Abstract
Simian immunodeficiency virus SIVrcm, which naturally infects red-capped mangabeys (RCMs), is the only SIV that uses CCR2 as its main coreceptor due to the high frequency of a CCR5 deletion in RCMs. We investigated the dynamics of SIVrcm infection to identify specific pathogenic mechanisms associated with this major difference in SIV biology. Four pigtailed macaques (PTMs) were infected with SIVrcm, and infection was monitored for over 2 years. The dynamics of in vivo SIVrcm replication in PTMs was similar to that of other pathogenic and nonpathogenic lymphotropic SIVs. Plasma viral loads (VLs) peaked at 107 to 109 SIVrcm RNA copies/ml by day 10 postinoculation (p.i.). A viral set point was established by day 42 p.i. at 103 to 105 SIVrcm RNA copies/ml and lasted up to day 180 p.i., when plasma VLs decreased below the threshold of detection, with blips of viral replication during the follow-up. Intestinal SIVrcm replication paralleled that of plasma VLs. Up to 80% of the CD4+ T cells were depleted by day 28 p.i. in the gut. The most significant depletion (>90%) involved memory CD4+ T cells. Partial CD4+ T-cell restoration was observed in the intestine at later time points. Effector memory CD4+ T cells were the least restored. SIVrcm strains isolated from acutely infected PTMs used CCR2 coreceptor, as reported, but expansion of coreceptor usage to CCR4 was also observed. Selective depletion of effector memory CD4+ T cells is in contrast with predicted in vitro tropism of SIVrcm for macrophages and is probably due to expansion of coreceptor usage. Taken together, these findings emphasize the importance of understanding the selective forces driving viral adaptation to a new host.
Simian immunodeficiency viruses (SIVs) share a number of biological and structural features with human immunodeficiency virus (HIV), making SIVs powerful tools for studying HIV infection. SIVs are the sources of the HIV pandemic (71), and SIV infection of nonhuman primates (NHPs) is at the origin of animal models of AIDS research (16, 28, 70, 74). Rhesus macaques (RMs; Macaca mulatta) infected with SIVmac, SIVsmm, and several chimeric simian-human immunodeficiency viruses (SHIVs) are the most widely used animal models of AIDS and provide essential insights into HIV/SIV immunopathogenesis and vaccine research (43, 65). The Indian RM (IndRM) is the best-characterized and most commonly studied model; however, significant expansion of AIDS research in recent years has resulted in limited availability of IndRM and a renewed interest in alternative animal models for pathogenic SIV infection (15, 47), such as Chinese RMs, cynomolgus macaques (Macaca fascicularis), and pigtailed macaques (PTMs; Macaca nemestrina) (15, 65). Chinese RMs and cynomolgus macaques show somewhat less pathogenic courses of SIVmac239, SIVmac251, and SHIV89.6P infections than IndRMs (36, 61). SIV and SHIV strains, as well as HIV type 2 (HIV-2), have also been used to infect PTMs (2, 12, 38, 48). Attempts have been also made to infect PTMs with HIV-1, but no persistent infection resulted (1, 18, 19). Several groups now primarily study PTMs as an animal model for HIV (28), and there are growing data on the common major histocompatibility complex molecules expressed by PTMs and the SIV epitopes they present (2, 12, 38, 48), thus enabling detailed immunopathogenic studies in PTMs similar to those originally pioneered with IndRMs.
One of the most important applications of the use of PTMs as animal models for AIDS research is the study of cross-species SIV transmission from the natural hosts. It is widely acknowledged that African NHPs that are natural hosts of SIV generally do not progress to AIDS when infected with their species-specific virus despite high levels of viral replication (51, 54, 55, 62). In striking contrast to the pacific SIV-host interactions observed in natural hosts, HIV-1 and HIV-2 infections in humans are highly pathogenic, being characterized by progression to AIDS in a variable time frame (27). Both HIV-1 and HIV-2 originated by cross-species transmission of the SIVcpz from chimpanzees (Pan troglodytes troglodytes) and SIVsmm from sooty mangabeys (Cercocebus atys), respectively (71). SIVs have a high propensity for cross-species transmission, which is not a rare event, having been documented in the wild (8, 31, 72). However, unlike cross-species transmission of SIVs to humans and macaques that resulted in the emergence of highly pathogenic viruses, cross-species transmission of SIVs to new African NHP hosts generally does not result in increases in SIV pathogenicity (71). Therefore, understanding the mechanisms behind the spectacular increase in pathogenicity of SIVs transmitted across species to humans is a high priority in the field, especially considering the high exposure of humans to a plethora of viruses naturally infecting African NHP hosts in sub-Saharan Africa (59).
A systematic approach of the events driving the increase in pathogenicity upon SIV cross-species transmission will need appropriate NHP models. For these types of studies RMs are probably not an appropriate host as studies carried out thus far demonstrated that RMs can control the majority of cross-species-transmitted SIV infections. Thus, SIVmnd-2 from mandrills (Mandrillus sphinx) (68), SIVrcm from red-capped mangabeys (RCMs; Cercocebus torquatus torquatus) (37, 64), SIVagm from African green monkeys (genus Chlorocebus) (52), SIVsyk from Syke's monkey (Cercopithecus albogularis) (26), and SIVtal from talapoins (Myopithecus talapoin) (49) are minimally pathogenic or controlled when transmitted to RMs. The only exception is the cross-species transmission of SIVsmm from sooty mangabeys (Cercocebus atys), which is pathogenic in RMs upon direct cross-species transmission (41, 62). Note that the emergence of SIVmac and SIVsmm reference strains currently used for pathogenesis and vaccine studies occurred through accidental transmission of SIVsmm from sooty mangabeys to different species of macaques in primate centers in the United States (4, 5, 34, 44) and that the high pathogenicity of these reference strains might have resulted as an effect of serial passages (5, 24). This conclusion is also supported by the recent observation that the intrinsic pathogenicity of primary SIVsmm isolates is significantly lower than previously believed (C. Apetrei, unpublished observations).
In contrast to RMs, PTMs appear to be more susceptible to cross-species-transmitted SIV, with cases of AIDS reported to occur in PTMs after experimental infection with SIVsmm (17), SIVagm from different species of African green monkeys (SIVagm.ver and SIVagm.sab) (22, 25) (I. Pandrea, unpublished observations), SIVlhoest from l'hoest monkeys (Cercopithecus lhoesti) (6), and SIVsun from sun-tailed monkeys (Cercopithecus solatus) (6). This higher susceptibility of PTMs to cross-species-transmitted viruses may be explained by peculiarities of the TRIM5 genes in PTMs. The TRIM5 gene encodes Trim5α, a protein that blocks infection of the cell by retroviruses through the inhibition of reverse transcription immediately after viral entry into the cell (67). An aberrant splicing of TRIM5 mRNA resulting in TRIM5α isoform transcripts was described in PTMs (46). These isoforms (TRIM5η or TRIM5θ) are incapable of restricting either HIV-1 or SIV infection (11), thus explaining the high susceptibility of PTMs to different retroviral pathogens.
The RCMs are found along the Atlantic forest coastal area of west and central Africa (9) and are naturally infected with a specific lentivirus, SIVrcm (7, 13, 21, 64). Since the RCMs were identified by genus and species previously (two paragraphs above), the identification was deleted from the sentence beginning “The RCMs are found.”− SIVrcm is a unique primate lentivirus in that it uses chemokine receptor CCR2 as a coreceptor for entry (13, 76) in contrast to the majority of HIV and SIV strains that use CCR5 and CXCR4 as the main coreceptors (14, 35). This peculiar coreceptor usage pattern is due to a deletion of 24 bp in the RCM CCR5 (Δ24 CCR5) gene that prevents expression of CCR5 on the surface of CD4+ T cells. This Δ24 CCR5 allele has a high frequency in RCM populations (13). Thus, SIVrcm is an example of a primate lentivirus that has adapted its coreceptor usage to allow productive replication and persistence in its natural host species.
Given the unique coreceptor usage, along with the observation that the CCR2 coreceptor is expressed mainly on macrophages and only at very low levels on CD4+ T cells, SIVrcm may be ideal for pathogenesis studies of macrophage-tropic viruses. To date, there is no available data on SIVrcm replication in its natural host, but it is believed that SIVrcm rarely causes disease in RCMs, suggesting that it has been associated with its natural host for a long time. Previous studies on experimental infection of cynomolgus macaques and RMs reported that SIVrcm replicated in both macaque species during acute infection, but the replication was completely controlled by day 60 postinfection (p.i.) (21, 64). This controlled pattern of infection was maintained even after serial passage of SIVrcm in RMs (37).
In order to develop an animal model for the in vivo study of SIVrcm pathogenesis and to confirm that this virus can be used as a model of macrophage-tropic SIV infection, we infected PTMs with SIVrcm and monitored the dynamics of viral loads (VLs), changes in lymphocyte subsets, antibody responses, and clinical and pathological features over a period of 27 months. Surprisingly, the patterns of SIVrcm replication and memory CD4+ T-cell depletion in PTMs were similar to those observed in lymphotropic SIV infections. Following inoculation in PTMs, SIVrcm expanded its coreceptor usage, becoming able to use CCR4 in vivo in addition to CCR2. This result indicates that lentiviral adaptation to a new host may occur rapidly through strain selection and involve new pathogenic pathways, pointing to the need for in vivo studies to characterize the pathogenesis of primate lentiviruses.
MATERIALS AND METHODS
Animals.
Four PTMs were included in this study. All animals were negative for serum antibodies to herpes B virus, SIV, type D retrovirus, and simian T-lymphotropic virus type 1. The animals were fed and housed according to regulations set forth by the Guide for the Care and Use of Laboratory Animals (45) and the Animal Welfare Act at the Tulane National Primate Research Center (TNPRC), which is an Association for Assessment and Accreditation of Laboratory Animal Care International facility. For all procedures, animals were anesthetized with 10 mg/kg ketamine. The Tulane University Institutional Animal Care and Use Committee approved all protocols and procedures for these studies.
Virus.
Animals were inoculated intravenously with 100 time the 50% tissue culture infective dose of SIVrcmGB1, a challenge stock of a primary isolate from Gabon (21) propagated and titrated in human peripheral blood mononuclear cells (PBMCs).
Sample collection and processing.
Blood was collected two times before infection (days −7 and 0), biweekly for the first 2 weeks p.i., weekly for the next 2 weeks, bimonthly up to 2 months p.i, monthly up to 4 months p.i., and finally, every 2 months during chronic infection (between 4 and 27 months p.i.).
Lymph node (LN) biopsies were sampled using sterile surgical procedures on days 0, 28, 42, 90, 180, 360, 420, 480, 540, 600, 720, and 820 p.i., as described previously (51, 58). Intestinal endoscopies (proximal jejunum) consisting of approximately 10 to 15, 1- to 2-mm2 pieces were obtained by endoscopic-guided biopsy on days 0, 28, 42, 90, 180, 240, 300, 360, 420, 480, 540, 600, 720, and 820 p.i., as described previously (56, 58). Intestinal resections (5 to 10 cm) were performed at days 0, 42, and 180 p.i., as previously described (56, 58). Additional intestine samples were collected at necropsy.
Isolation of lymphocytes.
Mononuclear cells were separated from the blood using Ficoll density gradient centrifugation. Lymphocytes from the intestine and LNs were isolated and stained for flow cytometry as previously described (51, 56, 58). Briefly, lymphocytes were isolated from intestinal biopsies using EDTA, followed by collagenase digestion and Percoll density gradient centrifugation (51, 56, 58). Lymphocytes were isolated from the axillary LNs by gently mincing and pressing tissues through nylon mesh screens.
Dynamics of anti-SIVrcm antibodies.
In order to detect anti-SIVrcm-specific antibodies, we designed an indirect enzyme-linked immunosorbent assay (ELISA) using peptides mapping sequences of gp41 immunodominant epitopes of the transmembrane protein of the SIVrcm.GB1. Wells of polyvinyl microtiter plates (Falcon) were coated at 100 μl/well with antigen diluted in 0.05 M bicarbonate buffer, pH 9.6, by incubation for 20 h at 37°C. The wells were washed twice with phosphate-buffered saline (PBS) containing 0.5% Tween 20 (PBS-TW). Incubation for 45 min at 37°C was followed by washing in PBS-TW. Each serum sample was tested at 1:100 dilution in 0.01 M sodium phosphate buffer, pH 7.4, containing 0.75 M NaCl, 10% newborn calf serum (NBCS), and 0.5% Tween 20 (PBS-TW-NBCS). One hundred microliters of diluted serum was added to the wells and incubated for 30 min at room temperature. The wells were washed four times with PBS-TW, and peroxidase-conjugated goat F(ab9)2 anti-human immunoglobulin (Sigma, St. Louis, MO) (100 μl of a 1:2,000 dilution in PBS-TW-NBCS) was added and incubated for 30 min at room temperature. The wells were washed four times with PBS-TW, and the reaction was revealed by incubation with hydrogen peroxide (H2O2)-o-phenylendiamine for 15 min at room temperature. Color development was stopped with 2 N H2SO4, and the absorbance value (optical density [OD]) was read at 492 nm. Based on our previous experience (63), the cutoff was arbitrarily established at 0.15.
SIVrcm RNA quantification.
RNA was extracted from 420 μl of EDTA plasma by using a QIAamp Viral RNA kit (Qiagen, Valencia, CA) and eluted in 40 μl of nuclease-free water. For tissue quantification, viral RNA was extracted from 5 × 105 to 5 × 106 cells from PBMCs, LNs, and intestine with RNeasy (Qiagen), as described elsewhere (56, 58). Viral RNA was quantified in plasma and tissues using a real-time PCR assay specific for SIVrcm. Briefly, total RNA was retrotranscribed into cDNA using a TaqMan Gold RT-PCR kit and random hexamers (PE-Applied Biosystems, Foster City, CA). The quantification was based on the amplification of a 163-bp fragment located in the gag region (positions 2723 to 2886). The primers and probe were the following: forward primer, 5′-ATGAGCTGCACCCAAAAAGT-3′; reverse primer, 5′-TGCTCCCCTTATCAACTTGC-3′; and probe, 5′-FAM-CACTGTAAATTTGACTTGCCCA-TAMSp-3′ (where FAM is 6-carboxyfluorescein and TAMSp is 6-carboxytetramethylrhodamine). Optimal conditions for PCR were as follows: 7 mM MgCl2; 200 μM each of dATP, dCTP, and dGTP; 400 μM dUTP; a 900 nM concentration of each primer; and 300 nM probe. The amplification protocol consisted of incubation for 2 min at 50°C, followed by 10 min at 95°C and subsequently 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. Amplification and detection were carried out using an ABI Prism 7700 Sequence Detection System (Applied Biosystems). All real-time PCRs were carried out in duplicate. Absolute viral RNA copy numbers were deduced by comparing the relative signal strength to corresponding values obtained for eight 10-fold dilutions of a standard RNA that was reverse transcribed and amplified in parallel. The target copy numbers of SIVrcm were determined by using an SIVrcm standard which was constructed as follows: a larger SIVrcm gag fragment (693 bp; positions 2494 to 3187 of the SIVrcm genome) encompassing the fragment targeted by real time PCR was PCR-amplified with primers SF1 (5′-TCTCCCGCCATCTTTCA-3′) and SR1 (5′-GCAGCCTTCCCCAAATA-3′). This PCR product was cloned into a pCR2.1 vector (Invitrogen, Carlsbad, CA), and the recombinant plasmid was transformed into Escherichia coli. The insert was sequenced to confirm the size and identity of the template. The plasmid was linearized using HindIII enzyme, and RNA transcripts were prepared from the linearized plasmid by use of a Megascript high-yield transcription kit (Ambion, Austin, TX). The transcripts were purified by phenol-chloroform extractions, resuspended in nuclease-free water, and quantified by spectrophotometer at an OD at 260 nm (OD260) by use of an extinction coefficient of 40 g/ml/OD unit.
For tissue SIVrcm RNA load quantification, simultaneous quantification of RNase P, a single copy gene with two copies per diploid cell, was done to normalize sample variability and allow accurate quantification of cell equivalents (53). RNase P primers, VIC probe, reagents, and standards were components of a commercially available kit (RNase P detection kit; Applied Biosystems).
The detection limits of the SIVrcm quantification assays were 102 SIVrcm RNA copies per ml of plasma and 10 RNA copies/105 cells.
SIVrcm proviral DNA quantification.
Genomic DNA was isolated from PBMCs by using a QIAamp DNeasy tissue Kit (Qiagen, Valencia, CA) in accordance with the manufacturer's instructions. Real-time PCR components and conditions for quantifying SIVrcm DNA were the same as described for viral RNA quantification but with the cDNA step omitted. A plasmid DNA standard was constructed based on the same gag region as for the RNA standard. Plasmid DNA was diluted to 1010 copies/ml, and subsequent dilutions were used for real-time PCR assays. Simultaneous quantification of RNase P was done to normalize sample variability and allow accurate quantification of cell equivalents (39). The detection limit of the proviral SIVrcm DNA quantification was 10 proviral DNA copies/105 PBMCs.
CCR2 gene expression quantification.
mRNA expression of the CCR2 gene in PBMCs was quantified at different time points p.i. by real-time PCR. cDNAs were generated by random hexamers using TaqMan reverse transcription reagents (Applied Biosystems). TaqMan assays for quantifying CCR2 mRNA were done with primers CCR2F1 (5′-CCTCCTGACAATCGATAGATACC-3′) and CCR2R1 5′-TTCCTGGCATTTAGTAAAGATGA-3′ and probe 5′-FAM-TGGCTGTGTTTGCTTCTGTC-TAMSp-3′. As a template, 2.5 μl cDNA was added to TaqMan reagents (Applied Biosystems), resulting in a total volume of 25 μl. PCR conditions were as follows: incubation for 2 min at 50°C, followed by 10 min at 95°C and subsequently 40 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. Amplification and detection were carried out using an ABI Prism 7700 Sequence Detection System (Applied Biosystems). A CCR2 RNA standard was prepared by targeting a 209-bp region of the CCR2 gene, and the amplicon was synthesized by using primers CCR2F2 (5′-GGTTATTTGGGCGGAATCTT-3′) and CCR2R2 (5′-GGGCCGCAGATATAAACAGA-3′). The PCR product was cloned into a pCR2.1 vector (Invitrogen), and the recombinant plasmid was transformed into E. coli. The insert was sequenced to confirm the size and identity of the template. The plasmid was linearized using HindIII enzyme, and RNA transcripts were prepared by use of a Megascript high-yield transcription kit (Ambion). The transcripts were purified by phenol-chloroform extractions, resuspended in nuclease-free water, quantified by spectrophotometer, and diluted to 1010 copies/ml. Aliquots were stored at −80°C. Subsequent 10-fold dilutions were made in nuclease-free water. The detection limit of the CCR2 mRNA quantification assay was 10 copies/105 cells.
Antibodies and flow cytometry.
Mononuclear cells derived from peripheral blood, intestinal biopsies, and LNs were stained for flow cytometric analysis using a four-color method with the following combinations of monoclonal antibodies: CD3 (clone SP34)-fluorescein isothiocyanate (FITC), CD8 (clone Leu2a)-phycoerythrin (PE), CD4 (clone L200)-peridinin chlorophyll A protein (PerCP), CD20 (clone 2H7)-allophycocyanin (APC), CD3-FITC, CCR5 (clone 3A9)-PE, CD4-PerCP, CD95 (clone DX2)-APC, CD95 (clone DX2)-FITC, CD3 (clone SP34)-PE, CD4-PerCP, CD28 (clone CD28.2)-APC, CD3-FITC, CXCR4 (clone 12G5)-PE, CD4-PerCP, CD95-APC, CD14 (clone M5E2)-FITC, CCR2 (clone 48607)-PE, CD4-PerCP, and CD3 (SP34-2)-APC (Becton Dickinson Biosciences-Pharmingen, San Diego, CA). Cells were incubated with an excess amount of monoclonal antibodies at 4°C for 30 min, followed by a PBS wash (400 × g for 7 min) and fixation in 2% paraformaldehyde. All antibodies were validated and titrated using PTM PBMCs. Flow-cytometric acquisition and analysis of samples were performed on at least 100,000 events on a FACSCalibur flow cytometer driven by the Cell Quest software package (Becton Dickinson). Analysis of the acquired data was performed using Flowjo software (Tree Star, Inc., Ashland, OR). Cell blood counts were performed for each animal and each time point and were used to determine the absolute numbers of CD4+ T cells.
Cell sorting.
Frozen PBMCs were thawed in a 37°C water bath with continuous agitation until completely thawed and then placed on ice for 2 min. Each 1 ml of thawed cell suspension was slowly diluted with 10 ml of RPMI 1640 medium supplemented with 20% fetal bovine serum and 25 mM HEPES buffer at room temperature, incubated for 10 min, pelleted by centrifugation, and washed with 10 ml of medium. Recovered cells were resuspended in staining buffer and stained for flow-cytometric analysis using combinations of the following fluorochrome-conjugated monoclonal antibodies: CD4-APC, CD14-FITC, CD8-PE-Texas Red, and CD3 Pacific Blue. Cell sorting was done using a fluorescence-activated cell sorting Aria instrument (BDIS), and at least 10,000 cells were sorted. Quantification of SIVrcm RNA and DNA in sorted CD4+ T cells and CD14+ cells was performed by quantitative PCR by means of a 5′ nuclease (TaqMan) assay with an ABI 7700 system (Applied Biosystems), as described above. Samples were run in duplicate, and template copies were calculated using ABI 7700 software.
Coreceptor usage.
Coreceptor usage was determined as described previously (76). Human osteosarcoma (GHOST) cells expressing CD4 and one of the following coreceptors were obtained through the NIH AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, contributed by Dan Littman and Vineet KewalRamani: CCR1, CCR2, CCR3, CCR4, CCR5, CCR8, CXCR4, BOB, and Bonzo. These cells were cultured in complete Dulbecco's minimal essential medium containing G418 (5 μg/ml), hygromycin (1 μg/ml), and puromycin (1 μg/ml). GHOST cells expressing only CD4 (GHOST-CD4 cells) served as controls; these cells were cultured in the same medium without puromycin. GHOST cells (105/ml; 500 μl per well) were maintained in 24-well plates for 24 h. The medium was then removed, and 200 μl of fresh medium was added, along with a viral inoculum at a multiplicity of infection of 10. On the next day, residual virus was removed, and the cells were washed once with 1 ml of medium. Then, 750 μl of fresh complete medium containing the selection antibiotics was added. Productive viral replication was monitored by measuring SIV p27 Gag antigen in the culture supernatants on days 0, 2, 4, 6, and 9 by ELISA (Zeptometrix Corp., Buffalo, NY). In all cases, the amount of antigen produced in control GHOST-CD4 cells was subtracted from the amount produced in coreceptor-transfected GHOST-CD4 cells.
Immunohistochemical staining.
Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded intestinal tissues using an avidin-biotin complex horseradish peroxidase technique (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA) and mouse monoclonal anti-human CD4 (NCL-CD4-1F6; Novocastra, Newcastle, United Kingdom), as previously described (58). CD4+ T cells were manually counted in the lamina propria of normal and SIVrcm-infected PTMs during acute and chronic infection.
SIVrcm env sequencing.
In order to understand the mechanisms behind the rapid expansion of coreceptor usage by SIVrcm in PTMS, SIVrcm was reisolated from infected PTMs (7 to 42 days p.i.) and subjected to env gene sequencing. cDNA was prepared using random hexamers, and a 2-kb fragment was obtained by PCR using primers envF (5′-GAAAATAGAATAAGATAAGATGGAT-3′) and envR (5′-TGCCTCCTCTAATTACAAAGTCTCT-3′). PCR conditions included an initial denaturation of 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 45°C for 45 s, and 68°C for 3 min, with a final extension for 10 min at 68°C. PCR products were visualized by agarose gel electrophoresis. PCR products were cloned using a TOPO TA Cloning Kit (Invitrogen). Plasmids with the correct insert were sequenced with the universal primer, and env sequences were aligned using the Clustal W (69) profile alignment option, translated, and analyzed.
In vitro replicative capacity of SIVrcm on T-cell and monocyte cell lines.
In vitro characterization of SIVrcm strains isolated from PTMs was performed as previously described (20) to identify discernible differences in the in vitro replication capacities compared to SIVmac. Human CD4+ T-cell lines (MT2, MT4, M8166, HUT78, C8166, U937.p11, U87.CD4, H9 SupT1, CEMss, CEMx174, PM1, and Molt4clone8) were maintained in RPMI 1640 medium, supplemented with 10% heat-inactivated fetal bovine serum, and 1% penicillin-streptomycin (1 mg/ml). For infection studies, 106 cells were infected with parental SIVrcm and SIVrcm strains isolated from the infected PTMs, corresponding to 1,000 pg of p27 antigen in 12-well plates (Costar, Corning, NY). Cells were incubated for 4 h and then washed extensively to remove cell-free virus. Virus production in culture supernatants was monitored biweekly by SIV p27 antigen capture assay (Zeptometrix Corp., Buffalo, NY).
Statistical analysis and data presentation.
The dynamics of different parameters of SIVrcm infection in PTMs (VLs, immunophenotypic markers, and gene expression) were analyzed for significant differences (P < 0.05) by using the Mann-Whitney test. Correlations were calculated and are expressed as the Spearman coefficient of correlation.
Nucleotide sequence accession numbers.
The nucleotide sequences of the env sequences from SIVrcm-infected PTMs were deposited in GenBank under accession numbers GQ267521 to GQ267527.
RESULTS
CCR2 expression on different immune cell populations.
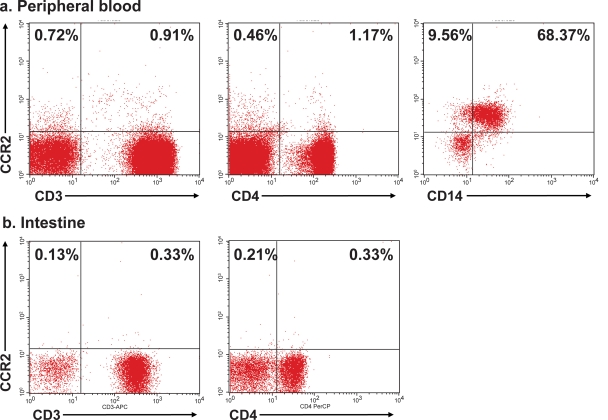
In order to confirm that SIVrcm, a CCR2-tropic virus, can be used as a model of macrophage-tropic SIV, we examined the surface expression of CCR2 on lymphocytes and monocytes in blood and intestine of uninfected PTMs using flow-cytometric analysis. High CCR2 expression was observed on monocytes, in contrast to the very low expression on CD4+ T cells in periphery as well as intestine (Fig. 1). These results indicate that CCR2 is mainly expressed on cells belonging to monocyte/macrophage lineages. Therefore, due to its tropism to CCR2-expressing cells (7, 13), we expected SIVrcm to be a macrophage-tropic virus, and we infected the PTMs to define the in vivo pathogenesis of this infection with a putative macrophage-tropic virus.
FIG. 1.
Flow-cytometric identification of CCR2 expression on blood and intestine of pigtailed macaques. CCR2 expression is restricted to CD14+ monocytes in blood, with the T-cell population (CD3+) and CD4+ T cells (gated on CD3+) in periphery (a) and intestine (b) expressing low levels of CCR2.
Clinical follow-up.
All four PTMs included in this study became infected with SIVrcm. None of the infected animals showed any clinical signs of SIV infection, weight loss, opportunistic infection, or increase in the size of LNs during or after acute SIVrcm infection. No clinical signs of AIDS were observed at the end of the follow-up, 820 days p.i. One of the animals (EC01) died at 132 days p.i. due to causes unrelated to SIV infection (abscess and adhesions originating from abdominal surgical incision). All four SIVrcm-infected PTMs seroconverted between days 21 and 42 p.i., as shown by the dynamics of anti-Gp36 SIVrcm binding antibodies (Fig. 2).
FIG. 2.
Dynamics of anti-SIVrcm gp41 binding antibodies by SIVrcm-specific ELISA. SIVrcm-infected PTMs seroconverted between days 21 and 42 p.i. Transmembrane ELISA results are presented as crude OD values. Cutoff was arbitrarily established at 0.15. Animal EC01, •; BU62, ▾; EC23, ▪; EC25, ♦.
Dynamics of SIVrcm replication.
To identify potential differences in the pattern of SIVrcm infection due to the peculiar biology of SIVrcm, which, in the context of coreceptor usage and CCR2 expression on macrophages rather than lymphocytes (Fig. 1), we monitored the dynamics of SIVrcm replication on both plasma and tissue samples from the four SIVrcm-infected PTMs (Fig. 3a to d) . Plasma VLs peaked as early as 7 to 10 days p.i. (ranging from 107 to 109 copies per ml) and established a set point at 103 to 105 copies/ml for a limited period of time (between days 42 p.i. and 180 p.i.); then, starting from day 180 p.i., virus was undetectable by our assay at most time points (Fig. 3a). However, control of viral replication was not complete, as demonstrated by blips of viral replication during the follow-up. This pattern of acute SIVrcm replication is similar to the patterns observed in infections with lymphotropic SIV strains in pathogenic or nonpathogenic infections (33, 57, 62). SIVrcm RNA in PBMCs peaked at day 10 (106 to 108 copies/106 PBMCs) and declined rapidly 10- to 100-fold by day 28 (Fig. 3b), and then at 4 months p.i., the RNA copy numbers in PBMCs varied from undetectable to 105 copies/106 cells (Fig. 3b). SIVrcm RNA became undetectable in PBMCs in all four PTMs after 240 days p.i. Since the gastrointestinal tract is known as a primary site of HIV/SIV replication, viral RNA loads in the intestine were also measured. Intestinal SIVrcm RNA loads paralleled those in blood but persisted longer and at high levels (104 to 106 SIVrcm RNA copies/106 cells). Intestinal SIVrcm RNA loads were still detectable at day 300 p.i. in all SIVrcm-infected PTMs (Fig. 3c). From there on, intestinal SIVrcm RNA loads became undetectable, with blips of viral replication. Finally, high levels of viral replication were detected in the LNs during both acute and chronic infection up to 180 days p.i., when VLs decreased below the limits of detection (Fig. 3d). Note that for both intestine and LN SIVrcm VLs, the sampling timing probably missed the peak of viral replication, which explains the lack of discernible VL peaks in both these tissues.
FIG. 3.
Dynamics of SIVrcm replication and burden in SIVrcm-infected PTMs. Viral replication was assessed by quantifying SIVrcm RNA loads in plasma (a), PBMCs (b), intestine (c), and LNs (d). The dynamics of proviral DNA burden was quantified in PBMCs (e). Plasma viral loads are expressed as SIVrcm RNA copies/ml of plasma; detection limit of the assay is 102 copies/ml. SIVrcm RNA/proviral DNA levels in tissues are expressed per 106 cells with a detection limit of 10 copies. Animal EC01, •; BU62, ▾; EC23, ▪; EC25, ♦.
We also measured the proviral SIVrcm DNA loads in PBMCs and showed that during the first 8 months of infection, they paralleled the dynamics of SIVrcm plasma VLs. Thus, the proviral DNA burden increased rapidly from day 7 to 10 p.i., reaching 105 to 106 SIVrcm DNA copies/106 PBMCs (Fig. 3e). Then, the postacute decline of proviral SIVrcm loads was of 2 to 3 logs (Fig. 3e). Proviral SIVrcm DNA loads were detectable at every time point of the follow-up, even after viral replication was controlled (Fig. 3e). This detection pattern is different from patterns observed in completely controlled SIV infections (58), suggesting that there is tissue reseeding with SIVrcm and thus ongoing cryptic replication below detection limits in SIVrcm-infected PTMs.
Changes in immune cell populations during SIVrcm infection of PTMs.
Surprisingly, immunophenotyping of immune cell populations did not show any significant changes of CCR2+ monocytes during acute or chronic SIVrcm infection of PTMs (Fig. 4a). Moreover, transient increases of the monocytes were observed during the acute SIVrcm infection in PTMs. The kinetics of CD4+ T cells in blood (Fig. 4b), LNs, and intestine (Fig. 4c) of PTMs showed that SIVrcm infection induced a transient but significant CD4+ T-cell depletion in periphery and a significant and more persistent CD4+ T-cell depletion in the intestine, suggesting that CD4+ T-cell lymphocytes are the major target cells supporting viral replication in SIVrcm-infected PTMs. Thus, in periphery, up to 64% of the CD4+ T cells were depleted during acute SIVrcm infection of PTMs (19.75% at day 28 p.i., compared to 35.75% ± 6.25% at the baseline). Although there was a significant (P < 0.05) CD4+ T-cell rebound in peripheral blood starting from day 28 p.i. (Fig. 4b), the CD4+ T-cell counts did not reach the preinfection levels during the follow-up, which is surprising in the context of control of viral replication at late time points. CD4+ T-cell depletion was even more pronounced in the intestine, where up to 80% of the total population of CD4+ T cells was depleted by 28 days p.i. in the lamina propria, and depletion of CD4+ T cells persisted up to day 300 p.i. (Fig. 4c). A significant immune restoration was then observed at later time points in the context of the control of viral replication. However, at the end of the follow-up, the restoration of intestinal CD4+ T cells was still partial, with only 60% of cells being restored (Fig. 4c). No significant changes in CD4+ T cells were observed in the LNs, with the exception of a moderate and transient depletion during the acute SIVrcm infection (Fig. 4d). Finally, expansion of CD8+ T cells was observed in SIVrcm-infected PTMs (Fig. 4e).
FIG. 4.
Impact of SIVrcm replication on major blood populations in PTMs. No change in CCR2+ monocyte population in periphery during acute or chronic SIVrcm infection of PTMs (a). Significant declines in CD4+ T cells in periphery (b), intestine (c), and LNs (d) were observed after the peak of viremia. Partial restoration of CD4+ T cells occurred at later time points. CD4+ T cells never reached preinfection levels. Expansion of CD8+ T cells was observed during chronic SIVrcm infection (e). Animal EC01, •; BU62, ▾; EC23, ▪; EC25, ♦.
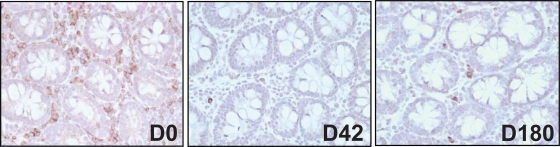
Flow cytometry data showing CD4+ T-cell depletion were further confirmed by immunohistochemistry (Fig. 5), which confirmed CD4+ T-cell depletion in the intestine during acute infection (42 day p.i.), followed by partial restoration during the chronic phase of infection (day 300 p.i.).
FIG. 5.
Immunohistochemical assessment of CD4+ T-cell depletion during SIVrcm infection of pigtailed macaques. Massive CD4+ T-cell depletion compared to baseline levels was observed after acute SIVrcm infection, followed by partial restoration during chronic infection. D, day.
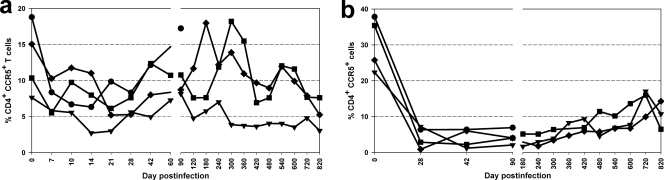
We then investigated the dynamics of specific CD4+ T-cell subsets during the course of SIVrcm infection in PTMs. Circulating CD4+ CCR5+ T cells in SIVrcm-infected PTMs dropped to significant levels during the acute phase of infection (i.e., day 14 to 21) (Fig. 6a) and recovered during the follow-up. At the end of the study, the recovery of peripheral CD4+ CCR5+ T cells was still incomplete. In the intestine, CCR5-expressing CD4+ T cells were markedly depleted within 28 days p.i. in all four animals, and restoration of this population was minimal during the course of infection (Fig. 6b).
FIG. 6.
Changes in CD4+ CCR5+ T cells in response to SIVrcm infection. Modest declines of CD4+ CCR5+ T cells in periphery (a) and a near total loss of CD4+ CCR5+ T cells in intestine (b) by day 28 p.i. Minimal restoration was noticed in intestine during the course of infection. Animal EC01, •; BU62, ▾; EC23, ▪; EC25, ♦.
Finally, we investigated the impact of SIVrcm infection on CD4+ T-cell subsets, and we observed a massive depletion of those CD4+ T cells expressing a memory phenotype in both periphery and intestine, of which most significant depletion (>90%) involved effector memory cells (Fig. 7). At later time points, a partial restoration of naive cells was observed in the intestine (Fig. 7b); however, restoration of memory CD4+ T cells was minimal (Fig. 7d and f). This observation is striking, as this pattern of SIVrcm infection is very similar to other lymphotropic viruses and not predicted by the in vitro data that showed an exclusive usage of the CCR2 coreceptor by SIVrcm.
FIG. 7.
Dynamics of CD4+ T-cell subsets in periphery and intestine during SIVrcm infection. Significant depletion with partial restoration of naïve CD4+ T cells (CD28+ CD95−) (a and b) was observed in periphery and intestine. Memory CD4+ T cells (CD28− CD95+) showed massive depletion (c to f), with the least restoration being observed for the effector memory CD4+ T-cell population in periphery as well as intestine of SIVrcm-infected PTMs (e and f). Animal EC01, •; BU62, ▾; EC23, ▪; EC25, ♦.
CCR2 mRNA expression.
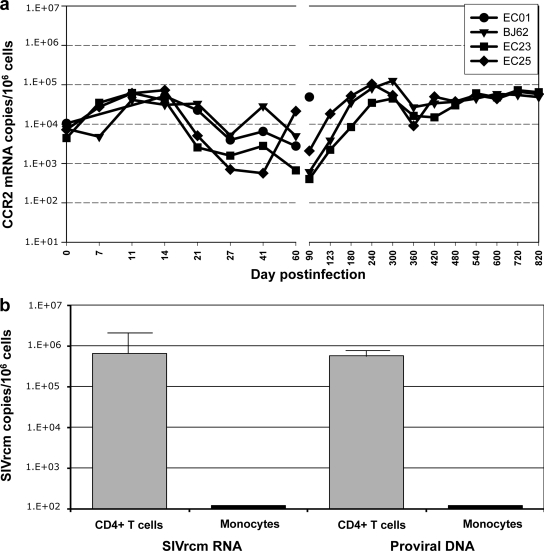
We further confirmed the flow cytometry data showing no major impact of SIVrcm replication on CCR2 expression by quantifying the dynamics of CCR2 mRNA expression in PBMCs. Although infection with a CCR2-tropic virus should have resulted in a CCR2+ cell depletion and thus in a decrease in CCR2 gene expression, we observed a slight increase in CCR2 mRNA expression in PBMCs during the acute SIVrcm infection. This upregulation in the PBMCs was rapid, marked, and sustained in the first 2 weeks of infection, paralleling the increases in immune activation, and was followed by transient downregulation up to day 90 p.i. (Fig. 8a). At later time points, CCR2 mRNA expression was at the preinfection levels in all animals.
FIG. 8.
Changes in the expression levels of CCR2 mRNA in PBMCs during SIVrcm infection of PTMs, as assessed by quantitative real-time PCR (a). The gene expression is shown as the absolute number of CCR2 mRNA copies per 106 cells. Quantification of SIVrcm RNA levels as well as that of proviral DNA showed that SIVrcm preferentially replicates in CD4+ T cells and not in monocytes (b).
SIVrcm replication in different cell subsets.
In order to confirm that CD4+ T cell depletion in SIVrcm-infected PTMs is a result of virus replication in lymphocytes and not in macrophages, we sorted CD4+ T cells and CD14+ monocytes from PBMCs collected at day 21 p.i. and quantified both SIVrcm RNA and proviral DNA in these cells. As shown in Fig. 8b, the bulk of viral replication occurred in CD4+ T cells, with negligible levels detected in monocytes. Note, however, that the lack of virus detection in monocytes may be due to the relatively small number of sorted cells and that monocyte infectivity is not predictive for that of tissue macrophages.
Coreceptor studies.
All the results presented thus far suggest that SIVrcm does not specifically deplete CCR2 monocytes in vivo (Fig. 4a), which is in striking contrast to what is expected for CCR2 coreceptor usage in vitro by SIVrcm (7, 13, 76). Moreover, our results suggest that SIVrcm specifically depletes CD4+ T cells in vivo, which would imply that the replicative virus has an expanded tropism to include lymphocytes. Such a property could only be achieved if the virus is able to use a broader range of coreceptors. We therefore assessed the coreceptor usage of the SIVrcm strains isolated from PBMCs of PTMs collected at day 10 p.i. Our results confirmed previous observations that SIVrcm uses CCR2 and not CCR5 as its major coreceptor (7, 13, 76) (Fig. 9). Consistent with the previous reports, SIVrcm also used Bonzo/STRL33 and CX3CR1 (V28) efficiently (76). Interestingly, expansion of coreceptor usage to CCR4 usage was also observed for the viruses isolated from PTMs but not for the parental virus (Fig. 9). This finding was consistent for the four PTMs included in this study.
FIG. 9.
Coreceptor usage of SIVrcm isolated from acutely infected PTMs. Coreceptor usage of SIVrcm involves CCR2, as previously reported, but expanded to CCR4 in all four infected PTMs. SIVmac239 served as a control. Ag, antigen.
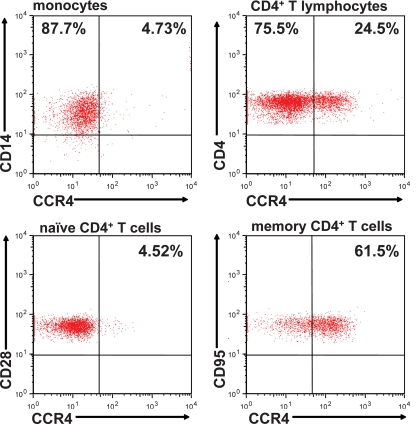
As we documented CCR4 coreceptor usage by SIVrcm, we assessed the expression of CCR4 on different cell populations in uninfected PTMs. CCR4 expression was abundant on CD4+ T cells and scarce on monocytes (Fig. 10). CCR4 expression varied among the CD4+ T-cell subsets, with low levels on naïve cells and high levels on memory cells (Fig. 10). Also, CCR4+ CD4+ T cells appeared to be significantly, albeit not preferentially, depleted during SIVrcm infection, as opposed to the minor CD4+ T-cell population that expresses CCR2, which was relatively well conserved during SIVrcm infection (data not shown).
FIG. 10.
Comparison of CCR4 expression on CD14+ monocytes and CD4+ T cells in blood from an uninfected PTM. CCR4 expression was significantly lower in CD14+ monocytes than in CD4+ T cells. Naïve CD4+ T cells (CD28+ CD95−) showed lower CCR4 expression while memory (CD95+) cells expressed significantly higher levels of CCR4 on their surfaces.
In vitro replication of SIVrcm from PTMs.
We next compared the replicative capacity and cytopathic properties of the parental SIVrcm and SIVrcm isolates from PBMCs of infected PTMs, and we infected different human T-cell lines with these viruses. SIVmac239 was tested in parallel and served as a control. Both parental SIVrcm and the SIVrcm strains isolated from PTMs showed similar replicative potentials on human cell lines (Table 1). All these strains replicated at high levels on MT-4, U937.p11, and CEMss cell lines. Moderate replication of SIVrcm was observed in MT-2 and H9 cells. In contrast, SIVrcm did not replicate in HUT78 or CEMx174 cells, compared with the relatively efficient replication of SIVmac239 (Table 1). Moreover, SIVrcm strains showed efficient replication in the human monocytic cell line U937.p11.
TABLE 1.
Cellular tropism of SIVrcm strains isolated from PTMs compared to that of parental SIVrcm and SIVmac
| Cell type | Replication potential by strain (source)a
|
|||
|---|---|---|---|---|
| SIVrcm (parental) | SIVrcm (PTM BJ62) | SIVrcm (PTM EC23) | SIVmac239 (parental) | |
| MT-2 | + | + | + | ++ |
| MT-4 | ++ | ++ | ++ | − |
| HUT78 | ± | − | ± | ++ |
| U937.p11 | ++ | ++ | ++ | − |
| CEMx174 | ± | ± | + | ++ |
| Molt4clone8 | + | ± | ++ | − |
| SupT1 | ± | − | ± | ± |
| H9 | + | + | + | ± |
| CEMss | + | ++ | ++ | ± |
+, replication; ++, efficient replication; −, partial/undetectable replication.
env sequence analysis.
In order to understand the mechanisms behind the rapid expansion of coreceptor usage by SIVrcm in PTMs, we cloned and sequenced the env genes of the SIVrcm strains isolated at serial time points from PTMs. The sequences of env genes were then compared to the sequence of the parental SIVrcmGB1 strain (GenBank accession no. AF382829). Although analysis of env sequences revealed accumulation of random mutations over the course of SIVrcm infection of PTMs, the strains from PTMs harbored 23 signature changes compared to the parental strain. The number of N-glycosylation sites was similar between parental SIVrcm strains and the strains isolated from PTMs; however, two glycosylation sites occurred at different positions in the SIVrcm isolates from PTMs. Two distinct populations were identified in the PTMs, and both were different from the parental strain (Fig. 11). Thus, the majority of SIVrcm isolates were very similar and formed a dominant population. Two strains (BJ62 D14 and EC25 D7) had a different profile in the 5′ part of the env gene and were similar to the parental virus. However, in the 3′ half of the env gene, these two sequences bore the same signature patterns as the dominant population. We therefore concluded that these two strains were recombinant. At those two time points, the sequence clones contained both the recombinant sequence and the dominant population, but in Fig. 11 only the recombinant sequences are shown. Altogether, sequence analysis strongly suggests that strain selection upon transmission rather than virus evolution may account for the coreceptor expansion of SIVrcm. Viral populations transmitted to PTMs were probably minor in the parental SIVrcm quasispecies as none of the clones amplified from the parental swarm harbored the signature patterns of the strains isolated from PTMs.
FIG. 11.
Alignment of envelope protein sequences from multiple clones of SIVrcm isolated from all four infected PTMs at different time points postinfection. PCR-derived env nucleotide sequences were translated and aligned with previously reported SIVrcm (NCBI database reference sequence AF382829). V1/V5 regions showed distinct changes in clones derived from infected PTMs compared to parental SIVrcm.
DISCUSSION
In this study we performed a systematic examination of the adaptation of an SIV to a new NHP species upon cross-species transmission. By infecting PTMs with SIVrcm that naturally infects RCMs and monitoring the virologic and immunologic parameters of this infection, we showed (i) that dynamics of viral replication and decline of CD4+ T cells during infection with the “CCR2-tropic” SIVrcm are similar to those observed in lymphotropic SIV infections and (ii) that viral adaptation to a new host occurs through strain selection and expansion of coreceptor usage. SIVrcm strains isolated from PTMs use CCR4 as the coreceptor for viral entry, in addition to CCR2, the only coreceptor used by the parental strain (7, 14, 76). Although our study was carried out on a limited number of PTMs, the results were highly consistent in all animals and therefore significant.
In order to study the mechanisms of SIV adaptation to a new host, SIVrcm is an ideal model because its coreceptor usage is unique among the different SIVs in that it uses CCR2B rather than CCR5 as its primary coreceptor for viral entry (7, 13, 76). This peculiarity is due to a deletion in the CCR5 gene that occurs in most of the RCMs in the wild and prevents CCR5 expression on the cell surface (13). Thus, SIVrcm is exemplary of SIV adaptation for persistence in NHP hosts, and one may expect that cross-species transmission of SIVrcm to new hosts that normally express chemokine receptors may result in a reversion of coreceptor usage as a mechanism of SIV adaptation for viral persistence.
A second reason for using SIVrcm in pathogenesis studies is that although there is no information available on the pathogenesis of SIVrcm in its natural host, the predictive tropism of the virus, based on the coreceptor usage, is on macrophages. Indeed, as illustrated in Fig. 1, CCR2 is mainly expressed on monocytes, and there is virtually no expression of this chemokine receptor on T lymphocytes. This putative macrophage tropism of the virus would have significant impact on the patterns of viral replication as monocytes/macrophages are long-lived cells compared with the short life span of lymphocytes (60). Note that previous in vivo studies employing SIVmac strains that were macrophage-tropic in vitro failed to confirm exclusive infection of macrophages in vivo (10).
However, PTM infection with SIVrcm resulted in a pattern of acute and chronic infection which was very similar to patterns observed in pathogenic and nonpathogenic infections with SIV lymphotropic strains, which were characterized by a peak of viral replication occurring at 10 days p.i., with a set point of viral replication established by day 42 p.i. and maintained for up to 240 days p.i., when the infection was controlled. This pattern of viral replication is characteristic for lentiviral infections in which the bulk of viral replication occurs in short-lived cells (23, 53, 75). In addition, the lack of significant depletion of the macrophage population, together with the significant loss of CD4+ T cells in intestine, and, to a lesser extent, in blood, was also highly suggestive of in vivo infection of lymphocytes. Depletion of memory CD4+ T cells and, to a lesser extent, of naïve CD4+ T cells from peripheral blood and intestine contributed to the overall depletion of CD4+ T cells. During the chronic infection, when viral replication was controlled, a partial immune restoration of the CD4+ T-cell population was observed, with memory CD4+ T cells being the least restored.
Altogether, these results suggest that, contrary to the in vitro predicted SIVrcm preferential tropism for macrophages due to CCR2 coreceptor usage, the in vivo replication of SIVrcm occurred in lymphocytes. This conclusion is supported by our sorting experiments, which demonstrated that, in peripheral blood, viral replication occurs in lymphocytes and not in monocytes. Note, however, that monocyte infectivity does not necessarily predict infectivity of activated tissue macrophages (66) and that limited availability of cells isolated from tissues, the lack of CD14 expression by tissue macrophages, and the lack of available reagents for SIVrcm detection in situ preclude a definitive conclusion with regard to SIVrcm ability to infect macrophages in vivo.
In order to understand the mechanism behind the ability of SIVrcm to infect CD4+ T cells, we have reisolated the virus from acutely infected PTMs and tested the coreceptor usage of these SIVrcm isolates. While this experiment confirmed that the SIVrcm isolates are using CCR2, as previously reported (7, 13, 76), the strains isolated from the four PTMs were also able to use CCR4. The usage of CCR4 occurred as an expansion of virus tropism as the parental SIVrcm strain exclusively used CCR2 as a coreceptor (Fig. 9). This expanded chemokine coreceptor tropism of SIVrcm isolates from PTMs could have promoted the dissemination of SIVrcm to T cells as CCR4 is mainly expressed on lymphocytes and only at a low level on monocytes/macrophages. Also, memory CD4+ T cells express CCR4 at higher levels than naïve cells, which explains the pattern of CD4+ T-cell depletion observed in our study. The use of CCR4 is unexpected, and very few HIV strains have been reported thus far to use this coreceptor (42). CCR4 binds two chemokines, thymus and activation-regulated chemokine (29) and macrophage-derived chemokine (30) that has a suppressive activity on HIV-1, independent from strain tropism (50), suggesting a role for CCR4 in HIV-1 entry (32). It is also being reported that CCR4+ T cells act at preferential sites of HIV replication and interact with other T cells to promote their activation for productive HIV infection (3). Note, however, that CCR4 expression on CD4+ T cells in peripheral blood and the intestine is lower than the overall magnitude of CD4+ T-cell depletion observed during SIVrcm infection. There are two potential explanations for this observation: first, it is possible that, similar to what was described for CCR5 expression on CD4+ T cells (40), CCR4 expression on CD4+ T cells is higher than detected by flow cytometry; second, in keeping with the extreme plasticity of SIVrcm, which adapted for the use of CCR2 and then, upon cross-species transmission, expanded the coreceptor usage to CCR4, one cannot exclude the possibility that, in spite of the in vitro coreceptor usage data showing the use of CCR2 and CCR4, the virus is actually using CCR5 as a coreceptor in vivo. This last hypothesis may be supported by the pattern of CD4+ T-cell subset depletion in the intestine which is very similar to that of other CCR5-tropic SIVs. Alternatively, the depletion of CD4+ T cells expressing CCR5 may reflect the depletion of activated CD4+ T cells, which were reported to express CCR5 (73). Finally, one should keep in mind that the coreceptor usage data are based on strains isolated early during the infection and that the virus might have evolved during the follow-up.
In order to understand the mechanism of this consistent rapid expansion of coreceptor usage of the SIVrcm strains in the PTMs, we have sequenced the envelopes of the viruses isolated from the PTMs at different time points during acute SIVrcm infection. Interestingly, sequence analysis revealed that although the PTMs were exposed to SIVrcm through the intravenous route, a stringent strain selection occurred during transmission. Thus, we were able to identify only two major virus populations in the SIVrcm-infected PTMs. The dominant population was very different from the major population in the swarm, with >23 substitutions in the Env proteins. These substitutions were consistently present in the strains isolated from the PTMs, suggesting that they did not occur randomly as point mutations but, rather, that they were present in the inoculum. Therefore, we hypothesize that the dominant SIVrcm population replicating in the PTMs preexisted in the transmitted quasispecies inocula and expanded upon transmission due to selective advantage in the new host. It is likely that this transmitted population accounted for a very limited number of strains in the original quasispecies as we did not amplify it from the parental swarm.
Therefore, our results identified a strain selection mechanism that contributes to evolutionary advantage for the cross-species-transmitted SIVs.
Previous studies identified SIVrcm as an example of a primate lentivirus that adapted its coreceptor usage to allow productive replication in a natural host resistant to infection with CCR5-tropic viruses (13, 76). Here, we report that upon cross-species transmission to a new host susceptible to various SIV infections, SIVrcm showed a remarkable plasticity and expanded coreceptor usage to infect new cell targets and induce successful cross-species transmission. This rapid adaptation occurred through effective strain selection rather than virus evolution. Our results thus demonstrate that lentiviruses can exploit numerous mechanisms in order to generate successful cross-species transmission, and our findings indicated the need for continuous monitoring of new cross-species transmission of SIVs to humans in central Africa. Finally, our results reinforce the value of in vivo studies for understanding HIV/SIV pathogenesis.
Acknowledgments
This work was supported by funds from grants R01 AI065325 and P20 RR020159 (C.A.) and RO1 AI064066 (I.P.) from the National Institute of Allergy and Infectious Diseases and from development grants G20 RR016930, RR018397, RR019628, RR013466, RR05169, C06 RR12112, and P51 RR000164 (TNPRC) from the National Center for Research Resources.
We thank Vanessa Hirsch, Petronela Ancuta, and Andrew Lackner for helpful discussions and Robin Rodriguez for help with manuscript preparation. We also thank the veterinary and animal care staff of TNPRC for their service and expertise.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Agy, M. B., L. R. Frumkin, L. Corey, R. W. Coombs, S. M. Wolinsky, J. Koehler, W. R. Morton, and M. G. Katze. 1992. Infection of Macaca nemestrina by human immunodeficiency virus type-1. Science 257103-106. [DOI] [PubMed] [Google Scholar]
- 2.Ambrose, Z., V. Boltz, S. Palmer, J. M. Coffin, S. H. Hughes, and V. N. Kewalramani. 2004. In vitro characterization of a simian immunodeficiency virus-human immunodeficiency virus (HIV) chimera expressing HIV type 1 reverse transcriptase to study antiviral resistance in pigtail macaques. J. Virol. 7813553-13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ancuta, P., P. Autissier, A. Wurcel, T. Zaman, D. Stone, and D. Gabuzda. 2006. CD16+ monocyte-derived macrophages activate resting T cells for HIV infection by producing CCR3 and CCR4 ligands. J. Immunol. 1765760-5771. [DOI] [PubMed] [Google Scholar]
- 4.Apetrei, C., A. Kaur, N. W. Lerche, M. Metzger, I. Pandrea, J. Hardcastle, S. Fakelstein, R. Bohm, J. Kohler, V. Traina-Dorge, T. Williams, S. Staprans, G. Plauche, R. S. Veazey, H. McClure, A. A. Lackner, B. Gormus, D. L. Robertson, and P. A. Marx. 2005. Molecular epidemiology of SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J. Virol. 798991-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apetrei, C., N. W. Lerche, I. Pandrea, B. Gormus, M. Metzger, G. Silvestri, A. Kaur, R. Bohm, D. L. Robertson, J. Hardcastle, A. A. Lackner, and P. A. Marx. 2006. Kuru experiments triggered the emergence of pathogenic SIVmac. AIDS 20317-321. [DOI] [PubMed] [Google Scholar]
- 6.Beer, B. E., C. R. Brown, S. Whitted, S. Goldstein, R. Goeken, R. Plishka, A. Buckler-White, and V. M. Hirsch. 2005. Immunodeficiency in the absence of high viral load in pig-tailed macaques infected with simian immunodeficiency virus SIVsun or SIVlhoest. J. Virol. 7914044-14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. St. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 7512014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibollet-Ruche, F., A. Galat-Luong, G. Cuny, P. Sarni-Manchado, G. Galat, J. P. Durand, X. Pourrut, and F. Veas. 1996. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J. Gen. Virol. 77773-781. [DOI] [PubMed] [Google Scholar]
- 9.Blois-Heulin, C., and B. Girona. 1999. Patterns of social visual attention in the red-capped mangabey (Cercocebus torquatus torquatus) in the context of food competition. Folia Primatol. (Basel) 70180-184. [DOI] [PubMed] [Google Scholar]
- 10.Borda, J. T., X. Alvarez, I. Kondova, P. Aye, M. A. Simon, R. C. Desrosiers, and A. A. Lackner. 2004. Cell tropism of simian immunodeficiency virus in culture is not predictive of in vivo tropism or pathogenesis. Am. J. Pathol. 1652111-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan, G., Y. Kozyrev, T. Kodama, and S. L. Hu. 2007. Novel TRIM5 isoforms expressed by Macaca nemestrina. J. Virol. 8112210-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carruth, L. M., M. C. Zink, P. M. Tarwater, M. D. Miller, M. Li, L. A. Queen, J. L. Mankowski, A. Shen, R. F. Siliciano, and J. E. Clements. 2005. SIV-specific T lymphocyte responses in PBMC and lymphoid tissues of SIV-infected pigtailed macaques during suppressive combination antiretroviral therapy. J. Med. Primatol. 34109-121. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Z., D. Kwon, Z. Jin, S. Monard, P. Telfer, M. S. Jones, C. Y. Lu, R. F. Aguilar, D. D. Ho, and P. A. Marx. 1998. Natural infection of a homozygous Δ24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J. Exp. Med. 1882057-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, Z., P. Zhou, D. D. Ho, N. R. Landau, and P. A. Marx. 1997. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J. Virol. 712705-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen, J. 2000. AIDS research. Vaccine studies stymied by shortage of animals. Science 287959-960. [DOI] [PubMed] [Google Scholar]
- 16.Fauci, A. S., M. I. Johnston, C. W. Dieffenbach, D. R. Burton, S. M. Hammer, J. A. Hoxie, M. Martin, J. Overbaugh, D. I. Watkins, A. Mahmoud, and W. C. Greene. 2008. HIV vaccine research: the way forward. Science 321530-532. [DOI] [PubMed] [Google Scholar]
- 17.Fultz, P. N., H. M. McClure, D. C. Anderson, and W. M. Switzer. 1989. Identification and biologic characterization of an acutely lethal variant of simian immunodeficiency virus from sooty mangabeys (SIV/SMM). AIDS Res. Hum. Retrovir. 5397-409. [DOI] [PubMed] [Google Scholar]
- 18.Gartner, S., Y. Liu, M. G. Lewis, V. Polonis, W. R. Elkins, P. M. Zack, J. Miao, E. A. Hunter, J. Greenhouse, and G. A. Eddy. 1994. HIV-1 infection in pigtailed macaques. AIDS Res. Hum. Retrovir. 10(Suppl. 2)S129-S133. [PubMed] [Google Scholar]
- 19.Gartner, S., Y. Liu, V. Polonis, M. G. Lewis, W. R. Elkins, E. A. Hunter, J. Miao, K. J. Corts, and G. A. Eddy. 1994. Adaptation of HIV-1 to pigtailed macaques. J. Med. Primatol. 23155-163. [DOI] [PubMed] [Google Scholar]
- 20.Gautam, R., A. C. Carter, N. Katz, I. F. Butler, M. Barnes, A. Hasegawa, M. Ratterree, G. Silvestri, P. A. Marx, V. M. Hirsch, I. Pandrea, and C. Apetrei. 2007. In vitro characterization of primary SIVsmm isolates belonging to different lineages. In vitro growth on rhesus macaque cells is not predictive for in vivo replication in rhesus macaques. Virology 362257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georges-Courbot, M. C., C. Y. Lu, M. Makuwa, P. Telfer, R. Onanga, G. Dubreuil, Z. Chen, S. M. Smith, A. Georges, F. Gao, B. H. Hahn, and P. A. Marx. 1998. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J. Virol. 72600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein, S., I. Ourmanov, C. R. Brown, R. Plishka, A. Buckler-White, R. Byrum, and V. M. Hirsch. 2005. Plateau levels of viremia correlate with the degree of CD4+-T-cell loss in simian immunodeficiency virus SIVagm-infected pigtailed macaques: variable pathogenicity of natural SIVagm isolates. J. Virol. 795153-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon, S. N., R. M. Dunham, J. C. Engram, J. Estes, Z. Wang, N. R. Klatt, M. Paiardini, I. V. Pandrea, C. Apetrei, D. L. Sodora, H. Y. Lee, A. T. Haase, M. D. Miller, A. Kaur, S. I. Staprans, A. S. Perelson, M. B. Feinberg, and G. Silvestri. 2008. Short-lived infected cells support virus replication in sooty mangabeys naturally infected with simian immunodeficiency virus: implications for AIDS pathogenesis. J. Virol. 823725-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gormus, B. J., L. N. Martin, and G. B. Baskin. 2004. A brief history of the discovery of natural simian immunodeficiency virus (SIV) infections in captive sooty mangabey monkeys. Front. Biosci. 9216-224. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch, V. M., G. Dapolito, P. R. Johnson, W. R. Elkins, W. T. London, R. J. Montali, S. Goldstein, and C. Brown. 1995. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J. Virol. 69955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirsch, V. M., G. A. Dapolito, S. Goldstein, H. McClure, P. Emau, P. N. Fultz, M. Isahakia, R. Lenroot, G. Myers, and P. R. Johnson. 1993. A distinct African lentivirus from Sykes' monkeys. J. Virol. 671517-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch, V. M., and P. R. Johnson. 1994. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 32183-203. [DOI] [PubMed] [Google Scholar]
- 28.Hu, S. L. 2005. Non-human primate models for AIDS vaccine research. Curr. Drug Targets Infect. Disord. 5193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imai, T., M. Baba, M. Nishimura, M. Kakizaki, S. Takagi, and O. Yoshie. 1997. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J. Biol. Chem. 27215036-15042. [DOI] [PubMed] [Google Scholar]
- 30.Imai, T., D. Chantry, C. J. Raport, C. L. Wood, M. Nishimura, R. Godiska, O. Yoshie, and P. W. Gray. 1998. Macrophage-derived chemokine is a functional ligand for the CC chemokine receptor 4. J. Biol. Chem. 2731764-1768. [DOI] [PubMed] [Google Scholar]
- 31.Jin, M. J., J. Rogers, J. E. Phillips-Conroy, J. S. Allan, R. C. Desrosiers, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J. Virol. 688454-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato, H., N. Tsuchiya, S. Izumi, M. Miyamasu, T. Nakajima, H. Kawasaki, K. Hirai, and K. Tokunaga. 1999. New variations of human CC-chemokine receptors CCR3 and CCR4. Genes Immun. 197-104. [DOI] [PubMed] [Google Scholar]
- 33.Kaur, A., R. M. Grant, R. E. Means, H. McClure, M. Feinberg, and R. P. Johnson. 1998. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J. Virol. 729597-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan, A. S., T. A. Galvin, L. J. Lowenstine, M. B. Jennings, M. B. Gardner, and C. E. Buckler. 1991. A highly divergent simian immunodeficiency virus (SIVstm) recovered from stored stump-tailed macaque tissues. J. Virol. 657061-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchhoff, F., S. Pohlmann, M. Hamacher, R. E. Means, T. Kraus, K. Uberla, and P. Di Marzio. 1997. Simian immunodeficiency virus variants with differential T-cell and macrophage tropism use CCR5 and an unidentified cofactor expressed in CEMx174 cells for efficient entry. J. Virol. 716509-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling, B., R. S. Veazey, A. Luckay, C. Penedo, K. Xu, J. D. Lifson, and P. A. Marx. 2002. SIV(mac) pathogenesis in rhesus macaques of Chinese and Indian origin compared with primary HIV infections in humans. AIDS 161489-1496. [DOI] [PubMed] [Google Scholar]
- 37.Ling, B., R. S. Veazey, and P. A. Marx. 2008. Nonpathogenic CCR2-tropic SIVrcm after serial passage and its effect on SIVmac infection of Indian rhesus macaques. Virology 37938-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Looney, D. J., J. McClure, S. J. Kent, A. Radaelli, G. Kraus, A. Schmidt, K. Steffy, P. Greenberg, S. L. Hu, W. R. Morton, and F. Wong-Staal. 1998. A minimally replicative HIV-2 live-virus vaccine protects M. nemestrina from disease after HIV-2(287) challenge. Virology 242150-160. [DOI] [PubMed] [Google Scholar]
- 39.Luo, W., H. Yang, K. Rathbun, C. P. Pau, and C. Y. Ou. 2005. Detection of human immunodeficiency virus type 1 DNA in dried blood spots by a duplex real-time PCR assay. J. Clin. Microbiol. 431851-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattapallil, J. J., D. C. Douek, B. Hill, Y. Nishimura, M. Martin, and M. Roederer. 2005. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature 4341093-1097. [DOI] [PubMed] [Google Scholar]
- 41.McClure, H. M., D. C. Anderson, P. N. Fultz, A. A. Ansari, E. Lockwood, and A. Brodie. 1989. Spectrum of disease in macaque monkeys chronically infected with SIV/SMM. Vet. Immunol. Immunopathol. 2113-24. [DOI] [PubMed] [Google Scholar]
- 42.McKnight, A., M. T. Dittmar, J. Moniz-Periera, K. Ariyoshi, J. D. Reeves, S. Hibbitts, D. Whitby, E. Aarons, A. E. Proudfoot, H. Whittle, and P. R. Clapham. 1998. A broad range of chemokine receptors are used by primary isolates of human immunodeficiency virus type 2 as coreceptors with CD4. J. Virol. 724065-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan, C., M. Marthas, C. Miller, A. Duerr, C. Cheng-Mayer, R. Desrosiers, J. Flores, N. Haigwood, S. L. Hu, R. P. Johnson, J. Lifson, D. Montefiori, J. Moore, M. Robert-Guroff, H. Robinson, S. Self, and L. Corey. 2008. The use of nonhuman primate models in HIV vaccine development. PLoS Med. 5e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphey-Corb, M., L. N. Martin, S. R. Rangan, G. B. Baskin, B. J. Gormus, R. H. Wolf, W. A. Andes, M. West, and R. C. Montelaro. 1986. Isolation of an HTLV-III-related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature 321435-437. [DOI] [PubMed] [Google Scholar]
- 45.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 46.Newman, R. M., L. Hall, A. Kirmaier, L. A. Pozzi, E. Pery, M. Farzan, S. P. O'Neil, and W. Johnson. 2008. Evolution of a TRIM5-CypA splice isoform in Old World monkeys. PLoS Pathog. 4e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Connor, D. H., T. M. Allen, and D. I. Watkins. 2001. Where have all the monkeys gone? Evaluating SIV-specific CTL in the post-Mamu-A*01 era. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 48.O'Neil, S. P., S. P. Mossman, D. H. Maul, and E. A. Hoover. 1999. Virus threshold determines disease in SIVsmmPBj14-infected macaques. AIDS Res. Hum. Retrovir. 15183-194. [DOI] [PubMed] [Google Scholar]
- 49.Osterhaus, A. D., N. Pedersen, G. van Amerongen, M. T. Frankenhuis, M. Marthas, E. Reay, T. M. Rose, J. Pamungkas, and M. L. Bosch. 1999. Isolation and partial characterization of a lentivirus from talapoin monkeys (Myopithecus talapoin). Virology 260116-124. [DOI] [PubMed] [Google Scholar]
- 50.Pal, R., A. Garzino-Demo, P. D. Markham, J. Burns, M. Brown, R. C. Gallo, and A. L. DeVico. 1997. Inhibition of HIV-1 infection by the beta-chemokine MDC. Science 278695-698. [DOI] [PubMed] [Google Scholar]
- 51.Pandrea, I., C. Apetrei, J. Dufour, N. Dillon, J. Barbercheck, M. Metzger, B. Jacquelin, R. Bohm, P. A. Marx, F. Barre-Sinoussi, V. M. Hirsch, M. C. Muller-Trutwin, A. A. Lackner, and R. S. Veazey. 2006. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J. Virol. 804858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandrea, I., C. Apetrei, S. Gordon, J. Barbercheck, J. Dufour, R. Bohm, B. Sumpter, P. Roques, P. A. Marx, V. M. Hirsch, A. Kaur, A. A. Lackner, R. S. Veazey, and G. Silvestri. 2007. Paucity of CD4+ CCR5+ T cells is a typical feature of natural SIV hosts. Blood 1091069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandrea, I., T. Gaufin, J. M. Brenchley, R. Gautam, C. Monjure, A. Gautam, C. Coleman, A. A. Lackner, R. M. Ribeiro, D. C. Douek, and C. Apetrei. 2008. Cutting edge: experimentally induced immune activation in natural hosts of simian immunodeficiency virus induces significant increases in viral replication and CD4+ T cell depletion. J. Immunol. 1816687-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pandrea, I., C. Kornfeld, M. J. Ploquin, C. Apetrei, A. Faye, P. Rouquet, P. Roques, F. Simon, F. Barre-Sinoussi, M. C. Muller-Trutwin, and O. M. Diop. 2005. Impact of viral factors on very early in vivo replication profiles in simian immunodeficiency virus SIVagm-infected African green monkeys. J. Virol. 796249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pandrea, I., R. Onanga, C. Kornfeld, P. Rouquet, O. Bourry, S. Clifford, P. T. Telfer, K. Abernethy, L. T. White, P. Ngari, M. Muller-Trutwin, P. Roques, P. A. Marx, F. Simon, and C. Apetrei. 2003. High levels of SIVmnd-1 replication in chronically infected Mandrillus sphinx. Virology 317119-127. [DOI] [PubMed] [Google Scholar]
- 56.Pandrea, I., R. M. Ribeiro, R. Gautam, T. Gaufin, M. Pattison, M. Barnes, C. Monjure, C. Stoulig, J. Dufour, W. Cyprian, G. Silvestri, M. D. Miller, A. S. Perelson, and C. Apetrei. 2008. Simian immunodeficiency virus SIVagm dynamics in African green monkeys. J. Virol. 823713-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pandrea, I., G. Silvestri, R. Onanga, R. S. Veazey, P. A. Marx, V. Hirsch, and C. Apetrei. 2006. Simian immunodeficiency viruses replication dynamics in African non-human primate hosts: common patterns and species-specific differences. J. Med. Primatol. 35194-201. [DOI] [PubMed] [Google Scholar]
- 58.Pandrea, I. V., R. Gautam, R. M. Ribeiro, J. M. Brenchley, I. F. Butler, M. Pattison, T. Rasmussen, P. A. Marx, G. Silvestri, A. A. Lackner, A. S. Perelson, D. C. Douek, R. S. Veazey, and C. Apetrei. 2007. Acute loss of intestinal CD4+ T cells is not predictive of simian immunodeficiency virus virulence. J. Immunol. 1793035-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 2711582-1586. [DOI] [PubMed] [Google Scholar]
- 61.Reimann, K. A., R. A. Parker, M. S. Seaman, K. Beaudry, M. Beddall, L. Peterson, K. C. Williams, R. S. Veazey, D. C. Montefiori, J. R. Mascola, G. J. Nabel, and N. L. Letvin. 2005. Pathogenicity of simian-human immunodeficiency virus SHIV-89.6P and SIVmac is attenuated in cynomolgus macaques and associated with early T-lymphocyte responses. J. Virol. 798878-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Silvestri, G., A. Fedanov, S. Germon, N. Kozyr, W. J. Kaiser, D. A. Garber, H. McClure, M. B. Feinberg, and S. I. Staprans. 2005. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J. Virol. 794043-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simon, F., S. Souquiere, F. Damond, A. Kfutwah, M. Makuwa, E. Leroy, P. Rouquet, J. L. Berthier, J. Rigoulet, A. Lecu, P. T. Telfer, I. Pandrea, J. C. Plantier, F. Barre-Sinoussi, P. Roques, M. C. Muller-Trutwin, and C. Apetrei. 2001. Synthetic peptide strategy for the detection of and discrimination among highly divergent primate lentiviruses. AIDS Res. Hum. Retrovir. 17937-952. [DOI] [PubMed] [Google Scholar]
- 64.Smith, S. M., M. Makuwa, F. Lee, A. Gettie, C. Russo, and P. A. Marx. 1998. SIVrcm infection of macaques. J. Med. Primatol. 2794-98. [DOI] [PubMed] [Google Scholar]
- 65.Staprans, S. I., and M. B. Feinberg. 2004. The roles of nonhuman primates in the preclinical evaluation of candidate AIDS vaccines. Expert Rev. Vaccines 3S5-S32. [DOI] [PubMed] [Google Scholar]
- 66.Stebbing, J., B. Gazzard, and D. C. Douek. 2004. Where does HIV live? N. Engl. J. Med. 3501872-1880. [DOI] [PubMed] [Google Scholar]
- 67.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427848-853. [DOI] [PubMed] [Google Scholar]
- 68.Takehisa, J., Y. Harada, N. Ndembi, I. Mboudjeka, Y. Taniguchi, C. Ngansop, S. Kuate, L. Zekeng, K. Ibuki, T. Shimada, B. Bikandou, Y. Yamaguchi-Kabata, T. Miura, M. Ikeda, H. Ichimura, L. Kaptue, and M. Hayami. 2001. Natural infection of wild-born mandrills (Mandrillus sphinx) with two different types of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 171143-1154. [DOI] [PubMed] [Google Scholar]
- 69.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uberla, K. 2005. Efficacy of AIDS vaccine strategies in nonhuman primates. Med. Microbiol. Immunol. 194201-206. [DOI] [PubMed] [Google Scholar]
- 71.VandeWoude, S., and C. Apetrei. 2006. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin. Microbiol. Rev. 1972-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Rensburg, E. J., S. Engelbrecht, J. Mwenda, J. D. Laten, B. A. Robson, T. Stander, and G. K. Chege. 1998. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J. Gen. Virol. 791809-1814. [DOI] [PubMed] [Google Scholar]
- 73.Veazey, R. S., K. G. Mansfield, I. C. Tham, A. C. Carville, D. E. Shvetz, A. E. Forand, and A. A. Lackner. 2000. Dynamics of CCR5 expression by CD4+ T cells in lymphoid tissues during simian immunodeficiency virus infection. J. Virol. 7411001-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watkins, D. I., D. R. Burton, E. G. Kallas, J. P. Moore, and W. C. Koff. 2008. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 14617-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, L., P. J. Dailey, T. He, A. Gettie, S. Bonhoeffer, A. S. Perelson, and D. D. Ho. 1999. Rapid clearance of simian immunodeficiency virus particles from plasma of rhesus macaques. J. Virol. 73855-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 746893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]