Abstract
T20 (generic name, enfuvirtide; brand name, Fuzeon) is a first-generation human immunodeficiency virus (HIV) fusion inhibitor approved for salvage therapy of HIV-infected patients refractory to current antiretroviral drugs. However, its clinical use is limited because of rapid emergence of T20-resistant viruses in T20-treated patients. Therefore, T1249 and T1144 are being developed as the second- and third-generation HIV fusion inhibitors, respectively, with improved efficacy and drug resistance profiles. Here, we found that combinations of T20 with T1249 and/or T1144 resulted in exceptionally potent synergism (combination index, <0.01) against HIV-1-mediated membrane fusion by 2 to 3 orders of magnitude in dose reduction. Highly potent synergistic antiviral efficacy was also achieved against infection by laboratory-adapted and primary HIV-1 strains, including T20-resistant variants. The mechanism underlying the synergistic effect could be attributed to the fact that T20, T1249, and T1144 all contain different functional domains and have different primary binding sites in gp41. As such, they may work cooperatively to inhibit gp41 six-helix bundle core formation, thereby suppressing virus-cell fusion. Therefore, these findings strongly imply that, rather than replacing T20, combining it with HIV fusion inhibitors of different generations might produce synergistic activity against both T20-sensitive and -resistant HIV-1 strains, suggesting a new therapeutic strategy for the treatment of HIV-1 infection/AIDS.
In the early 1990s, a number of highly potent anti-human immunodeficiency virus type 1 (HIV-1) peptides derived from the C-heptad repeat (CHR) domain of the HIV-1 envelope glycoprotein (Env) transmembrane subunit gp41 were discovered (21, 22, 35, 59, 61). Biophysical and biochemical analyses suggest that the CHR peptides inhibit HIV-1 Env-mediated membrane fusion by interacting with the viral gp41 N-heptad repeat (NHR) domain to form heterologous trimer-of-heterodimer complexes, thus blocking gp41 six-helix bundle (6-HB) core formation, a critical step in virus-cell fusion (4, 5, 31, 52, 57).
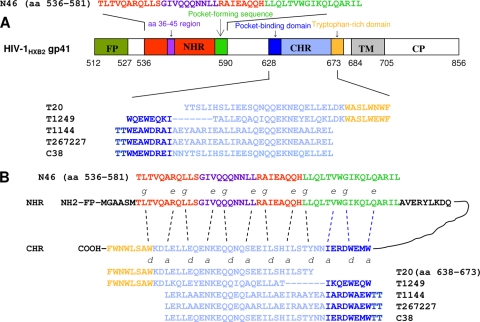
T20 (generic name, enfuvirtide; brand name, Fuzeon), a 36-mer CHR peptide (amino acids [aa] 638 to 673) containing a heptad repeat (HR) sequence-binding domain (HBD) and a tryptophan-rich domain (TRD) (Fig. 1) (30, 61), was licensed by the U.S. FDA as a first-generation HIV fusion inhibitor. T20 is very effective in inhibiting infection by HIV-1, especially the strains resistant to current antiretroviral therapies (24). However, many patients are now failing to respond to T20 because the viruses have developed T20 resistance (34, 51, 56, 62).
FIG. 1.
Functional domains of HIV fusion inhibitors and the interaction model. (A) Schematic view of the HIV-1HXB2 gp41 molecule and sequences of the first-, second-, and third-generation HIV fusion inhibitors. FP, fusion peptide; TM, transmembrane domain; CP, cytoplasmic domain. (B) Interaction between the NHR and CHR peptides. The dashed lines between the NHR and CHR domains indicate the interaction between the residues located at the e and g and a and d positions in the NHR and CHR, respectively. The PBD, HBD, and TRD in the CHR peptides are shown in blue, light blue, and orange, respectively. The HR sequence, the region of aa 36 to 45 (determinant for T20 resistance and the primary binding site for T20), and the pocket-forming sequence in the NHR are shown in red, purple, and green, respectively. The interaction between the PBD and pocket-forming sequence is critical for stabilization of the 6-HB (3).
T1249, a second-generation HIV fusion inhibitor, is a 39-mer peptide consisting of a pocket-binding domain (PBD), an HBD, and a TRD (Fig. 1). T1249 was shown to have a longer half-life than T20 in primates (7) and greater anti-HIV-1 potency than T20 in clinical studies and to be active against some T20-resistant HIV-1 variants (7, 14, 27, 38). However, the clinical development of T1249 was discontinued due to formulation difficulties (37).
T1144, a third-generation HIV fusion inhibitor, is a 38-mer peptide containing a PBD and an HBD (Fig. 1). T1144 was designed by modifying the amino acid sequence of T651 (peptide C38; aa 626 to 673) to increase α-helicity and 6-HB stability and to improve pharmacokinetic properties (10). T1144 and its analog peptides are effective against viruses that are resistant to T20 (11).
Sifuvirtide, a new generation of HIV fusion inhibitor, is a 34-mer peptide analogue of C34 containing a PBD and an HBD. Our previous studies have shown that sifuvirtide is more effective than T20 against both primary and laboratory-adapted HIV-1 strains. Pharmacokinetic studies of sifuvirtide demonstrated longer decay half-lives than T20 (19). Sifuvirtide is under phase II clinical trial (www.fusogen.com). Most recently, we found that the combination of sifuvirtide with T20 resulted in potent synergistic effect against T20-sensitive and -resistant HIV-1 strains (43). These findings encouraged us to test whether combining T20 with T1249 and/or T1144 would also have synergistic anti-HIV-1 activity since next-generation HIV fusion inhibitors, like C34 and sifuvirtide, also contain a PBD that can interact with pocket-forming sequence in the gp41 NHR. In this study, we were also motivated to address the mechanism(s) underlying a synergic effect. Once this effect is confirmed, a novel combination therapy could be designed for the treatment of HIV/AIDS patients who have failed to respond to T20 or other antiretroviral drugs.
MATERIALS AND METHODS
Peptide synthesis.
Peptides T20, T1249, T1144, T267227, C38, and N46 (Fig. 1) were synthesized by a standard solid-phase fluorenylmethoxycarbonyl method using an Applied Biosystems model 433A peptide synthesizer. All peptides were acetylated at the N termini and amidated at the C termini. The peptides were purified to homogeneity (>95% purity) by high-performance liquid chromatography and identified by laser desorption mass spectrometry (PerSeptive Biosystems, Framingham, MA). The peptide concentration was determined, according to Edelhoch's method (12), by measuring UV absorbance of the peptide in 6 M guanidinium-HCl solution and calculating the molar extinction coefficient ɛ (at 280 nm) of 5,500 mol/liter−1·cm−1 and 1,490 mol/liter−1·cm−1 based on the number of tryptophan (Trp) residues and tyrosine (Tyr) residues (all the peptides tested contain Trp and/or Tyr), respectively.
Assessment of inhibition of HIV-1-mediated cell-cell fusion.
A dye transfer assay was used for detection of HIV-1-mediated cell-cell fusion as previously described (21, 33). Briefly, calcein-acetoxy-methyl ester-labeled H9/HIV-1IIIB-infected cells were incubated with MT-2 cells (ratio, 1:5) at 37°C for 2 h in the presence or absence of the test peptide. The fused and unfused calcein-labeled HIV-1-infected cells were counted under an inverted fluorescence microscope (Zeiss, Germany) with an eyepiece micrometer disc. The percent inhibition of cell-cell fusion and the 50% inhibitory concentrations (IC50s) were calculated as described before (23).
Measurement of inhibition of infection by laboratory-adapted HIV-1 strains.
The inhibitory activity of the peptidic HIV fusion inhibitors on infection by laboratory-adapted HIV-1 strains and T20-resistant viruses was determined as previously described (23). In brief, 1 × 104 MT-2 cells were infected with HIV-1 isolates at 100 times the 50% tissue culture infective dose overnight in 200 μl of culture medium in the presence or absence of the test peptide. Then, the culture supernatants were removed, and fresh medium was added. On the fourth day postinfection, 100 μl of the culture supernatants was collected from each well, mixed with equal volumes of 5% Triton X-100, and assayed for p24 antigen by enzyme-linked immunosorbent assay (ELISA) as previously described (23).
Determination of inhibition of infection by primary HIV-1 isolates.
The inhibitory activity of the peptidic HIV-1 fusion inhibitors against a primary HIV-1 isolate was determined as previously described (23). Briefly, the peripheral blood mononuclear cells were isolated from the blood of healthy donors using a standard density gradient (Histopaque-1077; Sigma) centrifugation. After incubation at 37°C for 2 h, the nonadherent cells were collected and resuspended at 5 × 105/ml in RPMI 1640 medium containing 10% fetal bovine serum, 5 μg of phytohemagglutinin/ml, and 100 U of interleukin-2/ml, followed by incubation at 37°C for 3 days. The phytohemagglutinin-stimulated cells were infected with a primary HIV-1 isolate at a multiplicity of infection of 0.01 in the absence or presence of a peptide at graded concentrations. The supernatants were collected at 7 days postinfection and tested for p24 antigen by ELISA.
CD spectroscopic analysis.
Circular dichroism (CD) measurements were performed as previously described (32). Briefly, N46 and each of the CHR peptides were dissolved in phosphate-buffered saline (PBS) solution, pH 7.2. Individual peptides at 8 μM or mixtures of 8 μM of each peptide in PBS were incubated at 37°C for 30 min. The CD spectrum of each sample was acquired on a Jasco spectropolarimeter (Model J-715; Jasco Inc., Japan) at 20°C using a 5-nm bandwidth, 0.5-nm resolution, 0.1-cm path length, and an average time of 5.0 s. Spectra were corrected by the subtraction of a blank corresponding to the solvent composition of each sample. Peptide interactions were determined according to Lawless's protocol by comparing the spectrum of the peptide mixture (experimental spectrum) to the sum of the individual spectra of the peptides at the same concentration and identical experimental condition (calculated noninteracting spectrum) (28).
Detection of inhibition of 6-HB formation by ELISA.
Inhibitory activity of the peptides on the 6-HB core formation between N46 and biotinylated C34 (C34-biotin) was determined by ELISA, as previously described (45), using the conformation-specific monoclonal antibody (MAb) NC-1 (20). Briefly, a testing peptide at graded concentration was preincubated with an equal amount of N46 (0.5 μM) at 37°C for 30 min, followed by the addition of C34-biotin (0.5 μM). The mixture was added to a 96-well polystyrene plate (Costar; Corning Inc., Corning, NY) coated with MAb NC-1 immunoglobulin G (2 μg/ml in 0.1 M Tris, pH 8.8) and blocked with 2% nonfat milk in PBS. The plate was then incubated for 30 min and added to horseradish peroxidase labeled with streptavidin (Zymed Laboratories, S. San Francisco, CA). The plate was washed with the washing buffer (PBS containing 0.01% Tween 20) six times to remove any unbound peptide. The substrate TMB (3,3′,5,5′-tetramethylbenzidine; Sigma) was added sequentially. Absorbance at 450 nm (A450) was measured using an ELISA reader (Ultra 384; Tecan, Research Triangle Park, NC). The percent inhibition of 6-HB formation and the IC50s were calculated (23).
Synergy analysis.
Peptides were tested individually and in combination at a fixed molar ratio, which was optimized to give the greatest synergism over a range of serial dilutions. The inhibition data were analyzed for cooperative effects by using the method of Chou and Talalay (8, 9). The analysis was conducted in a stepwise fashion by calculating IC50 (or 75, 90, and 95% IC values) values based on the dose-response curves of (i) single drugs (T20, T1249, and T1144) that were tested separately, (ii) two drugs tested in combination (T20-T1249 and T20-T1144), and (iii) three drugs tested in combination (T20-T1249-T1144). Then, the combination index (CI) was calculated by using the median effect equation with the CalcuSyn program (kindly provided by T. C. Chou at the Memorial Sloan-Kettering Cancer Center) to assess the synergistic effect of combinations. A CI of <1 indicates synergism (CI values are interpreted as follows: <0.1, very strong synergism; 0.1 to 0.3, strong synergism; 0.3 to 0.7, synergism; 0.7 to 0.85, moderate synergism; and 0.85 to 0.90, slight synergism), a CI of 1 or close to 1 indicates additive effects, and a CI of >1 indicates antagonism (9). Dose reduction was calculated by dividing the IC50 value of a peptide when it was tested alone by that of the same peptide tested in combination with other peptide(s).
RESULTS
Combining T20 with T1249 and/or T1144 produced exceptionally potent synergism against HIV-1-mediated cell-cell fusion.
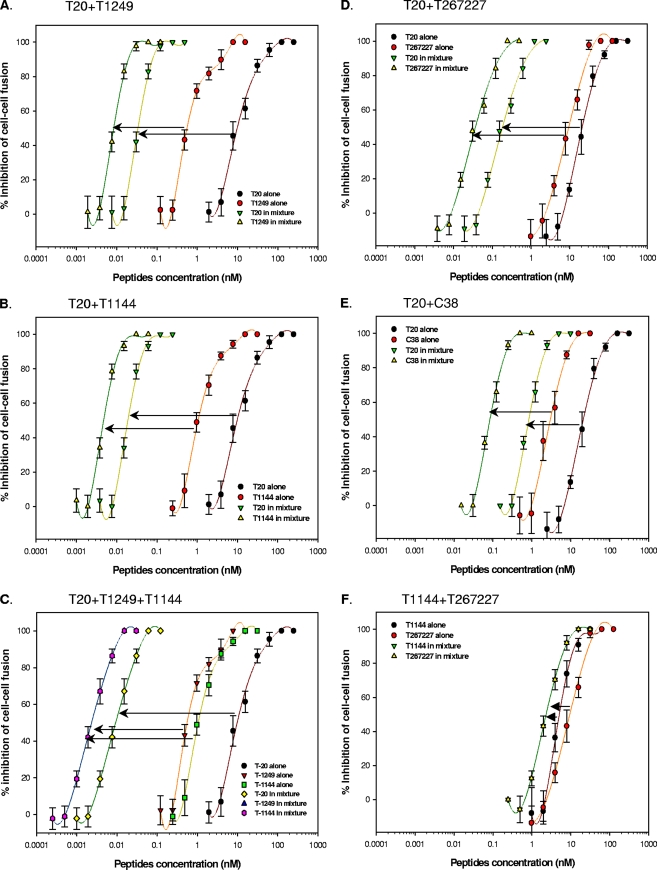
We first investigated the potential cooperative effects of various combinations of T20 with T1249 and/or T1144 on HIV-1-mediated cell-cell fusion. Very surprisingly, the combinations of T20 and T1249 (Fig. 2A) or of T20 and T1144 (Fig. 2B) resulted in exceptionally potent synergism, with a CI of <0.01, as well as reduction of the IC50 from nanomolar to picomolar levels. A triple combination (T20-T1249-T1144) also exhibited very strong synergism (CI of 0.008) (Fig. 2C), with a dose reduction (IC50 of a peptide when tested alone/IC50 of the peptide in combination) of 2 to 3 orders of magnitude. To elucidate the possible causation of synergism, we then synthesized two T1144 analogous peptides, T267227 and C38 (11), which are expected to have the same primary binding sites and mechanisms of action as T1144, and tested their inhibitory activity on HIV-1-mediated cell-cell fusion in combination with T1144 and T20, respectively. Similar to the T20-T1144 combination, the combinations of T20-T267227 and T20-C38 also exhibited potent synergism on inhibition of HIV-1-mediated cell-cell fusion (Fig. 2D and E). However, no significant synergism was observed when the T1144-T267227 combination was tested (Fig. 2F). These results suggest that the combination of T20 and T1144 demonstrates synergism because the peptides have different primary binding sites in the gp41 NHR region.
FIG. 2.
Synergistic effect of combinations of T20 with T1249 and/or T1144 and T1144 analogues T267227 and C38 on inhibition of HIV-1-mediated cell-cell fusion as determined by a dye transfer assay (21, 33). Ratios are as follows: T20-T1249, 4:1 (A); T20-T1144, 4:1 (B); T20-T1249-T1144, 4:1:1 (C); T20-T267227, 5:1 (D); T20-C38, 10:1 (E); and T1144-T2672227, 1:1 (F). Each sample was tested in quadruplicate, the experiment was repeated twice, and a representative set of data is shown.
Combining T20 with T1249 and/or T1144 leads to a potent synergistic effect against infection by laboratory-adapted and primary HIV-1 strains.
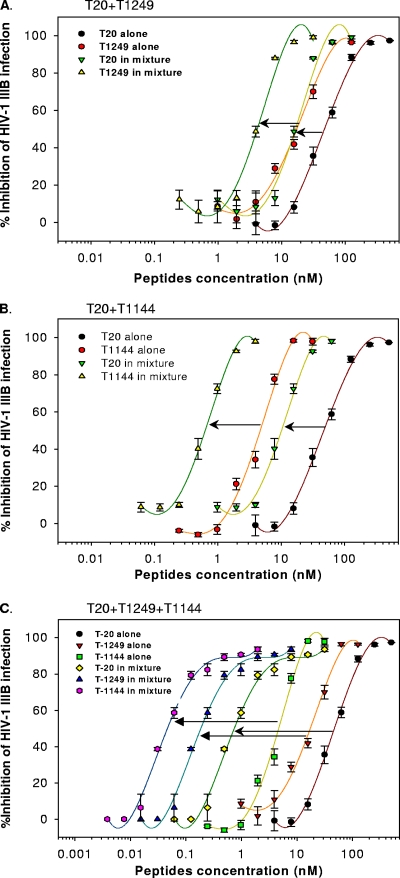
Next, we determined the potential synergistic effect against infection by two laboratory-adapted HIV-1 strains, IIIB (subtype B, X4) and BaL (subtype B, R5), and two primary HIV-1 isolates, 93IN101 (subtype C, R5) and RU570 (clade G, R5). Synergism was observed for all virus strains tested. The combination of T20 with T1249 or with T1144 resulted in a dose reduction of about 3- to 12-fold or 5- to 20-fold, respectively, to inhibit infection by laboratory-adapted HIV-1 strains. Strikingly, a triple combination (T20-T1249-T1144) caused the greatest synergism, with a 71- to 281-fold dose reduction to inhibit laboratory-adapted HIV-1 infection (Table 1 and Fig. 3). Potent synergism was also observed against infection by the primary HIV-1 isolates 93IN101 and RU570 with double and triple combinations of T20 with T1249 and/or T1144 (Table 1). Although combinations of T20 with T1249 and with T1144 exhibited strong synergism against infection by both laboratory-adapted and primary HIV-1 strains, these data confirm that triple combination leads to even stronger synergism.
TABLE 1.
CI and dose reduction in inhibition of infection by laboratory-adapted and primary HIV-1 strains by combining T20 with T1249 and/or T1144a
| Peptide combination and virus (subtype, tropism)b | CI | T20
|
T1249
|
T1144
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (nM)
|
Dose reduction (n-fold) | IC50 (nM)
|
Dose reduction (n-fold) | IC50 (nM)
|
Dose reduction (n-fold) | |||||
| Alone | In mixture | Alone | In mixture | Alone | In mixture | |||||
| T20 and T1249 | ||||||||||
| IIIB (B, X4) | 0.44 | 50.44 | 15.21 | 3.32 | 19.20 | 3.80 | 5.05 | |||
| Bal (B, R5) | 0.13 | 8.42 | 0.73 | 11.53 | 3.91 | 0.36 | 10.86 | |||
| 93IN101 (C, R5) | 0.16 | 39.89 | 3.66 | 10.90 | 13.58 | 0.91 | 14.92 | |||
| RU570 (G, R5) | 0.23 | 38.44 | 5.98 | 6.43 | 19.15 | 1.50 | 12.77 | |||
| T20 and T1144 | ||||||||||
| IIIB (B, X4) | 0.31 | 50.44 | 9.88 | 5.11 | 4.95 | 0.62 | 7.98 | |||
| Bal (B, R5) | 0.06 | 8.42 | 0.42 | 20.05 | 4.18 | 0.21 | 19.90 | |||
| 93IN101 (C, R5) | 0.18 | 39.89 | 4.29 | 9.30 | 15.47 | 1.07 | 14.46 | |||
| RU570 (G, R5) | 0.19 | 38.44 | 5.19 | 7.41 | 24.20 | 1.30 | 18.62 | |||
| T20, T1249, and T1144 | ||||||||||
| IIIB (B, X4) | 0.06 | 50.44 | 0.71 | 71.04 | 19.20 | 0.18 | 106.67 | 4.95 | 0.05 | 99.00 |
| Bal (B, R5) | 0.01 | 8.42 | 0.03 | 280.67 | 3.91 | 0.02 | 195.50 | 4.18 | 0.02 | 209.00 |
| 93IN101 (C, R5) | 0.15 | 39.89 | 2.54 | 15.70 | 13.58 | 0.64 | 21.22 | 15.47 | 0.64 | 24.17 |
| RU570 (G, R5) | 0.22 | 38.44 | 4.54 | 8.47 | 19.15 | 1.14 | 16.80 | 24.20 | 1.14 | 21.23 |
Data are representative of two separate experiments. Each sample was tested in triplicate, and the mean values are presented. Ratios of the peptides T20/T1249/T1144 in combinations are 16:4:1 for IIIB, 2:1:1 for Bal, and 4:1:1 for 93IN101 and RU650.
93IN101 and RU570 are primary HIV-1 isolates.
FIG. 3.
Synergistic effect of combinations of T20 with T1249 and/or T1144 on inhibition of HIV-1 IIIB infection. Ratios are as follows: T20-T1249, 4:1 (A); T20-T1144, 16:1 (B); and T20-T1249-T1144, 16:4:1 (C). Each sample was tested in triplicate, the experiment was repeated twice, and a representative set of data is shown.
Combining T20 with T1249 and/or T1144 exhibited a strong synergistic effect against infection by T20- and T1249-resistant HIV-1 strains.
The rapid emergence of T20-resistant viruses in T20-treated patients is one of the major causes for the failure of T20 therapy (34, 51). Here, we investigated whether combining T20 with T1249 and/or T1144 had a synergistic effect against T20- and T1249-resistant HIV-1 strains. We compared the antiviral activity of these peptides separately or in combination against one T20-sensitive strain, NL4-3D36G, and three T20-resistant strains, NL4-3(36G)V38A, NL4-3(36G)V38A/N42D, and NL4-3(36G)V38E/N42S, which contain a single or double mutation in the principal determinant of T-20 resistance (aa 36 to 45: GIVQQQNNLL) in the gp41 NHR domain (13, 16, 26, 34, 36, 39, 46, 47, 50, 51, 56), including V38A, V38A/N42D, and V38E/N42S. As shown in Table 2, when tested separately, T20, T1249, and T1144 were effective against the T20-sensitive strain NL4-3D36G, with an IC50 ranging from 6 to 49 nM. However, T20 could inhibit infection by these three T20-resistant variants only at a high concentration (IC50s of 313, 2,646, and 9,894 nM to inhibit infection by NL4-3(36G)V38A, NL4-3(36G)V38A/N42D, and NL4-3(36G)V38E/N42S, respectively), while T1144 was highly effective against all three T20-resistant viruses, with an IC50 of about 4 to 6 nM. Interestingly, both NL4-3(36G)V38A and NL4-3(36G)V38A/N42D were sensitive to T1249 (IC50 of 4 to 10 nM), but NL4-3(36G)V38E/N42S was resistant to T1249 (IC50 of 358 nM). This is consistent with the report by Eggink et al. (13) who have shown that some T20-resistant variants with a V38E mutation are also resistant to T1249. However, the combination of T20 and T1249 or of T20 and T1144 resulted in significant synergistic activity against T20- and T1249-resistant strains, with a 2- to 26-fold dose reduction. Consistent with the results of testing laboratory-adapted HIV-1 strains, the synergism observed in a triple combination of these peptides was stronger against these T20- and T1249-resistant strains, with a 9- to 68-fold dose reduction. These results suggest that combining T20 with T1249 and/or T1144 results in highly potent synergistic activity against both T20- and T1249-resistant HIV-1 strains, suggesting a new therapeutic strategy for the treatment of patients who have failed to respond to T20 monotherapy.
TABLE 2.
Synergistic effect of combinations of T20 with T1249 and/or T1144 on inhibition of infection by T20-sensitive and -resistant HIV-1 strainsa
| Peptide combination and NL4-3 mutant | CI | T20
|
T1249
|
T1144
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 (nM)
|
Dose reduction (n-fold) | IC50 (nM)
|
Dose reduction (n-fold) | IC50 (nM)
|
Dose reduction (n-fold) | |||||
| Alone | In mixture | Alone | In mixture | Alone | In mixture | |||||
| T20 and T1249 | ||||||||||
| D36G | 0.46 | 48.76 | 7.52 | 6.48 | 17.45 | 5.37 | 3.25 | |||
| (36G)V38A | 0.06 | 313.04 | 13.59 | 23.03 | 4.35 | 0.17 | 25.59 | |||
| (36G)V38A/N42D | 0.38 | 2645.98 | 228.83 | 11.56 | 10.36 | 2.86 | 3.62 | |||
| (36G)V38E/N42S | 0.21 | 9894.46 | 732.6 | 13.51 | 358.01 | 48.84 | 7.33 | |||
| T20 and T1144 | ||||||||||
| D36G | 0.26 | 48.76 | 3.14 | 15.53 | 5.85 | 0.45 | 13.00 | |||
| (36G)V38A | 0.09 | 313.04 | 18.65 | 16.78 | 3.76 | 0.23 | 16.35 | |||
| (36G)V38A/N42D | 0.53 | 2645.98 | 200.89 | 13.17 | 4.57 | 2.51 | 1.82 | |||
| (36G)V38E/N42S | 0.62 | 9894.46 | 3749.09 | 2.64 | 5.21 | 1.25 | 4.17 | |||
| T20, T1249, and T1144 | ||||||||||
| D36G | 0.22 | 48.76 | 2.62 | 18.61 | 17.45 | 1.87 | 9.33 | 5.85 | 0.37 | 15.81 |
| (36G)V38A | 0.07 | 313.04 | 8.29 | 37.76 | 4.35 | 0.10 | 43.50 | 3.76 | 0.10 | 37.60 |
| (36G)V38A/N42D | 0.17 | 2645.98 | 38.97 | 67.90 | 10.36 | 0.49 | 21.14 | 4.57 | 0.49 | 9.33 |
| (36G)V38E/N42S | 0.14 | 9894.46 | 410.16 | 24.12 | 358.01 | 27.34 | 13.09 | 5.21 | 0.14 | 37.21 |
NL4-3D36G is a T20-sensitive strain, which is the parent strain used for generation of T20-resistant mutants, including NL4-3(36G)V38A, NL4-3(36G)V38A/N42D, and NL4-3(36G)V38E/N42S. NL4-3(36G)V38E/N42S is also resistant to T1249 (13). Ratios of the peptides T20/T1249/T1144 in combination were 7:5:1 for NL4-3D36G, 80:1:1 for NL4-3(36G)V38A), 80:1:1 for NL4-3(36G)V38A/N42D, and 3,000:200:1 for NL4-3(36G)V38E/N42S. Data are representative of two separate experiments. Each sample was tested in triplicate, and the mean values are presented.
CD spectra showed distinct interactions among N46, T1144, T20, and T1149.
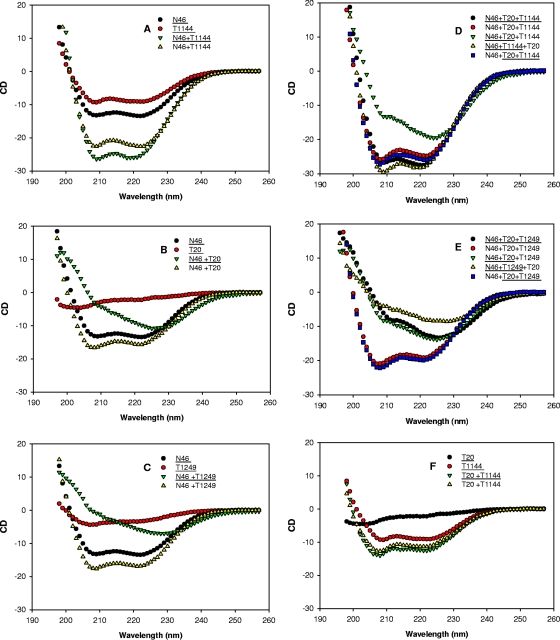
To delineate the putative mechanism of synergism resulting from the combinations of HIV fusion inhibitors, we used CD spectroscopy to study the gp41 NHR and CHR interactions involved in secondary structure changes. We first recorded CD spectra of single peptides and their mixtures under identical conditions. The spectra of the mixtures (experimental spectra) and the sum of the spectra of single peptides in the mixtures (calculated noninteracting spectra) were compared to determine the interactions. If no structural change occurs because of noninteraction in the mixture, identical experimental and calculated noninteracting spectra are expected (28). As shown in Fig. 4, the mixtures of N46 with T1144, T1249, and T20, all displayed large secondary structure changes, indicating the interaction between N46 and each of these CHR peptides. N46 and T1144, when mixed, formed a typical α-helical complex with increased α-helical content (Fig. 4A), which is consistent with CD spectral changes reported for N46 interactions with other CHR peptides containing the PBD, e.g., C34 and C36 (30, 31, 45). T20 and T1249 were unstructured in solution, with <20% helical content. When mixed with N46, instead of forming an α-helical complex with increased helical content, their interactions significantly disrupted α-helical conformation of N46 and resulted in a spectrum with a minimum at 228 nm (Fig. 4B and C). This is consistent with the T20-NHR interaction reported by Wild et al. (60) and Lawless et al. (28). This suggests that T1249 and T20 have a different interaction model with the NHR from that of T1144 with the NHR. Subsequently, we recorded CD spectra of mixtures of N46 or PBS with two CHR peptides, T20-T1144 or T20-T1249, respectively. The noninteracting spectra were calculated in three ways to dissect the interaction: (i) the sum of the spectra of three peptides (N46 and two CHR peptides) measured separately, (ii) the sum of the spectra of two CHR peptides measured separately, and (iii) the sum of the spectrum of one peptide measured separately and the spectra of two peptides measured in combination. As shown in Fig. 4D and E, the experimental spectra of the triple mixtures did not overlap with the calculated noninteracting spectra of the corresponding mixtures, indicating that interactions occur between N46 and the double combinations of the CHR peptides. Interestingly, the calculated noninteracting spectra of N46-T1144-T20 and N46-T20-T1249 are similar to those of the experimental spectra of N46-T1144-T20 and N46-T20-T1249, respectively. These data suggest that the N46 and T1144 interaction predominated in the N46-T1144-T20 mixture, while the N46 and T20 interaction was predominant in N46-T20-T1249 mixture. The CD signal changes from the T20 and T1144 interaction is not significant (Fig. 4F). This further distinguishes the role of that the different peptide fusion inhibitors played in combination.
FIG. 4.
Analysis of the interaction between N46 and the CHR peptide(s) by CD spectroscopy. All peptides and their complexes were measured at 8 μM in PBS. For peptide(s) underlined or not underlined data are presented as the experimental spectra (underlined) and calculated theoretical noninteracting spectra (not underlined), as previously described (28, 40). Combinations are indicated on the panels.
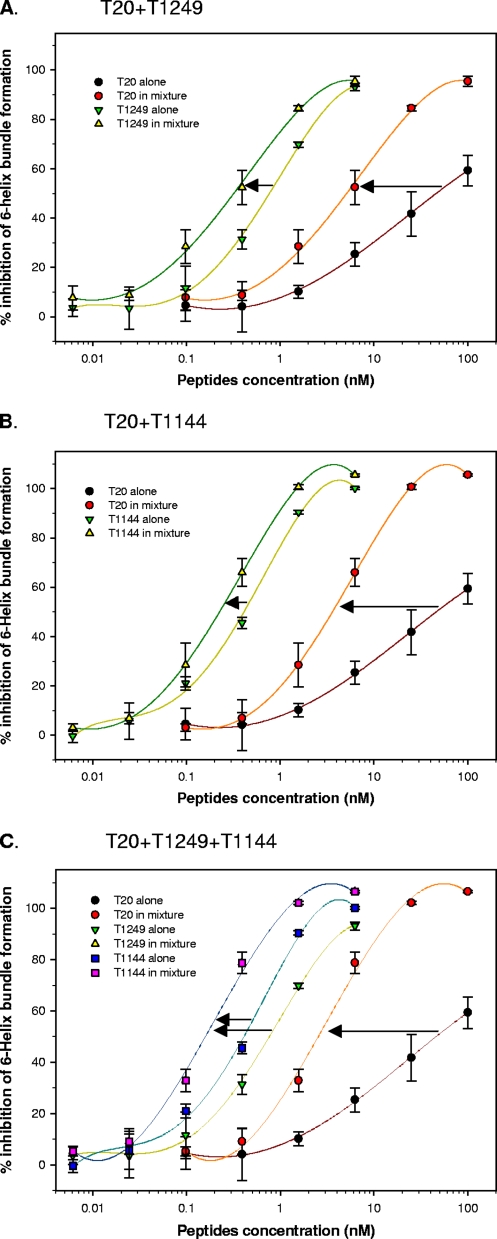
Combining T20 with T1249 and/or T1144 resulted in synergistic effect on 6-HB core formation.
Subsequently, we determined the potential synergism resulting from the combination of T20 with T1249 and/or T1144 against 6-HB formation between N46 and C34-biotin. Consistent with our previous observation (29), T20 alone could only weakly inhibit 6-HB formation, with an IC50 of 59 μM, while T1249 and T1144 alone significantly blocked 6-HB formation in a dose-dependent manner, with IC50s of 0.8 and 0.3 μM, respectively. Combining T20 and T1249 (Fig. 5A) or T20 and T1144 (Fig. 5B) resulted in a synergistic effect on inhibition of 6-HB formation, with CIs of 0.4 and 0.5, respectively. A triple combination also showed synergism, with a dose reduction for T20, T1249, and T1144 of about 26-, 4-, and 2-fold, respectively (Fig. 5C). These results suggest that the increased potency of these CHR peptides in combination against HIV-1 Env-mediated membrane fusion derives from their synergistic effect on inhibition of the gp41 6-HB core formation.
FIG. 5.
Synergistic effect on inhibition of 6-HB formation resulting from combinations of T20 with T1249 and/or T1144 as measured by ELISA (45). Ratios are as follows: T20-T1249, 16:1 (A); T20-T1144, 16:1 (B); T20-T1249-T1144, 16:1:1 (C). Each sample was tested in triplicate. The results shown are a representative set of data from two independent experiments.
DISCUSSION
In general, if a first-generation antiviral drug becomes ineffective against resistant viruses, it is replaced by a next-generation drug with improved efficacy and drug-resistant profile. In the case of T20, however, our study shows that it is preferable to use a next-generation HIV fusion inhibitors, T1249 or T1144, in combination with T20 rather than to replace it by either of the next-generation drugs studied. Specifically, our results showed that the combination of T20 with T1249 or T1144 leads to exceptionally potent synergism, with a dose reduction of 2 to 3 orders of magnitude in the inhibition of HIV-1-induced cell-cell fusion (Fig. 2), particularly since all of these peptides when used separately are already highly potent (at nM level). Similarly, strong synergism was also observed in the combination of T20 with T1249 or T1144 against infection by both laboratory-adapted strains and primary HIV-1 isolates. A triple combination exhibited even greater synergism (Table 1). Most importantly, combining T20 with T1249 and/or T1144 also exhibited strong synergism against T20- and T1249-resistant viruses, with a dose reduction as high as 68-fold (Table 2). These findings suggest a new strategy for treatment of patients who have failed to respond to first-generation drugs.
It has been reported that the combination of T20 with PRO 542 (a CD4-based HIV-1 entry inhibitor targeting gp120) or AMD3100 (a CXCR4 antagonist) or SCH-C (a CCR5 antagonist) results in strong synergistic anti-HIV-1 activity (42, 53, 54). Since it is well known that the combination of two drugs with different mechanisms of action or target sites may lead to synergism (9), it is understandable that combining T20 with other HIV entry inhibitors targeting gp120 or coreceptors would have synergistic effects. However, it seems difficult to explain why the combinations of different generations of HIV fusion inhibitors, all targeting gp41, also exhibit synergism. We attribute the mechanism of synergism resulting from T20, T1249, and T1144 combinations to the fact that these peptidic HIV fusion inhibitors have different primary binding sites in the gp41 NHR.
Both in vitro and in vivo studies (13, 16, 26, 34, 36, 39, 46, 47, 50, 51, 56) have shown that T20 resistance is associated with single or double mutations in the region of aa 36 to 45 in the gp41 NHR domain (e.g., G36D, I37V, V38A, V38E, V38M, N42D, N42S, and N43D) (Table 2), assuming that these mutations impact the binding of T20 and, hence, its potency and suggesting that this region is the primary binding site for T20. Using a turbidity clearance assay and CD analysis, Trivedi and colleagues have shown that the LLSGIV (aa 33 to 38) motif in the gp41 NHR is critical for the binding of T20 to NHR peptides (55). Besides the HR-binding sequence that can interact with the region of aa 36 to 45, T1144 also contains the pocket-binding sequence (Fig. 1). Through the pocket-binding sequence, T1144 is able to bind to the NHR hydrophobic pocket, which plays a critical role in stabilization of the gp41 6-HB core (3), to form a highly stable 6-HB with the viral gp41 NHR domain. Therefore, the pocket-forming sequence in NHR is regarded as the primary binding site for T1144. This may explain why T1144 and other CHR peptides with pocket-binding sequences, e.g., C34, C37, and C38, bind to the gp41 NHR domain much more strongly than T20 and are more effective than T20 in blocking gp41 6-HB formation (2, 6, 25, 29, 30). Since the region of aa 36 to 45 is not the primary binding site for T1144, the T20-resistant viruses with mutations in this region are sensitive to T1144 (Table 2). We have recently demonstrated that a peptide containing the pocket-binding sequence, but lacking the binding sequence in the region of aa 36 to 45, is exceptionally potent against T20-resistant variants (18), further confirming that the pocket-binding sequence is critical for the CHR peptides against T20-resistant viruses. T1249 contains both the pocket- and HR-binding sequences as well as the TRD (Fig. 1). Therefore, it is expected to function either like T1144 by binding to the pocket region and HR sequence in the NHR domain (Fig. 6, model I) or like T20 by interacting with the HR sequence in the gp41 NHR domain and lipid membrane (Fig. 6, model II). However, our results from CD analysis suggest that T1249 functions more like T20 than T1144 (Fig. 4C), perhaps because the modified pocket-binding sequence in T1249 may not function as well as the unmodified pocket-binding sequence in T1144. This may explain why some T20-resistant viruses are also resistant to T1249 (Table 2) (13).
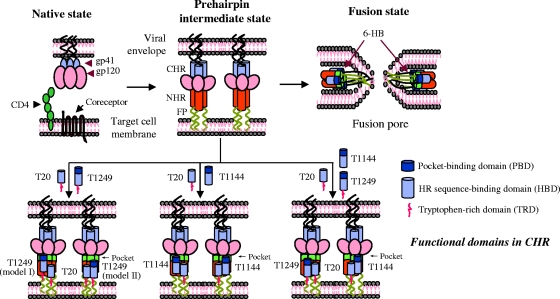
FIG. 6.
Putative models of action of different generations of HIV fusion inhibitors, T20, T1249, and T1144, which contain distinct functional domains. T1144 binds to the HR sequence and pocket-forming sequence in the NHR domain via its PBD and HBD, respectively, to form a stable heterologous 6-HB. T20 interacts with HR sequence in the NHR and lipid membranes through its HBD and TRD, respectively (1, 29, 30, 49, 58). T1249 may function either like T1144 through the PBD and HBD (model I) or like T20 via the HBD and TRD (model II). Binding of T20 to one of the three grooves on the NHR trimer may prolong the exposure of the fusion intermediate, which may promote T1249 or T1144 to interact with the unoccupied grooves, resulting in a synergistic effect against gp41-mediated membrane fusion.
Using the CD spectroscopy to analyze the secondary structure change of the complexes formed between the NHR peptide N46 and the individual CHR peptides as well as their combinations, we found that all the three CHR peptides, T20, T1249, and T1144, could interact with N46 in solution, but different outcomes occurred. Addition of T1144 to N46 resulted in formation of typical α-helical complex with increased α-helical content, while the interaction of T20 or T1249 with N46 led to the disruption of the α-helical conformation of N46 (Fig. 4), which is consistent with the CD spectrum change when an NHR peptide is mixed with a CHR peptide with or without the PBD (e.g., T20 and C34) (28, 30, 31, 45, 60). This result suggests that the model of the interaction of T1144 with the gp41 NHR differs from that of T1249 or T20 with the NHR. In the mixture of N46 with the T20-T1144 combination, the N46-T1144 interaction predominated over the N46-T20 interaction while in the mixture of N46 with the T20-T1249 combination, the N46-T20 interaction is predominant (Fig. 4). This indicates that different generations of HIV fusion inhibitors may play different roles in combination. However, the synergistic mechanism resulting from the complicated interactions between the multiple peptides and the corresponding regions in viral gp41 in the presence of the virus and the target cell may not be readily interpreted by using biophysical analysis. Particularly, since the binding of T20 to the HR sequence in the NHR domain is not strong enough to compete with the interaction between the viral gp41 CHR and NHR regions, T20 may have to use its C-terminal TRD to interact with the target cell membrane in order to stabilize its interaction with the viral gp41 NHR region (Fig. 6) (25, 29, 30, 44).
HIV-1 Env-mediated membrane fusion is a kinetics-limited process (41). Suboptimal temperature (31.5°C) and other influencing factors that slow the fusion kinetics and prolong exposure of the gp41 fusion intermediate could increase the sensitivity of the virus to the HIV fusion inhibitors targeting the gp41 NHR domain and make some nonneutralizing MAbs become neutralizing (15). Other investigators have shown that introduction of T20 resistance-associated mutations into the gp41 NHR region results in prolonged fusion processes and increased sensitivity of the virus to the neutralizing antibodies targeting gp41 (e.g., 2F5 and 4E10) (47, 48). Gustchina and coworkers (17) have demonstrated that combining the NHR peptide N36Mut(e,g) with a gp41 NHR-specific neutralizing MAb, Fab 3674, results in synergism, rescuing neutralizing activity of this MAb against resistant virus strains. This is because binding of N36Mut(e,g) to the viral gp41 NHR results in prolongation of the temporal window during which the virus is susceptible to neutralization by the MAbs. By a similar logic, binding of one peptide fusion inhibitor (e.g., T20) to the gp41 NHR may prolong the half-life of the fusion intermediate so that other fusion-inhibitory peptides (e.g., T1144 and T1249) in the mixture can bind more efficiently to the NHR domain. At low concentration, T1144 may bind to one of three grooves on the NHR trimer, which allows T20 to interact with other unoccupied grooves on the NHR trimer. The prolonged exposure of the fusion intermediate resulting from the mutations in the region of aa 36 to 45 (47, 48) may therefore benefit T1144 binding to the viral gp41 NHR trimer, which would, in turn, promote the interaction of T20 with the gp41 NHR domain (Fig. 6).
In summary, highly potent synergistic activity against both laboratory-adapted and primary HIV-1 strains, including those resistant to T20, is achieved by combining T20 with T1249 and/or T1144 because these peptidic HIV fusion inhibitors contain different functional domains and have distinct primary binding sites in the gp41 NHR domain. Binding of one fusion inhibitor to the viral gp41 NHR domain may extend the temporal window of the fusion intermediate, which thus becomes more accessible to other fusion inhibitor(s) targeting the NHR domain, resulting in synergistic anti-HIV-1 activity. Therefore, the synergism and the resulting dose reduction of the anti-HIV drugs in combination may provide maximum efficacy as well as low-dose and low-cost options, thus overcoming the three major weaknesses of T20 monotherapy: (i) ineffectiveness against T20-resistant viruses, (ii) requirement of a high-dose injection intramuscularly that causes serious injection site reaction, and (iii) high cost to patients. This combination therapy strategy, if proven successful in clinical trials, could also be applied to other drugs of different generations that have distinct primary binding sites.
Acknowledgments
We thank T. C. Chou at the Memorial Sloan-Kettering Cancer Center for providing the CalcuSyn program and consultancy on data analysis. We are grateful to the NIH AIDS Research and Reference Reagent Program for providing laboratory-adapted and primary HIV-1 isolates as well as T20-resistant HIV-1 strains and HIV-infected cells. We thank Veronica Kuhlemann for editorial assistance.
This work was supported by an NIH grant (AI46221) to S.J. and a scholarship from the China Scholarship Council to C.P.
There were no conflicts of interest.
S.J. designed the research; C.P., L.C., H.L., and Z.Q. performed the research; and C.P., L.C., and S.J. analyzed data and wrote the paper.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Bellamy-McIntyre, A. K., C. S. Lay, S. Baar, A. L. Maerz, G. H. Talbo, H. E. Drummer, and P. Poumbourios. 2007. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J. Biol. Chem. 28223104-23116. [DOI] [PubMed] [Google Scholar]
- 2.Champagne, K., A. Shishido, and M. J. Root. 2009. Interactions of HIV-1 inhibitory peptide T20 with the gp41 N-HR coiled coil. J. Biol. Chem. 2843619-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 9515613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89263-273. [DOI] [PubMed] [Google Scholar]
- 5.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93681-684. [DOI] [PubMed] [Google Scholar]
- 6.Chang, D. K., and C. S. Hsu. 2007. Biophysical evidence of two docking sites of the carboxyl heptad repeat region within the amino heptad repeat region of gp41 of human immunodeficiency virus type 1. Antivir. Res. 7451-58. [DOI] [PubMed] [Google Scholar]
- 7.Chinnadurai, R., D. Rajan, J. Munch, and F. Kirchhoff. 2007. Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J. Virol. 816563-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou, T. C., and P. Talalay. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 2227-55. [DOI] [PubMed] [Google Scholar]
- 9.Chou, T. C. 2006. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58621-681. [DOI] [PubMed] [Google Scholar]
- 10.Delmedico, M., B. Bray, N. Cammack, D. Davison, J. Dwyer, L. Frick, N. Tvermoes, S. Wring, H. Zhang, and M. Greenberg. 2006. Next generation HIV peptide fusion inhibitor candidates achieve potent, durable suppression of virus replication in vitro and improved pharmacokinetic properties, abstr. 48. Abstr. 13th Conf. Retrovir. Oppor. Infect., 5 to 8 February 2006. Foundation for Retrovirology and Human Health, Alexandria, VA.
- 11.Dwyer, J. J., K. L. Wilson, D. K. Davison, S. A. Freel, J. E. Seedorff, S. A. Wring, N. A. Tvermoes, T. J. Matthews, M. L. Greenberg, and M. K. Delmedico. 2007. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc. Natl. Acad. Sci. USA 10412772-12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelhoch, H. 1967. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 61948-1954. [DOI] [PubMed] [Google Scholar]
- 13.Eggink, D., C. E. Baldwin, Y. Deng, J. P. M. Langedijk, M. Lu, R. W. Sanders, and B. Berkhout. 2008. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J. Virol. 826678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eron, J. J., R. M. Gulick, J. A. Bartlett, T. Merigan, R. Arduino, J. M. Kilby, B. Yangco, A. Diers, C. Drobnes, R. DeMasi, M. Greenberg, T. Melby, C. Raskino, P. Rusnak, Y. Zhang, R. Spence, and G. D. Miralles. 2004. Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J. Infect. Dis. 1891075-1083. [DOI] [PubMed] [Google Scholar]
- 15.Golding, H., M. Zaitseva, E. de Rosny, L. R. King, J. Manischewitz, I. Sidorov, M. K. Gorny, S. Zolla-Pazner, D. S. Dimitrov, and C. D. Weiss. 2002. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J. Virol. 766780-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg, M. L., and N. Cammack. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54333-340. [DOI] [PubMed] [Google Scholar]
- 17.Gustchina, E., C. A. Bewley, and G. M. Clore. 2008. Sequestering of the prehairpin intermediate of gp41 by peptide N36Mut(e,g) potentiates the human immunodeficiency virus type 1 neutralizing activity of monoclonal antibodies directed against the N-terminal helical repeat of gp41. J. Virol. 8210032-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, Y., J. Cheng, H. Lu, J. Li, J. Hu, Z. Qi, Z. Liu, S. Jiang, and Q. Dai. 2008. Potent HIV fusion inhibitors against enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. USA 10516332-16337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, Y., Y. Xiao, H. Song, Q. Liang, D. Ju, X. Chen, H. Lu, W. Jing, S. Jiang, and L. Zhang. 2008. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 28311126-11134. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the HIV-1 envelope glycoprotein. J. Virol. 7210213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365113. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. Inhibition of HIV-1 infection by a fusion domain binding peptide from HIV-1 envelope glycoprotein gp41. Biochem. Biophys. Res. Commun. 195533-538. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, S., H. Lu, S. Liu, Q. Zhao, Y. He, and A. K. Debnath. 2004. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. Antimicrob. Agents Chemother. 484349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 3482228-2238. [DOI] [PubMed] [Google Scholar]
- 25.Kliger, Y., S. A. Gallo, S. G. Peisajovich, I. Munoz-Barroso, S. Avkin, R. Blumenthal, and Y. Shai. 2001. Mode of action of an antiviral peptide from HIV-1. Inhibition at a post-lipid mixing stage. J. Biol. Chem. 2761391-1397. [DOI] [PubMed] [Google Scholar]
- 26.Labrosse, B., L. Morand-Joubert, A. Goubard, S. Rochas, J. L. Labernardiere, J. Pacanowski, J. L. Meynard, A. J. Hance, F. Clavel, and F. Mammano. 2006. Role of the envelope genetic context in the development of enfuvirtide resistance in human immunodeficiency virus type 1-infected patients. J. Virol. 808807-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalezari, J. P., N. C. Bellos, K. Sathasivam, G. J. Richmond, C. J. Cohen, R. A. Myers, Jr., D. H. Henry, C. Raskino, T. Melby, H. Murchison, Y. Zhang, R. Spence, M. L. Greenberg, R. A. Demasi, and G. D. Miralles. 2005. T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J. Infect. Dis. 1911155-1163. [DOI] [PubMed] [Google Scholar]
- 28.Lawless, M. K., S. Barney, K. I. Guthrie, T. B. Bucy, Jr., S. R. Petteway, and G. Merutka. 1996. HIV-1 membrane fusion mechanism: structural studies of the interactions between biologically active peptides from gp41. Biochemistry 3513697-13708. [DOI] [PubMed] [Google Scholar]
- 29.Liu, S., W. Jing, B. Cheung, H. Lu, J. Sun, X. Yan, J. Niu, J. Farmar, S. Wu, and S. Jiang. 2007. HIV gp41 C-terminal heptad repeat contains multifunctional domains: relation to mechanisms of action of anti-HIV peptides. J. Biol. Chem. 2829612-9620. [DOI] [PubMed] [Google Scholar]
- 30.Liu, S., H. Lu, Y. Xu, S. Wu, and S. Jiang. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 28011259-11273. [DOI] [PubMed] [Google Scholar]
- 31.Liu, S., S. Wu, and S. Jiang. 2007. HIV entry inhibitors targeting gp41: from polypeptides to small-molecule compounds. Curr. Pharm. Design. 13143-162. [DOI] [PubMed] [Google Scholar]
- 32.Liu, S., Q. Zhao, and S. Jiang. 2003. Determination of the HIV-1 gp41 postfusion conformation modeled by synthetic peptides: applicable for identification of the HIV-1 fusion inhibitors. Peptide 241303-1313. [DOI] [PubMed] [Google Scholar]
- 33.Lu, H., Q. Zhao, Z. Xu, and S. Jiang. 2003. Automatic quantitation of HIV-1 mediated cell-to-cell fusion with a digital image analysis system (DIAS): application for rapid screening of HIV-1 fusion inhibitors. J. Virol. Methods 107155-161. [DOI] [PubMed] [Google Scholar]
- 34.Lu, J., S. G. Deeks, R. Hoh, G. Beatty, B. A. Kuritzkes, J. N. Martin, and D. R. Kuritzkes. 2006. Rapid emergence of enfuvirtide resistance in HIV-1-infected patients: results of a clonal analysis. J. Acquir. Immune Defic. Syndr. 4360-64. [DOI] [PubMed] [Google Scholar]
- 35.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 21075-1082. [DOI] [PubMed] [Google Scholar]
- 36.Matthews, T., M. Salgo, M. Greenberg, J. Chung, R. DeMasi, and D. Bolognesi. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3215-225. [DOI] [PubMed] [Google Scholar]
- 37.Melby, T., R. DeMasi, N. Cammack, G. D. Miralles, and M. L. Greenberg. 2007. Evolution of genotypic and phenotypic resistance during chronic treatment with the fusion inhibitor T-1249. AIDS Res. Hum. Retrovir. 231366-1373. [DOI] [PubMed] [Google Scholar]
- 38.Menzo, S., A. Castagna, A. Monachetti, H. Hasson, A. Danise, E. Carini, P. Bagnarelli, A. Lazzarin, and M. Clementi. 2004. Genotype and phenotype patterns of human immunodeficiency virus type 1 resistance to enfuvirtide during long-term treatment. Antimicrob. Agents Chemother. 483253-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mink, M., S. M. Mosier, S. Janumpalli, D. Davison, L. Jin, T. Melby, P. Sista, J. Erickson, D. Lambert, S. A. Stanfield-Oakley, M. Salgo, N. Cammack, T. Matthews, and M. L. Greenberg. 2005. Impact of human immunodeficiency virus type 1 gp41 amino acid substitutions selected during enfuvirtide treatment on gp41 binding and antiviral potency of enfuvirtide in vitro. J. Virol. 7912447-12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morton, T. A., D. G. Myszka, and I. M. Chaiken. 1995. Interpreting complex binding kinetics from optical biosensors: a comparison of analysis by linearization, the integrated rate equation, and numerical integration. Anal. Biochem. 227176-185. [DOI] [PubMed] [Google Scholar]
- 41.Munoz-Barroso, I., S. Durell, K. Sakaguchi, E. Appella, and R. Blumenthal. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagashima, K. A., D. A. Thompson, S. I. Rosenfield, P. J. Maddon, T. Dragic, and W. C. Olson. 2001. Human immunodeficiency virus type 1 entry inhibitors PRO 542 and T-20 are potently synergistic in blocking virus-cell and cell-cell fusion. J. Infect. Dis. 1831121-1125. [DOI] [PubMed] [Google Scholar]
- 43.Pan, C., H. Lu, Z. Qi, and S. Jiang. 2009. Synergistic efficacy of combination of enfuvirtide and sifuvirtide, the first- and next-generation HIV-fusion inhibitors. AIDS 23639-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peisajovich, S. G., S. A. Gallo, R. Blumenthal, and Y. Shai. 2003. C-terminal octylation rescues an inactive T20 mutant: implications for the mechanism of HIV/SIV-induced membrane fusion. J. Biol. Chem. 27821012-21017. [DOI] [PubMed] [Google Scholar]
- 45.Qi, Z., W. Shi, N. Xue, C. Pan, W. Jing, K. Liu, and S. Jiang. 2008. Rationally designed anti-HIV peptides containing multifunctional domains as molecule probes for studying the mechanisms of action of the first and second generation HIV fusion inhibitors. J. Biol. Chem. 28330376-30384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ray, N., J. E. Harrison, L. A. Blackburn, J. N. Martin, S. G. Deeks, and R. W. Doms. 2007. Clinical resistance to enfuvirtide does not affect susceptibility of human immunodeficiency virus type 1 to other classes of entry inhibitors. J. Virol. 813240-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ray, N., L. A. Blackburn, and R. W. Doms. 2009. HR-2 mutations in human immunodeficiency virus type 1 gp41 restore fusion kinetics delayed by HR-1 mutations that cause clinical resistance to enfuvirtide. J. Virol. 832989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves, J. D., F. H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 794991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reeves, J. D., J. L. Miamidian, M. J. Biscone, F. H. Lee, N. Ahmad, T. C. Pierson, and R. W. Doms. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 785476-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sista, P. R., T. Melby, D. Davison, L. Jin, S. Mosier, M. Mink, E. L. Nelson, R. DeMasi, N. Cammack, M. P. Salgo, T. J. Matthews, and M. L. Greenberg. 2004. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS 181787-1794. [DOI] [PubMed] [Google Scholar]
- 52.Tan, K., J. Liu, J. Wang, S. Shen, and M. Liu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 9412303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tremblay, C. L., F. Giguel, C. Kollmann, Y. Guan, T. C. Chou, B. M. Baroudy, and M. S. Hirsch. 2002. Anti-human immunodeficiency virus interactions of SCH-C (SCH 351125), a CCR5 antagonist, with other antiretroviral agents in vitro. Antimicrob. Agents Chemother. 461336-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tremblay, C. L., C. Kollmann, F. Giguel, T. C. Chou, and M. S. Hirsch. 2000. Strong in vitro synergy between the fusion inhibitor T-20 and the CXCR4 blocker AMD-3100. J. Acquir. Immune Defic. Syndr. 2599-102. [DOI] [PubMed] [Google Scholar]
- 55.Trivedi, V. D., S. F. Cheng, C. W. Wu, R. Karthikeyan, C. J. Chen, and D. K. Chang. 2003. The LLSGIV stretch of the N-terminal region of HIV-1 gp41 is critical for binding to a model peptide, T20. Protein Eng. 16311-317. [DOI] [PubMed] [Google Scholar]
- 56.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic Structure of the ectodomain from HIV-1 gp41. Nature 387426-428. [DOI] [PubMed] [Google Scholar]
- 58.Wexler-Cohen, Y., and Y. Shai. 2007. Demonstrating the C-terminal boundary of the HIV 1 fusion conformation in a dynamic ongoing fusion process and implication for fusion inhibition. FASEB J. 213677-3684. [DOI] [PubMed] [Google Scholar]
- 59.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retrovir. 91051-1053. [DOI] [PubMed] [Google Scholar]
- 60.Wild, C., T. Greenwell, D. Shugars, L. Rimsky-Clarke, and T. Matthews. 1995. The inhibitory activity of an HIV type 1 peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res. Hum. Retrovir. 11323-325. [DOI] [PubMed] [Google Scholar]
- 61.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 919770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, L., S. Hue, S. Taylor, D. Ratcliffe, J. A. Workman, S. Jackson, P. A. Cane, and D. Pillay. 2002. Minimal variation in T-20 binding domain of different HIV-1 subtypes from antiretroviral-naive and -experienced patients. AIDS 161684-1686. [DOI] [PubMed] [Google Scholar]