Abstract
Members of the Paramyxoviridae such as measles, mumps, and parainfluenza viruses have pleomorphic, enveloped virions that contain negative-sense unsegmented RNA genomes. This is encapsidated by multiple copies of a viral nucleocapsid protein N to form a helical ribonucleoprotein complex (termed the nucleocapsid), which acts as the template for both transcription and replication. Structure analysis of these viruses has proven challenging, owing to disordered regions in important constituent proteins, conformational flexibility in the nucleocapsid and the pleomorphic nature of virus particles. We conducted a low-resolution ultrastructural analysis of Sendai virus, a prototype paramyxovirus, using cryo-electron tomography. Virions are highly variable in size, ranging approximately from 110 to 540 nm in diameter. Envelope glycoproteins are densely packed on the virion surface, while nucleocapsids are clearly resolved in the virion interior. Subtomogram segmentation and filament tracing allowed us to define the path of many nucleocapsids and in some cases to determine the number of putative genomes within a single virus particle. Our findings indicate that these viruses may contain between one and six copies of their genome per virion and that there is no discernible order to nucleocapsid packaging.
The Paramyxoviridae include many medically important viruses such as measles, mumps, and parainfluenza viruses and the emerging zoonotic agents nipah and hendra viruses. There are also numerous animal pathogens within the Paramyxoviridae including Newcastle disease, rinderpest, peste-des-petits-ruminants, and canine distemper viruses. The Paramyxoviridae are divided into two subfamilies, the Paramyxovirinae and the Pneumovirinae. The Paramyxovirinae includes the above-mentioned animal and human pathogens and is divided into five genera: Avulavirus, Henipavirus, Morbillivirus, Respirovirus, and Rubulavirus. The Pneumovirinae comprise human and bovine respiratory syncytial viruses, pneumonia virus of mice, and the metapneumoviruses. Paramyxoviruses have a negative-sense nonsegmented RNA genome that is encapsidated by multiple copies of a virally encoded nucleocapsid protein (N) to form a helical ribonucleoprotein complex known as the nucleocapsid. This is associated with the virally encoded RNA-dependent RNA polymerase, which is made up of the large protein (L) and the phosphoprotein (P). During virion morphogenesis, the nucleocapsid/RNA-dependent RNA polymerase complex is packaged within a lipid envelope at the host-cell plasma membrane. The envelope is studded with glycoproteins necessary for virus egress, attachment, and entry and is lined internally by the matrix protein (M). A number of species-specific accessory proteins are also found within the virion (13).
Structural investigation of paramyxovirus biology has proven difficult, owing to disordered regions in some of their proteins, conformational flexibility in the nucleocapsid, and the pleomorphic nature of the paramyxovirus virion. For example, measles virus N has been found to bind several viral and host proteins. Many of these interactions involve the hypervariable C-terminal domain Ntail, which is intrinsically disordered and undergoes an unstructured to structured transition upon binding to P (15). In Sendai virus (SeV), Ntail has been shown to have a residual and dynamic structure that is stabilized by P binding (9). Ntail is also highly susceptible to proteolytic digestion, which leads to major conformational changes in the nucleocapsid, suggesting that it plays an important role in modulating nucleocapsid structure (7, 15, 19). Three-dimensional (3D) helical reconstructions of paramyxovirus nucleocapsids have revealed considerable structural variability with pitch values ranging from 46 to 66 Å and twists ranging from 12.35 to 13.6 subunits per turn (2, 4, 24).
Paramyxovirus virions are highly pleomorphic and have been shown to range in diameter from approximately 120 to 450 nm (25). We have investigated the structure of these particles by cryo-electron tomography of purified SeV (a member of the Respirovirus genus). Our results confirm the pleomorphic nature of these viruses, although we have found some larger particles measuring up to 540 nm in diameter. Within many particles, nucleocapsids were readily visualized in three dimensions, allowing us to trace the path of individual genomes through the virion. We show that paramyxovirus particles may contain multiple copies of their genome and that there is no obvious order to genome packaging.
MATERIALS AND METHODS
Virus growth and purification.
Five 9-day-old embryonated chicken eggs were injected with Sendai virus stock at 1,000 PFU/egg in the allantoic cavity. To minimize defective-interfering (DI) particles, the virus stock was prepared by inoculating eggs with 104 PFU of thrice-plaque-purified virus/egg. After 3 days of incubation at 33°C, the allantoic fluid was collected and clarified by centrifugation for 30 min at 3,500 rpm (4°C) in a refrigerated Megafuge 2.0R (Heraeus Instruments). Clarified allantoic fluid was loaded onto a 4-ml 20% sucrose cushion in Beckman SW28 tubes. The virus was then pelleted at 26,000 rpm and 4°C for 90 min. The pellet was resuspended in 5 ml of 20 mM Tris-HCl-20 mM NaCl-5 mM EDTA buffer (pH 7.0) by gentle Dounce homogenization. After purification, virus was stored at 4°C or on ice and prepared for cryo-electron microscopy within 3 days of production.
Cryo-electron tomography.
Purified SeV was mixed with 10-nm colloidal gold and prepared for electron microscopy by loading 5-μl samples onto 200 mesh R2/2 Quantifoil grids (Quantifoil Micro Tools GmbH, Jena, Germany) and plunge freezing the samples in liquid nitrogen-cooled liquid ethane as previously described (1). Vitrified grids were imaged at liquid nitrogen temperature in a JEOL 2200FSC transmission electron microscope equipped with a Gatan 914 cryo-holder and Ultrascan 4000 camera. Energy filtered imaging was performed with a slit-width of 30 eV and at a magnification of ×20,000. Images were binned by a factor of two giving a pixel size of 10.6 Å in the specimen. Low-dose tilt series were acquired at 2-μm underfocus by using SerialEM (17). Specimens were tilted in 2° steps from approximately 66° to −66° and subjected to a total electron dose of between 86 and 98 e/Å2 per tilt series.
Image reconstruction and analysis.
Tomograms were calculated by using the IMOD software package (12). Subtomograms were analyzed by using Amira (Visage Imaging GmbH, Berlin, Germany) and the Electron Tomography Toolbox (22). Individual virion reconstructions were first denoised by using the bilateral filter algorithm as implemented in EMAN (five iterations, σ1 = 1, σ2 = 2× standard deviation, kernel size 5 for segmenting and two iterations, σ1 = 1, σ2 = 2× standard deviation, kernel size 1 for two-dimensional visualization) (10, 16). The reconstructions were then segmented into interior, membrane, and glycoprotein regions. Membrane and glycoproteins were only segmented in areas where the membrane was clearly visible. After masking, virion interiors were low-pass filtered to 15 to 20 nm to facilitate visualization and tracing of the nucleocapsid. To estimate the proportion of the particle interior taken up by nucleocapsid, the volume was calculated for the masked interior after thresholding to optimally show the nucleocapsid with minimal noise; this value was then divided by the volume of the interior mask. The filament-tracing algorithm in Amira was used to determine the path of nucleocapsids within each virion and to measure their length. Two scientists independently traced the nucleocapsid paths, either using an auto-skeleton algorithm, followed by manual depletion of branches and nodes that were judged to be incorrect, or by manual placement of nodes. Segmented denoised virions were visualized by using UCSF Chimera (21) for isosurface rendering and Amira for volume rendering with filament traces. Subtomograms were viewed by using the slicer module of 3dmod (IMOD).
RESULTS
Paramyxovirus morphology.
Purified SeV was imaged in the cryo-electron microscope under low-electron dose conditions. Four tilt series of images were recorded and processed to calculate three-dimensional reconstructions from which 11 subtomograms of individual virus particles were excised for further analysis.
Virions were found to range in size from approximately 110 to 540 nm. Particles were mostly circular in projection, although some were elliptical or distorted into irregular shapes. Measurements of virions in the z-axis revealed that larger particles were significantly flattened in the vitreous ice layer (Fig. 1). Indeed, interpretation of flattened particles embedded in thinner ice (∼250 nm) was easier than for those that were broadly spherical but in thicker ice (ca. 300 to 400 nm).
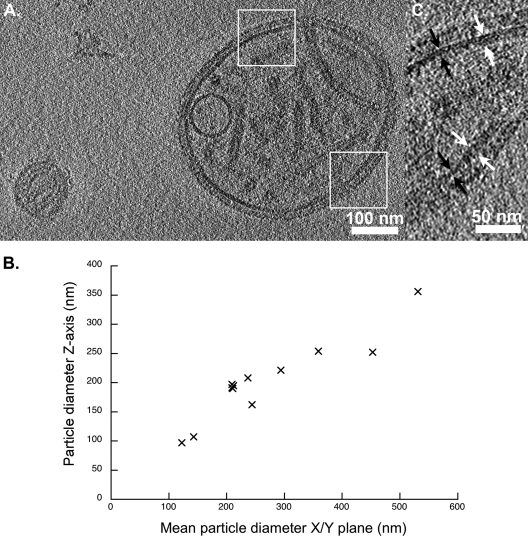
FIG. 1.
(A) Projection image of a 10-nm-thick section through a tomogram that shows two virus particles. Sendai virions range in diameter from approximately 110 to 540 nm; the particle to the left measures ∼140 nm in diameter, while the larger particle is 490 by 410 nm. Measurements of virion diameter in the x, y, and z axes show a degree of flattening in the z axis, particularly in the larger particles. Dimensions for 11 particles are plotted (B). Measurements in the x/y plane were taken along longest and shortest dimensions for nonspherical particles and averaged to give the “mean particle diameter.” (C) Close-up views of the SeV envelope (i.e., the boxed areas from panel A) show variations in membrane thickness. White arrows indicate thicker regions measuring ∼12 nm; black arrows indicate thinner regions measuring ∼7 nm.
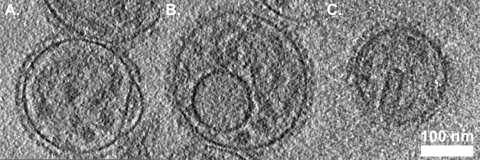
Measurements of the thickness of the viral envelope varied from 6 to 12 nm (Fig. 1C), thicker regions of the envelope sometimes appearing as two distinct layers. In some reconstructions, thinner regions of the envelope also appeared as a bilayer; however, this feature was lost when denoising was applied to improve image contrast (see Fig. S1 in the supplemental material). In some cases the viral envelope had a “blister” (Fig. 2A). Several virus particles were also found to contain a vesicle-like structure (Fig. 1A and 2B). The putative phospholipid bilayer in these vesicles measured approximately 8 to 12 nm thick. We also saw cylindrical or rectangular objects within some virions, measuring approximately 90 nm long and 30 nm in diameter (Fig. 2C, 4A, and 5A).
FIG. 2.
Several unanticipated features were seen in tomograms of SeV, including blisters in the viral envelope (A), vesicles inside the virions (B), and cylindrical and/or rectangular structures (C).
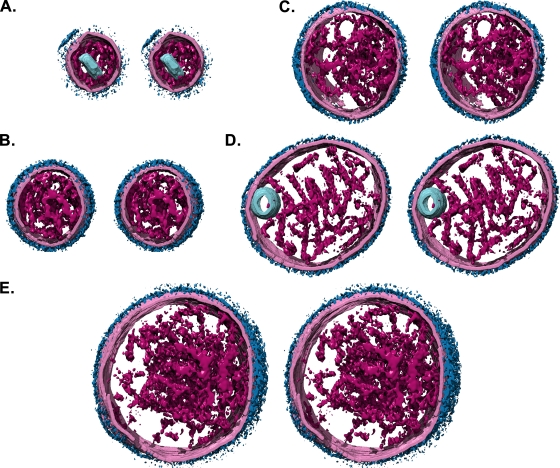
FIG. 4.
Stereo images (divergent) of segmented and isosurface rendered Sendai virion tomograms. The viral envelope is colored pale pink, envelope glycoproteins dark blue, the virion interior (nucleocapsid) is magenta, and inclusions are pale blue. A range of particle sizes is shown, approximately to scale. (A) 210-nm diameter particle containing a cylindrical or rectangular feature. (B) 290-nm diameter particle. (C) 350-by-370-nm particle. (D) 490-by-410-nm particle containing a vesicle. (E) The largest particle recorded was 540 by 510 nm.
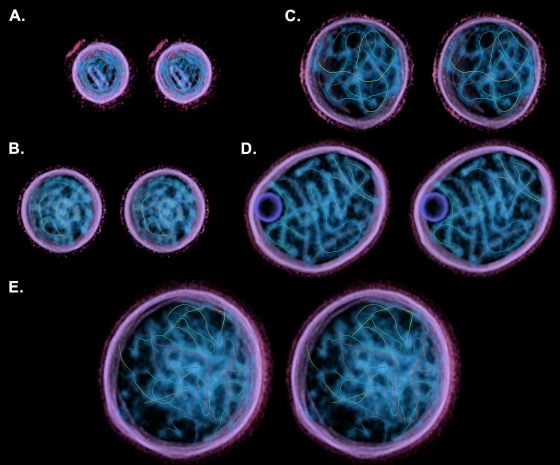
FIG. 5.
Stereo images (divergent) of volume-rendered, segmented tomograms showing the paths of traced sections of nucleocapsid. The particles are the same as those shown in Fig. 4. Each path is represented as a uniquely colored filament. The particle in panel A contains two fragments measuring 1,124 nm in total (482 nm, green; 642 nm, red), possibly representing a single copy of the viral genome incompletely traced. The total lengths for the remaining images are 1,840 nm (B), 3,033 nm (C), 3,649 nm (D), and 5,829 nm (E).
On the outer surface of the virus particles the envelope glycoproteins were densely packed and, in some tomograms, it was difficult to discern individual molecules. Where individual glycoproteins could be seen they appeared to have a thin stalk terminating in a membrane-distal globular domain (Fig. 3A).
FIG. 3.
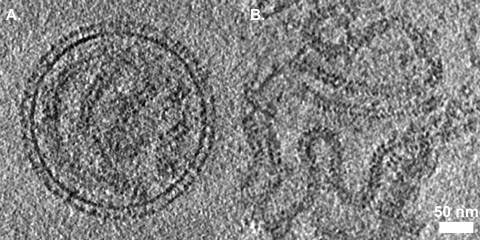
(A) Section through the center of a virus particle. Individual envelope glycoproteins are visible as thin stalks terminating in a membrane distal globular domain. Several sections of nucleocapsid are also visible, although finer features such as the helical pitch are not clearly resolved. (B) A section through a tomogram of free nucleocapsid shows such features more clearly. The 7-nm central cavity and helical pitch are visible.
Nucleocapsid structure and packaging.
Nucleocapsid was visible within virions although the herringbone morphology was more clearly seen in free nucleocapsid that was visualized in the same tomograms (Fig. 3). Many features were discernible in these data, including the 7-nm-diameter channel that runs along the center of the nucleocapsid and the helical repeat (pitch), which has been shown to vary between 5 and 7 nm in purified SeV nucleocapsids and in nucleocapsidlike particles produced by the recombinant expression of measles virus N (2, 4). In these data the helical pitch measured ∼7 nm.
Segmenting and filtering to suppress noise allowed us to visualize the nucleocapsids within virions in three dimensions (Fig. 4 and see also Movie S1 in the supplemental material). In these images the virus interior was segmented to include all visible features, while the viral envelope and glycoproteins are only shown where the membrane had strong density. It is clear from these images that there is no consistent order to nucleocapsid packaging; rather, they appear as a tangled knot, wrapping around each other and themselves. We estimated the proportion of the virion interior taken up by nucleocapsid in each particle; values ranged between 9 and 24%, with a mean value of 14% (standard deviation = 5%, n = 11). Filament tracing algorithms were used to determine the paths of the nucleocapsids within each virion (Fig. 5 and see Movie S2 in the supplemental material). In some cases, we were able to trace paths for significant proportions of each nucleocapsid within a virion, providing an estimate of the number of genomes packaged. For example, the particle shown in Fig. 5D contains four distinct nucleocapsids. Three of these filaments measure approximately 1 μm in length (indicated by color in Fig. 5: blue, 1,008 nm; green, 1,048 nm; and yellow, 923 nm), while the fourth measures ∼640 nm (red). These measurements are consistent with previously published estimates that SeV nucleocapsids are ∼1.1 μm in length (4, 5)}. In other particles, nucleocapsids were less well resolved, and we were only able to trace shorter sections, for example, where two nucleocapsid sections come into very close proximity, making it difficult to discern connectivity. The particle shown in Fig. 5B contains ∼1.8 μm of nucleocapsid in total, suggesting that the particle may contain two nucleocapsids and therefore two copies of the viral genome. Individual paths for each identified filament measured 393 nm (blue), 258 nm (dark green), 99 nm (light green), 639 nm (yellow), and 451 nm (red).
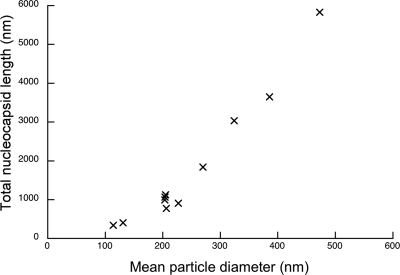
We calculated a total length for the nucleocapsid within each particle (Fig. 6), giving an estimate of the number of genomes packaged. Our measurements indicate that virions may contain between one and six genome copies. Five of the particles measured ∼200 nm in diameter and contained sufficient nucleocapsid to account for a single copy of the viral genome. Interestingly, smaller particles (those <200 nm in diameter) appear to contain <1 μm of nucleocapsid. Two particles were found to contain filaments measuring 341 and 406 nm. The mean diameters of these particles were 114 and 131 nm, respectively. Determining the path of the nucleocapsid in these particles was rather more challenging than in larger virions; however, the particles are significantly smaller than those found to contain nucleocapsid of the correct size to comprise a single genome.
FIG. 6.
Total nucleocapsid length measurements for each particle are plotted against the mean particle radius.
DISCUSSION
Features of the SeV envelope.
Our data provide the first three-dimensional view of paramyxovirus particles and confirm their pleomorphic nature. Some of our observations on the viral envelope and glycoproteins are consistent with those previously reported for parainfluenza virus type 5 (PIV5) or simian virus 5 (25). In that study, the discovery of variations in envelope thickness led to the suggestion that M may not completely line the inner surface of the envelope. A similar observation has been made for influenza virions (6). Freeze fracture experiments have shown the presence of crystalline arrays, thought to be M, covering the inner surface of budding SeV particles. These structures were less easily seen in virus particles after storage, suggesting some degree of rearrangement of M (3).
Internal structures: vesicles and filaments.
We have found that in addition to viral nucleocapsid, some Sendai virions contain vesicle-like structures or cylindrical objects. We have previously observed the presence of putative lipid vesicles in herpes virions (unpublished data). Such inclusions may therefore be a common feature of pleomorphic enveloped viruses. The cylindrical structures are similar in appearance and dimensions to helical assemblies found in purified preparations of SeV M stored under low-salt conditions (8). Filamentous virion formation is a feature of many negative-sense RNA containing viruses, including pneumovirus respiratory syncytial virus, influenza virus, and filovirus. We have not observed filamentous forms of SeV in our analysis, and members of the Respirovirus genus are thought not to adopt filamentous morphologies. However, such virions have been reported for PIV2 and PIV5; both are members of the Rubulavirus genus (26). The capacity to form helical tubules from M may be an important determinant of filamentous virus production. Our data indicate that SeV M may have the potential to form structures in vivo that could be compatible with filamentous virus assembly. Further studies, such as immunological staining, are required to ascertain whether the structures we observe are indeed composed of M however.
Nucleocapsid packaging.
We have demonstrated that SeV frequently packages more than one nucleocapsid per virion, suggesting that virus particles containing up to six genomes may be found. Polyploid measles viruses have previously been isolated, and calculations of virion volume have led to the suggestion that >30 genomes might be packaged (23). Our data provide the first structural evidence of multiple genome packaging and show that nucleocapsids are less well organized than in other negative-sense RNA viruses (6, 18, 20).
We have found that some particles appear to contain nucleocapsids too small to comprise a full-length viral genome. We cannot state categorically that we have traced the nucleocapsid in its entirety (imaging artifacts associated with tilt-series acquisition—the missing wedge—may be limiting); however, these data are consistent with previous studies that demonstrated the presence of subgenomic RNA in SeV particles. In common with many negative-sense RNA viruses, paramyxoviruses are susceptible to the accumulation of DI RNA-containing particles after serial passage. DI particles contain deleted genomes of mainly two types: internal deletions, in which the two genome RNA ends have been conserved while large sections of the internal sequence has been lost, and “copy-back.” The latter type results from the RNA polymerase leaving its template during replication of the genome to resume synthesis using the nascent RNA as the template. As a consequence, the two ends of these subgenomic RNAs are complementary over tens of nucleotides. Functionally, the copy-back RNAs see the weak replication promoter replaced with a copy of the strong replication promoter. This feature explains the interference that these RNAs exert on replication of the genomic full-length nondefective RNA. (14). An early investigation of the physical properties of subgenomic RNAs in SeV described the presence of slowly sedimenting virions (400S) that were ∼150 nm in diameter and did not contain full-length genomes, packaging two distinct populations of smaller RNAs instead (19S and 25S) (11). To reduce the presence of DI particles in the present study, stock virus was plaque purified prior to inoculation; nonetheless, such particles seem to have persisted. It is also likely that some of the shorter nucleocapsids we have traced in larger particles may also contain DI RNA.
In many cases, however, we have managed to clearly visualize and trace the path of several distinct full-length nucleocapsids. Flattening of particles aided this task somewhat, the particle shown in Fig. 4D and 5D and in Movies S1 and S2 in the supplemental material being the most straightforward to analyze. In this case, we found and traced the path of four discrete filaments, three of which had lengths of approximately 1 μm, the recorded length of an intact SeV nucleocapsid. In other cases, we measured multiple fragments of nucleocapsid, allowing us to estimate the likely genome content. We found sufficient nucleocapsid in four of the virus particles investigated to suggest a polyploid virion, while five particles appeared to contain a single copy of the genome. The capacity to form polyploid virions might be seen as an advantageous property in these viruses, whereby delivery of more than one nucleocapsid will increase the probability of a single particle initiating a successful infection by ameliorating the risk of releasing a single defective genome into the cytoplasm. Conversely, a more efficient mechanism of segregating single genomes into virions should lead to production of greater numbers of particles. The fact that many virions studied were found to be haploid suggests that there is not a strong selective pressure to favor incorporation of multiple genomes. Given the tangled arrangement of nucleocapsid that we have observed in larger paramyxovirus virions, it seems plausible that the presence of multiple genomes may simply reflect a poorly ordered assembly pathway.
Supplementary Material
Acknowledgments
We thank Duncan McGeoch and Frazer Rixon for critical readings of the manuscript.
Footnotes
Published ahead of print on 3 June 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adrian, M., J. Dubochet, J. Lepault, and A. W. McDowall. 1984. Cryo-electron microscopy of viruses. Nature 30832-36. [DOI] [PubMed] [Google Scholar]
- 2.Bhella, D., A. Ralph, and R. P. Yeo. 2004. Conformational flexibility in recombinant measles virus nucleocapsids visualized by cryo-negative stain electron microscopy and real-space helical reconstruction. J. Mol. Biol. 340319-331. [DOI] [PubMed] [Google Scholar]
- 3.Buechi, M., and T. Bachi. 1982. Microscopy of internal structures of Sendai virus associated with the cytoplasmic surface of host membranes. Virology 120349-359. [DOI] [PubMed] [Google Scholar]
- 4.Egelman, E. H., S. S. Wu, M. Amrein, A. Portner, and G. Murti. 1989. The Sendai virus nucleocapsid exists in at least four different helical states. J. Virol. 632233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finch, J. T., and A. J. Gibbs. 1970. Observations on the structure of the nucleocapsids of some paramyxoviruses. J. Gen. Virol. 6141-150. [DOI] [PubMed] [Google Scholar]
- 6.Harris, A., G. Cardone, D. C. Winkler, J. B. Heymann, M. Brecher, J. M. White, and A. C. Steven. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA 10319123-19127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heggeness, M. H., A. Scheid, and P. W. Choppin. 1981. the relationship of conformational changes in the Sendai virus nucleocapsid to proteolytic cleavage of the NP polypeptide. Virology 114555-562. [DOI] [PubMed] [Google Scholar]
- 8.Hewitt, J. A., and M. V. Nermut. 1977. A morphological study of the M-protein of Sendai virus. J. Gen. Virol. 34127-136. [DOI] [PubMed] [Google Scholar]
- 9.Jensen, M. R., K. Houben, E. Lescop, L. Blanchard, R. W. Ruigrok, and M. Blackledge. 2008. Quantitative conformational analysis of partially folded proteins from residual dipolar couplings: application to the molecular recognition element of Sendai virus nucleoprotein. J. Am. Chem. Soc. 1308055-8061. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, W., M. L. Baker, Q. Wu, C. Bajaj, and W. Chiu. 2003. Applications of a bilateral denoising filter in biological electron microscopy. J. Struct. Biol. 144114-122. [DOI] [PubMed] [Google Scholar]
- 11.Kingsbury, D. W., A. Portner, and R. W. Darlington. 1970. Properties of incomplete Sendai virions and subgenomic viral RNAs. Virology 42857-871. [DOI] [PubMed] [Google Scholar]
- 12.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 11671-76. [DOI] [PubMed] [Google Scholar]
- 13.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 14.Lazzarini, R. A., J. D. Keene, and M. Schubert. 1981. The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26145-154. [DOI] [PubMed] [Google Scholar]
- 15.Longhi, S., V. Receveur-Brechot, D. Karlin, K. Johansson, H. Darbon, D. Bhella, R. Yeo, S. Finet, and B. Canard. 2003. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 27818638-18648. [DOI] [PubMed] [Google Scholar]
- 16.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 12882-97. [DOI] [PubMed] [Google Scholar]
- 17.Mastronarde, D. N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 15236-51. [DOI] [PubMed] [Google Scholar]
- 18.Mavrakis, M., L. Kolesnikova, G. Schoehn, S. Becker, and R. W. Ruigrok. 2002. Morphology of Marburg virus NP-RNA. Virology 296300-307. [DOI] [PubMed] [Google Scholar]
- 19.Mountcastle, W. E., R. W. Compans, H. Lackland, and P. W. Choppin. 1974. Proteolytic cleavage of subunits of the nucleocapsid of the paramyxovirus simian virus 5. J. Virol. 141253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noda, T., H. Sagara, A. Yen, A. Takada, H. Kida, R. H. Cheng, and Y. Kawaoka. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439490-492. [DOI] [PubMed] [Google Scholar]
- 21.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 22.Pruggnaller, S., M. Mayr, and A. S. Frangakis. 2008. A visualization and segmentation toolbox for electron microscopy. J. Struct. Biol. 164161-165. [DOI] [PubMed] [Google Scholar]
- 23.Rager, M., S. Vongpunsawad, W. P. Duprex, and R. Cattaneo. 2002. Polyploid measles virus with hexameric genome length. EMBO J. 212364-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoehn, G., M. Mavrakis, A. Albertini, R. Wade, A. Hoenger, and R. W. Ruigrok. 2004. The 12 Å structure of trypsin-treated measles virus N-RNA. J. Mol. Biol. 339301-312. [DOI] [PubMed] [Google Scholar]
- 25.Terrier, O., J. P. Rolland, M. Rosa-Calatrava, B. Lina, D. Thomas, and V. Moules. 2009. Parainfluenza virus type 5 (PIV-5) morphology revealed by cryo-electron microscopy. Virus Res. 142200-203. [DOI] [PubMed] [Google Scholar]
- 26.Yao, Q., and R. W. Compans. 2000. Filamentous particle formation by human parainfluenza virus type 2. J. Gen. Virol. 811305-1312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.