Abstract
Rabies virus infection induces the formation of cytoplasmic inclusion bodies that resemble Negri bodies found in the cytoplasm of some infected nerve cells. We have studied the morphogenesis and the role of these Negri body-like structures (NBLs) during viral infection. The results indicate that these spherical structures (one or two per cell in the initial stage of infection), composed of the viral N and P proteins, grow during the virus cycle before appearing as smaller structures at late stages of infection. We have shown that the microtubule network is not necessary for the formation of these inclusion bodies but is involved in their dynamics. In contrast, the actin network does not play any detectable role in these processes. These inclusion bodies contain Hsp70 and ubiquitinylated proteins, but they are not misfolded protein aggregates. NBLs, in fact, appear to be functional structures involved in the viral life cycle. Specifically, using in situ fluorescent hybridization techniques, we show that all viral RNAs (genome, antigenome, and every mRNA) are located inside the inclusion bodies. Significantly, short-term RNA labeling in the presence of BrUTP strongly suggests that the NBLs are the sites where viral transcription and replication take place.
Viral infection can cause extensive cellular rearrangements leading to the formation of cytoplasmic inclusions that concentrate viral components. These inclusions are often considered as side-products of the infectious process due to the passive accumulation of large quantities of proteins produced in excess during infection. Nevertheless, in some cases, it has been demonstrated that these inclusions are in fact viral factories in which essential events of the viral cycle take place (30, 38). Indeed, in these compartments, the high concentration of the viral components increases the efficiency of many steps of the viral infection (including transcription and/or replication of the genome and assembly of subviral particles) such that they represent “viral factories.” Such factories have been identified for a variety of unrelated viruses, including large DNA viruses (such as Poxviridae, Iridoviridae, and the closely related African swine fever virus), small DNA viruses (such as adenovirus and poliomavirus), double-stranded RNA viruses (such as rotaviruses), and positive-stranded RNA viruses (such as togavirus) (39).
Similar inclusions called aggresomes are formed in response to protein misfolding (7, 20). Proteins aggregates of misfolded proteins are toxic to cells and are driven along microtubules to aggresomes for immobilization and subsequent degradation by proteasomes. The similarity between aggresomes and viral inclusions raises the possibility that viruses hijack the aggresome pathway to facilitate their own replication and assembly (14, 39). Alternatively, aggresomes may be part of the innate cellular response that recognizes viral components as foreign or misfolded and targets them for storage and degradation.
Rabies virus, the prototype of the lyssavirus genus that belongs to the Rhabdoviridae family (Mononegavirales order), causes a fatal disease associated with intense viral replication in the central nervous system. Its single-stranded negative-sense RNA genome (∼12 kb), which encodes five viral proteins, is encapsidated by the nucleoprotein N (50 kDa) to form the nucleocapsid that is associated with the RNA-dependent RNA polymerase L (220 kDa) and its cofactor the phosphoprotein P (33 kDa). Inside the viral particle, this nucleocapsid has a tightly coiled helical structure that is associated with the matrix protein M (22 kDa) and surrounded by a membrane containing a unique glycoprotein G (62 kDa). The virus enters the host cell through the endosomal transport pathway via a low-pH-induced membrane fusion process catalyzed by glycoprotein G (9). The nucleocapsid released into the cytoplasm serves as a template for transcription and replication processes that are catalyzed by the L-P polymerase complex. During transcription, a positive-stranded leader RNA and five capped and polyadenylated mRNAs are synthesized. The replication process yields nucleocapsids containing full-length antigenome-sense RNA, which in turn serve as templates for the synthesis of genome-sense RNA. During their synthesis, both the nascent antigenome and the genome are encapsidated by N proteins. The neosynthesized genome either serves as a template for secondary transcription or is assembled with M proteins to allow budding of the neosynthesized virion at a cellular membrane. Some electron microscopy studies suggest that the budding step occurs at the level of internal cellular compartments (27, 28).
Rabies virus infection induces the formation of cytoplasmic inclusion bodies (IBs) that are called Negri bodies (29). Since these structures are typical of rabies infection of the brain, they have a diagnostic value and have been used as definite histological proof of such infection. These spherical structures having a diameter of a few micrometers (2 to 10 μm) are found in the cytoplasm of some infected nerve cells. By electron microscopy observations, the Negri bodies were found to be composed by a matrix of granular or filamentous material consisting of viral nucleoprotein. Viral particles were also seen budding from this structure (21, 22, 24). Cytological stainings have shown that they contain nucleic acids, suggesting that they may be replication complexes. However, RNA labeling (with [3H]thymidine or [3H]uridine) failed to detect the IBs, arguing against this conclusion (23). Rabies virus RNA was detected by in situ hybridization in human brain, but no colocalization of viral RNA and Negri bodies have been reported (17, 18).
In cultured cells from a neuronal or nonneuronal origin, IBs are also formed during viral infection. Based on their size and shape, it has been considered that these inclusions might be Negri body-like structures (NBLs).
In the present study, we have shown that the IBs formed in infected cells do indeed have similar characteristics to those of Negri bodies. Using this model, we have been able to study the mechanisms involved in the formation of NBL structures in infected cells and to investigate the role of these IBs during viral infection, revealing that they may be factories where viral RNAs synthesis occurs.
MATERIALS AND METHODS
Cells and viruses.
Human neuroblastoma SK cells, human glioblastoma astrocytoma U373-MG cells and BSR cells cloned from BHK 21 (baby hamster kidney) were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum.
The CVS strain of rabies virus was grown in BSR cells. Virus titers were determined by standard plaque assays into BSR cells.
Antibodies and drugs.
The mouse polyclonal anti-P antibody has been described previously and the anti-P monoclonal antibodies (MAbs) 25C2 and 30F2 have been previously described (35). The rabbit polyclonal anti-N antibody conjugated to fluorescein isothiocyanate (FITC) was kindly provided by Pierre Perrin (Institute Pasteur); anti-N MAb 62B5 was produced in mice immunized with the virus. Rabbit polyclonal anti-P antibody and mouse polyclonal anti-M were obtained by repeated injection of purified recombinant protein produced in Escherichia coli. Anti-G MAb 30AA5 has been previously described (8). Anti-HSp70 MAb (SPA-810) and anti-ubiquitin (SPA-203) were obtained from Stressgen. Anti-bromodeoxyuridine (anti-BrdU) MAb (clone B1C 9318) from Roche and anti-α tubulin MAb (N356) from Amersham were used. Fluorescent secondary antibodies were purchased from Molecular Probes, and anti-vimentin (V6630) and phalloidin (P1951) from Sigma. MitoTracker Red was obtained from Invitrogen (M7513). Nocodazole (M1404), paclitaxel (T1402), cytochalasin D (catalog no. 30385), latrunculin A (L5163), and actinomycin D (ActD; A9415) were obtained from Sigma; jasplakinolide (420107) was purchased from Calbiochem.
Plasmids.
The Plasmids pCDM8-P encoding the P protein (CVS strain) and pCDM8-N encoding the N protein (CVS strain) have been described previously (3). The plasmid pP-RFP has been generated by cloning first the P gene in fusion with the red fluorescent protein (RFP) gene in the plasmid pDsRedM (Clontech; gift of David Pasdeloup, MRC Virology Unit, Glasgow, United Kingdom), and then the fusion product P-RFP was inserted into pcDNA-3 vector between NheI and XbaI restriction sites. The plasmid pPΔ143AQ147A-RFP was obtained from the previous construct pCDM8 encoding pPΔ143AQ147A (33) by using the strategy described above. The dynamitin-green fluorescent protein (GFP)-expressing construct (p50-GFP) was a generous gift from Richard Vallee (Columbia University, New York, NY) (5).
Cell infection and transfection.
Monolayers of U373-MG cells or BSR cells were grown on sterile glass coverslip in six-well plates (from 50 to 80% confluence) and infected at different multiplicity of infection (MOI) of rabies virus (CVS strain) and at different times postinfection (p.i.).
U373-MG cells or BSR cells were grown on sterile glass coverslip in six-well plates (from 50 to 80% confluence) and were transfected with 2.5 μg or 5 μg of plasmid DNA by the calcium phosphate coprecipitation procedure. For N and P coexpression, the T7 vaccinia virus system was used as described previously (3).
Treatments of cells with drugs.
Cells were kept in Dulbecco modified Eagle medium containing 2 μM nocodazole, 1.25 nM paclitaxel, 2.5 μM cytochalasin D, 1 μM latrunculin A, or 0.5 μM jasplakinolide for 1 h before virus inoculation and during infection. Since all drugs were reconstituted in dimethyl sulfoxide (DMSO), untreated cells were maintained in medium containing the same concentration of DMSO. Cells were fixed and processed for microscopy.
Immunofluorescence staining and confocal microscopy.
Cells monolayers grown on glass coverslip were fixed 20 min with 4% (wt/vol) paraformaldehyde in phosphate-buffered saline (PBS) and permeabilized for 5 min in 0.1% Triton X-100 in PBS. Cells were then prepared for double-immunofluorescence staining and analyzed by confocal microscopy. The intracellular distribution of viral antigen P, N, M, and G were analyzed by using the specific antibody at the dilution of 1/500 to 1/1,000, followed by incubation with the corresponding immunoglobulin G (IgG) antibody conjugated to Alexa 568 or Alexa 488 at the dilution of 1/1,000. The cells were mounted onto glass slides by using Immu-Mount (Shandon) containing DAPI (4′,6′-diamidino-2-phenylindole) to stain nuclei.
Confocal laser microscopy was performed on a Leica SP2 microscope (×63 oil-immersion objective lens) using UV excitation at 351 nm (DAPI) and blue laser excitation at 488 nm (Alexa 488) and green laser excitation at 545 nm (Alexa 568) in sequential recording mode.
Sellers' staining.
Infected cells grown on coverslips were stained with a mixture of saturated solution of basic fuschin and methylene blue dissolved in methanol as described by Sellers in 1927 (40). Cells were fixed and permeabilized with methanol at −20°C, dipped into the staining solution for several seconds, and then washed with water. Cells were observed under a light microscope.
Quantification of Negri body-like structures.
BSR cells were infected with CVS at different MOIs (0.5, 1, 3, 5, and 10) in the presence or not of nocodazole (2 μM). Cells were fixed and permeabilized at different times p.i. (4 to 16 h). The viral N protein was stained by a rabbit polyclonal anti-N conjugated to FITC. Quantification of IBs was performed by counting the viral inclusions (containing the N protein) present in infected cells. An average of 400 cells from three independent experiments were counted at each time p.i. and for each MOI. The results are expressed as a percentage of uninfected cells and cell containing one, two, three, four, and more IBs.
Poisson distribution predicts that the relative frequency (p) of one cell to be infected by n virus at an MOI of λ is p (n) = (e−λ·λn)/n!.
FISH.
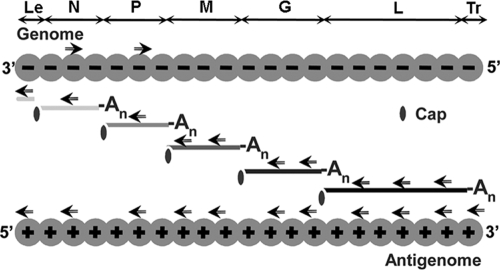
Fluorescence in situ hybridization (FISH) was performed by using 3′ biotinylated oligonucleotides (biotin 569.61, obtained from Eurogentec) (42 nucleotides) complementary to specific sequences of viral RNA (Table 1 and Fig. 1). According to the amount of viral RNA expected to be present during rabies virus infection, the number of probes mixed during hybridization was different: the NmRNA and PmRNA were detected by using one probe, the MmRNA and GmRNA were detected by using a mixture of two probes (M1 and M2 or G1 and G2), the LmRNA was detected by using three probes (L1, L2, and L3), and the genome was detected by using a mixture of two probes. The probe used to detect the antigenome was designed in order to avoid the detection of the mRNA, whereas the probe used to detect the mRNA hybridizes also to the antigenome.
TABLE 1.
Sequences of the probes used in the FISH experimentsa
| FISH probe | Sequence (5′-3′ biotinylated) | First nucleotide positionb |
|---|---|---|
| Leader RNA | GCTTTGCAATTGACGCTGTCTTTTTCTTCTTTGATCTGGTTG | 9 |
| N mRNA | CACCTGATTATTGACTTTGAACACAATCTTGTCGGCATCCAT | 71 |
| P mRNA | TATTTGCACTCCAACATTATCCAGGTACGATTGGAACAGGAG | 1784 |
| M1 mRNA | AGGAGAGGGCTTTTGAGTGTCCTCATCCCTACATTTTTTCAC | 2517 |
| M2 mRNA | CATAGTTGACATGCCCTCGAGTTCGAGTTGATACACCAGATC | 2996 |
| G1 mRNA | GCATCCTTCGTCCTCCACTACCAAATTGTTTGGGCAGCTGAG | 3438 |
| G2 mRNA | GGATCTCATTCCAAGTTCTGACTGACTTGTAGTGAGCATCGG | 4348 |
| L1 mRNA | TGGGTCAATAGGGTCATCATAGACCTCTCCAGGATCGAGCAT | 5417 |
| L2 mRNA | GTACTGATACAGAAACTCTAGGATTGCCTCCTCCTGTTGCTC | 8820 |
| L3 mRNA | CTGTTATACAGGTCGTGAAGTCTTGCGAAGCTAGGGACGTCA | 11470 |
| N genome | ATGGATGCCGACAAGATTGTGTTCAAAGTCAATAATCAGGTG | 71 |
| P genome | CTCCTGTTCCAATCGTACCTGGATAATGTTGGAGTCCAAATA | 1784 |
| Antigenome | GAGAAAAACAATCTCACACCAGAGGTTCGGATTCAAGATCTT | 11862 |
| Poly-A | d(T)x42 | |
| M mRNA VSV | CCTAATTTCTTAGATTTCTTAACCTTTCCCCTTCAGACCGAG | 2271 |
| GADPH mRNA | TGGCCAAGGTCATCCATGACAACTTTGGCATTGTGGAAGG |
The name of the probe corresponds to the viral RNA detected (the probe N mRNA is used to detect the N mRNA).
Nucleotides are numbered according to their distance from the 5′ end of rabies virus and VSV virus antigenome (positive strand).
FIG. 1.
Rabies virus RNA species (genome, antigenome, and mRNAs encoding N, P, M, G, and L proteins) and location of sense and antisense primers (indicated by arrows) used in the FISH experiments. Abbreviations: Le, leader; Tr, trailer; An, poly(A).
Infected cells grown on coverslips were fixed with paraformaldehyde and permeabilized as described above. After treatment with 200 U of DNase I (RNase-free D7201; Sigma)/ml, cells were prehybridized in hybridization buffer (50% formamide, 10% dextran sulfate, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) for 30 min at 37°C before being incubated with 10 μl of hybridization mix containing 10 ng of biotinylated probe, 10 μg of herring testis DNA, and 8.5 μl of hybridization buffer for 2 min at 95°C in presence or not of 15 U of RNase A (catalog no. 10109142001; Roche)/ml. Coverslips were then put in a humidified hybridization chamber at 37°C overnight and washed three times at 42°C for 5 min with 50% formamide-2× SSC and then three times at 42°C with 0.5× SSC. Cells were then processed for immunofluorescence staining. The N protein was stained with a mouse MAb anti-N (62B5) antibody, followed by incubation with Alexa 488-goat anti-mouse IgG. Viral biotinylated RNA were detected by incubation with streptavidin conjugated to cyanin 3 (Cy3; catalog no. 01 61 600 81; Jackson Immunotech).
BrUTP labeling of viral RNA.
BrUTP incorporation of newly synthezised viral RNA was performed as described by Georgi et al. (11). Briefly, the infected cells (16 h p.i.) were treated with 15 μg of ActD/ml for 1 h and then transfected with BrUTP (B 7166; Sigma) at a final concentration of 10 mM by using Lipofectamine 2000 (Invitrogen) and maintained in the presence of ActD for 20 min. The cells were fixed and permeabilized as described above and processed for immunofluorescence. P protein was detected by using the rabbit polyclonal anti-P antibody, followed by incubation with Alexa 568-conjugated goat anti-rabbit IgG. The BrUTP-labeled RNAs were stained by using mouse MAb to BrdU at a dilution of 1/50, followed by the addition of goat anti-mouse Alexa 488-conjugated antibody.
Electron microscopy.
For structural analysis, infected BSR cells were fixed for 1 h at room temperature with 2% glutaraldehyde (Sigma) in 0.1 M cacodylate buffer (pH 7.4; Sigma). Cells were washed with 0.1 M sucrose (Sigma) in 0.1 M cacodylate buffer (pH 7.4). Cells were then postfixed for 1 h at room temperature with 1% aqueous osmium tetroxide (OsO4) and 1.5% potassium ferrocyanide (Sigma) in 0.1 M cacodylate buffer (pH 7.4) and stained with 2% uranyl acetate in water. The cells were then dehydrated in increasing concentrations of ethanol and embedded by Epon with 2,4,6-tris(dimethylaminomethyl) phenol (DMP30; Delta Microscopies). Polymerization was carried out for 48 h at 60°C. Ultrathin sections of Epon-embedded material were collected on copper palladium grids (200 mesh) and stored until use. These sections were stained with lead citrate (Delta Microscopies). Sections were examined with a Zeiss EM902 electron microscope operated at 80 kV, and images were acquired with a charge-coupled device camera (Megaview III) and analyzed with ITEM software (Eloïse, France; MIMA2 platform [INRA-CRJ]).
For immunogold detection of rabies virus protein, cells were fixed for 1 h at 4°C with 4% formaldehyde (Merck) in 0.1 M phosphate buffer (pH 7.3). During fixation, cells were scraped and centrifuged. Pellets were dehydrated in increasing concentrations of methanol and embedded in Lowicryl K4M (Chemische Werke Lowi) at a low temperature according to the method of Roth et al. (36). Polymerization was carried out for 5 days at −30°C under long-wavelength UV light (Phillips fluorescent tubes [TL 6W]). Ultrathin sections of Lowicryl-embedded material were collected on carbon-coated gold grids (200 mesh) and stored until use. For immunogold labeling, grids bearing Lowcryl thin sections of infected cells were first placed for 2 min over drops of bovine serum albumin (5% PBS). The grids were then floated for 2 h, at room temperature, on drops of affinity purified mouse monoclonal or polyclonal anti-rabies virus proteins antibodies (diluted 1/20 in PBS) or over normal mouse serum as control (diluted 1/20 in PBS). After 15-min washes with 3 drops of PBS, the grids were incubated for 30 min over drops of a 1/30 dilution in PBS of goat anti-mouse IgG conjugated to gold particles (10 nm in diameter; Biocell Research Laboratories). After 5-min passages on 3 drops of PBS, the grids were rapidly rinsed in jet-distilled water, air dried, and stained for 10 min with 5% aqueous uranyl acetate. Grids were observed with a Philips 400 transmission electron microscope, at 80 kV at magnifications of ×8,000 to 22,000.
RESULTS
Characterization of the IBs found in rabies virus-infected cells.
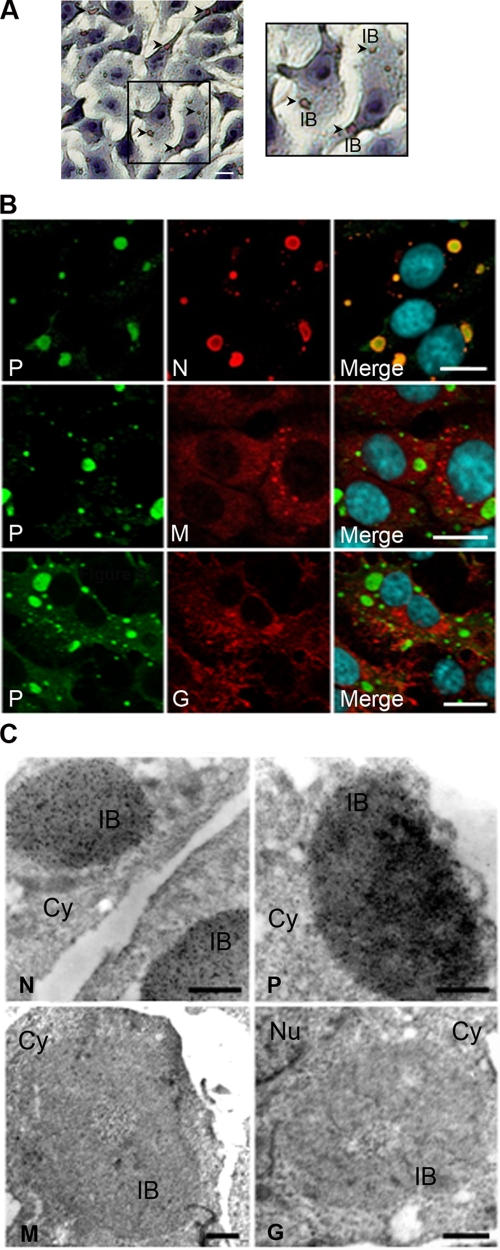
Rabies virus infection induces the formation of round IBs in the cytoplasm of neuronal and nonneuronal cells. We observed IBs in infected BSR cells (Fig. 2). Consistent with the known properties of Negri bodies, these inclusions could be stained with the Seller's solution containing the dye, basic fuschin (40) (Fig. 2A).
FIG. 2.
Characterization of the viral IBs. (A) BSR cells were infected with rabies virus (CVS strain) at an MOI of 3 for 16 h and stained by the Sellers' staining protocol as described in Materials and Methods. Viral IBs are indicated pointed by arrows (left panel). A higher magnification of an area is also presented (right panel). The scale bar corresponds to 12 μm. (B) BSR cells were infected as described above. After fixation and permeabilization, cells were processed for double-immunofluorescence staining with a rabbit polyclonal anti-P antibody (P) and the MAb 62B5 anti-N antibody (N), a mouse polyclonal anti-M antibody (M), or the MAb 30AA5 anti-G antibody (G). In all cases, DAPI (blue) was used to stain the nuclei (merge). Colocalization is apparent as yellow coloration in the merged panel. The scale bars correspond to 12 μm. (C) Infected BSR cells as described above were treated for immunogold labeling. P protein was detected by using two MAbs, 25C2 and 30F2; N protein was detected by using MAb 64B6; and M and G proteins were detected by using mouse polyclonal anti-M antibody and by anti-G MAb 30AA5, respectively. The scale bars correspond to 0.5 μm.
Unsurprisingly, confocal and electron microscopy analyses revealed that the IBs are full of N and P proteins (Fig. 2B and C). It has also been shown that the polymerase L is present in the IBs (6). In contrast and as expected, the M and G proteins were excluded from these structures, as indicated by the fact that IBs were neither labeled by a polyclonal serum against the M protein nor by an MAb directed against the G protein (Fig. 2B and C). Thus, the staining characteristics, the size (a few micrometers), and the shape, as well as the viral protein composition of the IBs formed in infected BSR cells, indicated that these structures share the defined features as Negri bodies and can thus be considered NBLs.
Morphogenesis of the NBLs during viral infection.
Little is known about the formation and evolution of Negri bodies during viral infection. Our model system permits us to perform such characterization. This is the first point that we have addressed.
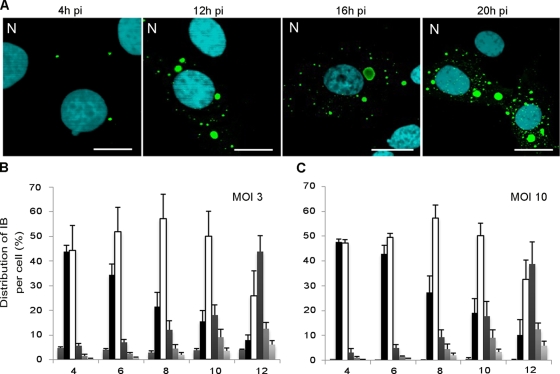
BSR cells or human neuroblastoma SK cells were infected at different MOIs (0.5 to 10) and analyzed at different times p.i. by immunofluorescence using anti-N or anti-P antibodies. At different times p.i., IBs in the cytoplasm of cells infected at different MOIs were counted. At an early stage of infection (4 h p.i.), infected cells presented one or two viral inclusions that then grew in size (10 to 12 h p.i.) (Fig. 3A). These values were observed regardless of the MOI used (between 3 and 10). Smaller IB structures appeared at later stage of infection (16 to 20 h p.i.) (Fig. 3A).
FIG. 3.
Evolution of the viral IBs during the viral cycle. (A) BSR cells were infected by CVS at an MOI of 3 for different times p.i. as indicated and stained with rabbit polyclonal anti-N conjugated to FITC antibody and DAPI. The scale bars correspond to 12 μm. (B and C) The numbers of IBs per cell were quantified at different times p.i. for an MOI of 3 (B) or an MOI of 10 (C). A histogram represents the average values from three independent experiments by counting 400 cells per experiment. The results are indicated as the percentages of cells that were uninfected or that contained one, two, three, four, and more IBs and are depicted by six variously shaded columns from left to right, respectively, for each time point. Error bars indicate the standard deviations.
The number of NBLs per cell was independent of the MOI and consequently was independent of the number of incoming virus per cell (Fig. 3B and C) that was expected to follow a Poisson distribution as mentioned in the Materials and Methods. These results suggest that there are a limited number of sites per cell that allow the formation of IBs.
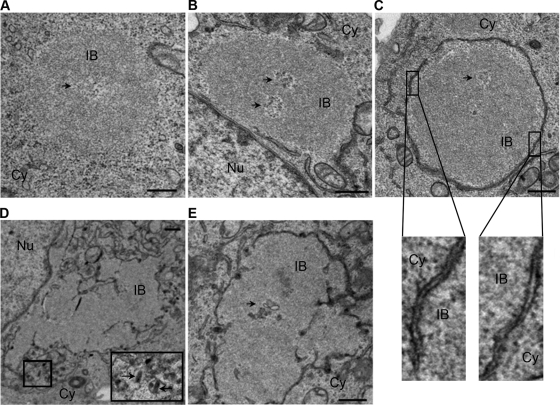
The morphogenesis and structural evolution of NBLs were also analyzed in infected BSR cells by electron microscopy (Fig. 4). NBLs are spherical structures (a few micrometers in size) that often contain a cavity. Although initially devoid of membranes (4 to 12 h p.i., Fig. 4A), NBLs progressively became associated with a double membrane (16 h p.i., Fig. 4B and C). The cytoplasmic granular surface of the membrane suggests that it might be derived from rough endoplasmic reticulum (Fig. 4C). Finally, NBLs were completely surrounded by these double membranes (16 to 24 h p.i.). This membrane organization is reminiscent of the organization of the double membrane around the cell nucleus. After 24 h p.i., the shape of the bodies appeared to be altered, and viral particles were also seen budding from some of these structures (Fig. 4D and E), most probably inside the lumen of the compartments that surround the bodies.
FIG. 4.
Morphology and evolution of viral IBs. Ultrastructural aspect of BSR cells infected for 12 h (A), 16 h (B and C), and 24 h (D and E) p.i. as indicated. Double fixation was performed with glutaraldehyde and osmium tetraoxide. Epon embedding and uranium-lead staining. IBs display a granular dense structure with a cavity (indicated by arrows in panels A, B, C, and E). Note the recruitment of membranes around IBs at 16 h p.i. (B and C) and the presence of virus particles close to the viral inclusion at 24 h p.i. (indicated by arrow in panel D). Nu, nucleus; Cy, cytoplasm. Bars correspond to 0.5 μm. Higher magnification (×13) of two membrane areas reveals a double-membrane with a granular surface are shown for panel C.
Comparison of the NBLs and aggresomes.
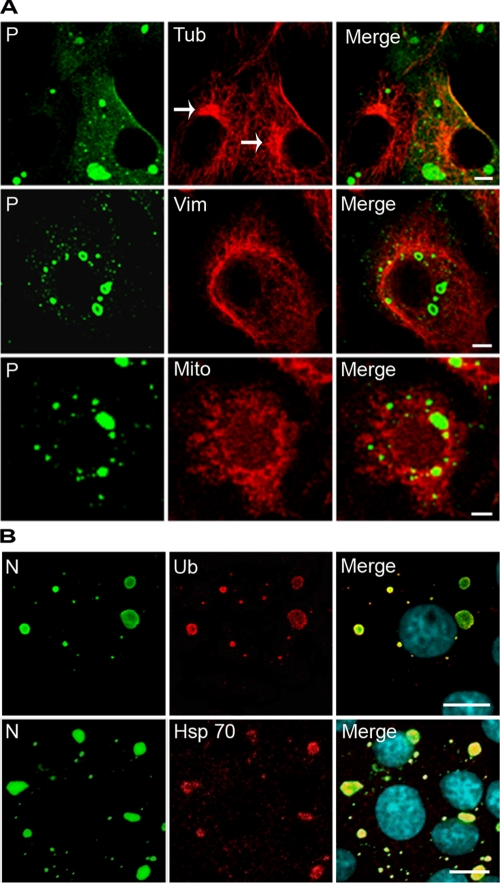
Aggresomes are misfolded protein aggregates that appear when the proteasome and chaperones can no longer deal with the aggregated proteins. They are driven on microtubules to the microtubule organizing center (MTOC), they are surrounded by vimentin cages to isolate and reduce the toxicity of misfolded proteins, and they recruit mitochondria that may provide the ATP required by chaperones and proteasomes for protein folding and degradation (7, 20). For many viral families, it has been proposed that the aggresome pathway is hijacked in order to concentrate viral components and to facilitate viral replication and assembly (14, 39). Therefore, we investigated whether NBLs share the characteristics of aggresomes described above. We observed that NBLs do not colocalize with the MTOC, as revealed by anti-α-tubulin staining (Fig. 5A). Costaining with anti-vimentin antibodies did not show any rearrangement of vimentin around the cytoplasmic inclusions at any stage of infection (Fig. 5A). Furthermore, we did not observe recruitment of mitochondria in close proximity to the NBLs (Fig. 5A). However, Hsp70 chaperone and ubiquitinylated proteins accumulate inside the IBs, as revealed by an anti-Hsp70 and anti-poly-ubiquitin staining, respectively (Fig. 5B). These results indicate that the NBLs do not share all of the features of aggresomes, although they contain Hsp70 and ubiquitinylated proteins.
FIG. 5.
Viral IBs exhibit some features of aggresomes but are distinct structures. (A) BSR cells were infected for 16 h and costained with a rabbit polyclonal anti-P antibody (P) and either mouse anti-α tubulin MAb (Tub), mouse anti-vimentin MAb (Vim), or the MitoTracker Red (Mito), followed by incubation with Alexa 488-goat anti-rabbit IgG and Alexa 568-goat anti-mouse IgG. Note that IBs are not localized at the MTOC (indicated by arrows) and that no vimentin cage or no recruitment of mitochondria (as would be expected for aggresomes) are observed around the IBs. (B) IBs contain Hsp70 and ubiquitinylated proteins. Infected BSR cells were immunostained with a rabbit polyclonal FITC-conjugated anti-N antibody (N) and a mouse MAb anti-ubiquitin antibody (Ub) or a mouse MAb anti-Hsp70 antibody (Hsp70), followed by incubation with Alexa 568-goat anti-mouse IgG. The scale bars correspond to 12 μm.
Role of the cytoskeleton in the morphogenesis of NBLs.
We next investigated the mechanism by which the different proteins gather and are recruited in NBL structures by analyzing the role of the cytoskeleton in the morphogenesis and evolution of the IBs.
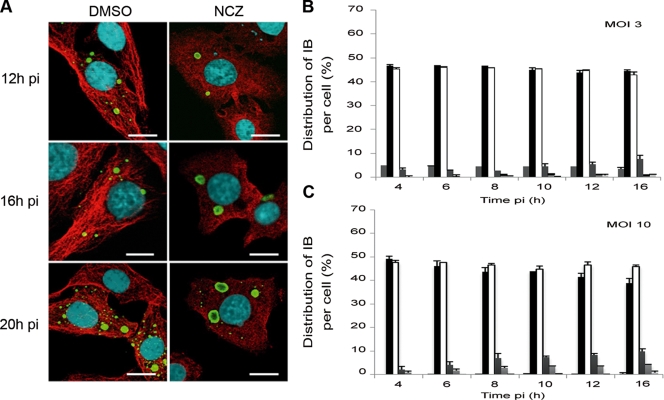
We studied the role of the microtubule network by using two different specific inhibitors of MT dynamics (nocodazole and paclitaxel). BSR or U373-MG infected cells (MOI = 3 and MOI = 10) were treated with nocodazole, which results in the disruption of microtubules, for 1 h before virus inoculation and during infection. In the presence of nocodazole, the “early” (large, one or two per cell) NBLs formed efficiently without any apparent delay (Fig. 6). These results indicate that microtubules are not needed for the IB formation.
FIG. 6.
Effect of nocodazole on formation and evolution of IBs during infection. (A) BSR cells were infected and treated with nocodazole (2 μM; NCZ) or mock treated (DMSO). At different times p.i. as indicated, the cells were costained with rabbit polyclonal anti-N conjugated to FITC antibody and with mouse MAb anti-α tubulin antibody, followed by incubation with Alexa 568-goat anti-mouse IgG. The scale bars correspond to 12 μm. (B and C) The numbers of IBs per cell were quantified in nocodazole-treated cells at different times p.i. as indicated for an MOI of 3 (B) or an MOI of 10 (C). A histogram represents the average values from three independent experiments by counting 400 cells per experiment. The results are expressed as the percentages of uninfected cells or cells containing one, two, three, four, and more IBs and are depicted by six variously shaded columns from left to right, respectively, for each time point. Error bars indicate the standard deviation.
In striking contrast, the formation of “late” (small) IBs was affected by nocodazole treatment since at later stages of infection (16 to 20 h p.i.), we observed NBLs that were much larger than those in the absence of the drug. Furthermore, at these late stages, no smaller structures were observed in nocodazole-treated cells (Fig. 6). This was in contrast to untreated cells where small IBs (more than 15 per cell) were detected. Thus, the drug inhibited the formation of additional IBs and/or the appearance of smaller structures.
We did not observe any effect of paclitaxel, a drug that stabilizes microtubules, on the formation and evolution of IBs (data not shown). This suggests that the mechanisms involved in these processes do not require dynamic microtubules.
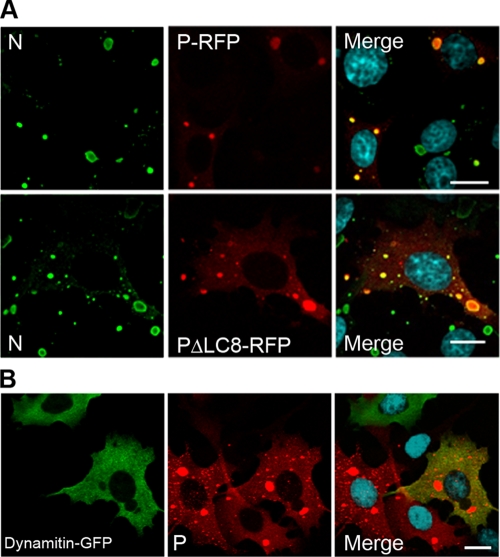
As P protein has been shown to interact with LC8, the light chain of the molecular motor dynein (19, 34), we checked whether this interaction plays a role in the targeting of the major P component to the viral inclusions. When cells were infected and transfected with the plasmid encoding the P protein mutant unable to interact with LC8 (PΔLC8-RFP) (33), RFP fluorescent viral inclusions were formed as in cells infected and transfected with the plasmid encoding the wild-type P-RFP protein (Fig. 7A). This indicates that LC8 is not required in the recruitment of P inside the IBs. This is consistent with the fact that overexpression of the inhibitor of dynein-mediated transport, dynamitin (dynamitin-GFP) during infection did not result in the inhibition of the formation of viral inclusions (Fig. 7B).
FIG. 7.
Formation of IBs does not require dynein-mediated transport. (A) BSR cells were transfected with a plasmid encoding the P-RFP or a P-RFP mutant that does not interact with LC8 (PΔLC8-RFP) and then infected for 24 h. Cells were stained with the MAb 62B5 anti-N antibody (N), followed by incubation with Alexa 488-goat anti-mouse IgG and DAPI (merge). The scale bars correspond to 12 μm. (B) BSR cells were transfected with a plasmid encoding dynamitin-GFP and then infected for 24 h. Cells were stained with a rabbit polyclonal anti-P antibody (P), followed by incubation with Alexa 568-goat anti-rabbit IgG and DAPI (merge). Colocalization is apparent as yellow coloration in the merged panel. The scale bars correspond to 12 μm.
The role of actin was also investigated in BSR cells or in U373-MG cells by using inhibitors resulting in depolymerization of actin (cytochalasin D and latrunculin A) or resulting in stabilization of actin (jasplakinolide). None of the inhibitors significantly affected the formation and evolution of NBLs, indicating that actin filaments are not required in these processes.
NBLs are potential sites of viral transcription and replication.
We then analyzed whether NBLs are functional structures where some essential steps of the virus cycle take place. Since the viral N, P, and L proteins involved in viral transcription and replication accumulate in IBs, we investigated the presence of viral RNA in these structures by using FISH. We therefore designed biotinylated oligonucleotide probes to detect specifically viral mRNAs, antigenomic RNA, and genomic RNA (Table 1 and Fig. 1).
After 16 h of infection, BSR cells were simultaneously prepared for FISH to detect the viral RNA and immunostained with anti-N antibody to visualize the Negri bodies.
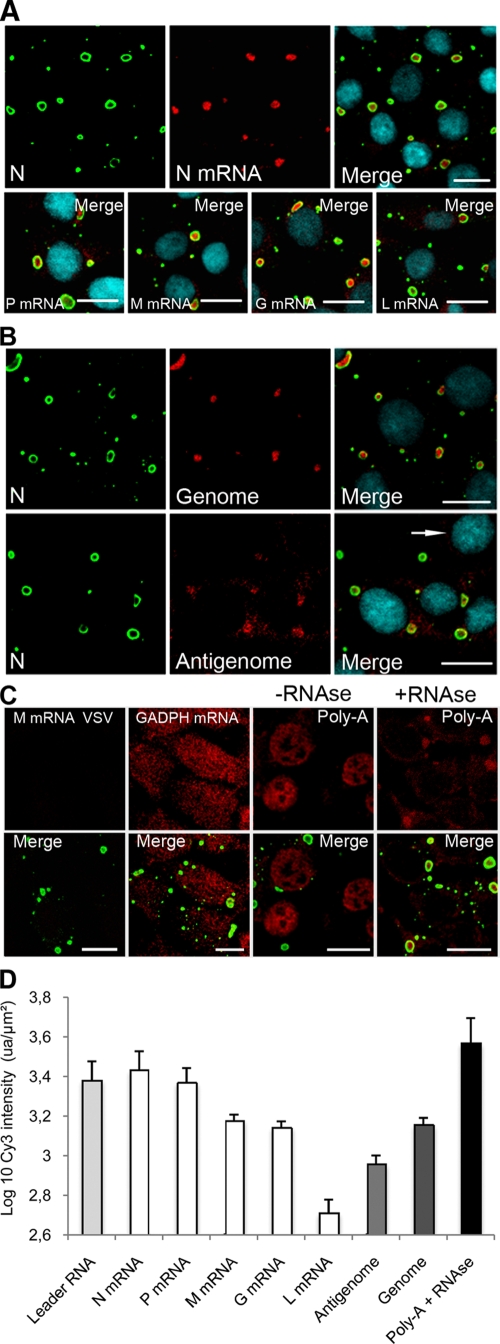
All viral mRNAs encoding the five viral proteins (N, P, M, G, and L) were detected inside the IBs with no viral mRNA readily detected elsewhere (i.e., nucleus/cytoplasm) outside of IBs (Fig. 8A). The relative quantities of the viral RNAs were determined (Fig. 8D). Although the FISH method does not allow a precise quantification, it clearly indicated that the N mRNA was more abundant than the L mRNA according to the known gradient of decreasing mRNAs concentration that reflects the gene order (N, P, M, G, and L) (1). These hybridizations were specific since no positive signal was observed in mock-infected cells (data not shown) or in infected cells treated with a probe detecting the mRNA of the M protein of vesicular stomatitis virus (VSV) (Fig. 8C). A positive control carried out in infected cells with a poly(dT) probe or a probe against the housekeeping GADPH mRNA gave rise to a signal diffusely distributed in the nucleus and the cytoplasm as expected (Fig. 8C). RNase A treatment of infected cells prior to FISH with poly(dT) probe prevented the hybridization signal in the nucleus and the cytoplasm but left intact the signal in the IBs (Fig. 8C). These results indicate that the viral mRNA accumulate inside the IBs and are protected against RNase digestion, most probably by the cage formed by the N and P proteins. Indeed, the N protein formed an apparent ringlike structure (cage) around the RNA (Fig. 2B and Fig. 8A).
FIG. 8.
Viral RNAs accumulate in viral IBs. BSR cells were infected (MOI = 3) for 16 h p.i. (A and B) FISH analyses were performed with biotinylated oligonucleotides (described in Materials and Methods), followed by incubation with streptavidin conjugated to Cy3 to detect NmRNA, PmRNA, MmRNA, GmRNA, and LmRNA as indicated (A) and to detect viral genomic and antigenomic RNA as indicated (B). Note that the probe used to detect the mRNA also allows the detection of the antigenome. In contrast, the probe used to detect the antigenome was designed in order to avoid the detection of the mRNA. Note that for the antigenomes, the photomultiplier tube (PMT) has been increased in order to clearly visualize the fluorescence signals resulting in an increase of the background signal. No positive signal was observed in mock-infected cells (indicated by an arrow). (C) FISH was also performed to detect the housekeeping GADPH mRNA or cellular mRNA (Poly-A) after treatment with RNase (+RNase) or without RNase (-RNase). No signal was observed with a control probe complementary to the MmRNA of VSV. In all cases, the N protein (N) was stained with the mouse MAb anti-N (62B5) antibody, followed by incubation with Alexa 488-goat anti-mouse IgG (A, B, and C). The scale bars correspond to 12 μm. (D) Relative quantification of the viral RNA accumulated inside the Negri bodies. FISH was performed as described above. Quantification of Cy3 fluorescent intensity (arbitrary units [ua]) was performed with the same PMT (the PMT adjusted with the N mRNA probe) on a Leica SP2 confocal microscope. The surface area and the Cy3 fluorescence intensity of each Negri body was calculated by using Leica confocal software. Two independent experiments were performed by counting an average of 300 IBs. The results were expressed as the log10 of Cy3's intensity per μm2 of Negri body and per probe (ua/μm2).
By using specific probes, the viral genomic RNA and antigenomic RNA were also detected exclusively in the IBs (Fig. 8B). The relative quantification of the viral RNAs indicated that the amount of genomic RNA is higher than the amount of antigenomic RNA (Fig. 8D).
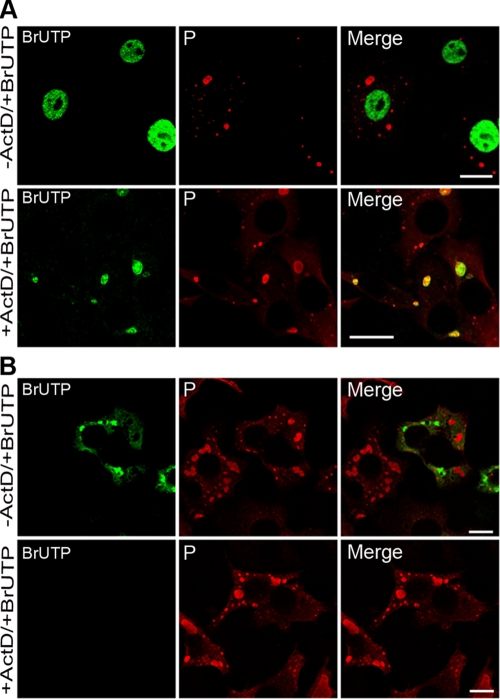
The experiments described above demonstrated that NBLs are structures where viral transcription and replication products accumulate. In order to establish whether the viral RNAs are synthesized inside IBs or synthesized outside IBs and then translocated into these structures, we performed short-term RNA labeling in the presence of BrUTP and ActD to inhibit cellular transcription. Cells were infected for 16 h before treatment without or with ActD for 1 h and transfected with BrUTP for 20 min. Cells were then examined by double-immunofluorescence staining using anti-BrdU and anti-P antibodies (Fig. 9). Without ActD, cellular RNA was mainly detected in the nucleus, as expected (Fig. 9A, upper panel). In the presence of the drug, newly synthesized RNA were detected inside the viral inclusions (Fig. 9A, lower panel). Control experiments were performed in which rabies virus N and P proteins were coexpressed (with the vaccinia virus T7 system) to mimic NBLs, as previously shown (3), and provide nonviral IBs. The fact that no BrUTP signal was detected in these nonviral IBs in the presence of the drug indicated that the BrUTP signals were dependent on the incorporation of BrUTP by the rabies virus RNA polymerase (Fig. 9B). These results provide evidence that NBs are functional structures where viral transcription and replication take place.
FIG. 9.
Newly synthesized viral RNAs are present inside the viral IBs. (A) BSR cells were infected at an MOI of 3 for 16 h p.i., transfected with BrUTP for 20 min in the presence (+ActD) or absence (-ActD) of ActD. P protein (as a marker of the Negri bodies) was detected by using the rabbit polyclonal anti-P antibody, followed by incubation with Alexa 568-goat anti-rabbit IgG (P). Newly synthesized RNAs were detected by using mouse MAb anti-BrdU antibody (BrUTP) and goat anti-mouse Alexa 488. Colocalization of the viral RNA (vRNA) with the IBs is shown in the merged panel in the presence of the drug (+ActD). In cells untreated with ActD, cellular mRNA is produced in vast excess over vRNA. Thus, for these images, the PMT gain of the confocal laser scanning microscope is lower, so the signal for vRNA is lower than for that shown in the presence of the drug, and specific colocalization is not readily observed. The scale bars correspond to 12 μm. (B) BrUTP signals are specific to RNA produced by rabies virus. BSR cells were infected with the VTF7-3 recombinant virus and cotransfected with plasmids expressing N and P proteins. Cells were then transfected with BrUTP for 20 min in the presence (+ActD) or absence (-ActD) of ActD. P protein and RNA were detected as in panel A. Note that in the absence of the drug, no BrUTP signal is detectable in the IBs formed by N and P proteins, but some RNA signals are detected in the cytoplasmic inclusions, most probably due to vaccinia virus infection (-ActD). In the presence of the drug, no RNA labeling is observed (+ActD).
DISCUSSION
The present study characterizes the morphogenesis and functions of the IBs that are found in the cytoplasm of cells infected by rabies virus. We show that these IBs have staining characteristics that are similar to those of the Negri bodies found in certain nerve cells infected by rabies virus. These structures are not canonical aggresomes containing misfolded proteins aggregates en route to degradation by the proteasome (consistent with the observations of Menager et al. [26]) but are functional structures with a central role in viral infection.
The detection of viral RNA in IBs indicates either that viral RNAs are synthesized within IBs or are synthesized elsewhere before import and accumulation within IBs. Our findings indicate that the former is the case and that Negri bodies are most probably specialized sites of viral transcription and replication. Significantly, the labeling period used in the BrUTP experiments is short compared to the duration of viral mRNA synthesis. Indeed, it has been reported that due to a quite slow elongation rate of transcription (3.1 to 4.3 nucleotides), it takes about 10 min for the VSV N mRNA and 1 to 2 h for the LmRNA to be synthesized (16). This is also consistent with the elongation rates recently reported for another Mononegavirales, the measles virus (three nucleotides) (32). Thus, the newly BrUTP-labeled RNAs are almost certainly synthesized within NBLs (during the 20-min labeling period), as indicated by the BrUTP labeling experiment previously used for VSV (4).
This would be consistent with the fact that proteins L, P, and N involved in viral RNA synthesis are accumulated in these structures. Interestingly, viral genomes, antigenomes, and mRNAs synthesized inside the IBs appear to be surrounded by a cage formed by N and P proteins that may protect them from degradation. We did not investigate whether NBLs are also sites of viral protein synthesis. Whether the viral proteins L, N, and P are translated inside or at the periphery of the viral inclusions or anywhere else, and in this latter case how these proteins are recruited in such IBs, remains an open question. Cellular proteins are also detected in these structures. Although the exact composition of the NBLs remains unknown, the Toll-like receptor 3 has been shown to be an important compound of the NBLs (26). In addition, the heat shock protein Hsp70 and ubiquitinylated proteins, which are also found associated with aggresomes, were accumulated inside these structures. This enrichment might be correlated with an intense production of viral proteins (particularly N and P) in these viral factories. However, in addition to continuously surveying the folding status of proteins as part of its quality control function, Hsp70 is also involved in many cellular processes such as cell cycle, apoptosis, or innate immunity (25). Similarly, ubiquitinylation of proteins is also involved in many cellular functions other than degradation, such as signalization events. Since viruses need to interfere with cellular processes to create a favorable environment for their multiplication, Hsp70 and ubiquitin ligases are frequently diverted and recruited by viruses (2, 25). In the case of rabies virus infection, Hsp70 might play a proviral role, as previously indicated by the presence of Hsp70 in purified rabies virus (37). In addition, NEDD4, an E3 ubiquitin ligase, is recruited by rabies virus matrix protein via a PPXY motif and is involved in the budding process (13, 15).
Electron-microscopy studies indicate that viral inclusions initially devoid of membranes recruit cellular membranes, suggesting that the budding step could occur inside intracellular compartment derived from the rough endoplasmic reticulum and wrapped around IBs as previously shown (24).
The fact that IBs may support viral transcription and replication leads us to consider that such structures may form around incoming genomes. Our results indicate that early after infection, infected cells contain one or two viral factories. This limited initial number of formation sites is independent of the MOI and consequently is independent of the number of incoming viruses per cell. The mechanism involved in this restriction is not known, but it might occur at the cell entry to limit the number of incoming viral particles and/or inside the cytoplasm to reduce the number of functional transcription and replication sites. It is also possible that a viral factory arises from many incoming genomes. The restriction would then be due to the requirement of specific cellular structures or organelles (present in a limited number in the cell) for the IBs to form.
We did not observe any preferential intracellular localization of the viral inclusions in the cells such as the MTOC, where aggresomes are typically located, as well as some other viral factories (31). The lack of a link between MTOC and rabies viral factories is consistent with the fact that microtubule networks are not involved in the morphogenesis of NBLs that are formed efficiently and without any delay in the presence of an inhibitor of microtubule polymerization. Furthermore, overexpression of dynamitin that blocks dynein/dynactin-mediated retrograde transport along microtubules has no effect on the morphogenesis of viral inclusions. All of these data indicate that the mechanism to gather the different viral and cellular components in viral inclusions does not require an active cellular transport along the microtubules.
Although microtubule depolymerization does not inhibit the formation of Negri bodies, it prevents the appearance of smaller IBs that otherwise form late in infection. Further experiments are required to establish whether these small inclusions are derived from the initial IBs or may be newly formed by infection at a later stage by new viral progeny or just by delayed formation. In contrast, the actin network does not play a prominent role in these processes.
It is possible that NBLs might be the result of a cytoplasmic compartmentalization due to a cellular defense mechanism involving components of the aggresome pathway, such as host factors involved in stress response, the storage or degradation of proteins to target virus components for storage, and degradation, as previously suggested for other viruses. These structures may be hijacked by the rabies virus to concentrate host and viral components in one place to facilitate viral replication, to immobilize and inactivate proteins that would otherwise inhibit infection, and also to mask rabies virus presence from the cellular components of the innate immune system. This is consistent with the findings of Menager et al. (26), who suggested that the sequestration of Toll-like receptor 3 in the Negri body could be a strategy whereby the virus prevents the antiviral or apoptotic effect of this cellular protein. Cytoplasmic inclusions have been reported for filoviruses (10) and some paramyxoviruses (12), suggesting that virus-induced formation of intracellular compartments might be general among viruses of the Mononegavirales order that contains numerous human and animal pathogens. It is probable that the mechanisms and the machineries hijacked by rabies virus are similarly diverted by these other viruses.
Acknowledgments
We are grateful to Greg Moseley (from Monash University, Clayton, Victoria, Australia) for helpful discussions and careful reading of the manuscript. We thank Manos Mavrakis for helpful discussions. We thank David Pasdeloup for providing the plasmid pDsRedM and for technical advice for the FISH. We acknowledge Richard Vallee for the gift of the plasmid encoding dynamitin-GFP. We acknowledge Christine Longin for helpful advice and assistance with the electron microscopy (Unité UR1196 Génomique et Physiologie de la Lactation, INRA, Plateau de Microscopie Electronique MIMA2, Jouy-en-Josas Cedex, France). Confocal microscopy was performed on the Plate-forme Imagerie et Biologie Cellulaire of the CNRS campus supported by the Institut Fédératif de Recherche 87, La Plante et Son Environnement, and the program ASTRE of the Conseil Général de l'Essonne.
We acknowledge support from the CNRS and INRA.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Ball, L. A., and C. N. White. 1976. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 73442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, M., and D. Gerlier. 2006. Viral hijacking of cellular ubiquitination pathways as an anti-innate immunity strategy. Viral Immunol. 19349-362. [DOI] [PubMed] [Google Scholar]
- 3.Chenik, M., K. Chebli, Y. Gaudin, and D. Blondel. 1994. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75(Pt. 11)2889-2896. [DOI] [PubMed] [Google Scholar]
- 4.Das, S. C., D. Nayak, Y. Zhou, and A. K. Pattnaik. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 806368-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dohner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 132795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finke, S., K. Brzozka, and K. K. Conzelmann. 2004. Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. J. Virol. 7812333-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Mata, R., Z. Bebok, E. J. Sorscher, and E. S. Sztul. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1461239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudin, Y. 1997. Folding of rabies virus glycoprotein: epitope acquisition and interaction with endoplasmic reticulum chaperones. J. Virol. 713742-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaudin, Y. 2000. Rabies virus-induced membrane fusion pathway. J. Cell Biol. 150601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisbert, T. W., and P. B. Jahrling. 1995. Differentiation of filoviruses by electron microscopy. Virus Res. 39129-150. [DOI] [PubMed] [Google Scholar]
- 11.Georgi, A., C. Mottola-Hartshorn, A. Warner, B. Fields, and L. B. Chen. 1990. Detection of individual fluorescently labeled reovirions in living cells. Proc. Natl. Acad. Sci. USA 876579-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldsmith, C. S., T. Whistler, P. E. Rollin, T. G. Ksiazek, P. A. Rota, W. J. Bellini, P. Daszak, K. T. Wong, W. J. Shieh, and S. R. Zaki. 2003. Elucidation of Nipah virus morphogenesis and replication using ultrastructural and molecular approaches. Virus Res. 9289-98. [DOI] [PubMed] [Google Scholar]
- 13.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 7510623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath, C. M., M. Windsor, and T. Wileman. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irie, T., J. M. Licata, J. P. McGettigan, M. J. Schnell, and R. N. Harty. 2004. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 782657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23477-484. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, A. C., D. L. Reimer, and W. H. Wunner. 1989. Detection of rabies virus RNA in the central nervous system of experimentally infected mice using in situ hybridization with RNA probes. J. Virol. Methods 251-11. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, A. C., and W. H. Wunner. 1991. Detection of rabies virus genomic RNA and mRNA in mouse and human brains by using in situ hybridization. J. Virol. 652839-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob, Y., H. Badrane, P. E. Ceccaldi, and N. Tordo. 2000. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 7410217-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1431883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manghani, D. K., D. K. Dastur, A. N. Nanavaty, and R. Patel. 1986. Pleomorphism of fine structure of rabies virus in human and experimental brain. J. Neurol. Sci. 75181-193. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto, S. 1962. Electron microscopy of nerve cells infected with street rabies virus. Virology 17198-202. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto, S. 1970. Rabies virus. Adv. Virus Res. 16257-301. [PubMed] [Google Scholar]
- 24.Matsumoto, S., L. G. Schneider, A. Kawai, and T. Yonezawa. 1974. Further studies on the replication of rabies and rabies-like viruses in organized cultures of mammalian neural tissues. J. Virol. 14981-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer, M. P. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 1531-46. [DOI] [PubMed] [Google Scholar]
- 26.Menager, P., P. Roux, F. Megret, J. P. Bourgeois, A. M. Le Sourd, A. Danckaert, M. Lafage, C. Prehaud, and M. Lafon. 2009. Toll-like receptor 3 (TLR3) plays a major role in the formation of rabies virus Negri bodies. PLoS Pathog. 5e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, F. A., S. P. Bauer, A. K. Harrison, and W. C. Winn, Jr. 1973. Comparative pathogenesis of rabies and rabies-like viruses. Viral infection and transit from inoculation site to the central nervous system. Lab. Investig. 28361-376. [PubMed] [Google Scholar]
- 28.Murphy, F. A., A. K. Harrison, W. C. Winn, and S. P. Bauer. 1973. Comparative pathogenesis of rabies and rabies-like viruses: infection of the central nervous system and centrifugal spread of virus to peripheral tissues. Lab. Investig. 291-16. [PubMed] [Google Scholar]
- 29.Negri, A. 1903. Contributo allo studio dell'eziologia della rabia. Bol. Soc. Med. Chir. Pavia 190388-114. [Google Scholar]
- 30.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nozawa, N., Y. Yamauchi, K. Ohtsuka, Y. Kawaguchi, and Y. Nishiyama. 2004. Formation of aggresome-like structures in herpes simplex virus type 2-infected cells and a potential role in virus assembly. Exp. Cell Res. 299486-497. [DOI] [PubMed] [Google Scholar]
- 32.Plumet, S., W. P. Duprex, and D. Gerlier. 2005. Dynamics of viral RNA synthesis during measles virus infection. J. Virol. 796900-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poisson, N., E. Real, Y. Gaudin, M. C. Vaney, S. King, Y. Jacob, N. Tordo, and D. Blondel. 2001. Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8: dissociation of dynein-binding properties and transcriptional functionality of P. J. Gen. Virol. 822691-2696. [DOI] [PubMed] [Google Scholar]
- 34.Raux, H., A. Flamand, and D. Blondel. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 7410212-10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raux, H., F. Iseni, F. Lafay, and D. Blondel. 1997. Mapping of monoclonal antibody epitopes of the rabies virus P protein. J. Gen. Virol. 78119-124. [DOI] [PubMed] [Google Scholar]
- 36.Roth, J., D. J. Taatjes, and K. T. Tokuyasu. 1990. Contrasting of Lowicryl K4M thin sections. Histochemistry 95123-136. [DOI] [PubMed] [Google Scholar]
- 37.Sagara, J., and A. Kawai. 1992. Identification of heat shock protein 70 in the rabies virion. Virology 190845-848. [DOI] [PubMed] [Google Scholar]
- 38.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312875-878. [DOI] [PubMed] [Google Scholar]
- 39.Wileman, T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61149-167. [DOI] [PubMed] [Google Scholar]
- 40.Young, C. C., and T. F. Sellers. 1927. Laboratory: a new method for staining Negri bodies of rabies. Am. J. Public Health 171080-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]