Abstract
The genome sequence of the giant virus Acanthamoeba polyphaga mimivirus revealed the presence of two putative cytochrome P450 (CYP) genes. The product of one of the two predicted CYP genes (YP_143162) showed low-level homology to sterol 14-demethylase (CYP51) and contained a C-terminal polypeptide domain of unknown function. YP_143162 expression (without an N-terminal membrane binding domain) in Escherichia coli yields a CYP protein which gives a reduced CO difference maximum at 448 nm and was formally demonstrated as the first viral cytochrome P450. Analysis of binding of lipid and sterol substrates indicated no perturbation in CYP heme environment, and an absence of activity was seen when 14-methyl sterols were used as a substrate. The function of the CYP protein and its C-terminal domain remain unknown.
Cytochromes P450 (CYP) are a superfamily of heme-thiolate enzymes that are distributed widely throughout Eukarya, Archaea, and Bacteria (http://drnelson.utmem.edu/CytochromeP450.html). Viruses are the most abundant biological entities in nature and are also responsible for many diseases in plants and animals. To date, 2,180 viral genome sequencing projects have been completed and annotated and no CYP open reading frames have been observed (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=10239&opt=Virus).
Acanthamoeba polyphaga mimivirus is the largest known virus, which grows in amoeba (5). In 2004, the 1.2-Mbp genome of mimivirus (GenBank accession no. AY653733) was sequenced (9). Its genome is larger than that of several bacteria and archaea and is predicted to encode 911 proteins, among which only 298 have predicted functions. Many atypical proteins are predicted to be encoded by the mimivirus genome, including key protein translation enzymes, a full complement of DNA repair pathway components, and the unique presence of three different topoisomerases (9). Interestingly, among genes never yet reported to occur in a virus, mimivirus contained two putative gene sequences predicted to encode cytochrome P450 enzymes (GenBank accession no. YP_142886 and YP_143162, also known as MIMI_L532 and MIMI_L808, respectively). First, YP_142886 is a putative protein of 468 amino acids in length. In a BLASTP search, this putative CYP protein showed homology to a range of bacterial P450 proteins, including a P450 protein from Chloroflexus aurantiacus (23% identity) and CYP171 from Streptomyces peucetius (23% identity). Additionally, YP_142886 also showed homology at the same level to nematode P450 proteins, including Caenorhabditis briggsae CYP37B1 (25% identity) and a P450 protein from the sea squirt Ciona intestinalis similar to the CYP4 family (24% identity). Efforts in our laboratory to express the YP_142886 gene and verify that it indeed encodes a cytochrome P450 have been unsuccessful, but additional attempts are in progress. The mimivirus protein YP_143886 was designated CYP5254A1 by David Nelson (http://drnelson.utmem.edu/CytochromeP450.html).
The second putative mimivirus CYP protein (YP_143162) showed in a BLASTP search the strongest homology to CYP51 proteins (7) from a variety of organisms, including protozoal CYP51 proteins from, e.g., Leishmania major (23% identity); plant CYP51 proteins from, e.g., Arabidopsis thaliana (22% identity); and fungal CYP51 proteins from, e.g., Aspergillus fumigatus (21% identity). This homology is low, strongly suggesting the absence of a functional link. Further analysis of the YP_143162 709-amino-acid sequence revealed this putative CYP protein to be approximately 200 residues longer than its closest CYP homologues, and this protein was proposed to comprise a fused protein domain of unknown function, with the best homologies to lipopolysaccharide core biosynthesis glycosyl transferase from Proteus mirabilis HI4320 (26% identity), integral membrane sensor signal transduction histidine kinase from Dinoroseobacter shibae DFL 12 (24% identity), a short region of dysferlin from Strongylocentrotus purpuratus (35% identity), and pierisin-1 (NAD-DNA ADP-ribosyltransferase) from Pieris rapae (24% identity). Interestingly, several putative posttranslational modifications, including one N glycosylation site, a protein kinase C phosphorylation site, four casein kinase II phosphorylation sites, and three myristoylation sites, were predicted to exist in this C-terminal extension peptide, representing the first time these specific modifications were present in a P450 molecule.
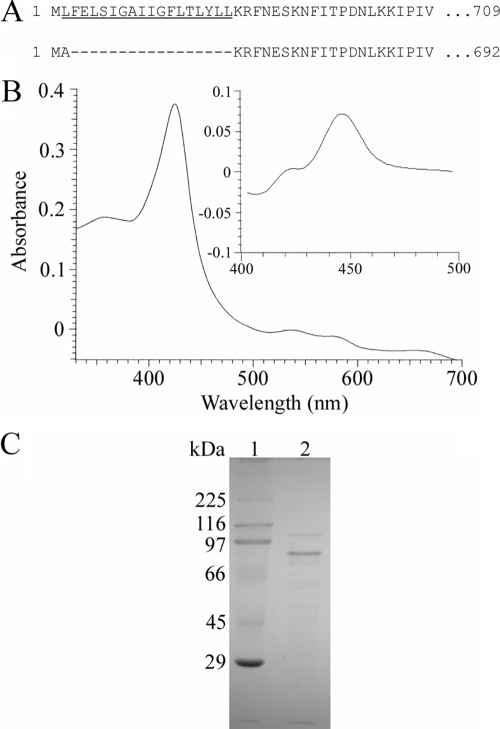
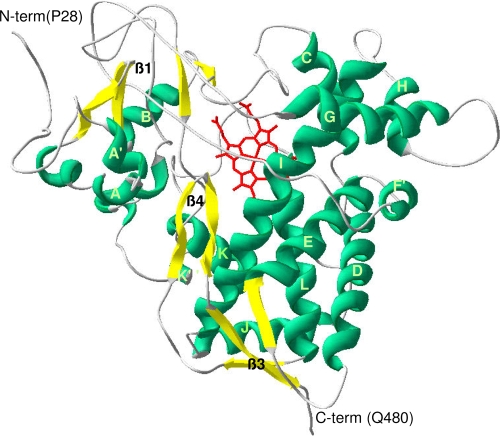
Historically, a protein can be identified as a cytochrome P450 through the production of the carbon monoxide (CO)-bound form of the reduced (sodium dithionite-treated) pigment, which has an intense absorption band at 450 nm (8). Following isopropyl β-d-thiogalactopyranoside-induced expression of the full-length YP_143162 gene, utilizing the T7 promoter of the Escherichia coli expression vector pET17b (Novagen), only a protein producing a Soret maximum at 420 nm, recognized as the misfolded, incorrect form of CYP, was detected in reduced-difference CO spectrophotometry. Alterations in temperature, coexpression with molecular chaperones GroES and GroEL of E. coli (which allow production of active and correctly folded human P450s [4]), and the use of different E. coli strains for recombinant protein expression did not produce correctly folded YP_143162 (data not shown). Analysis of the primary sequence revealed the presence of a putative membrane-spanning segment located from residue 2 to 19 which may interfere with the expression of correctly folded P450 (1). A modified YP_143162 gene sequence encoding an insertion of alanine at amino acid position 2 was generated by PCR. This N-terminally truncated enzyme (Fig. 1A), expressed as a correctly folded CYP protein, generated a characteristic reduced-CO-difference spectrum with a maximum at 448 nm (Fig. 1B). Cell fractionation revealed the truncated protein to be associated with the membrane fraction following ultracentrifugation at 100,000 × g, and no CYP protein was detected in the E. coli cytoplasm, thus necessitating the use of detergents to purify the enzyme. The truncated but membrane-bound enzyme was expressed at CYP levels of >1,000 nmol P450/liter of culture, with supplementation of the growth medium with the heme precursor δ-aminolevulinic acid increasing heme-incorporated CYP expression levels approximately twofold, to 2,000 to 3,000 nmol P450/liter of culture. Truncated YP_143162 was the major band observed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (Fig. 1C), with the molecular mass estimated to be 78 kDa, in agreement with the predicted molecular mass of the truncated protein. The absolute absorbance spectrum of the purified (oxidized, Fe3+) YP_143162 protein showed a Soret band at 419 nm and α and β bands at 572 and 536 nm, while reduction with sodium dithionite (Fe2+) resulted in a typical Soret peak shift to 417 nm. Most CYP enzymes are purified in a low-spin state with a water molecule hexacoordinated to the CYP heme iron, as indicated by a peak at 390 nm in the absolute spectrum. Substrate or inhibitor addition shifts the heme to the high-spin state, as indicated by a peak at 419 nm in the absolute spectrum, which can be the case when imidazole, used to purify the protein, binds to the heme iron. Continued dialysis for removal of the imidazole from the protein resulted in a shift from a high- to a low-spin state. Furthermore, quantification of the iron content (2) of YP_143162 (0.97 ± 0.06 atoms of iron per heme-containing molecule of YP_143162) indicated that there is one atom of iron per heme-containing molecule of YP_143162, confirming one atom of iron associated with the heme of this P450 protein. Given the weak homology of YP_143162 to sterol-metabolizing CYP proteins, the binding of the sterols lanosterol and obtusifoliol as well as the final A. polyphaga sterol end product ergosterol to purified enzyme was investigated as previously described (3). No evidence of sterol binding or metabolism was obtained (data not shown). Such data can be confirmed by the fact that the key motif aGQHTSs (which is involved in catalysis, includes an invariant H in all CYP51 proteins to date [6], and corresponds to a negatively charged residue [D/E] in other P450 families) is missing in YP_143162 (Fig. 2A). Additionally, YP_143162 did not cluster with any CYP51 proteins but mapped to a distinct and separate branch on the tree (Fig. 2B). A homology model was generated for mimivirus YP_143162 protein on the basis of the resolved P450 crystal structures of Mycobacterium tuberculosis CYP51 (Protein Data Bank accession no. 1E9X) and flavocytochrome CYP102A1 from Bacillus megaterium (Protein Data Bank accession no. 1JPZ). YP_143162 is predicted to adopt a typical P450 fold reflecting the similarity in helix assignment. It was possible to confirm the likely heme-coordinating residue (C425), and the EXXR motif, present in nearly all P450 proteins and involved in heme binding and P450 architecture, is also conserved in YP_143162 (Fig. 3). Consequently, the mimivirus protein YP_143162 was designated CYP5253A1 (http://drnelson.utmem.edu/CytochromeP450.html).
FIG. 1.
Purification and spectral characterization of mimivirus YP_143162. (A) N-terminal sequence of the full-length native enzyme, with the hydrophobic stretch that most likely forms a transmembrane α helix underlined. Below is shown the N-terminal sequence of the truncated CYP protein used to obtain the correctly folded P450 protein. (B) Absolute oxidized and reduced CO difference (inset) spectra at 1 μM P450 concentration. (C) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (10%). Lane 1, rainbow marker; lane 2, purified mimivirus CYP51-like protein.
FIG. 2.
Alignment of mimivirus YP_143162 with 143 CYP51 family members (only two representatives of each biological kingdom are shown). (A) The fragments shown are the BC loop (SRS1 region, helices F and G [SRS2 and SRS3] and helix I [SRS4]). (B) Representative phylogenetic tree of CYP51 sequences showing the position of YP_143162.
FIG. 3.
Mimivirus P450 model with marked secondary structural elements. The resultant homology models were validated by cross-reference to the secondary structure predictions. Homology models were generated with 10 iterations of the MODELLER program, and the model structure was clipped to the first 480 amino acids, for which the sequence identity with the resolved crystal structures of MTCYP51 and flavocytochrome P450-2 of Bacillus megaterium for the CYP domain was 16%. The energy-minimized model has very good ProsaII and Profiles 3D scores.
The presence of genes encoding CYP in the mimivirus genome is intriguing from the standpoint of P450 evolutionary discussion. Phylogenetically, mimivirus and other giant viruses are very old and are thought to have existed prior to cellular organisms (9). Furthermore, it was previously proposed that a form of cytochrome P450 has been present in life forms for billions of years and before the advent of free atmospheric oxygen (10). Subsequently P450 had protective value in detoxifying reactive oxygen species and was retained in aerobic organisms as a monooxygenase in biosynthetic processes and in the degradation of complex molecules. Consequently, it can be speculated that P450 may have been present in a viral genome prior to the establishment of the three domains of life (eukaryotes and prokaryotes, which consist of bacteria and archaea). It is possible that mimivirus acquired CYP genes from a more ancient progenitor. Conversely, Moreira and Brochier-Armanet (7) hypothesize that the diverse mimivirus genes, many with eukaryotic homology, including CYP, were obtained by horizontal gene transfer. The basis of this theory suggests that the mimivirus host, A. polyphaga, is also host to parasitic, bacterial endosymbionts, including many Mycobacteria spp. which have high numbers of CYP genes within their genomes. Consequently, mimivirus may have picked up genes from such endosymbionts or from the amoeba host itself. At present, no Acanthamoeba genome has been sequenced to allow comparisons, but Acanthamoeba genomes certainly contain CYP51 for synthesis of sterols. Finally, although mimivirus was first isolated from A. polyphaga, additional hosts acting as a source of new genetic material for this virus cannot be ruled out. For example, mimivirus has been implicated as a causative agent of influenza in mice and humans, suggesting mammalian cellular hosts for mimivirus. It can also be argued that mimivirus CYP has evolved into a gene encoding a protein with a totally different function, unrelated to its being a P450. It is probable that we will never know the nature of the ancient P450 ancestor or how it evolved into the superfamily that we see today (>8,500 genes). However our confirmation of a viral gene encoding a cytochrome P450 protein invigorates this continuing debate.
Acknowledgments
Support was provided by National Institutes of Health grants R01GM69970 and R01GM067871, a Leverhulme Trust Research Fellowship, and a Royal Society Travel Grant.
Footnotes
Published ahead of print on 10 June 2009.
REFERENCES
- 1.Barnes, H. J. 1996. Maximizing expression of eukaryotic cytochrome P450s in Escherichia coli. Methods Enzymol. 2723-14. [DOI] [PubMed] [Google Scholar]
- 2.Fish, W. W. 1988. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158357-364. [DOI] [PubMed] [Google Scholar]
- 3.Jefcoate, C. R. 1978. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 52258-279. [DOI] [PubMed] [Google Scholar]
- 4.Kagawa, N., H. Hori, M. R. Waterman, and S. Yoshioka. 2004. Characterization of stable human aromatase expressed in E. coli. Steroids 69235-243. [DOI] [PubMed] [Google Scholar]
- 5.La Scola, B., S. Audic, C. Robert, L. Jungang, X. de Lamballerie, M. Drancourt, R. Birtles, J.-M. Claverie, and D. Raoult. 2003. A giant virus in amoeba. Science 2992033. [DOI] [PubMed] [Google Scholar]
- 6.Lepesheva, G. I., and M. R. Waterman. 2007. Sterol 14alpha-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta 1770467-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreira, D., and C. Brochier-Armanet. 2008. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Omura, T., and R. Sato. 1964. The carbon monoxide binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 2392370-2378. [PubMed] [Google Scholar]
- 9.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J.-M. Claverie. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 3061344-1350. [DOI] [PubMed] [Google Scholar]
- 10.Wickramashighe, R. H., and C. A. Ville. 1975. Early role during chemical evolution for cytochrome P450 in oxygen detoxification. Nature 256509-510. [DOI] [PubMed] [Google Scholar]