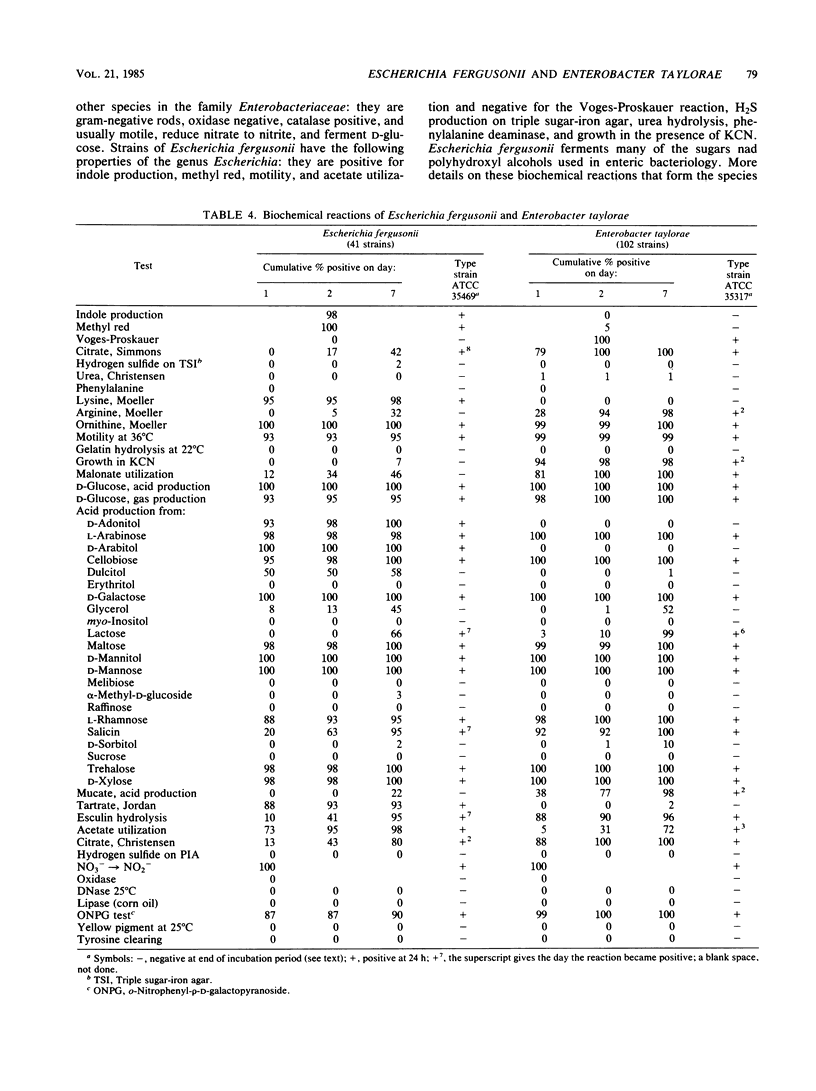
Abstract
Escherichia fergusonii (formerly known as Enteric Group 10) and Enterobacter taylorae (formerly known as Enteric Group 19) are proposed as new species in the family Enterobacteriaceae. By DNA hybridization (32P, 60 degrees C, hydroxyapatite), strains of E. fergusonii were 90 to 97% related to the type strain (holotype) ATCC 35469. They were most closely related to Escherichia coli and more distantly related to species in other genera. E. fergusonii strains are positive for indole production, methyl red, lysine decarboxylase, ornithine decarboxylase, and motility. They ferment D-glucose with gas production and also ferment adonitol, L-arabinose, L-rhamnose, maltose, D-xylose, trehalose, cellobiose, and D-arabitol. They are negative for Voges-Proskauer, citrate utilization (17% positive), urea hydrolysis, phenylalanine deamination, arginine dihydrolase, growth in KCN, and fermentation of lactose, sucrose, myo-inositol, D-sorbitol, raffinose, and alpha-methyl-D-glucoside. By DNA hybridization (32P, 60 degrees C, hydroxyapatite), strains of E. taylorae were 84 to 95% related to the type strain (holotype) ATCC 35317. Their nearest relative was E. cloacae, to which they were 61% related. Other named species were more distantly related. Strains of E. taylorae are positive for Voges-Proskauer, citrate utilization, arginine dihydrolase, ornithine decarboxylase, motility, growth in KCN medium, and malonate utilization. They ferment D-glucose with gas production and also ferment D-mannitol, L-arabinose, L-rhamnose, maltose, D-xylose, trehalose, and cellobiose. They are negative for indole production, methyl red, H2S production on triple sugar-iron agar, urea hydrolysis, phenylalanine deamination, lysine decarboxylase, gelatin hydrolysis, and fermentation of adonitol, i-inositol, D-sorbitol, and raffinose. Both new species occur in human clinical specimens. Two strains of E. fergusonii were isolated from blood. Five stains of E. taylorae were isolated from blood, and one was from spinal fluid. These blood and spinal fluid isolates suggest possible clinical significance, but this point requires further study.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Johnson K. E., Citarella R. V., Falkow S. Polynucleotide sequence relationships among members of Enterobacteriaceae. J Bacteriol. 1969 May;98(2):637–650. doi: 10.1128/jb.98.2.637-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. J., McWhorter A. C., Knutson J. K., Steigerwalt A. G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982 Jun;15(6):1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer J. J., 3rd, Davis B. R., Hickman-Brenner F. W., McWhorter A., Huntley-Carter G. P., Asbury M. A., Riddle C., Wathen-Grady H. G., Elias C., Fanning G. R. Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol. 1985 Jan;21(1):46–76. doi: 10.1128/jcm.21.1.46-76.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman F. W., Farmer J. J., 3rd Salmonella typhi: identification, antibiograms, serology, and bacteriophage typing. Am J Med Technol. 1978 Dec;44(12):1149–1159. [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]


