Abstract
In this study we examined whether human immunodeficiency virus type 1 (HIV-1) is equally susceptible to neutralization by a given antibody when the epitope of this antibody is introduced at different positions within the viral envelope glycoprotein (Env). To this end, we introduced two exogenous “epitope tags” at different locations within three major Env regions in two distinct HIV-1 isolates. We examined how the introduction of the exogenous epitopes affects Env expression, Env incorporation into virions, Env fusogenic potential, and viral susceptibility to neutralization. Our data indicate that even within the same Env region, the exact positioning of the epitope impacts the susceptibility of the virus to neutralization by the antibody that binds to that epitope. Our data also indicate that even if the same epitope is introduced in the exact same position on two different Envs, its exposure and, as a result, the neutralization susceptibility of the virus, can be very different. In contrast to the findings of previous studies conducted with HIV-1 isolates other than those used here, but in agreement with results obtained with simian immunodeficiency virus, we observed that tagging of the fourth variable region of Env (V4) did not result in neutralization by the anti-tag antibodies. Our data indicate that epitopes in V4 are not properly exposed within the functional HIV-1 trimeric Env spike, suggesting that V4 may not be a good target for vaccine-elicited neutralizing antibodies.
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) is expressed as a heavily glycosylated peptide of approximately 160 kDa (gp160), which is cleaved intracellularly into two noncovalently associated subunits: an extracellular subunit (gp120), responsible for CD4 and coreceptor (primarily CCR5 and/or CXCR4) binding, and a transmembrane subunit (gp41) that mediates fusion between viral and host cell membranes. Based on amino acid sequence homology analysis of gp120s derived from diverse HIV-1 isolates, gp120 is divided into five “constant” regions (C1 to C5) and five “variable” regions (also called “loops,” because most of them have cysteines in the N and C termini that form disulfide bonds). Despite their extensive amino acid variability, the variable loops of gp120 play central roles during the entry of the virus into the cell, for instance, by directly or indirectly modulating the interaction of Env with coreceptor molecules on the target surfaces during virus-cell fusion. They also offer protection from neutralizing antibodies (NAbs) by various mechanisms. The variable loops themselves are targets of NAbs, and during infection, the replicating virus accumulates mutations in the variable regions that allow it to escape the action of anti-variable loop-directed NAbs, while at the same time the variable loops are positioned within the Env trimer so that they prevent, or minimize, the binding of NAbs to more-conserved epitopes, such as the receptor and coreceptor binding sites (4, 5, 12, 15, 20, 23, 25, 27, 31).
HIV-1 strains display distinct neutralization phenotypes. Some isolates, such as SF162, are generally susceptible to NAbs that bind to many distinct regions of Env, including the variable regions, while other isolates, such as YU2 or JRFL, are generally resistant to neutralization by the same NAbs (1). It has been proposed that irrespective of the overall neutralizing phenotype of HIV-1 isolates, the binding of only a single antibody per Env trimer on the virion surface can lead to neutralization, when all Env trimers present on the virion surface are bound by at least one antibody (32). This important observation also implies that the epitope specificity of an antibody may not be as important for neutralization as its ability to bind to its target within the trimeric Env structure. In fact, antibodies to diverse regions of Env, such as V1, V2, V3, and the receptor and coreceptor binding sites, can all neutralize HIV-1 (1, 3, 6, 8, 10, 18, 20, 23, 25, 27, 29, 30).
In many cases, a given isolate will not be equally susceptible to neutralization by NAbs that bind to different Env regions, for example, the V3 loop and the CD4-binding site (CD4-BS). Whether differences in the neutralizing potentials of two antibodies that bind to distinct epitopes on HIV-1 Env are due to differences in the binding affinities of the two antibodies or whether they occur because the viruses are intrinsically more susceptible to NAbs that bind certain epitopes and not others (i.e., the relative importance of the various regions of Env in Env function and virus neutralization sensitivity differs) is not yet fully understood. One way to address these issues is to introduce small non-HIV Env amino acid sequences (tags) that are targets of known monoclonal antibodies (MAbs) at various positions within the viral Env and to examine how the placement of the same epitope at different positions within Env affects the neutralization phenotype of the virus.
Foreign epitopes have been introduced into the variable regions of HIV and simian immunodeficiency virus (SIV) Envs, and their effects on viral neutralization potential have been examined (14, 19, 22, 33). Yang and colleagues (33) introduced the FLAG epitope into the V4 regions of three HIV-1 isolates (YU2, JRFL, and HxB2) displaying distinct neutralization phenotypes in response to anti-HIV NAbs; they found that all three pseudotyped viruses were equivalently neutralized by an anti-FLAG MAb. One important implication of that study is that neutralization-resistant isolates, such as YU2 or JRFL, are not intrinsically more resistant to neutralization than more-susceptible isolates, such as HxB2, so long as the antibody binds to its epitope on the functional virion-associated Env spike. A second implication is that since the FLAG epitope was exposed in the V4 loops of all three isolates, the V4 loop could theoretically be a good target for vaccine-elicited antibodies. In contrast, Pantophlet et al. (19) introduced the HA tag into various regions of the JRCSF (neutralization-resistant) and HxB2 (neutralization-sensitive) isolates and reported that JRCSF was intrinsically more resistant than HxB2 to anti-HA antibodies. This observation implies, therefore, that some HIV-1 strains (primary, neutralization-resistant strains) have developed mechanisms that limit the accessibility of multiple Env regions, including variable regions, to antibodies developed during infection. Laird and Desrosiers (14) introduced the FLAG epitope into two positions within each of the V1, V2, and V4 loops of SIV239 and SIV316. They reported that the functionality of Env was differentially affected by the precise location of the exogenous tag sequence within the variable loops examined. Importantly, and in contrast to what was reported for the HIV-1 isolates mentioned above, the SIV239 variants containing a V4 FLAG epitope were not neutralized by an anti-FLAG MAb. It appeared, however, that the FLAG epitope was not well exposed on the trimeric Env when introduced into the V4 loop of SIV but was exposed when introduced into the V1 loop of the same virus. Potentially, this means that the V4 loop is differentially exposed in the context of the HIV-1 and SIV Envs.
The FLAG epitope (DYKDDDDK) is highly charged. Therefore, it is possible that the effect on Env function and epitope exposure could differ if a different exogenous epitope were inserted instead of FLAG. Here we examined the effect of variable loop tagging on the Env functions and viral neutralization phenotypes of two primary HIV-1 clade B isolates, SF162 (CCR5 tropic) and SF33 (CXCR4 tropic), using two exogenous epitopes (FLAG and hemagglutinin [HA] tags) positioned at multiple locations within the V1, V2, and V4 loops. By placing the same tag in several regions within each loop, we investigated the accessibilities of various parts of the same loop to a given NAb. By using two tags that differ significantly in amino acid composition (FLAG tag, DYKDDDDK; HA tag, YPYDVPDYA), we aimed at distinguishing between the effects of amino acid composition and the positioning of the tag on Env function and overall epitope exposure. Finally, identical evaluations of R5 and X4 Envs may provide information about the relative roles played in neutralization by variable loops in Envs displaying distinct coreceptor usage. We report that both the amino acid sequence and the position of the tag within and among the variable loops greatly affected the functionality of Env. In contrast to previous observations made with other HIV-1 Envs (33) but in agreement with what was reported for the SIV239 Env (14), we observed that tagging of the V4 loops of SF162 and SF33 did not render these isolates susceptible to neutralization by the corresponding anti-tag MAbs.
MATERIALS AND METHODS
Cell lines.
Human embryonic kidney (HEK) 293T cells, the human astroglioma cell line U87 (N. R. Landau, Salk Institute), stably expressing CD4 and CXCR4, and TZM-bl cells (David Montefiori, Duke University, Durham, NC), stably expressing CD4, CCR5, and CXCR4, were cultured as described previously (25).
Antibodies.
MAbs 2G12, 4E10, and 2F5 were purchased from Polymun Scientific. MAbs P3C8 and P3E1 were isolated from mice immunized with SF162 gp140 and SF162ΔV2 gp140, respectively (6). The former MAb is directed against the V1 loop, while the latter is directed against the crown of the V3 loop. The anti-FLAG MAb M2 (catalog no. F3165) and the anti-HA MAb HA-7 (catalog no. H3363) were purchased from Sigma-Aldrich. These MAbs recognize their respective epitopes when positioned internally as well as at the N and C termini of proteins. D7324 (an anti-carboxy terminus antibody) was purchased from Aalto Bio Reagents (Dublin, Ireland).
Engineering of FLAG- or HA-substituted HIV Env.
FLAG and HA epitope tags (FLAG tag, DYKDDDDK; HA tag, YPYDVPDYA) were introduced into the V1, V2, and V4 loops of the envelope genes of the R5-tropic isolate SF162 and the X4-tropic isolate SF33 by replacing the native amino acid sequences with those of either tag (see Fig. 1). The FLAG epitope is a synthetic peptide of eight amino acids. It is highly charged (seven of eight amino acids are charged at pH 7.0) and strongly hydrophilic. The HA epitope comprises nine amino acids from the human influenza virus HA1 fragment. Is largely uncharged (two of nine amino acids are charged at pH 7.0) and is considerably less hydrophilic than the FLAG epitope. Each tag was incorporated at three positions within each loop for a total of 18 tag-substituted Envs per isolate.
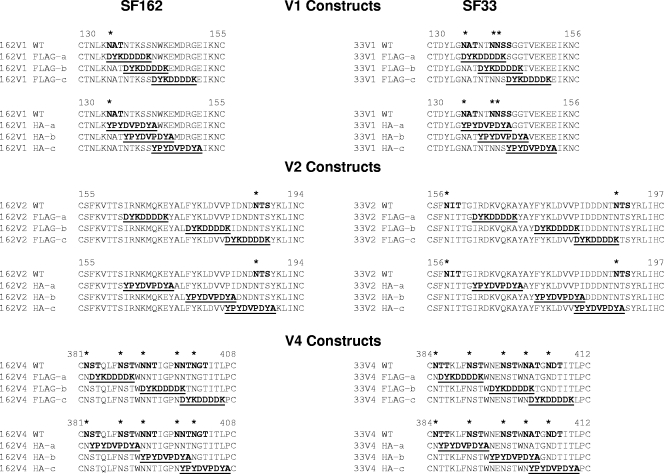
FIG. 1.
Amino acid sequences of the V1, V2, and V4 loops of SF162 and SF33 Envs, showing the positioning of the FLAG and HA tags. The amino acid sequence of each tag is shown in boldface and underlined. Potential N-linked glycosylation sites are indicated on the WT sequence with boldface letters and asterisks. Each Env uses its own numbering.
To generate epitope-tagged Envs, the gp160s of both SF162 and SF33 within the expression vector pEMC* were used as templates for mutagenesis reactions. Reactions were carried out using the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Following mutagenesis, gp160s were fully sequenced to verify the presence of the epitope as well as to confirm the absence of undesirable mutations elsewhere by using an Applied Biosystems 3730XL genetic analyzer with BigDye technology.
Generation of single-round competent virus.
Single-round replication-competent viral particles (pseudoviruses) were generated by cotransfecting HEK 293T cells with the pEnv and the pNL4-3 (Luc+ Vpr− Env−) backbone. Transfections were carried out using GeneJuice (Novagen) according to the manufacturer's instructions at a Δenv backbone/env ratio of 20:1. Seventy-two hours following transfection, supernatants were harvested, clarified by centrifugation at 860 × g for 10 min, aliquoted, and stored at 80°C until use. The p24 antigen concentration for each pseudovirus preparation was determined using the HIV type 1 p24 antigen capture assay kit (AIDS Vaccine Program, NCI—Frederick Cancer Research and Development Center).
Entry assays.
The abilities of pseudoviruses expressing epitope tag-substituted Env to mediate virus-cell fusion were examined using TZM-bl (for SF162) or U87 (CD4+ CXCR4+) (for SF33) target cells. Cells were plated at either 3 × 103 (TZM-bl) or 7 × 103 (U87) per well in a 96-well plate (Falcon) using Dulbecco's modified Eagle's medium (Mediatech, Inc.) containing 10% fetal bovine serum, 100 U/ml each of penicillin and streptomycin, and 2 mM glutamine (complete DMEM). Following Polybrene treatment (2 μg/ml), cells were incubated with serially diluted (fivefold) pseudovirus, and each dilution was tested in triplicate. After a 72-h incubation at 37°C, cells were lysed with SteadyLite (Perkin-Elmer), and cell-associated luciferase levels were measured in relative light units (RLU) using a Fluoroskan Ascent FL luminometer (Thermo Biosystems). All data analyses were carried out using GraphPad Prism software, version 4.03. The entry of each variant Env was tested at least twice. Fusogenicity was compared using the percentage of entry of each Env relative to the entry of its corresponding wild type (WT) after standardization for p24.
Neutralization assays.
Sensitivity to neutralization by a panel of MAbs was tested as described previously (5). Briefly, TZM-bl and U87 cells were plated and Polybrene treated as described above. Fifty micrograms of anti-Env MAbs per milliliter or 200 μg/ml of FLAG M2 and anti-HA MAbs was added to the top row of a separate 96-well plate (37.5 μl/well) and serially diluted fivefold in complete DMEM. For each virus, the equivalent of the amount of p24 (in nanograms) required for 2 × 105 RLU, as determined in an entry assay, was added to each well in 30 μl complete DMEM (60 μl total MAb-virus mixture/well). Plates were incubated at 37°C for 90 min, after which 50 μl was added to each well of a 96-well plate containing Polybrene-treated cells. Triplicate wells were used for each MAb concentration. Cells were incubated at 37°C for 72 h, after which cells were lysed and cell-associated luciferase levels were determined as described above. Results were analyzed using GraphPad Prism. The percentage of neutralization was calculated as [(RLUVA − RLUCA) − (RLUV+Ab − RLUCA)]/(RLUVA − RLUCA) × 100, where VA stands for the virus alone (without a MAb), CA stands for cells alone, and V+Ab stands for the virus preincubated with a MAb. All assays were performed at least twice.
Western blotting and Odyssey imaging.
To examine the relative expression and processing of the various tagged Envs, HEK 293T cells were transfected with GeneJuice and 12 μg of each Env plasmid in 100- by 20-mm plates (4 × 106 cells/plate) according to the manufacturer's instructions. Following a 72-h incubation at 37°C, cells were removed from the plate with cold phosphate-buffered saline (PBS) plus 1 mM EDTA, washed twice with cold PBS, and lysed with PBS containing 0.5% each NP-40 and deoxycholic acid. Cellular debris was removed by centrifugation (10 min at 17,000 × g in a microcentrifuge), and the resulting clarified supernatant was aliquoted and stored at −20°C until use.
Western blotting was carried out using NuPage 4-to-12% Tris-Bis gels and buffers according to the manufacturer's instructions. Following blotting, protein was transferred to a nitrocellulose membrane, which was then dried at room temperature for 30 min and finally blocked overnight at 4°C with Li-Cor Odyssey buffer (20 ml). Blocked membranes were probed with a 1:8,000 dilution of anti-Env rabbit polyclonal sera (raised against an SF162 gp140-gp120 mixture and kindly provided by Nancy Haigwood) in 10 ml Odyssey blocking buffer, 10 ml PBS, and 0.2% Tween 20 for 2 h, washed five times with 0.1% Triton X in PBS, and incubated with IRDye 700DX-conjugated goat anti-rabbit immunoglobulin G (IgG) at a 1:18,000 dilution in 10 ml Odyssey blocking buffer, 10 ml PBS, 0.2% Tween 20, and 0.02% sodium dodecyl sulfate. After a final set of washes, protein was visualized using a Li-Cor Odyssey infrared imager with Imaging System application software (version 2.1) to determine the amounts (in nanograms) of gp160 and gp120 per lane. Known amounts of purified SF162 gp140 were used to generate a standard curve. These values were then used to calculate a gp160/gp120 ratio for each Env.
To determine whether the replacement of specific HIV Env epitopes by non-HIV epitope tags altered the incorporation of gp120 into viral particles, pseudoviruses were produced as described above. Following clarification, virions were pelleted through a 20% glycerol-PBS cushion by centrifugation at ∼50,000 × g for 2 h. The viral pellets were resuspended and lysed in 1% Triton X-100-PBS; then the lysates were aliquoted, and aliquots were frozen at −80°C until use. The ratio of gp120 to p24 was evaluated using the Odyssey imaging system as described above with a MAb against HIV-1 p24 Gag (NIH AIDS Research and Reference Reagent Program) as the primary antibody (1:1,000) and IRDye 800CW-conjugated goat anti-mouse IgG (Li-Cor) at a 1:18,000 dilution as the secondary antibody for detection. The gp120/p24 ratio was calculated as (gp120 band intensity)/(p24 band intensity) for each sample. For intergel standardization, WT controls were run on each gel, and a standardized score for each sample was calculated as (gp120/p24 ratio for the sample)/(gp120/p24 ratio for the WT control).
Flow cytometry.
To determine the exposure of tags within the variable regions of HIV Env, HEK 293E cells (1 × 106/ml) were transfected with plasmids expressing either WT or variously tagged SF162 and SF33 Envs. Transfections were carried out using polyethylenimine. After a 4-h incubation, cells were diluted to 5 × 105/ml and grown in FreeStyle medium (Invitrogen) containing 0.1% Pluronic F-68 and 25 μg/ml G418. Following a 72-h incubation, either 10% goat serum (for 2G12) or 10% mouse serum (for anti-FLAG and anti-HA) was added to each sample. Samples were then incubated on ice with fluorescein isothiocyanate-conjugated MAb FLAG-M2 (5.5 μg/ml) or anti-HA (2.5 μg/ml) (Sigma) or with 2G12 (2 μg/ml) for 30 min and were washed twice with cold PBS plus 10% sera. Samples with 2G12 were incubated an additional 30 min with a fluorescein isothiocyanate-conjugated goat anti-human MAb (Invitrogen) at a 1:200 dilution, followed by two washes with cold PBS plus 10% goat serum. The percentage of binding was determined using a BD LSR II flow cytometer (BD Biosciences).
RESULTS
Generation of FLAG- and HA-tagged envelope glycoproteins.
A total of 36 tagged Envs (18 each for SF162 and SF33) were generated (Fig. 1). In order to keep the length of the Env constant, amino acid substitutions instead of insertions (which would have increased the size of Env) were used. Thus, our approach differed from that used by others who inserted the tags into the HIV envelope (19, 33), but it was similar to the substitution approach used by Laird and Desrosiers (14). The tags were introduced at three different positions (sometimes overlapping) within each of the V1, V2, and V4 loops: at the amino terminus (position “a”), in the central region (position “b”), and at the carboxy terminus (position “c”). Where possible, we tried to avoid disruption of naturally occurring potential N-linked glycosylation sites. This was not always possible, however; for example, several glycosylation sites within the V1 loop (for instance, insertions of tags in position “a” in the SF162 Env and in positions “a” and “b” in the SF33 Env) and within the V4 loop (in positions “a,” “b,” and “c” for either Env) were lost.
Effects of epitope tagging on the fusogenic potential of Env.
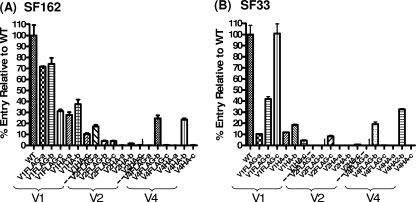
Tagged envelopes were tested for their abilities to mediate virus entry into the cell by using single-round competent viruses and either TZM-bl cells as targets for viruses expressing SF162 Env or U87 (CD4+ CXCR4+) cells for viruses expressing SF33 Env. The data from these studies are summarized in Table 1, and representative results are shown in Fig. 2. For both SF162 and SF33, tag substitutions within the V1 region were generally better tolerated than V2 or V4 substitutions, even though a wide range of entry potentials (∼10% and 100% of WT Env entry) was evident among the V1-tagged Envs. In contrast, for both Envs tested, substitutions in the V2 loop either completely abolished the fusogenic potential of Env or drastically reduced it (less than 10% of WT entry). In the case of V4, only the viruses with tag substitutions at position “b” (for either Env) remained functional (percentages of WT entry were ∼25% for SF162 V4FLAG-b, ∼23.5% for SF162 V4HA-b, 19% for SF33 V4 FLAG-b, and ∼32% for SF33 V4HA-b). V4 substitutions at position “a” or “c” resulted in nonfusogenic Envs, irrespective of the tag and the Env backbone.
TABLE 1.
Env processing, virion incorporation, and fusogenic potentials
| Env | Ratioa
|
% Entry relative to that of WT Envb | |
|---|---|---|---|
| gp160/gp120 | gp120/p24 | ||
| SF162 | |||
| WT | 0.52 | 1.00 | 100.00 |
| V1FLAG-a | 1.02 | 1.38 | 71.64 |
| V1FLAG-b | 0.62 | 2.33 | 74.08 |
| V1FLAG-c | 0.83 | 1.10 | 31.19 |
| V1HA-a | 0.69 | 0.53 | 27.72 |
| V1HA-b | 0.46 | 0.68 | 37.44 |
| V1HA-c | 0.63 | 1.73 | 10.34 |
| V2FLAG-a | 1.10 | 1.76 | 17.28 |
| V2FLAG-b | 1.91 | 1.55 | 3.97 |
| V2FLAG-c | 3.22 | 1.58 | 4.02 |
| V2HA-a | 2.41 | 0 | (−) |
| V2HA-b | 2.08 | 0.64 | 1.67 |
| V2HA-c | 3.39 | 2.69 | (−) |
| V4FLAG-a | 5.85 | 0.19 | (−) |
| V4FLAG-b | 1.28 | 3.74 | 24.75 |
| V4FLAG-c | 5.16 | 1.37 | (−) |
| V4HA-a | 11.38 | 0.30 | (−) |
| V4HA-b | 1.10 | 1.46 | 23.51 |
| V4HA-c | 9.96 | 0.15 | (−) |
| SF33 | |||
| WT | 0.73 | 1.00 | 100.00 |
| V1FLAG-a | 1.28 | 0.87 | 10.25 |
| V1FLAG-b | 1.45 | 0.58 | 41.95 |
| V1FLAG-c | 0.75 | 1.05 | 100.93 |
| V1HA-a | 0.96 | 1.00 | 11.80 |
| V1HA-b | 0.55 | 1.07 | 18.47 |
| V1HA-c | 0.86 | 1.25 | 4.42 |
| V2FLAG-a | 1.55 | 0.74 | (−) |
| V2FLAG-b | 1.66 | 0.78 | (−) |
| V2FLAG-c | 0.97 | 0.72 | 8.29 |
| V2HA-a | 14.13 | 0.13 | (−) |
| V2HA-b | 1.27 | 0.61 | (−) |
| V2HA-c | 1.68 | 0.32 | 0.80 |
| V4FLAG-a | >20 | 0.24 | (−) |
| V4FLAG-b | 0.65 | 0.73 | 19.25 |
| V4FLAG-c | >20 | 0.34 | (−) |
| V4HA-a | >20 | 0.31 | (−) |
| V4HA-b | 0.73 | 1.21 | 32.69 |
| V4HA-c | >20 | 0.21 | (−) |
gp160/gp120 ratios are for cell-associated Env; gp120/p24 ratios are for virion-associated gp120.
(−), <1% of WT entry.
FIG. 2.
Entry potentials of tagged Envs. The levels of entry of the tagged SF162 (A) and SF33 (B) Envs were determined as described in Materials and Methods and are presented here as percentages of the entries of the corresponding WT Envs. Results are averages and standard deviations from triplicate experiments.
Overall, FLAG tags were better tolerated than HA tags in both Envs. For example, SF162 V1 FLAG-tagged Envs entered with 30 to 74% of the efficiency of WT SF162, while SF162 V1 HA-tagged Envs entered with 10 to 37% of the efficiency of WT SF162. Similarly, SF33 V1 FLAG-tagged Envs entered with 10 to 100% of the efficiency of WT SF33, while SF33 V1HA Envs entered with less than 20% of the efficiency of WT SF33.
In agreement with the findings of previous studies (14, 19), we observed that the precise location of the tags within the variable loops (i.e., at the amino terminus, in the central region, or at the carboxy terminus [position “a,” “b,” or “c,” respectively]) differentially affected the fusogenic potentials of Envs in an Env background-dependent manner. Specifically, for SF162, Env FLAG tagged in the amino-terminal or central region of V1 (V1FLAG-a or -b, respectively) mediated entry at 70 to 74% of the WT level, while Env FLAG tagged at the C terminus (V1FLAG-c) mediated entry at only ∼30% of the WT level. Similarly, Envs tagged with HA at the amino terminus or in the central region of the V1 loop (V1 HA-a and -b) mediated entry at levels between 28% and 37% of the WT level, while HA tagging at the carboxy terminus of the V1 loop (V1HA-c) resulted in approximately 10% of WT entry. In contrast to what we observed with SF162, tagging the SF33 Env with FLAG at the V1 carboxy terminus (V1FLAG-c) did not affect the fusogenic potential of Env relative to that of the WT (i.e., 100% entry); however, FLAG tagging the central region of the V1 loop reduced the fusogenic potential of Env by 60%, and a FLAG tag at the amino terminus of that loop reduced the fusogenic potential of that Env by ∼90%. Tagging the V4 loop of either Env at the amino- or carboxy-terminal position abrogated the fusogenic potential, while tagging the V4 loop in its central region (“b”' constructs) did not abrogate fusogenic potential but reduced it approximately 75% to 80% from that of the corresponding WT Env.
Therefore, substitutions in the V4 of either virus affected the fusogenic potential of Env similarly irrespective of the Env backbone, while this was not the case for the V1 substitutions, where Env background dependence was recorded.
Effects of epitope tagging on Env processing and incorporation into virions.
Considering that improper Env processing has been observed in FLAG-tagged SIV Envs that failed to mediate entry (14), one possible explanation for the differences observed in the fusogenic potentials of the variously tagged Envs could be that the introduction of tags at different positions within the V1, V2, and V4 variable regions differentially affected the intracellular processing of Env, specifically the cleavage of the gp160 precursor into gp120 and gp41. To examine this, cells transfected with the various Env constructs were lysed; the cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis; and the gp160/gp120 ratios were determined (Table 1). Both WT Envs displayed gp160/gp120 ratios of <1, whereas this ratio for tagged Envs was much more variable. In some cases, ratios similar to that of WT Env were recorded, while in other cases (SF33 V4FLAG-a and -c and SF33 V4HA-a and -c), the ratios were >20, essentially indicating that the gp160 was unprocessed. A significant negative correlation was observed between the gp160/gp120 ratios and the relative fusogenic potentials of Envs for SF162 (Spearman's r, −0.8927; 95% confidence interval [95% CI], −0.9608 to −0.7229; P < 0.001) and for SF33 (Spearman's r, −0.6264; 95% CI, −0.8975 to −0.0969; P = 0.022). We note here that although some Envs were expressed at lower (or higher) levels than others, we did not find a significant relationship between the levels of Env expression, Env processing, and Env fusogenic potential.
Although in many cases, the poor fusogenic potential of a tagged Env could be associated with potential defects in the processing of gp160 into gp120 and gp41 (for example, SF33 V4FLAG-a and c and SF33 V4HA-a and c), in other cases the tagged Envs were processed as efficiently as the WT Env but nonetheless displayed significant reductions in their fusogenic potentials (for example, SF33 V1HA-b was processed more efficiently than the WT but displayed less than 80% of its fusogenic potential). Thus, factors other than the processing of gp160 into gp120 and gp41 are also contributing to the observed reduction in the fusogenic potentials of viruses expressing certain tagged Envs. It is possible that in some cases, although Env tagging does not reduce the processing of Env or its intrinsic ability to mediate fusion, it negatively affects the incorporation of Env molecules into virions. To examine this, we determined the relative gp120/p24 ratios of virions expressing the various Envs (Table 1).
No significant correlation was observed between the gp120/p24 ratios and the relative fusogenic potentials of the variously tagged SF162 Envs (r, 0.2921; 95% CI, −0.2168 to 0.6761; P = 0.2396), suggesting overall that for SF162, the extent of tagged-Env incorporation into virions was not associated with the virion fusion results. In contrast, for SF33 Envs, a significant correlation was observed (r, 0.6251; 95% CI, 0.2248 to 0.8449; P = 0.0042). Overall, therefore, the observed defect in the fusogenic potentials of several tagged SF162 Envs appears to be linked to defects in Env processing, while for tagged SF33 Envs, both processing and virion incorporation levels appear to be involved.
Neutralization sensitivities of viruses expressing tagged SF162 or SF33 envelopes.
Observations from previous studies using tagged Envs (14, 19), as well as the results described above, suggest that the introduction of exogenous tags into the variable regions of either the SF162 or the SF33 Env could result in changes in the overall conformation of the individual Env protomers participating in the formation of the trimer or in the overall association of the three Env protomers within the trimer. Such changes could affect the relative exposures of various neutralization epitopes. Although our goal was to examine how a specific antibody neutralizes a virus when its epitope is introduced at different locations of Env, the observations described above made us first examine whether the tagging of specific Env regions resulted in gross changes in the overall neutralization phenotypes of these two viruses (Table 2).
TABLE 2.
Neutralization phenotypes of SF162 and SF33 viruses expressing tagged Envs
| Env | Concn (μg/ml) of the following MAb resulting in 50% inhibition of infectiona:
|
||||||
|---|---|---|---|---|---|---|---|
| P3C8 (anti-V1) | P3E1 (anti-V3) | 2G12 (anti-gp120) | 2F5 (anti-gp41) | 4E10 (anti-gp41) | Anti-FLAG | Anti-HA | |
| SF162 | |||||||
| WT | 0.073 | 0.033 | 0.162 | 0.903 | 2.063 | (−) | (−) |
| V1FLAG-a | (−) | 0.034 | 0.138 | 0.873 | 1.623 | 2.474 | (−) |
| V1FLAG-b | (−) | 0.007 | 0.387 | 0.181 | 1.239 | 0.186 | (−) |
| V1FLAG-c | (−) | <0.001 | 0.152 | 0.196 | 1.632 | 0.016 | (−) |
| V1HA-a | (−) | 0.003 | 0.081 | 1.555 | 1.779 | (−) | 0.064 |
| V1HA-b | (−) | 0.026 | 0.058 | 1.997 | 2.946 | (−) | 0.053 |
| V1HA-c | (−) | <0.001 | 0.249 | 0.176 | 0.273 | (−) | 0.002 |
| V2FLAG-a | 0.014 | 0.004 | 0.120 | 0.033 | 0.218 | 29.700 | (−) |
| V4FLAG-b | 0.139 | 0.003 | 3.782 | 1.250 | 1.888 | (−) | (−) |
| V4HA-b | 0.165 | 0.004 | (−) | 0.502 | 3.724 | (−) | (−) |
| SF33 | |||||||
| WT | (−) | (−) | 0.041 | 0.103 | 0.024 | N/A | (−) |
| V1FLAG-a | NT | NT | 0.038 | 0.017 | 0.036 | 6.268 | (−) |
| V1FLAG-b | (−) | (−) | 0.024 | 0.065 | 0.019 | 0.902 | (−) |
| V1FLAG-c | NT | NT | 0.039 | 0.059 | 0.036 | (−) | (−) |
| V1HA-a | NT | NT | 0.650 | 0.014 | 0.019 | (−) | 2.104 |
| V1HA-b | NT | NT | 0.004 | 0.017 | 0.033 | (−) | 0.006 |
| V1HA-c | NT | NT | 0.074 | 0.005 | 0.012 | (−) | 1.008 |
| V2FLAG-a | NT | NT | 0.024 | 0.063 | 0.019 | 0.708 | (−) |
| V4FLAG2-b | NT | NT | 0.389 | 0.067 | 0.011 | (−) | (−) |
| V4HA2-b | NT | NT | 1.595 | 0.050 | 0.033 | (−) | (−) |
Values are means for at least two separate experiments. A value of <0.001 indicates that 50% neutralization was not reached at the lowest MAb concentration tested (0.001 μg/ml). Values in boldface indicate a >1 log10 difference in neutralization susceptibility from that of the WT. (−), the MAb did not achieve 50% neutralization even at the highest MAb concentration tested; NT, the indicated MAb was not tested against the particular virus.
The overall neutralization phenotypes of virions expressing tagged Envs were determined using known anti-gp120 and anti-gp41 neutralizing MAbs. Of course, only entry-competent viruses (nine SF162- and nine SF33-derived constructs) were used for these neutralization experiments (Table 1).
(i) Anti-gp120 MAbs.
All viruses were highly susceptible to neutralization by anti-CD4-BS reagents such as IgGCD4 and MAb B12 (50% inhibitory concentrations, <0.01 μg/ml), and thus we assume that the overall exposure of the CD4-BS was not affected by tagging.
MAb P3C8 was elicited in mice immunized with the SF162 Env, binds within the V1 loop, and strongly neutralizes the SF162 virus (6). This MAb did not neutralize the SF33-derived Envs. We are unaware of known anti-V1 MAbs that neutralize SF33. None of the SF162 viruses expressing V1-tagged Envs were neutralized by the anti-V1 MAb P3C8, since its epitope was destroyed by the introduction of the tags. In contrast, SF162 viruses expressing V4-tagged Envs were as susceptible (within twofold) as the WT to neutralization by P3C8.
MAb P3E1, which was also isolated from mice immunized with the SF162 Env, binds within the IGPGRAF crown motif of the V3 loop and displays limited cross-neutralizing activity (6). Since we did not tag the V3 loop, all SF162-derived viruses with tagged Envs were susceptible to neutralization by this MAb. The relative neutralization susceptibilities of V1-tagged SF162 viruses to P3E1 was dependent both on the epitope tag and on its location within the V1 loop. Although V1FLAG-a and V1HA-b were as susceptible to P3E1 as WT SF162, all other SF162 viruses expressing V1-tagged Envs were approximately 1 log10 unit more susceptible than the WT. These observations are consistent with a functional and structural interaction between the V3 loop and the V1V2 region of HIV-1 Env (9, 10, 13, 16, 20, 34). Interestingly, SF162 viruses expressing V4-tagged Envs were more susceptible to P3E1-mediated neutralization (by ∼1 log10 unit) than the WT. It appears, therefore, that the tagging of the V4 loop altered the accessibility of epitopes within the V3 loop.
MAb 2G12 binds to a conformational epitope formed by terminal mannose residues located on specific N-linked glycosylation sites on gp120 (24, 26). The most important N-linked glycosylation sites for the binding of MAb 2G12 to gp120 are at positions 295, 332, and 392 (based on HxB2 Env numbering), while glycans at positions 339, 386, and 448 also participate. Of these sites, only position 392 was eliminated when we introduced the tags in V4 at position “b” (Fig. 1). As a result, although the FLAG- or HA-tagged viruses expressing SF162 Env were as susceptible (within onefold) to MAb 2G12 as the corresponding virus expressing WT Env, viruses expressing the tags at position V4-b were significantly more resistant than the corresponding WT Env-expressing viruses. A similar observation was made for SF33, but here we also observed that tagging the V1 loop could render the virus less susceptible to 2G12 neutralization. These observations, again, suggest that any global structural alterations induced during Env tagging are dependent on the Env background. Interestingly, despite the relative resistance of V4-tagged Envs to MAb 2G12-mediated neutralization, these Envs were recognized by this MAb at levels similar to that of the corresponding WT (see Fig. 4, discussed below).
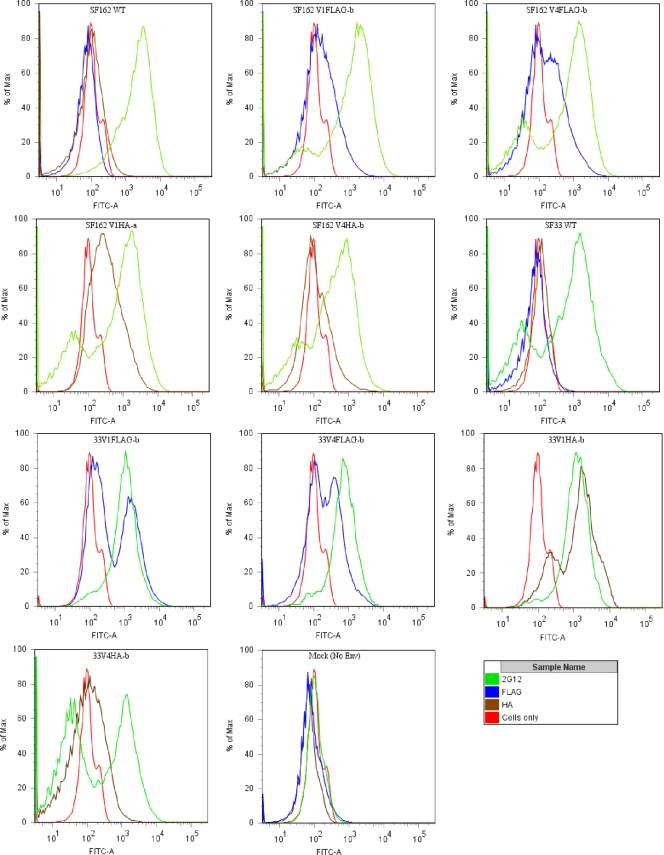
FIG. 4.
Cell surface staining of Env-expressing cells. The extents of binding of the anti-tag MAbs to WT, V1-tagged, and V4-tagged SF162 and SF33 Envs expressed on the cell surface were determined by fluorescence-activated cell sorting. MAb 2G12 was used as a positive control; as a negative control, we used cells expressing no Env (cells only).
(ii) Anti-gp41 MAbs.
The tags were introduced into gp120, and thus, in the vast majority of cases, they had no effect on the susceptibility of SF1262 or SF33 to neutralization by the anti-gp41 MAbs 2F5 and 4E10 (Table 2). However, SF162 V1HA-c and SF162 V2FLAG-a were more sensitive (by ∼1 log10 unit) to MAb 4E10, and SF162 V2FLAG-a was more sensitive to MAb 2F5, than WT SF162. Also, SF33 V1HA-c was more sensitive (by ∼1 log10 unit) to MAb 2F5 than WT SF33. It is known that changes in gp120 can modulate the exposure of epitopes in gp41 (15), and vice versa (2).
(iii) Anti-tag MAbs.
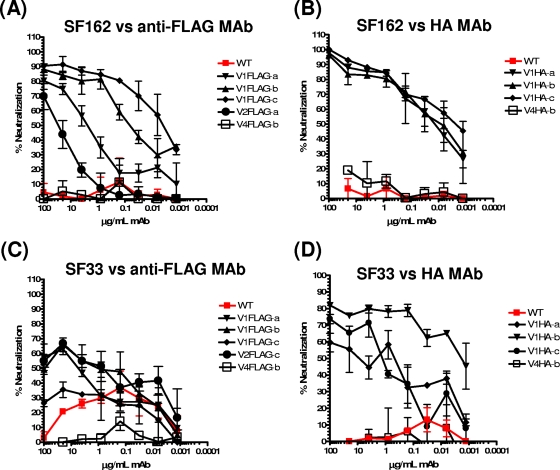
As expected, neither the WT SF162 nor the WT SF33 virus was susceptible to neutralization by anti-FLAG or anti-HA MAbs (Fig. 3 and Table 2). The V1-tagged SF162 viruses, however, were susceptible to anti-tag MAb-mediated neutralization. For FLAG-tagged Envs, neutralization susceptibility increased as the FLAG tag was moved from the amino-terminal (position “a”) to the carboxy-terminal (position “c”) region of the V1 loop (Fig. 3A). In contrast, this gradation in neutralization susceptibility was not observed for HA-tagged SF162 viruses (Fig. 3B). SF162 V2FLAG-a was also susceptible to neutralization by the anti-FLAG MAb, but less so than the V1 FLAG-tagged SF162 viruses. SF162 V2FLAG-a was less fusogenic than the SF162 V1 FLAG-tagged viruses (Table 1 and Fig. 2), and thus, no correlation between neutralization susceptibility and the entry potential of the viruses was observed here.
FIG. 3.
Neutralization of tagged SF162 and SF33 by anti-FLAG and anti-HA MAbs. The neutralizing potentials of anti-FLAG and anti-HA MAbs against SF162 and SF33 virions expressing WT or tagged Envs capable of mediating viral entry were evaluated as described in Materials and Methods. (A and C) Neutralization of FLAG-tagged fusogenic SF162 (A) and SF33 (C) Envs by anti-FLAG MAb M2. (B and D) Neutralization of HA-tagged fusogenic SF162 (B) and SF33 (D) Envs by an anti-HA MAb. Representative results are shown.
For SF33, the V1 HA-tagged Envs were susceptible to neutralization by the anti-HA MAb (Fig. 3D). In contrast to what we observed for SF162 (Fig. 3B), tagging the central region (position “b”) of the V1 loop of SF33 rendered the virus more susceptible to anti-HA MAb-mediated neutralization than tagging of either the amino (position “a”) or the carboxy (position “c”) terminus of that loop. Interestingly, the V1 FLAG-tagged SF33 viruses were more resistant to neutralization by the anti-FLAG MAb than were the V1 HA-tagged SF33 viruses to the anti-HA MAb (compare Fig. 3C and D). In fact, SF33 V1FLAG-c was as resistant as WT SF33 to anti-FLAG-mediated neutralization. The differences observed in the abilities of the anti-FLAG and anti-HA MAbs to neutralize the respectively tagged viruses (either SF162 or SF33) are most likely due to differences in the binding affinities of the two MAbs for their cognate epitopes. Interestingly, in most cases (with the exception of the HA-tagged SF162 viruses), 100% neutralization was not observed even at the highest MAb concentrations tested (100 μg/ml). Incomplete HIV-1 or SIV neutralization by anti-tag MAbs has been observed previously for both FLAG-tagged and HA-tagged Envs (14, 19), and although no definitive answer as to the exact reason(s) for this has yet been reported, it could potentially be attributed to heterogeneity in viral populations.
Overall, the neutralization results described above suggest that although the epitope tags were substituted at the same positions within the V1 loop of SF162 and that of SF33, their relative exposures within the context of the Env trimer differed between the two Envs. This observation, therefore, implies that structural differences exist between the envelope glycoproteins of SF162 and SF33.
Epitope tag exposure on Env trimers.
Introduction of either tag into the V4 loop of SF162 or SF33 did not result in virus neutralization by the corresponding anti-tag MAbs (Fig. 3 and Table 2). This result is similar to what has been reported for SIV (14) but different from what was reported for other HIV-1 strains (22, 33). Although the results of Pantophlet et al. (19) indicate that HIV-1 JRCSF with an HA tag in the V4 loop is marginally susceptible to neutralization by anti-HA MAbs, the inabilities of the two anti-tag MAbs to neutralize SF162 or SF33 viruses expressing V4-tagged Env could be explained if either the tag was not exposed on the surface of the Env trimer when the tag was introduced into V4 or the binding of the anti-tag MAbs to their epitopes when positioned within the V4 loop does not lead to inhibition of the Env-mediated virus-cell fusion process.
To address the first of these possibilities, we determined the binding of both anti-tag MAbs to cell surface-expressed Envs (Fig. 4). As a control, we used the anti-gp120 MAb 2G12, which bound equally all Env-expressing cells tested in this experiment. Binding of an anti-HA MAb to neutralization-sensitive V1HA Env-expressing viruses was observed for both SF162 (SF162 V1HA-a) and SF33 (SF33 V1HA-b), whereas the binding of the same MAb to neutralization-resistant V4HA Envs (SF162 V4HA-b and SF33 V4HA-b) was at or close to background levels, consistent with the hypothesis that the V4HA tag was occluded within these trimeric Envs. These results suggest that the SF162 and SF33 viruses that express V4 HA-tagged Envs evade neutralization by their cognate MAbs due to epitope occlusion within the Env trimeric spike. In contrast, the anti-FLAG MAb bound similarly to both the sensitive (SF162 V1FLAG-b or SF33 V1FLAG-b) and the resistant (SF162 V4FLAG-b or SF33 V4FLAG-b) Envs (Fig. 4). The differences in neutralization in these cases cannot, therefore, be explained by differences in epitope exposure within the Env trimeric spike. We speculated, therefore, that for the SF162 or SF33 V4FLAG-b Env, the anti-FLAG MAb could bind more efficiently to the unprocessed gp160 than to the processed gp120. Indeed, the anti-FLAG MAb bound SF162 V1FLAG-b gp120 very efficiently (endpoint binding titer by enzyme-linked immunosorbent assay [ELISA], 0.7 μg/ml) but did not bind SF162 V4FLAG-b gp120 in an ELISA format (Table 3). Similar observations were made for the SF33 Env.
TABLE 3.
Gp120 expression of FLAG and HA tags
| Env | Endpoint titer (μg/ml)a of the following MAb:
|
|||
|---|---|---|---|---|
| 2G12 | Anti-FLAG | Anti-HA | Anti-tag/2G12 | |
| SF162 | ||||
| WT | 0.0501 | NA | NA | NA |
| V1FLAG-b | 0.0404 | 0.7134 | — | 17.66 |
| V4FLAG-b | 1.8764 | NA | — | NA |
| V1HA-a | 0.1834 | — | NA | NA |
| V4HA-b | 1.0312 | — | NA | NA |
| SF33 | ||||
| WT | 0.0103 | 40.427 | NA | 3924.95 |
| V1FLAG-b | 0.0284 | 1.8337 | — | 64.57 |
| V4FLAG-b | 0.0134 | 13.91 | — | 1038.06 |
| V1HA-b | 0.0153 | — | 0.0169 | 1.10 |
| V4HA-b | 0.0181 | — | NA | NA |
NA, the highest titer was below the endpoint cutoff; —, ELISA was not done.
DISCUSSION
In this study we examined whether or not a given antibody can neutralize HIV to the same extent when its epitope is positioned at various regions of the HIV Env. To this end, we introduced exogenous tags (HA or FLAG) within the V1, V2, and V4 loops of two Envs, SF162 and SF33. We first examined how the introduction of these exogenous epitopes affects Env expression and fusogenic potential. Then we investigated whether or not the introduction of these tags globally altered the neutralization phenotypes of viruses expressing these Envs. Finally, we examined whether anti-tag MAbs could neutralize these viruses irrespective of the positioning of the tag, or whether neutralization occurred only when the tag was positioned at specific Env regions but not at others.
Our results are in general agreement with those reported by Laird and Desrosiers for SIV (14) and by Pantophlet et al. (19) for HIV-1, in that we observed that the functionality of the HIV envelope following tagging depended on the position of the tag. Thus, for both SF162 and SF33, the introduction of tags in the V2 or V4 loop abrogated or greatly reduced the fusogenic potentials of the Envs. In contrast, the V1-tagged Envs from both viruses were fusogenic (in most cases). In some cases, tagging of the V2 or V4 loop resulted in inefficient processing of the gp160 into gp120 and gp41, while in other cases, inefficient incorporation of Env into virions was observed. The observation that tagging of the V2 loop of SF162 Env had such a negative effect on Env processing and cell surface expression was unexpected, since this Env remains fusogenic even when major deletions are introduced into the V2 loop (28). Also, we observed that the potential of the HIV-1 Envs to mediate virus-cell fusion was differentially affected by the actual amino acid sequence of the tag. HA-tagged Envs were, in general, less fusogenic than FLAG-tagged Envs. The greater negative effect of HA tagging on Env fusogenic potential could be due to the fact that the HA tag contains two prolines, which would disrupt the secondary Env structure and affect Env expression as well as fusion potential.
In a previous study by Yang et al. (33), it was observed that the introduction of a FLAG tag into the V4 loops of three HIV-1 isolates with distinct neutralization phenotypes (YU2, JRFL, and HxB2) rendered all three viruses equally susceptible to neutralization by an anti-FLAG MAb. This neutralization was achieved without an obvious alteration of the overall neutralization phenotypes of these viruses in response to anti-HIV NAbs. A subsequent study by Pantophlet et al. (19), however, suggested that two HIV-1 isolates (JRCSF and HxB2) with distinct neutralization susceptibilities to anti-HIV-1 NAbs were also differentially neutralized by anti-tag MAbs when the appropriate tags were inserted into those Envs. In addition, it was noted that the neutralization susceptibilities of tagged viruses differed depending on the actual positioning of the tag within Env. This could potentially mean that not all neutralization epitopes are created equal and that the binding of the same antibody to the exact same epitope at two different regions of Env may result in significant differences in the neutralization of HIV-1. Similar observations were made for SIV239, in that neutralization of the tagged viruses was dependent on the positioning of the tag within the trimeric Env (14). In contrast to what was reported for HIV-1, V4-tagged SIVs were resistant to neutralization by anti-tag MAbs because the tag was not exposed within the SIV Env trimers. Thus, the V4 loop may be differentially exposed within the trimeric HIV-1 and SIV Envs.
Our results indicate that the V4 loops on SF162 and SF33 may not be well exposed (as observed for SIVmac239) on the functional trimeric Env spike, and as a result, the V4 HA-tagged SF162 and SF33 viruses were resistant to neutralization by the anti-HA MAb used here. We believe that the positions within the V4 loop where the tags were introduced are occluded within the functional, fully processed trimeric Env spike. In other cases (V4FLAG Envs), it appears that the anti-FLAG MAb bound preferentially to the “uncleaved” gp160. This is consistent with studies discussing antigenic differences between the properly cleaved and uncleaved versions of HIV Env (7, 17). Even though both processed and unprocessed Envs are present on the surfaces of viral particles, our results suggest that primarily the unprocessed Env forms were recognized by the anti-FLAG MAb and that this binding did not lead to neutralization. In this regard, our neutralization data would be in agreement with previous observations that only properly processed Envs are involved in the neutralization of HIV-1 (11, 21). We note, however, that another potential reason for the differences observed between the neutralization susceptibilities of the V4-tagged HIVs in the present study and those in the study by Yang et al. is that we replaced amino acids in the V4 loop with those of the tags, while Yang et al. inserted the tags into the V4 loop. Thus, in our case, the overall size of the V4 loop did not change as much as the size of the V4 loop in the previous publication.
Our results indicate that the extent of neutralization of HIV by antibodies that target the variable regions of Env depends on the exact location of the epitope within any given variable loop. They also indicate that the exposure of the exact same epitope positioned at the exact same region in two different Envs may actually differ depending on the Env backbone. This result also suggests that the orientation and exposure of the variable HIV-1 Env loops will be different for diverse HIV isolates. This diverse exposure of a given epitope among distinct isolates most likely results in the escape and predominance of certain viral species during infection. Our results also highlight the problem that a vaccine against HIV that is based on the elicitation of NAbs against specific epitopes within the variable regions of Env would be extremely difficult to develop, not only because of the variable nature of these regions, but also because of the differential exposure of specific epitopes in distinct Envs. The elicitation of broad NAb responses against conserved elements of the HIV-1 envelope may not face similar problems with regard to the “conservation” of these epitopes in diverse viruses but may face similar problems with regard to the “exposure” of such epitopes in diverse isolates.
Acknowledgments
This study was funded by NIH grant R01 AI47708 (to L.S.). We acknowledge the financial support of the M. J. Murdock Charitable Trust and the J. B. Pendleton Charitable Trust.
We also express our gratitude to those who contributed reagents for this study.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 7813232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blish, C. A., M. A. Nguyen, and J. Overbaugh. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton, D. R., R. L. Stanfield, and I. A. Wilson. 2005. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA 10214943-14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 719808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ching, L. K., G. Vlachogiannis, K. A. Bosch, and L. Stamatatos. 2008. The first hypervariable region of the gp120 Env glycoprotein defines the neutralizing susceptibility of heterologous human immunodeficiency virus type 1 isolates to neutralizing antibodies elicited by the SF162gp140 immunogen. J. Virol. 82949-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derby, N. R., S. Gray, E. Wayner, D. Campogan, G. Vlachogiannis, Z. Kraft, S. W. Barnett, I. K. Srivastava, and L. Stamatatos. 2007. Isolation and characterization of monoclonal antibodies elicited by trimeric HIV-1 Env gp140 protein immunogens. Virology 366433-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dey, A. K., K. B. David, M. Lu, and J. P. Moore. 2009. Biochemical and biophysical comparison of cleaved and uncleaved soluble, trimeric HIV-1 envelope glycoproteins. Virology 385275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 688312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 782394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, X. H. Wang, S. Burda, T. Kimura, F. A. Konings, A. Nadas, C. A. Anyangwe, P. Nyambi, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2006. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J. Virol. 806865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrera, C., C. Spenlehauer, M. S. Fung, D. R. Burton, S. Beddows, and J. P. Moore. 2003. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J. Virol. 771084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 762075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koito, A., L. Stamatatos, and C. Cheng-Mayer. 1995. Small amino acid sequence changes within the V2 domain can affect the function of a T-cell line-tropic human immunodeficiency virus type 1 envelope gp120. Virology 206878-884. [DOI] [PubMed] [Google Scholar]
- 14.Laird, M. E., and R. C. Desrosiers. 2007. Infectivity and neutralization of simian immunodeficiency virus with FLAG epitope insertion in gp120 variable loops. J. Virol. 8110838-10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCaffrey, R. A., C. Saunders, M. Hensel, and L. Stamatatos. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 783279-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nabatov, A. A., G. Pollakis, T. Linnemann, A. Kliphius, M. I. Chalaby, and W. A. Paxton. 2004. Intrapatient alterations in the human immunodeficiency virus type 1 gp120 V1V2 and V3 regions differentially modulate coreceptor usage, virus inhibition by CC/CXC chemokines, soluble CD4, and the b12 and 2G12 monoclonal antibodies. J. Virol. 78524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pancera, M., and R. Wyatt. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332145-156. [DOI] [PubMed] [Google Scholar]
- 18.Pantophlet, R., E. Ollmann Saphire, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantophlet, R., M. Wang, R. O. Aguilar-Sino, and D. R. Burton. 2009. The human immunodeficiency virus type 1 envelope spike of primary viruses can suppress antibody access to variable regions. J. Virol. 831649-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 785205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren, X., J. Sodroski, and X. Yang. 2005. An unrelated monoclonal antibody neutralizes human immunodeficiency virus type 1 by binding to an artificial epitope engineered in a functionally neutral region of the viral envelope glycoproteins. J. Virol. 795616-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rong, R., F. Bibollet-Ruche, J. Mulenga, S. Allen, J. L. Blackwell, and C. A. Derdeyn. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 811350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 767293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saunders, C. J., R. A. McCaffrey, I. Zharkikh, Z. Kraft, S. E. Malenbaum, B. Burke, C. Cheng-Mayer, and L. Stamatatos. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 799069-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 767306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization resistant, clade B HIV-1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 727840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatatos, L., M. Wiskerchen, and C. Cheng-Mayer. 1998. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV-1 isolate on viral envelope structure, cell-entry and replication. AIDS Res. Hum. Retrovir. 141129-1139. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan, N., Y. Sun, J. Li, W. Hofmann, and J. Sodroski. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 694413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 701100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 32.Yang, X., S. Kurteva, S. Lee, and J. Sodroski. 2005. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 793500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, X., I. Lipchina, S. Cocklin, I. Chaiken, and J. Sodroski. 2006. Antibody binding is a dominant determinant of the efficiency of human immunodeficiency virus type 1 neutralization. J. Virol. 8011404-11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye, Y., Z. H. Si, J. P. Moore, and J. Sodroski. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J. Virol. 7411955-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]