi) JE3ncrF+JEΔ473R and JEΔ473F+JE3ncrR for MUTΔ1-473. In all cases, two primary overlapping PCR products were gel purified and subsequently fused by a second round of PCR with the 5′ outside primer JE3ncrF and the 3′ outside primer JE3ncrR. The resulting amplicons were digested with NdeI and RsrII, generating fragments of the expected sizes: 887 bp for MUTΔ1-133 (i), 809 bp for MUTΔ1-211 (ii), 736 bp for MUTΔ1-284 (iii), 639 bp for MUTΔ1-381 (iv), 570 bp for MUTΔ1-450 (v), and 547 bp for MUTΔ1-473 (vi). Finally, each of the NdeI-RsrII fragments was cloned into the corresponding site in WT cDNA by a three-piece ligation, i.e., the 887-bp NdeI-RsrII fragment created to construct MUTΔ1-133 was ligated to the 7,449-bp RsrII-PacI and 10,096-bp PacI-NdeI fragments of the WT. In the case of the last (single-deletion) mutant, MUTΔ477-574, a fragment of the WT was first amplified by PCR with a pair of primers, JE3ncrF and JEΔ574R, and the 915-bp NdeI-NotI fragment of the resulting amplicons was then ligated with the 7,456-bp NotI-PacI and 10,096-bp PacI-NdeI fragments of the WT.
(ii) Reconstruction of MUTΔ1-450/83ntIns.
MUTΔ1-450/83ntIns, a derivative of MUTΔ1-450, contained an 83-nt sequence duplication that was discovered at the 3′-terminal region of the replicating genomic RNAs recovered from MUTΔ1-450-derived pseudorevertants. To construct MUTΔ1-450/83ntIns, a bacterial plasmid that we used for sequencing (see below) was utilized as a template for PCR with a pair of primers, JE3ncrF and JE3ncrR, to amplify a fragment containing the 83-nt sequence duplication. The 653-bp NdeI-RsrII fragment of the resulting amplicons was then ligated with the 7,449-bp RsrII-PacI and 10,096-bp PacI-NdeI fragments of MUTΔ1-450.
(iii) Reconstruction of MUTΔ1-381 derivatives.
A panel of 12 MUTΔ1-381 derivatives was constructed; each of these carried 1 of 12 point mutations that were identified at the 3′-terminal region of the genomic RNAs extracted from MUTΔ1-381-derived pseudorevertants. To reproduce these 12 derivatives of MUTΔ1-381, 12 fragments containing 1 of the 12 genetic changes each were PCR-amplified with primers JE3ncrF and JE3ncrR from the corresponding individual bacterial plasmids that we cloned for sequencing analysis (see below). The amplicons were digested with NdeI and RsrII, and the resulting NdeI-RsrII fragments each were ligated with two fragments of MUTΔ1-381 (the 7,449-bp RsrII-PacI and 10,096-bp PacI-NdeI fragments).
(iv) Reconstruction of MUTΔ1-284 derivatives.
A set of seven MUTΔ1-284 derivatives was constructed, each harboring one of seven sequence duplications of various sizes that were detected at the 3′-terminal region of the genomic RNAs recovered from MUTΔ1-284-derived pseudorevertants. In each case, a fragment containing one of the seven sequence duplications was PCR amplified with primers JE3ncrF and JE3ncrR from the corresponding individual clone that was used for sequencing (see below). The NdeI-RsrII fragment of the resulting variably sized amplicons was substituted for the corresponding region of MUTΔ1-284 by a three-piece ligation, as described above for the MUTΔ1-381 derivatives.
RNA transcription.
Template cDNA plasmids were linearized by initial digestion with XbaI and subsequent treatment with mung bean nuclease, and linearized plasmids were purified by phenol-chloroform extraction and ethanol precipitation. Runoff RNA transcripts were synthesized from linearized plasmid templates in vitro with SP6 RNA polymerase in a 25-μl reaction mixture containing ∼200 ng of template cDNA, 0.6 mM of cap analog [m7G(5′)ppp(5′)A; New England Biolabs, Beverly, MA], 1 mM each of the four nucleoside triphosphates (GE Healthcare, Piscataway, NJ), 0.5 μM of [3H]UTP (1.0 mCi/ml and 50 Ci/mmol; PerkinElmer, Waltham, MA), 15 U of SP6 RNA polymerase (New England Biolabs), 40 U of RNaseOUT (Invitrogen), 10 mM dithiothreitol (DTT), and the buffer supplied by the manufacturer. The reactions were carried out at 37°C for 1 h. Transcript yields were quantified on the basis of [3H]UTP-incorporated RNA adsorption to DE-81 (Whatman, Maidstone, United Kingdom) filter paper (59), and transcript integrity was verified by agarose gel electrophoresis and ethidium bromide staining. RNA transcripts were used for transfection without any additional purification.
RNA transfection and quantitation of RNA infectivity.
For RNA transfection, subconfluent BHK-21 cells were trypsinized, washed three times with cold phosphate-buffered saline (PBS), and resuspended at 2 × 107 cells/ml in PBS. Cell suspensions (400 μl each) were then transfected with 2 μg of RNA transcripts by electroporation under previously optimized conditions (980 V, 99-μs pulse length, and five pulses) with a model ECM 830 electroporator (BTX, San Diego, CA) (83) and then transferred to 10 ml of the fresh complete medium.
The specific infectivity of RNA transcripts was determined by infectious center assays. The RNA-transfected cells were serially diluted in 10-fold steps and plated onto monolayers of naïve BHK-21 cells (3 × 105 cells/well) in a 6-well plate. After incubation at 37°C for 4 to 6 h, the attached cells were overlaid with semisolid minimal essential medium (MEM) containing 0.5% SeaKem LE agarose (Lonza, Rockland, ME) and 10% fetal bovine serum (FBS) and then incubated for 4 days at 37°C under 5% CO2. The cells were overlaid with 2 ml of 7% formaldehyde per well and kept at room temperature for >4 h. After three 10-min washes with PBS, the cells were permeabilized with 0.25% Triton X-100 in PBS for 10 min and then incubated with the mouse anti-JEV antiserum (diluted 1:500 in PBS with 0.25% Triton X-100) at room temperature for 2 h. After three additional washes with PBS, the cells were incubated with HRP-conjugated goat anti-mouse IgG (1:1,000 dilution) at room temperature for 2 h. The cells were washed again with PBS, and the infectious centers of foci were stained by the addition of 3,3′-diaminobenzidine (DAB; Vector Laboratories, Burlingame, CA) according to the manufacturer's instructions.
Immunoblotting.
Total cell lysates were prepared by direct lysis of cell monolayers with 1× sample loading buffer (80 mM Tri-HCl [pH 6.8], 2.0% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1 M DTT, and 0.2% bromophenol blue). An equal amount of cell lysates was boiled for 5 min, separated on an SDS-12% polyacrylamide gel under reducing conditions, and electrotransferred to a methanol-activated polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA). The membrane was treated with 5% (wt/vol) nonfat dried milk in TBS-T buffer (20 mM Tris [pH 8.8], 137 mM NaCl, and 0.1% Tween 20) at room temperature for 1 h, followed by three 10-min washes with TBS-T buffer. The membrane was then incubated in TBS-T buffer containing 0.5% nonfat dried milk at room temperature for 2 h with a mouse anti-JEV (1:1,000 dilution), rabbit anti-JEV NS1 (1:1,000 dilution), or rabbit anti-GAPDH antiserum (1:10,000 dilution). The primary antibody-decorated membrane again was washed three times with TBS-T buffer and incubated with an AP-conjugated goat anti-mouse or anti-rabbit IgG, as appropriate, at a 1:5,000 dilution in TBS-T buffer containing 0.5% nonfat dried milk at room temperature for 2 h. Following three 10-min washes with TBS-T buffer, the membrane was stained with a mixture of 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) and nitroblue tetrazolium (NBT).
Real-time quantitative RT-PCR.
Total cellular RNAs were purified by direct lysis of cell monolayers with TRIzol reagent (Invitrogen) according to the manufacturer's instructions and used for the first-strand cDNA synthesis with two reverse primers (listed below), one specific for the JEV NS3 protein-coding region and the other specific for BHK-21 β-actin RNA (for use in normalizing the JEV RNA values). Each 20-μl reverse transcription (RT) reaction mixture contained 50 ng of template RNA, 5 pmol of the primers, 10 mM of deoxynucleoside triphosphate mix, 100 U of Superscript II reverse transcriptase (Invitrogen), 40 U of RNaseOUT (Invitrogen), 0.1 mM DTT, and the buffer supplied by the manufacturer. The RT reaction was carried out at 45°C for 30 min, followed by PCR amplification of 2-μl aliquots of the RT reaction mixture using the iQ Supermix Quantitative PCR system (Bio-Rad Laboratories) and the iCycler iQ Multicolor Real-Time PCR Detection system (Bio-Rad Laboratories). The reactions were carried out under the following conditions: 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The target sequences were amplified by using the following primer pairs and fluorogenic TaqMan probes (85): JEV RNA forward (5′-ATCCAACTCAACCGCAAGTC-3′, corresponding to nt 5,757 to 5,766), reverse (5′-TCTAAGATGGTGGGTTTCACG-3′, corresponding to nt 5,894 to 5,914), and probe (5′-6-carboxyfluorescein-CATCTCTGAAATGGGGGCTA-black hole quencher 1-3′, corresponding to nt 5,837 to 5,856); β-actin RNA forward (5′-ACTGGCATTGTGATGGACTC-3′), reverse (5′-CATGAGGTAGTCTGTCAGGTC-3′), and probe (5′-HEX-CCAGCCAGGTCCAGACGCAGG-black hole quencher 2-3′). Samples were run in duplicate; a reaction without an aliquot of the RT reaction mixture was used to establish baseline fluorescence levels. Data were based on a threshold cycle (CT) in which the signal was higher than that of the background. Relative changes in JEV RNA levels were estimated by using the 2−ΔΔCT method (60, 75).
Analysis of viral growth kinetics.
Naïve BHK-21, SH-SY5Y, or C6/36 cells were preseeded in 35-mm dishes at 3 × 105 cells/dish for 12 to 18 h before infection with RNA transcript-derived viruses at a multiplicity of infection (MOI) of 1. After incubation for 1 h at 37°C (for BHK-21 and SH-SY5Y) or 28°C (C6/36), the cells were washed twice and incubated with fresh complete medium for 4 days (BHK-21 and SH-SY5Y) or 6 days (C6/36). At the indicated time points, equal volumes of culture supernatants were collected and used for titration on naïve BHK-21 cells. Cells were preseeded onto 6-well plates at 3 × 105 cells/well for 12 to 18 h and then infected with serial 10-fold dilutions of the culture supernatants containing virus for 1 h at 37°C with frequent shaking. The cell monolayers were overlaid with semisolid MEM containing 0.5% SeaKem LE agarose (Lonza) and 10% FBS. After incubation for 4 days at 37°C with 5% CO2, the infectious centers of foci were immunostained with mouse anti-JEV antiserum and HRP-conjugated goat anti-mouse IgG and then visualized with DAB as described above. Virus yields were calculated and are presented as focus-forming units (FFU) per milliliter.
Serial passage of viruses.
WT virus and five mutant viruses (MUTΔ1-133, MUTΔ1-211, MUTΔ1-284, MUTΔ1-381, and MUTΔ1-450/83ntIns) obtained from the corresponding RNA-transfected BHK-21 cells were serially passaged in naïve BHK-21 cells for three consecutive rounds of infection. For each round, a monolayer of naïve BHK-21 cells (3 × 105) preseeded in a 35-mm dish was infected with 1 ml of virus suspension at an MOI of 1 and then gently agitated for 1 h at 37°C. After a rinse with 3 ml of serum-free MEM, the cells were incubated in 3 ml of the fresh complete medium containing 10% FBS at 37°C with 5% CO2. Once any JEV-induced cytopathic effects were noted, the culture supernatants containing progeny virions were clarified by centrifugation at 2,000 rpm for 10 min and kept at −80°C until the next round of infection.
Sequence analysis of the 3′-terminal region of JEV genomic RNAs.
Viral genomic RNAs were isolated from 200 μl of virus-containing culture supernatants with 600 μl of TRIzol LS reagent (Invitrogen), as recommended by the manufacturer, and resuspended in 20 μl of RNase-free distilled water. To sequence the 3′-terminal region of JEV genomic RNAs, we employed the 3′ rapid amplification of cDNA ends (3′RACE), as described previously (83). Synthetic oligonucleotide T (OligoT) was ligated to the 3′ end of the genomic RNA to serve as a specific primer binding site for cDNA synthesis and subsequent PCR amplification. OligoT was 5′ phosphorylated with T4 polynucleotide kinase (Takara, Shiga, Japan) and 3′ blocked by incorporating ddATP with terminal deoxynucleotidyltransferase (Takara) to prevent the intra- and intermolecular ligation of OligoT. The ligation of the modified OligoT to the 3′ end of the genomic RNA was carried out at 16°C for 12 h using T4 RNA ligase in a 20-μl reaction mixture containing 10 μl of extracted genomic RNAs, 10 pmol of OligoT, 10 U of T4 RNA ligase (New England Biolabs), 40 U of RNaseOUT (Invitrogen), and the buffer supplied by the manufacturer. The OligoT-ligated RNA was extracted with phenol-chloroform, precipitated with ethanol, and resuspended in 20 μl of RNase-free distilled water. Half of the ligated RNA was used for cDNA synthesis with oligonucleotide TR (OligoTR), which is complementary to OligoT. RT was carried out for 1 h at 45°C in a 20-μl reaction mixture containing 10 μl of the ligated RNA, 5 pmol of primer, 10 mM of deoxynucleoside triphosphate mix, 100 U of Superscript II reverse transcriptase (Invitrogen), 40 U of RNaseOUT (Invitrogen), 0.1 mM DTT, and the buffer supplied by the manufacturer. One-quarter of the RT reaction mixture was subsequently used for PCR amplification with Pfu-derived Pyrobest DNA polymerase (Takara) and a pair of primers, OligoTR and JEcircleF (complementary to nt 10,345 to 10,364). The PCRs consisted of 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C, followed by a final extension of 10 min at 72°C. cDNA amplicons were digested with SacI and EcoRI and subsequently cloned into the respective sites of pRS2 cloning vector. Approximately 20 randomly picked individual clones were sequenced to characterize the genetic changes in the 3′-terminal region of the viral genomes.
Secondary structure modeling.
Optimal RNA secondary structures were predicted by using the mfold, version 3.2, web server (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi) under default folding conditions (37°C, 1 M NaCl, no divalent ions, and no limit on distance between paired bases) (48, 88).
RESULTS
Mapping of the 3′ cis-acting RNA elements required for genomic RNA replication and infectious virus production.
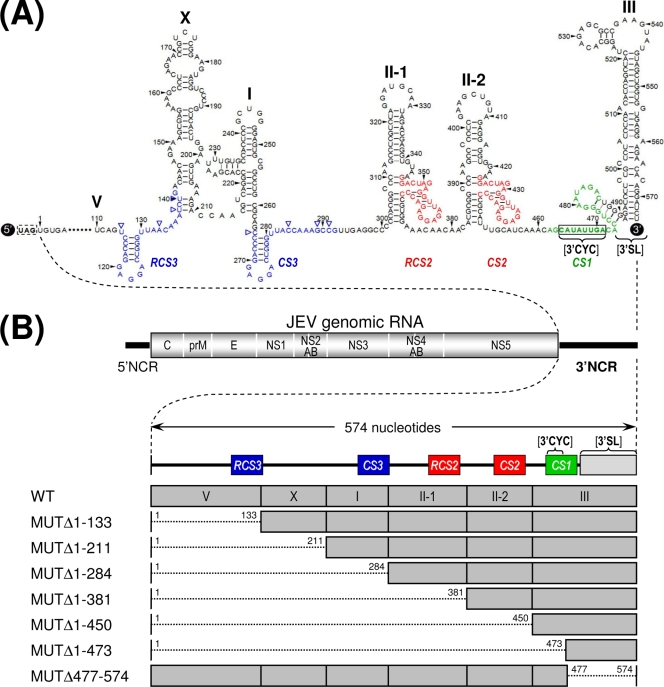
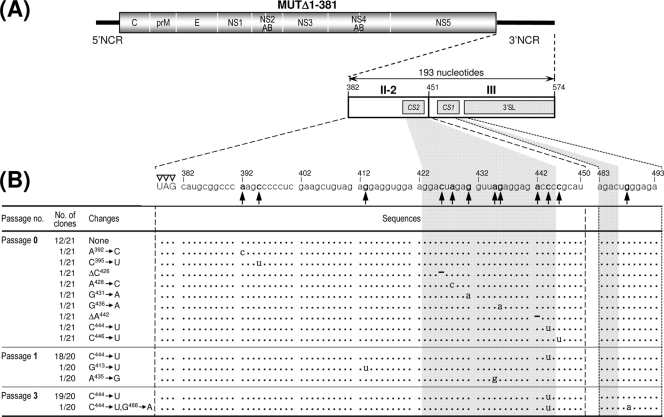
Comprehensive sequence comparisons and computer-based modeling previously revealed a pattern of RNA elements in the 3′NCR of JEV genomic RNA that are highly conserved among all members of JEV-related mosquito-borne flaviviruses (30, 52, 55, 56, 68). Based on these previous reports and our own comparative studies here, we have subdivided the 574-nt JEV 3′NCR into six domains (V, X, I, II-1, II-2, and III in the 5′-to-3′ direction). With the exception of the X domain, all of these domains include one of the ∼20- to 30-nt conserved sequences (CSs) or repeated conserved sequences (RCSs) that were originally recognized by Hahn et al. (30); domain V contains RCS3, I contains CS3, II-1 contains RCS2, II-2 contains CS2, and III contains CS1, which incorporates the 8-nt core sequence of 3′CYC (Fig. 1A). Downstream sequences of the CS1 motif in the 3′-terminal domain III correspond to the ∼90-nt 3′SL.
FIG. 1.
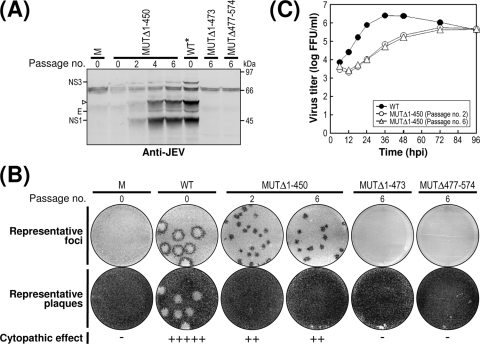
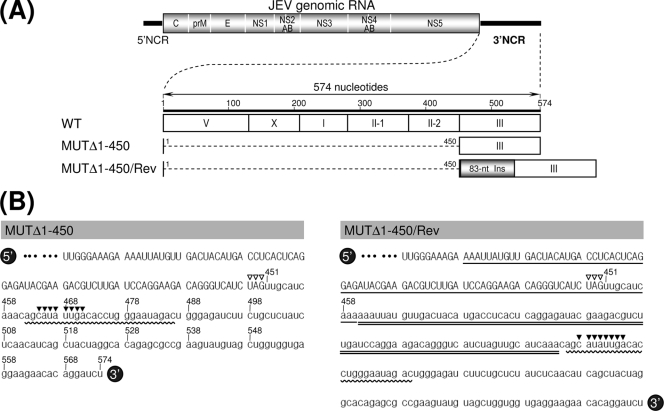
Schematic presentation of JEV 3′NCR mutants constructed in the context of a full-length infectious JEV cDNA molecular clone. (A) Detailed view of short CSs and predicted secondary structures within the 574-nt 3′NCR of JEV genomic RNA (30, 52, 55, 56, 68) from the translation stop codon (UAG; box outlined with a dotted line) of the single ORF to the 3′ end of the genome. Nucleotides are numbered from the first base of the 3′NCR, excluding the translation stop codon, toward its 3′ end. The 574-nt JEV 3′NCR was divided into six domains (55): V, nt 1 to 133; X, nt 134 to 211; I, nt 212 to 284; II-1, nt 285 to 381; II-2, nt 382 to 450; and III, nt 451 to 574, as ordered in the 5′ to 3′ direction. Highlighted in color are the five ∼20- to 30-nt CS or RCS motifs originally described by Hahn et al. (30): CS1, CS2, RCS2, CS3, and RCS3. Also indicated at the 3′ end of the genome are two well-documented RNA motifs, the 3′CYC (8-nt core sequence; box outlined with a solid line) embedded in the CS1 motif and the ∼90-nt 3′SL. (B) Schematic diagram of JEV 3′NCR deletion mutants. Shown at the top is the organization of the ∼11,000-nt JEV genomic RNA; viral proteins are indicated with the thick solid lines at both termini representing the 5′NCR and 3′NCR of the viral genome. Shown below is an expanded view of the six domains of the 574-nt JEV 3′NCR, in conjunction with the relative location of the five CSs (CSs or RCSs), 3′CYC, and 3′SL. Shown at the bottom are seven JEV 3′NCR deletion mutants, representing 5′- or 3′-truncated versions of various lengths within the 3′NCR, compared to full-length infectious JEV cDNA (WT). The designations of the seven mutants indicate the regions deleted from each of the individual constructs (shown by dotted lines).
To investigate the functional importance of these six domains of JEV 3′NCR in viral replication, particularly focusing on the 5′-proximal region, we introduced a series of progressive deletions into the 3′NCR in the context of the viral genome by using a full-length infectious JEV cDNA (WT), generating a total of seven mutant cDNAs. For discussion purposes, the nucleotides of the 574-nt WT 3′NCR are numbered consecutively from 1 to 574 in the 5′-to-3′ direction in this report; the designations of the seven mutants indicate the regions deleted from the corresponding mutant constructs. The first five mutant cDNAs (MUTΔ1-133, MUTΔ1-211, MUTΔ1-284, MUTΔ1-381, and MUTΔ1-450) were generated by progressively deleting from one to five of the 5′-proximal five domains in the 5′-to-3′ direction, the sixth (MUTΔ1-473) by removing all five domains plus the 3′CYC at the 5′ end of the 3′-proximal domain III, and the seventh (MUTΔ477-574) by eliminating only the 3′SL at the 3′ end of the genome (Fig. 1B).
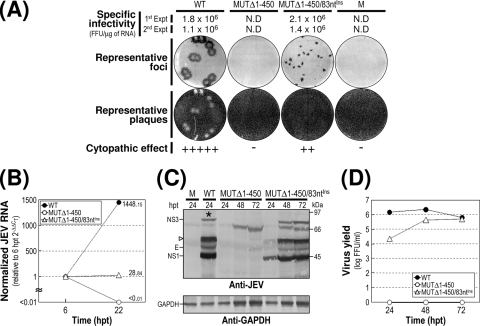
We first analyzed the effect of these deletions on the specific infectivity and viability of JEV genomic RNA by transfecting naïve BHK-21 cells with 2 μg of RNA transcripts synthesized from the WT or one of the seven 3′NCR mutant cDNA templates. At 4 days after transfection, monolayers of the RNA-transfected cells were first immunostained with a mouse JEV-specific hyperimmune antiserum to determine the number and size of infectious focus centers; subsequently, the same monolayers were restained with crystal violet to visualize the number and size of the infectious plaques. This sequential staining strategy allowed us to accurately quantitate the specific infectivity of our 3′NCR mutant RNAs, in case any of them failed to form the visible plaques under our experimental conditions, even though they were actually replication competent.
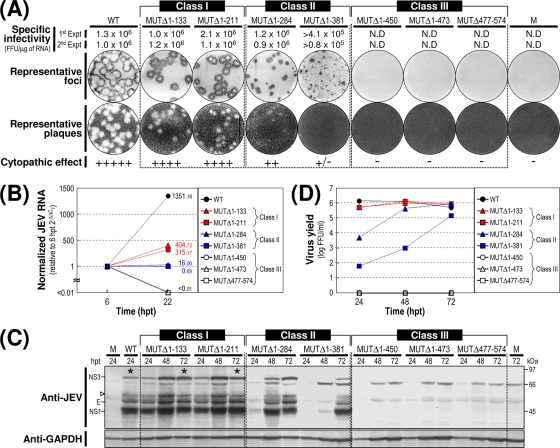
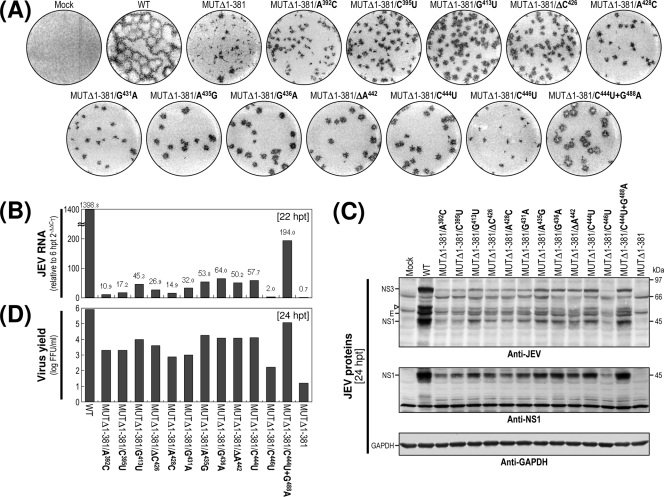
Based on our findings from this assay, we grouped the seven mutants into three classes (Fig. 2A). Class I mutants (MUTΔ1-133 and MUTΔ1-211) produced RNAs with specific infectivities similar to that of WT-derived RNA (1.0 × 106 to 2.1 × 106 FFU/μg or PFU/μg versus 1.0 × 106 to 1.3 × 106 FFU/μg or PFU/μg for the WT). The infectious foci and plaques displayed by these two mutant RNAs were as homogeneous as those of WT-derived RNA. In both cases, however, the average size was invariably ∼25% smaller than that of WT-derived RNA. Class II mutants (MUTΔ1-284 and MUTΔ1-381) generated RNAs with specific infectivities that were similar to or only moderately lower than that of WT-derived RNA (0.9 × 106 to 1.2 × 106 FFU/μg and >0.8 × 105 to 4.1 × 105 FFU/μg, respectively). However, the average size of their infectious foci varied significantly, ranging from an ∼2.5-fold reduction in the case of the MUTΔ1-284-derived RNA to nearly undetectable in the case of the MUTΔ1-381-derived RNA. Also, only a few or no plaques were seen in cells transfected with either of these two mutant RNAs. Class III mutants (MUTΔ1-450, MUTΔ1-473, and MUTΔ477-574) produced RNAs that generated no detectable infectious foci or plaques; this loss of infectivity was confirmed by the confocal immunofluorescence microscopy of single RNA-transfected cells (data not shown).
FIG. 2.
Functional analysis of JEV 3′NCR mutants. Naïve BHK-21 cells were mock transfected (M) or transfected with 2 μg of the RNA transcripts synthesized from WT cDNA or each of the seven JEV 3′NCR mutant cDNAs. (A) Specific infectivity and representative focus (or plaque) morphology. The specific infectivities of RNA transcripts were determined by infectious center assays, as described in Materials and Methods. At 4 days after transfection, the cell monolayers were first immunostained with a mouse anti-JEV antiserum (representative foci) and then restained with crystal violet (representative plaques). RNA infectivities are given as FFU/microgram of RNA. The number of plus signs indicates the magnitude of the cytopathic effect produced by the corresponding mutant RNAs relative to that induced by the WT RNA; a minus sign indicates no cytopathic effect. N.D, not detectable. (B) Production of JEV RNA. Equal amounts of total cellular RNAs extracted at 6 and 22 h posttransfection (hpt) were used for real-time quantitative RT-PCR with a JEV-specific probe complementary to a sequence in the NS3 protein-coding region; a β-actin RNA-specific probe was used to normalize total RNA levels. Changes in JEV RNA levels in real-time quantitative RT-PCR assays relative to those detected at 6 hpt were calculated using the 2−ΔΔCT method. (C) Accumulation of JEV proteins. Equal volumes of total cell lysates collected at the indicated time points were separated by SDS-polyacrylamide gel electrophoresis, and the levels of JEV proteins were visualized by immunoblotting with a mouse anti-JEV antiserum (anti-JEV). GAPDH, used as a loading and transfer control, was detected with a rabbit anti-GAPDH antiserum (anti-GAPDH). An asterisk indicates that ∼80% of RNA-transfected cells displayed a cytopathic effect. The positions of the viral proteins (E, NS1, and NS3) and a cleavage-related intermediate (open arrowhead) are shown on the left; molecular size markers (in kilodaltons) are shown on the right. (D) Yield of infectious virions. The amount of the infectious virus particles released into the culture supernatants was monitored by virus titration on naïve BHK-21 cells. The seven JEV 3′NCR mutants examined in our study were grouped into three classes (I to III) based on their phenotypes. See the text for detailed descriptions.
We next examined the production levels of JEV RNA and proteins in RNA-transfected cells and assessed the accumulation of infectious virus particles released into the culture medium. At 22 h posttransfection, the amount of JEV RNA produced was quantified by real-time quantitative RT-PCR using a JEV-specific probe complementary to a sequence in JEV NS3 and compared to those at 6 h posttransfection (Fig. 2B). The first class I and II mutants displayed levels of RNA production that were significantly lower than that of WT-derived RNA (∼3-fold, MUTΔ1-133; ∼4-fold, MUTΔ1-211; ∼84-fold, MUTΔ1-284; and ∼1,958-fold, MUTΔ1-381) and were consistent with the average size of the infectious foci observed (Fig. 2A). As expected, however, the remaining three mutant RNAs (class III; MUTΔ1-450, MUTΔ1-473, and MUTΔ477-574) showed no detectable level of JEV RNA production. In all cases, the production level of JEV RNA was proportional to the accumulation level of JEV proteins (such as E, NS1, and NS3) during the first 3 days after transfection, as demonstrated by immunoblotting with a JEV-specific hyperimmune antiserum (Fig. 2C), and to the level of infectious virus particles released into the medium during the same period (Fig. 2D).
We then analyzed the 3′-terminal nucleotide sequences of the viral genomic RNAs recovered from RNA-transfected cells at 72 h posttransfection by 3′RACE, followed by cDNA cloning into a bacterial plasmid and the sequencing of ∼20 independent clones containing inserts. In the case of the class I mutants (MUTΔ1-133 and MUTΔ1-211), all of the recovered genomic RNAs had 3′NCRs identical to those of their original transfected RNAs (data not shown). In the case of the class II mutants (MUTΔ1-284 and MUTΔ1-381), however, some of the genomic RNAs acquired a variety of secondary mutations in the vicinity of the corresponding primary deletion sites in their 3′NCRs (see Fig. 7 to 11 for a detailed description). The emergence of these secondary mutations was consistent with the heterogeneity in focus morphology observed in the cells transfected with these RNAs. As expected, we obtained no PCR products from the culture supernatants of BHK-21 cells transfected with any of the three class III mutant RNAs (data not shown).
FIG. 7.
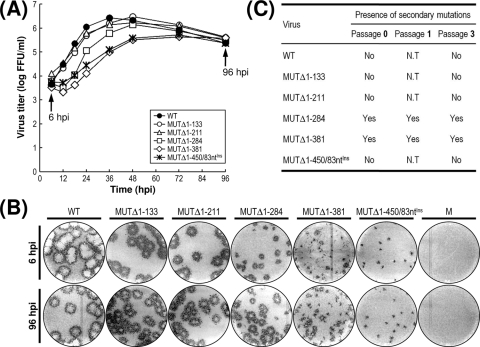
Characterization of recombinant viruses generated from five replication-competent JEV 3′NCR mutant cDNAs. (A and B) Viral growth and focus morphology. Subconfluent monolayers of BHK-21 cells were infected at an MOI of 0.1 with the WT or one of the five 3′NCR mutant viruses derived from the respective cDNAs, as indicated. (A) At the indicated hour postinfection (hpi), aliquots of culture supernatants were collected during a period of 4 days and used for virus titration on naïve BHK-21 cells. (B) The same monolayers were immunostained at 6 and 96 hpi (indicated by arrows) for infectious foci with the anti-JEV antiserum. The data shown are from one of two independent experiments, which produced similar results. M, mock infected. (C) Acquisition of spontaneous secondary mutations during three consecutive rounds of viral passage in BHK-21 cells. To examine the genetic stability of the five mutant viruses, each of the viruses collected from RNA-transfected cells (passage 0) was passaged three times in naïve BHK-21 cells at an MOI of 0.1. At the indicated passages, the 3′-terminal sequences of the replicating genomic RNAs were determined by 3′RACE, the cloning of the cDNA amplicons, and the sequencing of ∼20 independent clones containing the insert. Indicated is the presence (Yes) or absence (No) of secondary mutations for each mutant virus. N.T, not tested.
FIG. 11.
Effects of seven sequence insertions (duplications) discovered in MUTΔ1-284-derived pseudorevertants on RNA replication efficiency. Naïve BHK-21 cells were mock transfected (Mock) or transfected with the synthetic RNAs transcribed from WT cDNA or one of the reconstructed MUTΔ1-284 derivative cDNAs, as indicated. (A) Representative focus morphology. (B) Production of JEV RNA. (C) Accumulation of JEV proteins. (D) Production of infectious virions. Experiments were performed as described in the legend to Fig. 9.
Of the seven JEV 3′NCR mutants, four (MUTΔ1-133, MUTΔ1-211, MUTΔ1-284, and MUTΔ1-381) showed little or no loss of RNA infectivity, with a spectrum of average focus sizes that was correlated with their levels of RNA production, protein accumulation, and virus production. In contrast, the other three mutants showed an undetectable level of RNA infectivity: those in which the deletion extended further toward the 3′ end of the genome and included the II-2 domain (MUTΔ1-450) or the 3′CYC motif embedded in domain III (MUTΔ1-473), or in which only the 3′SL at the 3′ end of the genome was removed (MUTΔ477-574). These results suggested that in the BHK-21 cells we used for RNA transfection, the presence of the two 3′-proximal domains (II-2 and III) of the JEV 3′NCR was sufficient to maintain the replication competency of the genomic RNA, whereas the presence of the four remaining 5′-proximal domains (V, X, I, and II-1) was necessary to achieve a maximal efficiency of RNA replication. Also, we noted that domain X, which was missing from MUTΔ1-211 but not MUTΔ1-133, had only a minimal effect on RNA replication; this result is consistent with the fact that the X domain, unlike the other domains, does not contain any recognized CS motifs.
Recovery of pseudorevertants originating from the replication-incompetent MUTΔ1-450-derived RNA.
Initially, we did not detect any replication-competent pseudorevertants from BHK-21 cells transfected with any of the three class III mutant RNAs (MUTΔ1-450, MUTΔ1-473, and MUTΔ477-574) (Fig. 2). In an attempt to obtain pseudorevertants from these mutants, we serially passaged the undiluted culture supernatants from the RNA-transfected cells in naïve BHK-21 cells to amplify any very small amounts of replication-competent pseudorevertants that might have been present. After six passages, 1 ml of each undiluted culture medium was used to infect naïve BHK-21 cells, and the production of JEV-specific proteins and the formation of JEV-induced foci was assessed by immunoblotting with cell lysates obtained at 72 h postinfection with the mouse anti-JEV antiserum (Fig. 3A) and visualizing the infectious foci by the immunostaining of monolayers of the infected cells at 96 h postinfection with the same antiserum (Fig. 3B).
FIG. 3.
Recovery of MUTΔ1-450-derived pseudorevertants and their phenotypic characteristics. Undiluted culture supernatants of BHK-21 cells that had been transfected with 2 μg of the RNA synthesized from one of three mutant cDNAs (MUTΔ1-450, MUTΔ1-473, or MUTΔ477-574) were passaged six times in naïve BHK-21 cells. (A and B) Emergence of MUTΔ1-450-derived pseudorevertants. Culture medium (1 ml) collected at passage 0, 2, 4, and/or 6, as indicated, was used to infect naïve BHK-21 cells. (A) Equal amounts of the total cell lysates were analyzed for JEV protein expression at 72 h postinfection by immunoblotting with the mouse anti-JEV antiserum; the lysate from WT virus-infected cells at 24 h postinfection (WT*) was used as a reference to show the viral proteins E, NS1, and NS3 and a cleavage intermediate (open arrowhead). (B) To visualize representative foci (or plaques), cell monolayers at 4 days postinfection were immunostained first with the anti-JEV antiserum and then with crystal violet. The cytopathic effect produced by the individual mutant viruses relative to the WT level is indicated by plus or minus signs as described in the legend to Fig. 2. (C) Growth of MUTΔ1-450-derived pseudorevertants. Naïve BHK-21 cells were infected at an MOI of 0.1 with the WT or one of two MUTΔ1-450-originated pseudorevertants obtained at passages 2 and 6. Culture supernatants were harvested at the indicated hour postinfection (hpi) and used for virus titration on naïve BHK-21 cells. The data shown are from one of two independent experiments yielding similar results.
In the case of MUTΔ1-450, to our surprise, we noted the production of a significant quantity of JEV-specific proteins (i.e., E, NS1, and NS3), along with a homogeneous population of infectious foci, at passage 2 and thereafter, indicating the emergence of replication-competent pseudorevertants. These foci produced at passages 2 to 6 were invariably homogeneous but consistently ∼4.0-fold smaller than those produced by WT virus. Furthermore, the growth properties of these pseudorevertants were identical from passages 2 to 6, although they displayed a ∼36-h delay in growth rate and an ∼10-fold reduction in peak titer compared to those of the WT virus (Fig. 3C). In the case of the other two mutants (MUTΔ1-473 and MUTΔ477-574), neither viral proteins (Fig. 3A) nor infectious foci (Fig. 3B) were detectable in any of the six serial passages of the culture supernatants, indicating no evidence of reversion. Overall, these data suggest that although all three mutant RNAs of class III had no detectable infectivity, some replication-competent pseudorevertants could be recovered after the serial passage of the culture supernatants from BHK-21 cells transfected with MUTΔ1-450-derived RNA but not with the mutant RNAs derived from either MUTΔ1-473 or MUTΔ477-574.
Acquisition of a novel 83-nt sequence duplication capable of restoring the replicability of MUTΔ1-450-derived RNA.
To understand the genetic basis for the reestablishment of RNA replication in the MUTΔ1-450-derived pseudorevertants, we analyzed the 3′-terminal nucleotide sequences of the replicating genomic RNAs extracted from the pseudorevertants in the culture medium at passages 2 and 6. In both cases, the 3′-terminal sequence, including the 50-nt 3′-terminal region of the viral ORF and the entire 3′NCR, was amplified by 3′RACE; subsequently, the cDNA amplicons were cloned into a bacterial plasmid, and 20 independent clones containing the insert were sequenced. No differences were found between genomic RNAs prepared from the second and sixth culture passages. All of the clones lacked the 5′-proximal 450-nt sequence that we had originally deleted, and to our surprise, they all had acquired an additional copy of an 83-nt sequence duplicated from the 3′-terminal region of the viral ORF (MUTΔ1-450/Rev) (Fig. 4A). This newly acquired 83-nt sequence was located 10 nt downstream of the translation stop codon UAG in the forward direction, generating a new direct repeat sequence of two identical 83-nt sequences just 1 nt upstream of the CS1 motif (3 nt upstream of the 8-nt core sequence of the 3′CYC motif) in domain III without disturbing the single viral ORF (MUTΔ1-450/Rev) (Fig. 4B).
FIG. 4.
Identification of a novel 83-nt sequence duplication in the 3′-terminal region of the genomic RNAs extracted from MUTΔ1-450-derived pseudorevertants (MUTΔ1-450/Rev). (A) Relative location of a newly acquired 83-nt sequence (83-nt Ins; gray box) duplicated from the 3′-terminal region of the viral ORF in the 3′-terminal region of the MUTΔ1-450/Rev RNAs. (B) Nucleotide sequences of the 3′-terminal region of the original MUTΔ1-450 RNA (left) and its pseudorevertant MUTΔ1-450/Rev RNA (right). The uppercase sequences indicate the 3′-terminal sequences of the viral ORF; the lowercase sequences represent the entire 3′NCR of the corresponding RNAs. The 5′-end nucleotide of the 3′NCRs is numbered nt 451, and bases extending downstream are assigned consecutively according to the WT sequence. Also indicated are the translation stop codons UAG (open arrowheads), the 25-nt CS1 motif (curved lines), and the 8-nt core sequence of the 3′CYC motif (solid arrowheads). Highlighted only in MUTΔ1-450/Rev RNA is the newly discovered 83-nt sequence (double straight lines) originating from the corresponding 3′-terminal region (single straight lines) of the viral ORF.
To address the functional importance of this duplicated 83-nt sequence in MUTΔ1-450-derived RNA replication, we constructed a derivative of MUTΔ1-450 (designated MUTΔ1-450/83ntIns) with an additional copy of the 83-nt sequence and determined the specific infectivity of its RNA transcripts as an indication of its potential replicability after RNA transfection into BHK-21 cells (Fig. 5A). As expected, no infectivity was detectable in cells transfected with the original MUTΔ1-450-derived RNA. In contrast, the specific infectivity of the MUTΔ1-450/83ntIns-derived RNA was estimated to be in the range of 1.4 × 106 to 2.1 × 106 FFU/μg, which is indistinguishable from that of WT-derived RNA (1.1 to 1.8 × 106 FFU/μg), and its infectious foci were also as homogeneous as those of WT-derived RNA. On the other hand, the average focus size was dramatically reduced by ∼4.0-fold compared to that of WT-derived RNA. The foci were detectable only by the immunostaining of RNA-transfected cells with the anti-JEV antiserum and not by staining with crystal violet. Also, the difference in focus size was proportional to their relative accumulation of JEV RNA (Fig. 5B) and JEV proteins (Fig. 5C) in RNA-transfected cells, parallelling the production levels of infectious virus particles (Fig. 5D) released over time into their culture supernatants.
FIG. 5.
Functional importance of the 83-nt sequence duplication in MUTΔ1-450 RNA replication. Naïve BHK-21 cells were mock transfected (M) or transfected with 2 μg of the synthetic RNA transcripts derived from the WT cDNA, the original MUTΔ1-450 cDNA, or its derivative MUTΔ1-450/83ntIns containing the 83-nt sequence duplication that was originally identified from MUTΔ1-450/Rev. (A) Specific infectivity, representative focus (or plaque) morphology, and cytopathic effect. At 4 days after transfection, the specific infectivity and representative focus morphology were assessed by immunostaining the cell monolayers with the anti-JEV antiserum; plaque morphology was then visualized by restaining with crystal violet. The cytopathic effect induced by MUTΔ1-450/83ntIns RNA replication is indicated relative to the WT level. N.D, not detectable. (B) Production of JEV RNA. The levels of JEV RNA production at 22 hpt relative to those at 6 hpt were evaluated by real-time quantitative RT-PCR using a JEV-specific probe. (C) Accumulation of JEV proteins. The levels of JEV protein accumulation at 24, 48, and 72 hpt were analyzed by immunoblotting with the anti-JEV antiserum (anti-JEV). GAPDH was used as a loading and transfer control (anti-GAPDH). An asterisk indicates that ∼80% of RNA-transfected cells displayed a cytopathic effect. The viral proteins E, NS1, and NS3 and a cleavage-related intermediate (open arrowhead) are shown on the left; on the right are molecular size markers in kilodaltons. (D) Production of infectious virions. Virus titers accumulated in the culture supernatants at the indicated time points were estimated on naïve BHK-21 cells. Experiments were carried out as described for Fig. 2.
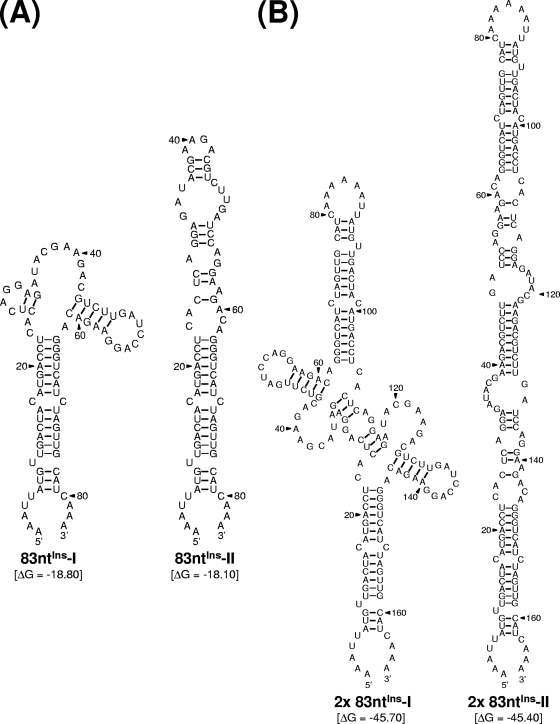
Taken together, these results indicate that the replicating genomic RNAs that emerged from MUTΔ1-450-derived RNA after amplification in BHK-21 cells acquired an additional copy of the viral 83-nt sequence, duplicated from the 3′-terminal region of the viral ORF, just upstream of domain III in its 3′NCR. The duplication of this 83-nt sequence was sufficient to restore the replication competence of the MUTΔ1-450-derived RNA to a level similar to that of WT-derived RNA, although the replication efficiency was significantly compromised. In addition, we noted that the duplication of this 83-nt sequence did not disrupt the virus's single ORF, suggesting that the reestablishment of the replication competence was attributable to an RNA element(s) of the primary sequences and/or secondary structures embedded in the duplicated 83-nt sequence, acting alone or in combination with other RNA elements present within the genome. We found that, consistent with this notion, a single copy of the 83-nt sequence was predicted to form two alternative stem-loop structures that are identical to each other except for their upper parts, which can assume either a cruciform-like structure (83ntIns-I) or extend the basal stem region further (83ntIns-II) (Fig. 6A). Also, a tandem repeat of the 83-nt sequence, identical to the one identified in MUTΔ1-450/83ntIns-derived RNA, was predicted to form two other alternative structures (designated 2× 83ntIns-I and 2× 83ntIns-II), both of which are arranged in a head-to-head configuration of 83ntIns-I and 83ntIns-II, respectively (Fig. 6B). The molecular basis for the role of this duplicated 83-nt sequence in MUTΔ1-450/83ntIns-derived RNA replication remains to be determined.
FIG. 6.
Predicted RNA secondary structures of the 83-nt sequence duplicated in the 3′-terminal region of the genomic RNAs of MUTΔ1-450-derived pseudorevertants. Shown are two pairs of four predicted stem-loop structures: one for a single copy of the 83-nt sequence (A) and the other for a tandem repeat of the 83-nt sequence (B). Also indicated are the initial thermodynamic free energies (ΔG, in kilocalories/mole) of all four potential structures, as determined by the RNA folding program mfold.
Genetic instability of progeny virions derived from class II mutants MUTΔ1-284 and MUTΔ1-381.
Of the seven 3′NCR mutant cDNAs constructed in this study, five produced mutant viruses that were competent for propagation in BHK-21 cells: two from class I (MUTΔ1-133 and MUTΔ1-211), two from class II (MUTΔ1-284 and MUTΔ1-381), and MUTΔ1-450/83ntIns cDNA (a MUTΔ1-450 derivative). These five mutant viruses produced infectious foci (or plaques) that were phenotypically distinct from each other, in terms of the magnitude of their reduction in focus size and the extent of their heterogeneity in focus morphology, compared to those of the WT virus. To further characterize their phenotypic differences, we examined the growth properties of these five mutant viruses during multiple cycles of viral replication, comparing not only their ability to grow and develop a productive infection but also the focus morphology produced by their progeny virions over time, as an indication of genetic stability. For this purpose, monolayers of naïve BHK-21 cells were infected at an MOI of 0.1 with the WT and each of the five mutant viruses in parallel. The yield of progeny virions released into the culture medium during the course of the 4-day incubation was monitored by titration on naïve BHK-21 cells.
The five mutant viruses revealed three different patterns of growth rate and virus yield, which is consistent with the phenotypic classification of the seven 3′NCR mutant cDNAs into three groups (Fig. 2). The first pattern was exhibited by the two mutant viruses derived from class I mutant cDNAs (MUTΔ1-133 and MUTΔ1-211); their growth property was essentially identical to that of the WT virus (peak titer of 1.8 × 106 FFU/ml at 36 h postinfection) (Fig. 7A); they achieved a peak titer comparable to that of the WT virus, albeit with an ∼12-h delay, peaking at 48 h postinfection. The second pattern was exhibited by the viruses derived from the other two class II mutant cDNAs (MUTΔ1-284 and MUTΔ1-381); this pattern consisted of two distinct stages, a noticeable eclipse-like stage during the first 18 h postinfection and a sharp exponential stage thereafter. The growth of MUTΔ1-284-derived mutant virus lagged behind that of the WT and two class I-derived mutant viruses for the first 36 h postinfection, but by 48 h postinfection and thereafter, this mutant achieved maximal titers slightly lower than those of WT virus, peaking at 48 to 72 h postinfection. Similarly, the growth of MUTΔ1-381-derived mutant virus was by far the most delayed of all four mutant viruses during the entire incubation period, but it yielded maximal titers that were ∼10-fold lower than those of WT virus at 72 h postinfection. The third pattern was exemplified by MUTΔ1-450/83ntIns-derived recombinant virus; the growth rate and virus yield were similar to those of the MUTΔ1-381-derived mutant at all time points, but the eclipse-like stage observed during the first 18 h postinfection was not as noteworthy as that seen for the MUTΔ1-381-derived virus during the same period of time.
The focus morphologies (size and heterogeneity) observed during multiple cycles of viral replication for the five viruses were consistent with three patterns of viral growth (Fig. 7B): (i) a homogeneous population of medium-sized foci, with the two class I-derived mutant viruses MUTΔ1-133 and MUTΔ1-211 representing the upper end of the size range; (ii) a heterogeneous population of medium-to-small foci, exemplified by the two class II-derived mutant viruses MUTΔ1-284 and MUTΔ1-381; and (iii) a homogeneous population of small foci, exemplified by the MUTΔ1-450/83ntIns-derived recombinant virus. Thus, the diversity in focus morphology and growth property described above provided additional evidence for the functional importance of the 5′-proximal four domains of JEV 3′NCR in terms of achieving a maximal level of replication efficiency. Of particular note was the fact that two of the class II-derived mutant viruses, unlike the other three, formed infectious foci of rather heterogeneous sizes; furthermore, this heterogeneity became more evident at the later time points during this 4-day experiment (Fig. 7B, compare the focus morphologies at 6 and 96 hpi), suggesting a rapid emergence of pseudorevertants.
To further investigate the genetic stability of the five mutant viruses, we serially passaged each of these viruses three times in naïve BHK-21 cells at an MOI of 0.1 and then determined the 3′-terminal sequences of the genomic RNAs extracted from progeny virions at passages 0, 1, and 3 by 3′RACE, cDNA cloning, and the sequencing of ∼20 independent clones for each mutant (Fig. 7C). Viruses derived from two class I mutant cDNAs (MUTΔ1-133 and MUTΔ1-211) and from MUTΔ1-450/83ntIns cDNA yielded genomic RNAs whose 3′NCR sequences were identical to their original transfected RNAs at passages 0 and 3. However, in the case of the remaining two viruses derived from class II mutant cDNAs (MUTΔ1-284 and MUTΔ1-381), a wide variety of spontaneous mutations emerged in their 3′NCRs with different ratios depending on the passage number (Fig. 8 to 11). Again, these findings supported our premise that the replicating genomic RNAs derived from two class II mutant cDNAs, MUTΔ1-284 and MUTΔ1-381, are genetically unstable.
FIG. 8.
Identification of 12 point mutations acquired within the 3′-terminal region of the genomic RNAs of MUTΔ1-381-derived pseudorevertants. The original recombinant viral stock was generated by transfecting BHK-21 cells with RNA transcripts derived from MUTΔ1-381 cDNA (passage 0); this viral stock was serially passaged three times in naïve BHK-21 cells at an MOI of 0.1 (passages 1 to 3). The 3′-terminal sequences of the genomic RNAs at passages 0, 1, and 3 were determined as described in Materials and Methods. The relative locations (A) and nucleotide sequences (B) acquired at the 3′-terminal region of the genomic RNAs of MUTΔ1-381-derived pseudorevertants are shown. (A) Illustrated at the top is the organization of the genomic RNA derived from the MUTΔ1-381 cDNA; shown below is an expanded view of the 193-nt 3′NCR, including only the two 3′-proximal domains (II-2 and III), with the CS1 and CS2 motifs and the 3′SL indicated by gray boxes. (B) Twelve point mutations (single-nucleotide substitutions or deletions are in boldface type) acquired in MUTΔ1-381-derived pseudorevertants. Dots indicate the conserved nucleotide sequences in adapted pseudorevertants, and hyphens indicate the deleted nucleotide sequences. The number of clones containing each identified nucleotide sequence is given over the total number of clones containing the insert that were sequenced. Open arrowheads indicate the translation stop codon UAG. The 5′-end nucleotide of the 3′NCR is numbered nt 382, and downstream sequences are consecutively assigned according to the WT sequence.
Acquisition of a collection of 12 point mutations capable of increasing the replication efficiency of MUTΔ1-381-derived RNA.
The sequence analysis of the 3′-terminal region of the MUTΔ1-381-derived viruses at passages 0, 1, and 3 identified a total of 12 point mutations predominantly within the 3′-penultimate domain II-2 of its 3′NCR, with the remaining 3′-terminal domain III left intact (Fig. 8A and B). At passage 0, 12 of 21 independent clones had the same sequence as the original MUTΔ1-381, whereas the remaining nine clones each contained a unique point mutation within the II-2 domain. Of these nine point mutations, seven were single-nucleotide substitutions (viz. A392→C, C395→U, A428→C, G431→A, G436→A, C444→U, and C446→U); the remaining two were single-nucleotide deletions (ΔC426 and ΔA442). At passage 1, none had a sequence identical to that of the original MUTΔ1-381, whereas a majority of the sequenced clones (18/20 clones) had the single C444→U substitution that had been found at passage 0 in 1 of 21 clones; the remaining two clones had a single G413→U or A435→G substitution in domain II-2. Of note was the finding that the single C444→U substitution appeared to be predominant at passage 1. This observation became more evident at passage 3, when all the sequenced clones had the same single C444→U substitution either alone (19/20 clones) or in combination with a single-nucleotide substitution of G488→A (1/20 clones) immediately downstream of the CS1 motif in the 5′ portion of domain III. In all, ∼60% of the MUTΔ1-381-derived viruses directly recovered from RNA-transfected cells acquired a group of single point mutations exclusively within domain II-2 of their 3′NCRs. Of these mutations, the single C444→U substitution was predominant, most likely because it provided a growth advantage for MUTΔ1-381-derived virus.
To address the functional significance of these 12 point mutations in MUTΔ1-381-derived RNA replication, we reconstructed 12 derivatives of MUTΔ1-381 with each of these genetic changes and then characterized the RNAs synthesized from these reconstructed cDNAs after transfection into BHK-21 cells with respect to (i) specific infectivity, (ii) focus morphology, (iii) viral RNA production, (iv) viral protein accumulation, and (v) virus yield. In all 12 cases, the specific infectivities were in the range of 1.0 × 106 to 2.1 × 106 FFU/μg, which is similar to that of WT-derived RNA (1.1 × 106 to 1.9 × 106 FFU/μg) and slightly higher than that of the original MUTΔ1-381-derived RNA (>5.6 × 105 to 0.5 × 105 FFU/μg). Thus, the RNAs generated from each of the 12 MUTΔ1-381 derivative cDNAs were as infectious as those produced from WT cDNA (data not shown). However, the foci formed in RNA-transfected cells at 4 days posttransfection, while consistently homogeneous, showed a noticeable variation in size compared to those in WT-derived RNA-transfected cells (Fig. 9A). The differences in average focus sizes were proportional to the relative levels of the accumulation of JEV RNA (Fig. 9B) and proteins (Fig. 9C) in RNA-transfected cells during the first 22- to 24-h incubation period and largely paralleled the levels of infectious virions released into the culture supernatants during the same period (Fig. 9D).
FIG. 9.
Effects on RNA replication efficiency of 12 point mutations discovered in MUTΔ1-381-derived pseudorevertants. Naïve BHK-21 cells were mock transfected (Mock) or transfected with a synthetic RNA transcribed from the WT cDNA or one of the reconstructed MUTΔ1-381 derivative cDNAs, as indicated. (A) Representative focus morphology. Infectious foci were visualized at 4 days posttransfection by immunostaining with the anti-JEV antiserum. (B) Production of JEV RNA. The levels of JEV RNA production at 22 hpt relative to those at 6 hpt were estimated by real-time quantitative RT-PCR using a JEV-specific probe. (C) Accumulation of JEV proteins. The levels of JEV protein accumulation at 24 hpt were examined by immunoblotting with the anti-JEV antiserum (anti-JEV) or a rabbit antiserum specific for JEV NS1 (anti-NS1). The positions of the viral proteins E, NS1, and NS3 and a cleavage-related intermediate (open arrowhead) are shown on the left; molecular size markers are shown on the right. (D) Production of infectious virions. Virus yields in the culture supernatants at 24 hpt were determined by virus titration on naïve BHK-21 cells.
Collectively, these results indicate that the acquisition of these 12 point mutations within or close to domain II-2 in MUTΔ1-381 is functionally important for increasing RNA replication efficiency (by ∼3-fold to ∼90-fold). In particular, we noted that a higher level of RNA replication was associated with the acquisition of four point mutations, A435→G, G436→A, ΔA442, and C444→U; all of these mutations converged on the ∼10-nt 3′-proximal region of the II-2 domain in MUTΔ1-381, corresponding to the 3′ half of the CS2 motif. Also, the enhanced replication efficiency of the MUTΔ1-381-derived RNA with a single C444→U substitution was increased further by the acquisition of an additional G488→A substitution just downstream of the CS1 motif at the 5′-proximal region of domain III. Thus, our findings demonstrated that although the original MUTΔ1-381-derived RNA was replication competent, the replication efficiency was severely impaired (near the limit of detection); nevertheless, various single point mutations acquired within the II-2 domain, particularly the CS2 motif in this domain, were able to enhance the RNA replication efficiency significantly, to the point of generating readily detectable levels of viral RNA and proteins, in parallel with infectious virus particles.
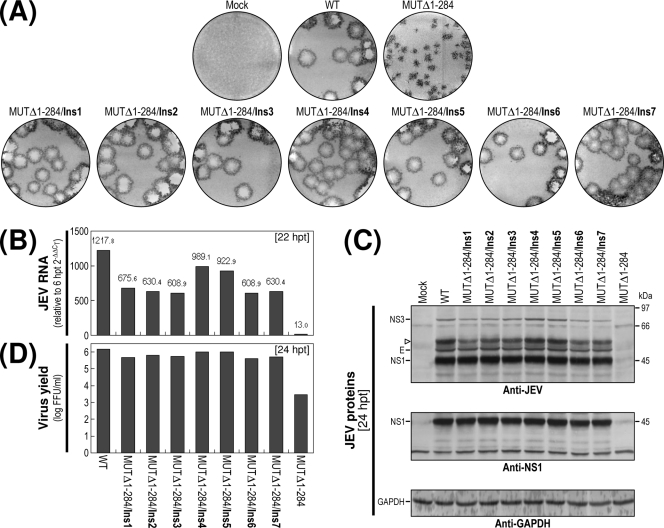
Acquisition of a selection of seven sequence duplications capable of enhancing the replication efficiency of MUTΔ1-284-derived RNA.
We also determined the 3′-terminal sequences of MUTΔ1-284-derived viruses that had been collected directly from RNA-transfected cells (passage 0), in parallel with those from the first (passage 1) and third (passage 3) passages. Remarkably, we found a total of seven nucleotide insertions of various sizes (designated Ins1 to Ins7), ranging from 40 to 168 bases; all of these insertions appeared to be located at the junction between domains II-1 and II-2 (Fig. 10A and B). Whereas 18 out of 20 independent clones had no insertions at passage 0, the remaining two clones had insertions of 155 and 168 nt (Ins2 and Ins3, respectively). At passage 1, the size of the inserted sequences became more heterogeneous. We saw no insertion in 11/21 clones; however, in addition to Ins2 in 2/21 clones and Ins3 in 1/21 clones, we also identified four other insertions of various sizes: 162 nt (Ins1, 4/21 clones), 145 nt (Ins4, 1/21 clones), another of 168 nt (Ins5, 1/21 clones), and 149 nt (Ins7, 1/21 clones). At passage 3, the 162-nt Ins1 became predominant, being found in 16/19 clones; we also saw Ins2 (2/19 clones) and a new 40-nt insertion (Ins6) in 1/19 clones. Of the seven inserted sequences, six (Ins1 to Ins5 and Ins7) corresponded to the 3′-terminal region of the viral ORF followed by the entire (or nearly entire) domain II-1; the remaining sequence (Ins6) corresponded to the 3′ half of domain II-1, including the complete RCS2 motif (Fig. 10C). More importantly, in all cases the newly inserted sequences perfectly matched a sequence immediately upstream of their respective insertion sites (Fig. 10C), indicating that these sequences were introduced by the duplication of the corresponding individual sequences just upstream of domain II-2.
FIG. 10.
Identification of seven sequence insertions (duplications) of various sizes at the junction between domains II-1 and II-2 in the genomic RNAs of MUTΔ1-284-derived pseudorevertants. The original recombinant viral stock was obtained after transfection of BHK-21 cells with the RNA transcripts derived from MUTΔ1-284 cDNA (passage 0); this virus stock was then passaged three times in naïve BHK-21 cells at an MOI of 0.1 (passages 1 to 3). The 3′-terminal sequences of the genomic RNAs at passages 0, 1, and 3 were determined as described in Materials and Methods. (A) Relative locations of the seven sequence insertions (Ins1 to Ins7) duplicated from upstream sequences of domain II-2 in the genomic RNAs of MUTΔ1-284-derived pseudorevertants. At the top is a schematic presentation of the genomic RNA derived from the MUTΔ1-284 cDNA; given below is an expanded view of the 290-nt 3′NCR containing only the three 3′-proximal domains (II-1, II-2, and III), with the three CSs (CS1, CS2, and RCS2) and the 3′SL indicated. (B and C) Nucleotide sequences of the seven sequence insertions (Ins1 to Ins7) of various sizes. (B) The number of clones containing each identified nucleotide sequence/the total number of clones containing the insert that we examined is indicated. (C) The nucleotides of the seven sequence insertions (duplications) were aligned; the RCS2 motif is highlighted with a gray box. Open arrowheads indicate the translation stop codon UAG.
To assess the functional importance of these duplicated sequences in MUTΔ1-284-derived RNA replication, we constructed seven derivatives of MUTΔ1-284 with all of these duplicated sequences and examined their phenotypes after the transfection of BHK-21 cells with the RNAs synthesized from the corresponding cDNAs. In all cases, the specific infectivities were essentially the same as those estimated for the original MUTΔ1-284-derived RNA, which were comparable to those of the WT-derived RNA (1.3 × 106 to 2.2 × 106 FFU/μg; data not shown). There were, however, significant differences between the phenotypes of the seven MUTΔ1-284 derivatives and that of the original MUTΔ1-284. At 4 days posttransfection, all seven derivatives always produced homogeneous foci/plaques that were essentially indistinguishable from those produced by WT-derived RNA. These foci/plaques were, on average, ∼2.5-fold larger than the rather heterogeneous foci produced by the original MUTΔ1-284-derived RNA (Fig. 11A). Consistent with these results, the levels of JEV RNA produced by the cells transfected with any of the seven MUTΔ1-284 derivative RNAs during the first 22 h of the incubation period were invariably ∼50 to 70-fold higher than that of the original MUTΔ1-284-derived RNA, ranging from ∼50% of the WT level to nearly the same as that of the WT (Fig. 11B). This increase in JEV RNA production was reflected in their levels of viral proteins (Fig. 11C) and virus yield (Fig. 11D).
Taken together, our findings indicate that MUTΔ1-284-derived RNA duplicated a viral sequence of various sizes (40 to 168 nt) just upstream of domain II-2 to generate a direct repeat sequence. All of these duplications dramatically increased the RNA replication efficiency of the MUTΔ1-284-derived RNAs to a level close to that of WT-derived RNA. Of the seven duplicated sequences, the shortest was surprisingly only 40 nt long, corresponding to the 3′ half of domain II-1, which includes the complete RCS2 motif. Therefore, our data suggest that the duplication of a sequence as short as the RCS2 motif in MUTΔ1-284 is sufficient to achieve RNA replication levels that are nearly indistinguishable from WT levels.
Cell type-specific replication of MUTΔ1-381-derived viruses, which are capable of replicating in BHK-21 and SH-SY5Y cells but not in C6/36 cells.
The five 3′NCR mutant viruses (MUTΔ1-133, MUTΔ1-211, MUTΔ1-284, MUTΔ1-381, and MUTΔ1-450/83ntIns) that were competent for replication in BHK-21 cells were examined further for their growth properties in two other cell types, human neuroblastoma SH-SY5Y and mosquito C6/36 cells, which are potentially important for JEV pathogenesis and transmission. Monolayers of each of these two cell lines were infected at an MOI of 1 with the WT or each of the five mutant viruses, and during an incubation period of 4 days (SH-SY5Y) or 6 days (C6/36), equal volumes of the culture fluids were harvested for progeny virions at the indicated time points. The production levels of the progeny virions were determined by titration on naive BHK-21 cells.
As shown in Fig. 12A, we found that the five mutant viruses all replicated successfully in SH-SY5Y cells, with the differential patterns of growth rate and virus yield being analogous to those observed in BHK-21 cells (Fig. 7A). In C6/36 cells, however, these characteristic growth properties were noted for only four (MUTΔ1-133, MUTΔ1-211, MUTΔ1-284, and MUTΔ1-450/83ntIns) of the five mutant viruses, although their growth kinetics were significantly delayed compared to those seen in BHK-21 and SH-SY5Y cells. In the case of the remaining MUTΔ1-381-derived virus, no signs of viral growth were observed during a 6-day period of incubation. To confirm the inability of the MUTΔ1-381-derived viruses to replicate in C6/36 cells, we performed immunoblotting to assess the production of JEV-specific proteins by the infected cells. Consistent with the lack of viral growth that we observed, we were unable to detect any JEV-specific proteins in the lysates of MUTΔ1-381-infected C6/36 cells at 48 or even 144 h postinfection, whereas significant quantities of JEV-specific proteins (i.e., E, NS1, and NS3) were readily detectable at the same time points in the lysates of C6/36 cells infected with the WT or MUTΔ1-284-derived viruses (Fig. 12B).
FIG. 12.
Cell type-dependent replication of MUTΔ1-381-derived mutant virus. (A) Viral growth properties in human neuroblastoma SH-SY5Y and mosquito C6/36 cells. Monolayers of SH-SY5Y or C6/36 cells were infected at an MOI of 1 with the WT or one of the five 3′NCR mutant viruses derived from the respective cDNAs, as indicated. At various time points postinfection, aliquots of culture supernatants were collected and used for virus titration on naïve BHK-21 cells. Data were consistent in two independent experiments. (B) Viral protein accumulation in infected C6/36 cells. Monolayers of C6/36 cells were mock infected or infected at an MOI of 1 with WT, MUTΔ1-284-derived, or MUTΔ1-381-derived virus, or at an MOI of 10 with MUTΔ1-381-derived virus in parallel. At 48 and 144 h postinfection (hpi), the levels of JEV-specific proteins or JEV NS1 protein were analyzed by immunoblotting with the anti-JEV antiserum (anti-JEV) or a rabbit antiserum specific for JEV NS1 (anti-NS1), respectively. The positions of the viral proteins (E, NS1, and NS3) and a cleavage-related intermediate (open arrowhead) are indicated on the left. Molecular size markers (in kilodaltons) are shown on the right.
Taken together, these data demonstrate that of the five 3′NCR mutant viruses recovered from BHK-21 cells after RNA transfection, four (MUTΔ1-133, MUTΔ1-211, MUTΔ1-284, and MUTΔ1-450/83ntIns) are replication competent in not only BHK-21 cells but also in human neuroblastoma SH-SY5Y and mosquito C6/36 cells, with a progressive negative effect on viral growth that reflects the size of the deletion introduced in the 5′-proximal region of the JEV 3′NCR. On the other hand, the replication of the remaining mutant virus derived from MUTΔ1-381 was cell type dependent (capable of replicating in BHK-21 and SH-SY5Y cells but not in C6/36 cells), suggesting that one or more of the four 5′-proximal domains (V, X, I, and II-1) of the JEV 3′NCR is (are) involved in maintaining the replication competence of the genomic RNA in a cell type-specific fashion.
DISCUSSION
In the present study, we have used an infectious cDNA technology previously developed in our laboratory (83) to elucidate the role in the virus life cycle of 3′ cis-acting RNA elements scattered throughout the 574-nt JEV 3′NCR, with a particular focus on the ∼450-nt 5′-proximal region. One of the most interesting findings from this study is that only a relatively short region of the JEV 3′NCR is required for genomic RNA replication. We have demonstrated that MUTΔ1-381-derived mutant RNA, consisting of the 3′-proximal 193 nt (domains II-2 and III) but not the other most 5′-proximal 381 nt (domains V, X, I, and II-1), is competent for replication in two mammalian cell types (BHK-21 and SH-SY5Y cells) and can produce infectious virions; however, two mutant RNAs (derived from MUTΔ1-450 and MUTΔ1-473) with the shorter 3′-terminal sequences were not replication competent, and neither was an MUTΔ477-574-derived mutant RNA lacking only the 3′SL at the 3′ end of the genome (Fig. 1, 2, 7, and 12). By comparison, the minimal 3′ region required for genomic RNA replication in DENV is shorter than that in JEV: a full-length mutant RNA (designated 3′d 384-183/172-113), bearing only a sequence that is structurally equivalent to domain III of the JEV 3′NCR, was able to produce infectious virions, although the RNA infectivity was severely impaired (49). In the case of the tick-borne encephalitis virus, on the other hand, the minimal 3′ region for genomic RNA replication was designated a core region (46, 54) that appears to be analogous to the two 3′-proximal domains (II-2 and III) of the JEV 3′NCR.
The requirement for the 3′-terminal domain III of the JEV 3′NCR for genome replication is consistent with previously published work. In the case of other mosquito-borne flaviviruses, the functionality of 3′CYC and 3′SL in domain III has been well documented; both have been shown to be essential for genome cyclization and RNA replication, either by using an in vivo genetic approach employing full-length or subgenomic replicon RNAs in the case of KUNV (34, 35), WNV (18, 42, 70, 82), YFV (8, 14), and DENV (2, 22, 49, 86), or by using an in vitro biochemical approach employing short exogenous viral RNAs in the case of WNV (51) and DENV (1, 3, 79, 80). The 3′SL of domain III also includes a loop of conserved pentanucleotides (5′-CACAG-3′) at its top and a pseudoknot structure at the bottom, both of which have been shown to be involved in efficient viral replication (35, 61, 62, 70, 80). Although these RNA elements are also found in the 3′-terminal domain III of JEV genomic RNA, the identification of the major determinants of viral replication within this region will require further investigation.
We recently described a novel complex RNA motif in JEV that is defined by three 5-nt-long discontinuous complementary sequences: an L strand corresponding to a sequence upstream of the translation start codon AUG close to the 5′ end of the genome, and M and R strands corresponding to the bottom stem of the 3′SL at the 3′ end (64). This complex motif has been shown to participate in two base-pairing interactions that are capable of forming L-M and M-R duplexes, which are critical for the 5′-3′ long-distance interaction and genome replication of JEV and possibly other mosquito-borne flaviviruses. However, definitive experimental evidence to define the molecular mechanism(s) by which this complex motif regulates JEV genome replication, in connection with other cis-acting replication signals present within the genome, still is lacking. Several studies have indicated that a pair of complementary sequences (named 5′UAR/3′UAR, for upstream AUG region), along with the well-documented 5′CYC/3′CYC, are functionally important for the genome cyclization and replication of WNV and DENV (3, 4, 17, 87); interestingly, this 5′UAR/3′UAR motif appears to be analogous to the L/M strands of JEV. For tick-borne encephalitis virus, a pair of complementary sequences (designated 5′-CS-A/3′-CS-A), which are different from the 5′CYC/3′CYC but similar to the L/M strands of JEV with respect to their locations but not their sequences, have been shown to be involved in genome cyclization and replication (37). It is still unclear how these cyclization sequence motifs found in mosquito- and tick-borne flaviviruses coordinate the multiple interactions of the 3′-terminal region of the genome with the 5′-terminal region, although these interactions most likely occur in association with viral and cellular proteins. Thus far, a panel of mammalian and insect proteins that interact with the 5′- and/or 3′-terminal regions has been identified (6, 7, 15, 16, 19, 20, 39, 53, 66, 77). Further studies are needed to elucidate the molecular mechanism(s) by which these host factors contribute to RNA replication and other virus-induced cellular processes.
Unlike the 3′-terminal domain III, the 3′-penultimate domain II-2 appears to have differential effects on RNA replication competence, largely depending on the type of constructs used (full-length versus replicon viral RNAs, for which the sensitivity of the assays is different), and the type of deletions (internal versus progressive deletions, in which the region and size of deletions introduced in the individual viral systems is varied). In the context of full-length viral RNA, DENV mutants lacking either domain II-2 or II-1 or both domains are viable and produce infectious virions, although the viral growth is compromised (2, 49); likewise, one YFV mutant lacking its CS2 motif (embedded in domain II-2) is also viable, although its level of RNA replication is reduced (8). In the case of luciferase-expressing replicon RNA, DENV mutants lacking either domain II-2 or II-1 have a drastically lower level of RNA replication than does WT virus, but a mutant lacking both domains shows no detectable RNA replication; in all three cases, the level of translation efficiency is not significantly affected (2). Similarly, WNV replicon mutant RNAs lacking the motif CS2 or RCS2 (embedded in domain II-1) are also viable, but their RNA replication is severely affected (42); however, no RNA replication can be detected in the case of mutants that have a 3′-terminal region corresponding to domain II-2 and the downstream domain III but lack the remaining most 5′-proximal region, although the translation efficiency is not compromised (42, 70). In the present study, we have demonstrated that the 3′-penultimate domain II-2, along with the 3′-terminal domain III, is sufficient for JEV genome replication in BHK-21 and SH-SY5Y cells, although the replication efficiency was dramatically reduced (Fig. 1, 2, 7, and 12). Also, as has been shown previously for other flavivirus systems (2, 42, 70), the mutant RNAs generated in the present study appear to have no deleterious effect on the translation efficiency, as assayed by transfecting BHK-21 cells with the luciferase-expressing replicon versions of their respective mutant RNAs, in which the luciferase gene was fused in frame to the 35 N-terminal amino acids of the JEV C protein, and subsequently monitoring luciferase activity at 4 to 6 h posttransfection (data not shown). Thus, it appears that domains II-2 and III at the 3′ end of JEV genomic RNA play an important role in RNA synthesis rather than translation.
Remarkably, we were able to recover infectious pseudorevertants from the replication-incompetent JEV 3′NCR mutant RNA (MUTΔ1-450), which lacks the entire 3′NCR but preserves the 124-nt 3′-terminal domain III (Fig. 3). It was particularly striking that all of these pseudorevertants acquired an additional copy of a viral 83-nt sequence duplicated from the 3′-terminal region of the viral ORF, which was subsequently shown to be sufficient for restoring the viability of this mutant RNA (Fig. 4 to 6). This is, to the best of our knowledge, the first example of a novel recombinant flavivirus generated by sequence duplication that is capable of regulating RNA replication even with such a short 3′NCR. However, the mechanism of how the 83-nt duplicated sequence is able to restore the replicability of the MUTΔ1-450-derived mutant RNA is as yet unknown. Considerable effort has been made to explore the possible phylogenetic conservation of the 83-nt sequence in the full-length genomes of other flaviviruses, including one YFV (vaccine strain 17D [X03700]), two WNVs (lineage I strain NY99 [NC_009942] and lineage II strain 956 [NC_001563]), and four DENVs (one for each serotype, DENV-1 [FJ850077], DENV-2 [FJ850088], DENV-3 [FJ850094], and DENV-4 [FJ850095]). We found that the nucleotide composition and predicted secondary structures of the 83-nt sequence found at the 3′-terminal region of the JEV ORF was distinct from those found at their corresponding loci in other flaviviruses, since a low percentage of sequence identities (i.e., ∼16.8% for YFV 17D, ∼33.1 to 38.7% for two WNVs, and ∼11.7 to 45.3% for four DENVs) was noted, with no significant similarity in the predicted secondary structures being identified (data not shown). Interestingly, an ∼50-nt cis-acting element (5BSL3.2) required for the replication of hepatitis C virus (HCV), a nonflavivirus in the family Flaviviridae, has been shown to be a part of a larger cruciform-like structure (5BSL3) located at the 3′-terminal region of the HCV ORF (81). Subsequently, the element 5BSL3.2 has been indicated to form a kissing-loop tertiary structure with part of the 3′NCR (78). Comparisons of the HCV 5BSL3 sequence to the JEV 83-nt sequence discovered in this study revealed no recognizable similarity in the primary sequences or predicted secondary structures, although both share their loci within their respective genomes (data not shown).
Although it is not yet clear how this 83-nt sequence becomes duplicated, we can envision three possible scenarios. In the first scenario, SP6 RNA polymerase involved in the in vitro transcription reaction duplicates the 83-nt sequence during RNA synthesis from MUTΔ1-450 cDNA, albeit only rarely and by chance, and the preexisting replication-competent mutant RNA is selected over the vast majority of other replication-incompetent mutant RNAs. The other two scenarios involve viral RNA polymerase-mediated RNA recombination involving template switching or copy choice, as has been described for many other RNA viruses (5, 38, 65, 76). Thus, the viral RNA polymerase could duplicate the 83-nt sequence intramolecularly during negative-strand RNA synthesis, given a situation in which the viral polymerase pauses during the elongation step at a given locus and reinitiates the polymerization upstream of the pause site on the same RNA template. Alternatively, the sequence duplication could be generated by template switching between two different RNA templates during negative-strand RNA synthesis, creating a direct repeat sequence.
While the 193-nt 3′-proximal region of the JEV 3′NCR includes the two CS motifs (CS1 in domain III and CS2 in domain II-2) and a downstream 3′SL that are shared with all mosquito-borne flaviviruses, the remaining 381 nt of the 5′-proximal region contains the three other CS motifs (RCS2 in domain II-1, CS3 in domain I, and RCS3 in domain V) that differ not only between members of different serological groups but also within the same serological group (reviewed in references 23 and 47). Specifically, CS3 and RCS3 are seen only in the JEV-related group (JEV, WNV, and MVEV); RCS2 is found in the JEV- and DENV-related groups but not the YFV-related group, which instead contains another set of one to three tandem repeat sequences. In the present study, we have shown that the 381-nt 5′-proximal region of the JEV 3′NCR is biologically important for the replication of JEV genomic RNA; we also made the interesting observation that this region acts in a cell type-specific fashion (Fig. 7 and 12). In the case of two mammalian cell lines (BHK-21 and SH-SY5Y), the cis-acting elements of primary sequences and/or secondary structures present within the 381-nt 5′-proximal region displayed an enhancer-like function in promoting RNA replication efficiency but was dispensable for RNA replication competence; this observation is in agreement with previous findings with other flaviviruses despite their high degree of sequence divergence (2, 8, 31, 33, 42, 49, 58). In the case of mosquito C6/36 cells, however, one or more of the cis-acting element(s) in the 381-nt 5′-proximal region of the JEV 3′NCR, along with all of the elements found in the 193-nt 3′-proximal region, appeared to be required for RNA replication competence; thus, it appears that additional cellular/viral factors or different strategies for JEV RNA replication are also involved depending on the cell type.
We have also made the important observation that the replication efficiency of JEV genomic RNA can be enhanced by two types of adaptive mutations occurring in the 3′-terminal region: (i) point mutations (single-nucleotide substitutions or deletions), as exemplified by the MUTΔ1-381-derived RNA, and (ii) sequence duplications, as exemplified by the MUTΔ1-284-derived RNA. In the case of the MUTΔ1-381-derived RNA (Fig. 8 and 9), despite the significant effect of the 12 different point mutations on RNA replication efficiency, there was no obvious relationship between these acquired point mutations and the predicted secondary structures (data not shown). However, we noted that a higher level of RNA replication with the single C444→U substitution was further enhanced by ∼4-fold with an additional G488→A substitution in a region between the 3′CYC and the 3′SL (Fig. 1). This region is known to form a small stem-loop, with the loop forming a pseudoknot with the base stem of the 3′SL (61, 80). Based on this model, the G488→A substitution would change the U476-G488 base pairing in this stem-loop to U-A base pairing, in part stabilizing its secondary structure. How each of these point mutations is able to enhance RNA replication efficiency is currently unknown. We suggest that these point mutations are able to rearrange the local secondary RNA structure surrounding the CS2 motif, thereby facilitating the RNA-RNA interaction with other cis-acting RNA elements or the RNA-protein interaction with viral replicase proteins and/or other as-yet-unknown cellular protein(s). An alternative but less likely possibility is that these point mutations can increase the stability of the RNA or its translation rate.
In the case of MUTΔ1-284-derived RNA (Fig. 10 and 11), it was intriguing that the duplication of a viral sequence of various sizes (40 to 168 nt) took place exclusively at the junction between domains II-1 and II-2 and that this sequence duplication was sufficient to increase the level of RNA replication to levels nearly comparable to that of the WT. Of the seven duplicated sequences, the shortest was 40 nt long and matched the 3′ half of domain II-1; of the 40 nt, ∼30 nt were shared with the other duplicated sequences. Since this shared sequence incorporated the complete RCS2 motif, all of the duplicated sequences created a new direct repeat (RCS2/RCS2) separated by an interrupting sequence of various sizes. Therefore, the presence of this direct repeat sequence in the duplicated region most likely is responsible for the enhancement in RNA replication efficiency.
From an evolutionary standpoint, it is important to understand the molecular mechanism(s) undergirding the appearance of short direct repeats in the flavivirus 3′NCR and to identify their function(s) in viral replication and transmission. Recently, insights from rigorous comparative sequence alignments have suggested that the direct repeats have originated from a longer hypothetical precursor that includes an earlier version of all of the preexisting direct repeat sequences; subsequently, this longer precursor may have undergone a variety of mutagenetic events (i.e., deletions, insertions, and substitutions) that create the unique array of shorter direct repeats, varying in terms of number, size, and nucleotide composition, that currently are found in the 3′NCR of each serological group (recently reviewed in reference 28). This longer primeval precursor has not been identified in the genomes of mosquito-borne flaviviruses; in the case of tick-borne flaviviruses, however, the origins of this precursor are found in both the 3′NCR and the viral ORF (24-27). Remarkably, the existence of such a precursor was substantiated experimentally by the results of the present study, which have demonstrated that JEV genomic RNAs with an impaired capacity for RNA replication can undergo sequence duplications, thereby acquiring a new pair of short direct repeats that can drastically enhance the RNA replication efficiency. It is particularly interesting that this duplicated sequence originated not only from the 5′-proximal region of the 3′NCR (MUTΔ1-284) (Fig. 10 to 11) but also from the 3′-terminal region of the viral ORF (MUTΔ1-450) (Fig. 3 to 6). In addition, the acquisition of a variety of point mutations in one of the CS motifs led to increases in RNA replication efficiency to various degrees that proved to be advantageous for viral growth (MUTΔ1-381) (Fig. 8 and 9).
In summary, the findings described in this report not only provide new information regarding an array of 3′ cis-acting RNA elements that plays a vital, cell type-specific role in controlling JEV genomic RNA replication but also offer new insights into 3′-directed repeat sequences that are newly acquired by the sequence duplication of the coding region and the 3′NCR of JEV genomic RNA. Furthermore, we have generated recombinant JEVs harboring various 5′-truncated versions of the 3′NCR (i.e., MUTΔ1-381 and MUTΔ1-450/83ntIns), which we believe provide an initial platform for developing safe and effective live-attenuated vaccines and new antiviral strategies against JEV.
Acknowledgments
This work was supported by a grant (KRF-2006-312-C00312) from the Korea Research Foundation funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) and a grant (#0320110) from the National R&D Program for Cancer Control funded by the Ministry of Health and Welfare, Republic of Korea.
We thank Deborah McClellan for editorial assistance.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 27639926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, D. E., A. L. De Lella Ezcurra, S. Fucito, and A. V. Gamarnik. 2005a. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339200-212. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez, D. E., M. F. Lodeiro, S. J. Luduena, L. I. Pietrasanta, and A. V. Gamarnik. 2005b. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 796631-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez, D. E., C. V. Filomatori, and A. V. Gamarnik. 2008. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology 375223-235. [DOI] [PubMed] [Google Scholar]
- 5.Ball, L. A. 1997. Nodavirus RNA recombination. Semin. Virol. 895-100. [Google Scholar]
- 6.Blackwell, J. L., and M. A. Brinton. 1995. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol. 695650-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 716433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 841261-1268. [DOI] [PubMed] [Google Scholar]
- 9.Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153113-121. [DOI] [PubMed] [Google Scholar]
- 10.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162290-299. [DOI] [PubMed] [Google Scholar]
- 11.Calisher, C. H., and E. A. Gould. 2003. Taxonomy of the virus family Flaviviridae. Adv. Virus Res. 591-19. [DOI] [PubMed] [Google Scholar]
- 12.Cammisa-Parks, H., L. A. Cisar, A. Kane, and V. Stollar. 1992. The complete nucleotide sequence of cell fusing agent (CFA): homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology 189511-524. [DOI] [PubMed] [Google Scholar]
- 13.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44649-688. [DOI] [PubMed] [Google Scholar]
- 14.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. H. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 772265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, W. G., J. L. Blackwell, P. Y. Shi, and M. A. Brinton. 2007. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 8110172-10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Nova-Ocampo, M., N. Villegas-Sepúlveda, and R. M. del Angel. 2002. Translation elongation factor-1alpha, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295337-347. [DOI] [PubMed] [Google Scholar]
- 17.Dong, H., B. Zhang, and P. Y. Shi. 2008. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 381123-135. [DOI] [PubMed] [Google Scholar]
- 18.Elghonemy, S., W. G. Davis, and M. A. Brinton. 2005. The majority of the nucleotides in the top loop of the genomic 3′ terminal stem loop structure are cis-acting in a West Nile virus infectious clone. Virology 331238-246. [DOI] [PubMed] [Google Scholar]
- 19.Emara, M. M., and M. A. Brinton. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. USA 1049041-9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emara, M. M., H. Liu, W. G. Davis, and M. A. Brinton. 2008. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J. Virol. 8210657-10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endy, T. P., and A. Nisalak. 2002. Japanese encephalitis virus: ecology and epidemiology. Curr. Top. Microbiol. Immunol. 26711-48. [DOI] [PubMed] [Google Scholar]
- 22.Filomatori, C. V., M. F. Lodeiro, D. E. Alvarez, M. M. Samsa, L. Pietrasanta, and A. V. Gamarnik. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 202238-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gritsun, T. S., A. Tuplin, and E. A. Gould. 2006. Origin, evolution, and function of flavivirus RNA in untranslated and coding regions: implications for virus transmission, p. 47-99. In M. Kalitzky and P. Borowski (ed.), Molecular biology of the flavivirus. Horizon Scientific Press, Norwich, United Kingdom.
- 24.Gritsun, T. S., and E. A. Gould. 2006a. The 3′ untranslated regions of Kamiti River virus and cell fusing agent virus originated by self-duplication. J. Gen. Virol. 872615-2619. [DOI] [PubMed] [Google Scholar]
- 25.Gritsun, T. S., and E. A. Gould. 2006b. The 3′ untranslated region of tick-borne flaviviruses originated by the duplication of long repeat sequences within the open reading frame. Virology 354217-223. [DOI] [PubMed] [Google Scholar]
- 26.Gritsun, T. S., and E. A. Gould. 2006c. Direct repeats in the 3′ untranslated regions of mosquito-borne flaviviruses: possible implications for virus transmission. J. Gen. Virol. 873297-3305. [DOI] [PubMed] [Google Scholar]
- 27.Gritsun, T. S., and E. A. Gould. 2007a. Direct repeats in the flavivirus 3′ untranslated region; a strategy for survival in the environment? Virology 358258-265. [DOI] [PubMed] [Google Scholar]
- 28.Gritsun, T. S., and E. A. Gould. 2007b. Origin and evolution of 3′UTR of flaviviruses: long direct repeats as a basis for the formation of secondary structures and their significance for virus transmission. Adv. Virus Res. 69203-248. [DOI] [PubMed] [Google Scholar]
- 29.Gubler, D. J., G. Kuno, and L. Markoff. 2007. Flaviviruses, p. 1153-1252. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins Publishers, Philadelphia, PA.
- 30.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 19833-41. [DOI] [PubMed] [Google Scholar]
- 31.Hoenninger, V. M., H. Rouha, K. K. Orlinger, L. Miorin, A. Marcello, R. M. Kofler, and C. W. Mandl. 2008. Analysis of the effects of alterations in the tick-borne encephalitis virus 3′-noncoding region on translation and RNA replication using reporter replicons. Virology 377419-430. [DOI] [PubMed] [Google Scholar]
- 32.Khromykh, A. A., and E. G. Westaway. 1994. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J. Virol. 684580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 711497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 756719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khromykh, A. A., N. Kondratieva, J. Y. Sgro, A. Palmenberg, and E. G. Westaway. 2003. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 7710623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, J.-M., S.-I. Yun, B.-H. Song, Y.-S. Hahn, C.-H. Lee, H.-W. Oh, and Y.-M. Lee. 2008. A single N-linked glycosylation site in the Japanese encephalitis virus prM protein is critical for cell type-specific prM protein biogenesis, virus particle release, and pathogenicity in mice. J. Virol. 827846-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kofler, R. M., V. M. Hoenninger, C. Thurner, and C. W. Mandl. 2006. Functional analysis of the tick-borne encephalitis virus cyclization elements indicates major differences between mosquito-borne and tick-borne flaviviruses. J. Virol. 804099-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai, M. M. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 5661-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, W., Y. Li, N. Kedersha, P. Anderson, M. Emara, K. M. Swiderek, G. T. Moreno, and M. A. Brinton. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 7611989-12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindenbach, B. D., and C. M. Rice. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 5923-61. [DOI] [PubMed] [Google Scholar]
- 41.Lindenbach, B. D., H.-J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins Publishers, Philadelphia, PA.
- 42.Lo, M. K., M. Tilgner, K. A. Bernard, and P. Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 7710004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackenzie, J. S., C. A. Johansen, S. A. Ritchie, A. F. Van Den Hurk, and R. A. Hall. 2002. Japanese encephalitis as an emerging virus: the emergence and spread of Japanese encephalitis virus in Australasia. Curr. Top. Microbiol. Immunol. 26749-73. [DOI] [PubMed] [Google Scholar]
- 44.Mackenzie, J. S., D. J. Gubler, and L. R. Petersen. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10S98-S109. [DOI] [PubMed] [Google Scholar]
- 45.Mandl, C. W., H. Holzmann, C. Kunz, and F. X. Heinz. 1993. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology 194173-184. [DOI] [PubMed] [Google Scholar]
- 46.Mandl, C. W., H. Holzmann, T. Meixner, S. Rauscher, P. F. Stadler, S. L. Allison, and F. X. Heinz. 1998. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 722132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markoff, L. 2003. 5′- And 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 59177-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 49.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 703930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohan, P. M., and R. Padmanabhan. 1991. Detection of stable secondary structure at the 3′ terminus of dengue virus type 2 RNA. Gene 108185-191. [DOI] [PubMed] [Google Scholar]
- 51.Nomaguchi, M., T. Teramoto, L. Yu, L. Markoff, and R. Padmanabhan. 2004. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J. Biol. Chem. 27912141-12151. [DOI] [PubMed] [Google Scholar]
- 52.Olsthoorn, R. C., and J. F. Bol. 2001. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 71370-1377. [PMC free article] [PubMed] [Google Scholar]
- 53.Paranjape, S. M., and E. Harris. 2007. Y box-binding protein-1 binds to the dengue virus 3′-untranslated region and mediates antiviral effects. J. Biol. Chem. 28230497-30508. [DOI] [PubMed] [Google Scholar]
- 54.Pletnev, A. G. 2001. Infectious cDNA clone of attenuated Langat tick-borne flavivirus (strain E5) and a 3′ deletion mutant constructed from it exhibit decreased neuroinvasiveness in immunodeficient mice. Virology 282288-300. [DOI] [PubMed] [Google Scholar]
- 55.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 251194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rauscher, S., C. Flamm, C. W. Mandl, F. X. Heinz, and P. F. Stadler. 1997. Secondary structure of the 3′-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. RNA 3779-791. [PMC free article] [PubMed] [Google Scholar]
- 57.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229726-733. [DOI] [PubMed] [Google Scholar]
- 58.Romero, T. A., E. Tumban, J. Jun, W. B. Lott, and K. A. Hanley. 2006. Secondary structure of dengue virus type 4 3′ untranslated region: impact of deletion and substitution mutations. J. Gen. Virol. 873291-3296. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 60.Schmittgen, T. D., B. A. Zakrajsek, A. G. Mills, V. Gorn, M. J. Singer, and M. W. Reed. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 285194-204. [DOI] [PubMed] [Google Scholar]
- 61.Shi, P. Y., M. A. Brinton, J. M. Veal, Y. Y. Zhong, and W. D. Wilson. 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 354222-4230. [DOI] [PubMed] [Google Scholar]
- 62.Silva, P. A., R. Molenkamp, T. J. Dalebout, N. Charlier, J. H. Neyts, W. J. Spaan, and P. J. Bredenbeek. 2007. Conservation of the pentanucleotide motif at the top of the yellow fever virus 17D 3′ stem-loop structure is not required for replication. J. Gen. Virol. 881738-1747. (Erratum, 88:2361). [DOI] [PubMed] [Google Scholar]
- 63.Solomon, T. 2003. Recent advances in Japanese encephalitis. J. Neurovirol. 9274-283. [DOI] [PubMed] [Google Scholar]
- 64.Song, B.-H., S.-I. Yun, Y.-J. Choi, J.-M. Kim, C.-H. Lee, and Y.-M. Lee. 2008. A complex RNA motif defined by three discontinuous 5-nucleotide-long strands is essential for flavivirus RNA replication. RNA 141791-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strauss, J. H., and E. G. Strauss. 1997. Recombination in alphaviruses. Semin. Virol. 885-94. [Google Scholar]
- 66.Ta, M., and S. Vrati. 2000. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 745108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thiel, H.-J., M. S. Collett, E. A. Gould, F. X. Heinz, M. Houghton, G. Meyers, R. H. Purcell, and C. M. Rice. 2005. Family Flaviviridae, p. 981-998. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the international committee on taxonomy of viruses. Elsevier Academic Press, San Diego, CA.
- 68.Thurner, C., C. Witwer, I. L. Hofacker, and P. F. Stadler. 2004. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 851113-1124. [DOI] [PubMed] [Google Scholar]
- 69.Tilgner, M., and P. Y. Shi. 2004. Structure and function of the 3′ terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 788159-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tilgner, M., T. S. Deas, and P. Y. Shi. 2005. The flavivirus-conserved pentanucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 331375-386. [DOI] [PubMed] [Google Scholar]
- 71.Weaver, S. C., and A. D. Barrett. 2004. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wengler, G., and G. Wengler. 1981. Terminal sequences of the genome and replicative-from RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology 113544-555. [DOI] [PubMed] [Google Scholar]
- 73.Wengler, G., and E. Castle. 1986. Analysis of structural properties which possibly are characteristic for the 3′-terminal sequence of the genome RNA of flaviviruses. J. Gen. Virol. 671183-1188. [DOI] [PubMed] [Google Scholar]
- 74.Westaway, E. G., J. M. Mackenzie, and A. A. Khromykh. 2003. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 5999-140. [DOI] [PubMed] [Google Scholar]
- 75.Winer, J., C. K. Jung, I. Shackel, and P. M. Williams. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 27041-49. [DOI] [PubMed] [Google Scholar]
- 76.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 802535-2543. [DOI] [PubMed] [Google Scholar]
- 77.Yocupicio-Monroy, M., R. Padmanabhan, F. Medina, and R. M. del Angel. 2007. Mosquito La protein binds to the 3′ untranslated region of the positive and negative polarity dengue virus RNAs and relocates to the cytoplasm of infected cells. Virology 35729-40. [DOI] [PubMed] [Google Scholar]
- 78.You, S., and C. M. Rice. 2008. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J. Virol. 82184-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′ end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 27433714-33722. [DOI] [PubMed] [Google Scholar]
- 80.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 27615581-15591. [DOI] [PubMed] [Google Scholar]
- 81.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 781352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu, L., and L. Markoff. 2005. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J. Virol. 792309-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yun, S.-I., S.-Y. Kim, C. M. Rice, and Y.-M. Lee. 2003. Development and application of a reverse genetics system for Japanese encephalitis virus. J. Virol. 776450-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yun, S.-I., and Y.-M. Lee. 2006. Japanese encephalitis virus: molecular biology and vaccine development, p. 225-271. In M. Kalitzky and P. Borowski (ed.), Molecular biology of the flavivirus. Horizon Scientific Press, Norwich, United Kingdom.
- 85.Yun, S.-I., Y.-J. Choi, X.-F. Yu, J.-Y. Song, Y.-H. Shin, Y.-R. Ju, S.-Y. Kim, and Y.-M. Lee. 2007. Engineering the Japanese encephalitis virus RNA genome for the expression of foreign genes of various sizes: implications for packaging capacity and RNA replication efficiency. J. Neurovirol. 13522-535. [DOI] [PubMed] [Google Scholar]
- 86.Zeng, L., B. Falgout, and L. Markoff. 1998. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 727510-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang, B., H. Dong, D. A. Stein, P. L. Iversen, and P. Y. Shi. 2008. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 3731-13. [DOI] [PubMed] [Google Scholar]
- 88.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]