Abstract
Hypermutation, the introduction of excessive G-to-A substitutions by host proteins in the APOBEC family, can impair replication of the human immunodeficiency virus (HIV). Because hypermutation represents a potential antiviral strategy, it is important to determine whether greater hypermutation is associated with slower disease progression in natural infection. We examined the level of HIV-1 hypermutation among 28 antiretroviral-naive Kenyan women at two times during infection. By examining single-copy gag sequences from proviral DNA, hypermutation was detected in 16 of 28 individuals. Among individuals with any hypermutation, a median of 15% of gag sequences were hypermutated (range, 5 to 43%). However, there was no association between the level of gag hypermutation and the viral load or CD4 count. Thus, we observed no overall relationship between hypermutation and markers of disease progression among individuals with low to moderate levels of hypermutation. In addition, one individual sustained a typical viral load despite having a high level of hypermutation. This individual had 43% of gag sequences hypermutated and harbored a partially defective Vif, which was found to permit hypermutation in a peripheral blood mononuclear cell culture. Overall, our results suggest that a potential antiviral therapy based on hypermutation may need to achieve a substantially higher level of hypermutation than is naturally seen in most individuals to impair virus replication and subsequent disease progression.
Interactions between the human immunodeficiency virus type 1 (HIV-1) protein Vif (for viral infectivity factor) and antiviral host factors in the APOBEC family (for apolipoprotein B mRNA editing enzyme, catalytic polypeptide) have recently attracted interest as potential targets for antiviral therapy (10, 13, 16, 26, 28, 40). One of the mechanisms by which APOBEC3G, a cytidine deaminase, may impair HIV-1 replication is through the introduction of C-to-U lesions in the negative-sense DNA strand during reverse transcription (RT). This process, known as hypermutation, results in excessive G-to-A changes in the HIV-1 coding strand, leading to premature stop codons and other potentially disruptive mutations (24). APOBEC3G activity is typically counteracted by the HIV-1 protein Vif, which targets it for degradation by the proteasome (38). This prevents APOBEC from being incorporated into nascent virions and causing hypermutation. However, Vif activity in vivo is imperfect, as demonstrated by the observation of hypermutated sequences in HIV-infected individuals (9, 17, 18, 31, 42). Because hypermutation is expected to impair virus replication, recent efforts have focused on characterizing hypermutation in HIV-infected individuals and examining its relationship with disease progression.
The relationship between hypermutation and markers of disease progression has been addressed in two different ways. In three studies, markers of disease progression were compared between individuals with or without hypermutation detected by population sequencing (18, 31, 42). In this method, HIV-1 DNA is amplified and sequenced in bulk, and the predominant variants (i.e., those comprising more than ∼20% of the total sequence population [12, 44]) are identified in a single sequence chromatogram. Thus, studies using this method have focused on individuals with high levels of hypermutation, as illustrated by the fact that such studies have detected hypermutation in just 7 to 9% of HIV-infected individuals (18, 31). Using this approach, Pace et al. (31) found that individuals with detectable hypermutation had ∼0.7-log-lower viral loads than individuals without detectable hypermutation. Land et al. (18) found that individuals with detectable hypermutation had no difference in viral load, but their CD4 counts were approximately 150 to 200 cells/mm3 higher than individuals without detectable hypermutation. In contrast to these findings, Ulenga et al. found no association between G-to-A substitutions and viral load (42). Although these studies all detected hypermutation by population sequencing, it is important to note that they used different methods to define which of the sequences were hypermutated. Specifically, Pace et al. defined hypermutated sequences by cluster analysis, whereas Land et al. used a web program Hypermut 2.0 (34) and Ulenga et al. evaluated G-to-A changes without specifically identifying hypermutated sequences. Thus, it is difficult to precisely compare the results of these three studies. However, a possible explanation for the difference in their results is that the relatively low sensitivity of population sequencing necessitates studying a large number of individuals. Therefore, the Ulenga et al. study, which included 29 individuals, may have been too small to detect the magnitude of difference observed in the other two studies, which each included over a hundred individuals.
In contrast to examining population sequences, an alternative approach is to examine many independent sequences from each individual. The sensitivity of this method increases as more sequences are examined, so lower levels of hypermutation can be detected. As a result, studies that used this method have found hypermutated sequences in 43 to 100% of HIV-infected individuals (9, 15, 17). In addition to detecting lower levels of hypermutation, this method also allows quantitative assessment of the relationship between hypermutation and disease progression. Using this approach, Gandhi et al. did not find a greater amount of hypermutation in elite suppressors compared to HIV-infected individuals on highly active antiretroviral therapy (HAART) (17), suggesting that hypermutation does not contribute to slow disease progression in elite suppressors. However, there was limited virus replication in the individuals included in that study (elite suppressors and individuals on HAART). Thus, prior studies of hypermutation in natural infection have focused on select populations, either those with limited virus replication or those with high enough levels of hypermutation to be detected by population sequencing. It is not known whether there is an association between disease progression and HIV-1 hypermutation across the full spectrum of hypermutation that is likely to be present in HIV-infected individuals with ongoing virus replication.
We examined the extent of hypermutation among 28 HIV-infected, antiretroviral-naive Kenyan women monitored longitudinally from prior to infection through approximately 5 years of infection. We characterized hypermutation by examining multiple independent sequences, each from a separate single-copy PCR, from two times in infection. We identified a wide range of hypermutation within these individuals, but there were no associations between the level of hypermutation and markers of disease progression in this group. For a subset of individuals, we examined the role of Vif in contributing to hypermutation by measuring hypermutation in peripheral blood mononuclear cells (PBMC) culture. We found that one individual harbored a partially defective Vif but sustained a typical viral load, suggesting that in some cases even a high level of hypermutation does not confer a clinical benefit.
MATERIALS AND METHODS
Study population.
Individuals in the present study were part of a prospective seroincident cohort of high-risk women in Mombasa, Kenya (25). Methods for determining the timing of HIV-1 infection and measuring plasma viral load by the Gen-Probe HIV-1 viral load assay and CD4 count have been described previously (19). The 28 women included in the present study had a blood sample taken within the first year of infection, were antiretroviral naive at the time of sample collection, and were not dually infected (32). The study was approved by the ethical review committees of the University of Nairobi, the University of Washington, and the Fred Hutchinson Cancer Research Center.
Single-copy sequencing and detection of hypermutation.
Methods for obtaining single-copy sequences from patient PBMC samples have been described (32). Briefly, DNA was extracted from approximately 5 million frozen PBMC by using a QIAamp DNA blood minikit (Qiagen, Valencia, CA). The HIV-1 proviral copy number was estimated by using quantitative PCR (1, 35), and DNA samples were diluted to an estimated single proviral copy per PCR. A region encompassing gag p17-partial p24 (∼700 bp) was amplified in a nested PCR using previously published primers (32). A region encompassing envelope V1-V5 (∼1.2 kb) was also amplified using single-copy PCR and previously published primers (32). PCR products were treated with ExoSap-IT (USB, Cleveland, OH) and sequenced directly using the second round PCR primers. For each sample, at least 24 independent single-copy gag and env PCRs were performed and one-third to one-half were successful, as would be expected from an average of one copy per reaction. Sequences were aligned by using CLUSTALX (41) and manually edited using MacClade 4.0 (23). Regions that could not be unambiguously aligned between individuals (mainly regions of env V1/V2 and V4) were excluded from analysis. For each individual, the most recent common ancestor(s) (MRCA) of sequences from the early time point was reconstructed using maximum likelihood in GARLI 0.95 (48).
Sequences from each individual were aligned with their MRCA and uploaded to the Hypermut 2.0 tool on the Los Alamos National Laboratories website (http://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html) (34). In addition, all gag sequences from individuals with subtype A viruses were aligned with the gag subtype A consensus sequence from the Los Alamos HIV sequence database, and this alignment was uploaded to the Hypermut 2.0 program. Hypermutated sequences were defined as those with a Fisher exact P value of <0.05, as calculated by Hypermut 2.0.
In addition to the Hypermut 2.0 program, hypermutation was assessed by using a cluster analysis method similar to that introduced by Pace et al. (31), part of which was used by Ulenga et al. (42). Sequences from each individual were aligned with their MRCA and uploaded to the LANL Hypermut program, which determined the number of G-to-A changes, GG-to-AG changes, and GA-to-AA changes (Hypermut original output). For each sequence, the overall substitution frequency was calculated as the total number of nucleotide changes compared to the MRCA, divided by the sequence length. The G→A substitution frequency was calculated as the number of G→A changes divided by the number of Gs in the MRCA. Similarly, the GG→AG and GA→AA substitution frequencies were calculated as the number of GG→AG or GA→AA changes divided by the number of GG or GA positions in the MRCA, respectively. A general G→A substitution score was calculated as the G→A substitution frequency divided by the overall substitution frequency (as in reference 31). APOBEC3G- and APOBEC3F-specific substitution scores were calculated by dividing the GG→AG and GA→AA substitution frequencies by the overall substitution frequency, as in reference 31). In contrast to the Pace et al. study, the scores defined here were not log transformed because transformation may inappropriately affect clustering. A k-means cluster analysis with two groups was used to sort the sequences into two groups (hypermutated and not hypermutated) based on both the general and APOBEC3G-specific scores.
RNA isolation and gag amplification from plasma samples.
To obtain HIV-1 RNA sequences from frozen heparinized plasma samples, intact virions were first isolated by using a microMACS VitalVirus isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). For each sample, 200 μl of plasma was thawed and spun at 13,000 × g for 30 s to pellet debris. The supernatant was removed and passed through the microMACS VitalVirus isolation kit according to the manufacturer's instructions. Intact virions were eluted in 140 μl of phosphate-buffered saline, and viral RNA was immediately extracted by using a QIAamp viral RNA minikit according to the manufacturer's instructions. Viral RNA was eluted in 60 μl of RNase-free water and stored at −20°C. The plasma viral load as determined by Gen-Probe (7) was available for all plasma samples. The copy number in the final sample of extracted RNA was estimated as follows, including an assumed 80% loss during extraction, as determined by control experiments (not shown): plasma viral load (copies/ml)·0.2 ml·0.2 (loss)/60 μl (final volume) = copies/μl.
Based on this calculation, RNA was diluted to an estimated 10 copies/reaction, and RT-PCR was performed using the gag first-round primers described above (32). A SuperScript III One-Step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) was used according to the manufacturer's instructions, with a 60-min RT step at 45°C, followed by 30 cycles of PCR amplification with an annealing temperature of 55°C. A second round of PCR amplification was performed using the described second-round primers (32), and PCR products were sequenced directly as described.
vif amplification and cloning.
To determine the predominant vif variant in each individual in the present study, population sequencing of the full vif open reading frame was performed using undiluted DNA from PBMC (representing ca. 1 to 50 HIV-1 copies per sample, measured by quantitative PCR as described above). For some individuals with unique vif polymorphisms, the sequence was confirmed by single-copy sequencing. Sequences containing the vif open reading frame were amplified in a nested PCR using AmpliTaq (ABI, Foster City, CA) according to the manufacturer's instructions. In the first round, the outer primers pol15 (5′-TACAGTGCAGGGGAAAGAATA-3′) and vpu2 (5′-GCCACTGTCTTCTGCTCTTT-3′) were used to amplify a 1.4-kp product in 25 PCR cycles with an annealing temperature of 60°C. In the second round, the inner primers pol4r (5′-GGAAAGGTGAAGGGGCAG-3′) and vpr5r (5′-CATGAAGCCATGGCCTAGG-3′) were used to amplify a 731-bp product in 25 PCR cycles with an annealing temperature of 60°C. PCR products were treated with ExoSap-IT (USB) and sequenced directly using the second-round PCR primers. After amplification and sequencing, the vif genes from several individuals of interest (QA039, QA268, QB368, QC440, and QF307) were cloned into TOPO TA vectors for further analyses.
Construction of full-length infectious clones expressing different vif sequences.
A proviral clone defective for vif (Q23Δvif) was constructed from a full-length, CCR5-tropic, subtype A clone, Q23-17, which was originally isolated from a Kenyan individual early in infection (33). Q23Δvif was created according to methods described by Sakurai et al. (36). A SalI site was introduced immediately 3′ of the pol stop codon, resulting in a frameshift mutation in vif that prevented vif expression. To eliminate alternate initiation codons in the pol/vif overlapping region, four point mutations were introduced. Three of these were synonymous in the pol reading frame, and the other resulted in a methionine-to-valine change but did not affect viral replication (data not shown). In addition to the SalI restriction site, an MluI restriction site was introduced immediately 5′ of the vpr initiation codon so that vif variants of interest could be inserted into this clone (see below). All mutations were made by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
To insert vif variants of interest into the Q23Δvif clone, the vif open reading frame was amplified from TOPO TA plasmids (described above). A single round of PCR was performed by using TaqPrecisionPlus (Stratagene) according to the manufacturer's instructions, with an annealing temperature of 55°C. The forward primer was 5′-GAAGGTCGACATGGAAAACAGATGGCAGG-3′, and the reverse primer was 5′- GTACACGCGTAAGCTCTTCTAACAGTTCTA-3′. Underlined sequences in the forward and reverse primers indicate the SalI and MluI restriction sites, respectively. PCR products representing vif variants were digested using SalI and MluI and cloned into Q23Δvif. This resulted in the variants Q23/QA039vif, Q23/QB368vif, Q23/QA268vif, Q23/QC440vif, and Q23/QF307vif, which were verified by sequence analysis. To create the positive control Q23/Q23vif, the vif open reading frame was amplified from the original Q23 plasmid and reinserted into Q23Δvif. Amplification and cloning were performed as described for the other variants; however, the reverse PCR primer used to amplify the Q23 vif was 5′-GAAGACGCGTGATCATCTAACAGCTCTAAC-3′.
Viruses expressing the vif variants of interest were generated by transfecting 293T cells with the full-length clones by using previously described transfection methods (21). For each transfection, 2 × 106 293T cells were plated and, 24 h later, the cells were transfected with 6 μg of DNA and 18 μl of FuGENE 6 (Boehringer, Mannheim, Germany). Supernatants were collected 48 h after transfection, filtered, and frozen at −80°C until use. Infectious virus titers were obtained by using the previously described TZM-bl assay (4). Briefly, TZM-bl indicator cells (NIH AIDS Research and Reference Reagent Program) were infected for 48 h in the presence of 10 μg of DEAE-dextran/ml, fixed, and stained for β-galactosidase activation, and blue foci were counted (43).
PBMC culture of viruses with vif variants.
PBMC from HIV-negative donors were isolated by the Ficoll gradient method, activated for 48 h with 10 U of phytohemagglutinin M (Roche, Indianapolis, IN)/ml, and maintained in RPMI 1640 medium (Invitrogen) plus 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 10 U of interleukin-2 (Roche)/ml. Viruses with different vif variants were each mixed with 10 × 106 donor PBMC at a multiplicity of infection (MOI) of 0.02 or 0.1, as indicated. After 2 to 3 h of infection, cells were washed three times in PBS and resuspended in 3 ml of media in one well of a six-well dish. Cultures were maintained for ∼14 days, with the medium replaced every 3 to 4 days. The amount of virus in culture supernatants was measured by using a p24 ELISA kit (ZeptoMetrix, Buffalo, NY) every 3 to 4 days. After ∼14 days of culture, DNA was isolated from culture cells by using a Qiagen extraction method, as described above for PBMC samples. The HIV-1 copy number was estimated by using quantitative PCR as described above. To facilitate the examination of a larger number of sequences from culture samples, DNA samples were diluted to an estimated 10 copies per reaction, and gag and env amplification and sequencing was performed as described previously (referred to as “10-copy sequencing”). Hypermutation was defined in these 10-copy sequences by the presence of G-to-A changes or mixed G-A peaks occurring in a GG or GA dinucleotide context. Mixed G-A peaks were defined as positions at which two peaks were visible in the sequence chromatogram, and the smaller peak was substantially larger than any “background” visible across the chromatogram. In general, the smaller peak was greater than 20% of the peak height of the larger peak.
Statistical analyses.
Associations between the percentage of hypermutated sequences and the plasma viral load or CD4 count were determined by using the nonparametric Spearman rank correlation test. The Mann-Whitney test was used to compare plasma viral loads or CD4 counts between individuals with or without hypermutation. The Wilcoxon matched-pairs signed rank test was used to compare the amount of hypermutation in early versus late infection and in gag versus env. All analyses were performed in STATA 9 (Stata, College Station, TX).
GenBank accession numbers.
Sequences discussed in this study are available via GenBank under accession numbers FJ876002 to FJ876042 for gag, FJ876043 to FJ876047 for env, and FJ882081 to FJ882086 for vif.
RESULTS
Defining hypermutated sequences.
In order to characterize the extent of hypermutation among 28 individuals, we examined a total of 465 gag single-copy sequences encompassing a region from the beginning of p17 through the first half of p24. These single-copy sequences were obtained from two times in infection: a median of 107 days postinfection (“early infection”) and a median of 5.4 years postinfection (ypi; “chronic infection”). A median of eight gag single-copy sequences were examined per individual per time point (range, 4 to 17).
We measured the extent of HIV-1 hypermutation within each individual by using the LANL Hypermut 2.0 program. The analysis was first limited to individuals infected with subtype A viruses (n = 23/28) in order to compare the use of the subtype A consensus and individual MRCA as a reference sequence. Compared to the subtype A consensus, a total of 20 of 373 single-copy sequences were identified as hypermutated. Compared to the individual MRCA, 31 of 373 sequences were identified as hypermutated, including the 20 that were identified as hypermutated using the subtype A consensus and an additional 11 sequences. Because no sequences were identified as hypermutated by comparison to only the subtype A consensus, comparison to the individual MRCA allows greater sensitivity in detecting hypermutated sequences. In addition, using the MRCA also permits analysis of sequences from individuals infected with different subtypes. The Hypermut 2.0 analysis was repeated using the individual MRCA for all 28 individuals, including those with subtype C and D viruses, and a total of 43 of 465 single-copy sequences were found to be hypermutated.
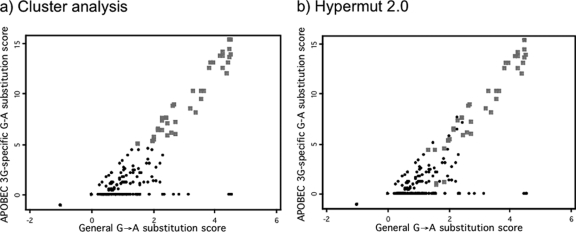
We also compared the Hypermut 2.0 method to a k-means cluster analysis similar to that used by Pace et al. (31). Whereas 43 of 465 hypermutated sequences were found by Hypermut 2.0, 45 of 465 hypermutated sequences were found by cluster analysis (Fig. 1). Of the six sequences identified as hypermutated by Hypermut 2.0 but not cluster analysis, five had Hypermut 2.0 P values between 0.03 and 0.05, suggesting that cluster analysis may be more conservative than the P < 0.05 cutoff we used with Hypermut 2.0. The eight sequences identified as hypermutated by cluster analysis but not Hypermut 2.0 had high GG→AG scores but relatively low G→A scores. This is consistent with the fact that Hypermut 2.0 does not take into account the context of G→A changes, whereas the cluster analysis gives more weight to changes in a GG→AG context. In general, however, there was good agreement between these methods, with 36 of 465 sequences identified as hypermutated by both methods and 412 of 465 sequences not identified as hypermutated by either method. The results using the Hypermut 2.0 definition of hypermutation are presented here, in order to better compare our results to those of other recent studies that have used this method.
FIG. 1.
Definition of hypermutation by two methods. In each plot, the APOBEC3G-specific score for each sequence is plotted against its general G→A score (see Materials and Methods). In panel a, sequences identified as hypermutated by k-means cluster analysis are indicated as gray squares, while those not hypermutated are black circles. In panel b, sequences identified as hypermutated by Hypermut 2.0 are indicated as gray squares, while those not hypermutated are black circles.
Characterization of hypermutation in infected individuals.
Using Hypermut 2.0 and comparison to the individual MRCA, a total of 9.2% of the gag single-copy sequences examined were hypermutated (43/465). As shown in Table 1, hypermutated sequences had a much higher G→A substitution frequency (median = 8.4% of Gs changed to As) than nonhypermutated sequences (median = 0.7%), although the overall substitution frequencies in these groups were similar. Hypermutated sequences also had a substantially higher GG→AG substitution frequency than nonhypermutated sequences (median = 22.5% versus 0%, respectively). The GA→AA substitution frequency was low in both groups, indicating that most G→A substitutions occurred in the APOBEC3G-preferred context (GG) rather than the APOBEC3F-preferred context (GA). Most of the hypermutated sequences (35/43) had at least one nonsense mutation within the ∼600-bp gag region examined.
TABLE 1.
Substitution frequencies in gag proviral sequences
| Substitution | Median frequency (%)a
|
|
|---|---|---|
| Not hypermutated | Hypermutated | |
| Overall | 1.0 (0-10) | 3.3 (1-6) |
| G→A | 0.7 (0-7) | 8.4 (3-9) |
| GG→AG | 0 (0-15) | 22.5 (3-50) |
| GA→AA | 0 (0-10) | 2.5 (0-10) |
The median frequencies and ranges (indicated in parentheses) are shown for 422 sequences defined as not hypermutated by Hypermut 2.0 and 43 sequences defined as hypermutated by Hypermut 2.0.
Overall, sixteen out of 28 individuals (57%) had at least one hypermutated gag sequence (Table 2) . Among those individuals with at least one hypermutated sequence, individuals had a median of 15% hypermutated sequences in gag (range, 5 to 43%). There was a trend for more hypermutation at the chronic infection time point than the early time point (median = 14 and 11.5%, respectively, of the sequences hypermutated; Wilcoxon P = 0.1).
TABLE 2.
Characterization of hypermutation in gag and env sequences from 28 individuals at two times during infection
| Sequence | No. of individuals (%) with any hypermutation
|
Median % of sequences hypermutated (range)a
|
||||
|---|---|---|---|---|---|---|
| Early infection | Chronic infection | Total | Early infection | Chronic infection | Total | |
| gag | 10 (35.7) | 13 (46.4) | 16 (57.0) | 11.5 (0-25) | 14 (0-54) | 15 (5-43) |
| env | 2 (7.1) | 3 (10.7) | 4 (14.3) | 4.4 (0-9.1) | 11.7 (0-20) | 7.6 (5.6-11) |
That is, among individuals with any sequences hypermutated.
We also examined hypermutation among a total of 437 env V1-V5 single-copy sequences sampled at the same time points. Only five hypermutated sequences (1.1%) were detected, as shown in Table 2. Of the 28 individuals, 3 had one hypermutated env sequence, and 1 had two hypermutated env sequences. Individuals had a significantly higher percentage of hypermutated sequences in gag compared to env (Wilcoxon P = 0.0003). The low level of detection of hypermutation in env is not likely to be due to the fact that highly variable regions were excluded from our analyses; the full V1-V5 sequences were visually inspected for a subset of sequences, and no additional hypermutation was detected (not shown). Because of the small number of hypermutated env sequences identified, further analyses are limited to gag sequences.
Relationships between hypermutation and markers of disease progression.
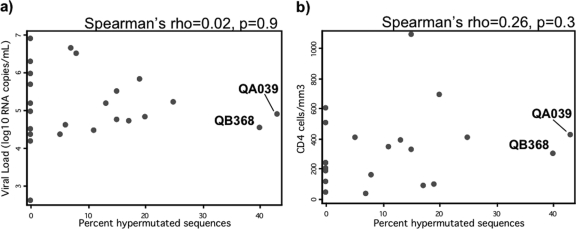
We investigated whether the level of hypermutation, which often leads to the production of defective proviruses, was associated with slower disease progression in these individuals. Plasma viral load measured at the chronic infection time point and CD4 count measured at or within 6 months of the chronic infection time point were used as markers of disease progression. As shown in Fig. 2, there was no relationship between the percentage of hypermutated gag single-copy sequences and either viral load (Fig. 2a, Spearman's rho = 0.02, P = 0.9) or CD4 count (Fig. 2b, Spearman's rho = 0.26, P = 0.3). We also compared these markers of disease progression between individuals with no hypermutated sequences (n = 12) and those with any hypermutated sequences (n = 16). The median viral loads in the no-hypermutation and any-hypermutation groups were 5.1 (range, 2.6 to 6.9) and 4.9 (range, 4.3 to 6.6), respectively, and there was no significant difference between these groups (Mann-Whitney P = 0.95). There was also no difference in CD4 counts, which had a median of 220 cells/mm3 in the no-hypermutation group (range, 44 to 599) and 343 cells/mm3 in the any-hypermutation group (range, 37 to 1,088) (Mann-Whitney P = 0.47). Therefore, our results indicate that greater gag hypermutation is not associated with slower disease progression among these individuals.
FIG. 2.
Associations between the level of gag proviral hypermutation and markers of disease progression. Each point represents one individual, with the percentage of gag sequences that were identified as hypermutated indicated on the x axis. Plasma viral load (a) and CD4 count (b) measured in chronic infection are indicated on the y axis. Associations were evaluated by using the Spearman rank correlation test, with results indicated above the plots. The two individuals with extensive hypermutation (at least 40%) are labeled.
In order to compare our results to those of prior studies that used population sequencing, we classified individuals in our study as having at least 20% hypermutated sequences or less than 20% hypermutated sequences (including those with no hypermutation). Only 4 of the 28 individuals (14%) in the present study had more than 20% hypermutated sequences. There was no difference in viral load between these groups, although there was a trend for higher CD4 count among individuals with >20% hypermutated sequences (median CD4 count = 418 versus 241, Mann-Whitney P = 0.07).
Extensive hypermutation in two individuals with unusual vif polymorphisms.
As illustrated in Fig. 2, there were two individuals with extensive hypermutation in gag (40% or more hypermutated sequences). To determine whether hypermutated sequences were present in actively replicating viruses in these individuals, we examined gag sequences from plasma RNA at the chronic infection time point. Using population sequencing starting from an estimated 10 HIV-1 RNA copies per reaction, we did not find evidence of hypermutation in any of eight sequencing reactions (corresponding to ∼80 total copies) from each individual. These results are consistent with prior studies showing evidence of hypermutation in proviruses but not in virions (17, 18).
We hypothesized that the extensive and persistent hypermutation observed in proviruses from these two individuals, QA039 and QB368, might be due in part to Vif defects. To investigate this, vif sequences were obtained from these individuals, as well as 25 other individuals from this group, and compared to vif sequences available in the LANL database. Two unusual polymorphisms were present in these individuals and are illustrated in Fig. 3. The first is a two-amino-acid (RP) insertion after position 161 of Vif (based on HXB2), which was observed in sequences from QA039 in both early and chronic infection. The second is a glutamine substitution at the C-terminal amino acid of Vif, which was found in both QA039 and QB368. These Vif variants were the only variants detected in both PBMC and plasma samples (results not shown).
FIG. 3.
Alignment of Vif sequences. This alignment includes sequences from the two individuals with extensive hypermutation (QA039 and QB368) at both early (E) and chronic (C) infection time points, three other randomly chosen individuals in the present study (QA268, QC340, and QF307), and a subtype A reference strain from Kenya (Q23). All sequences are subtype A based on phylogenetic analysis (not shown) and are aligned against the subtype A consensus sequence (A con). Amino acid substitutions are indicated with capital letters, and positions with a mixture of amino acids are indicated with lowercase letters. The two polymorphisms of interest are highlighted in gray. *, Stop codon.
To investigate whether these polymorphisms might have contributed to the extensive hypermutation observed in vivo, vif variants were cloned from QA039, QB368, and three other individuals in this group. These vif variants were inserted into a full-length, subtype A backbone virus defective for vif, Q23Δvif, and cultured in human PBMC. Hypermutation in culture DNA was assessed by examination of gag sequences representing an estimated 10 copies per reaction (“10-copy sequences”), which allowed us to examine a greater total number of sequences. Hypermutation was identified by the presence of G-A substitutions or mixed peaks containing both Gs and As in the sequence chromatogram (as illustrated in Fig. 4).
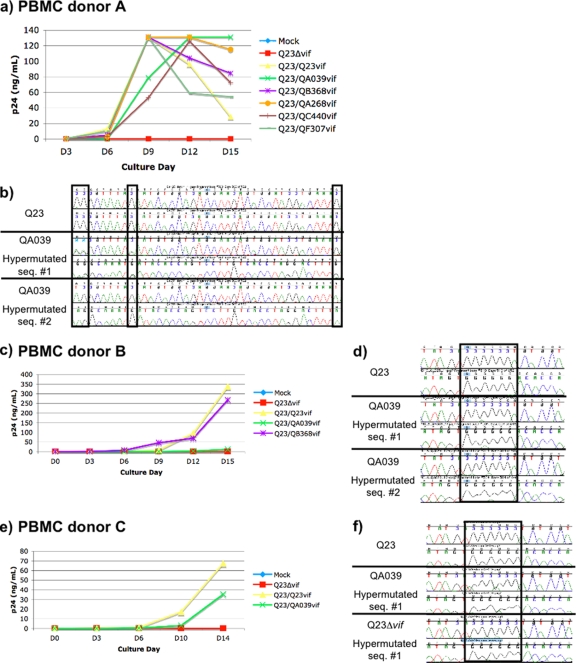
FIG. 4.
Virus replication and hypermutation in PBMC cultures of vif variants. (a) Plot of p24 pg/ml versus day of culture for each vif variant in the first set of cultures (PBMC from donor A, infected at an MOI of 0.02). Q23Δvif (red line) is the negative control; Q23/Q23vif (yellow) is the vif defective clone with its own vif reinserted; Q23/QA039vif (green) and Q23/QB368vif (purple) contain vif variants from individuals with extensive hypermutation; and Q23/QA268vif, Q23/QC440vif, and Q23/QF307vif are vif variants from other individuals without extensive hypermutation. (b) Example of a gag sequence from the Q23 culture (top) and two gag sequences from the QA039 culture (middle and bottom) for donor A. For each sequence, a region of both the forward and reverse sequencing chromatograms is shown. The positions in which G-to-A changes or mixed G-A peaks were identified are indicated in boxes. For QA039 hypermutated sequence 1, three positions with mixed peaks are indicated in the first two boxes. For QA039 hypermutated sequence 2, two G-to-A substitutions are indicated in the first and third boxes. Cultures were repeated for a subset of the vif variants in PBMC from two different donors infected at an MOI of 0.1, and plots of p24 versus day of culture are shown for donor B (c) and donor C (e). (d) Two hypermutated sequences from the QA039 culture for donor B are shown, with mixed peaks highlighted in the box. (f) One sequence each from the Q23, QA039, and Q23Δvif cultures for donor C. The box highlights a region in which two mixed peaks were found in QA039 and three in Q23Δvif.
In the first set of cultures, PBMC (from donor A) were infected with viruses containing vif from the two individuals of interest—Q23/QA039vif and Q23/QB368vif—and three other individuals who did not have evidence of extensive hypermutation: Q23/QA268vif, Q23/QC440vif, and Q23/QF307vif (whose sequences are shown in Fig. 3). The original Q23 with its own vif, Q23/Q23vif was included as a positive control, and Q23Δvif served as a negative control. As shown in Fig. 4a, Q23/QA039vif showed somewhat delayed replication relative to the other viruses, but by day 12 of culture, this virus had reached a peak p24 level similar to the other viruses. There was negligible replication of Q23Δvif virus, as expected given that PBMC are nonpermissive cells (8). vif sequences were examined from extracted culture DNA, and no reversion of vif polymorphisms was observed (data not shown).
The results of gag 10-copy sequencing from these cultures are summarized in Table 3. At least 10 gag 10-copy sequences were examined from each of the cultures except Q23Δvif, which did not replicate to a high enough level. The one sequence examined from the Q23Δvif culture showed evidence of hypermutation, with multiple mixed G-A peaks (results not shown). As shown in Table 3, there was no evidence of hypermutation in the cultures with Q23/Q23vif, Q23/QB368vif, Q23/QC440vif, or Q23/QF307vif. There was evidence of hypermutation in 2 of 11 sequencing reactions from the Q23/QA039vif culture (Table 3). One of these sequences had mixed G-A peaks at 8 positions (out of 125 total guanine positions), while the second was dramatically hypermutated, with G-to-A substitutions at 13 positions. Figure 4b shows a sequence chromatogram of a ∼40-bp region from these two hypermutated gag sequences, with positions containing mixed G-A peaks (sequence 1) and G-to-A substitutions (sequence 2) indicated in boxes. The low level of background peaks in these chromatograms is representative of the sequences as a whole.
TABLE 3.
Detection of hypermutation in PBMC cultures of vif variants
| PBMC donor | vif variant | No. of 10-copy sequencing reactions
|
No. of G-to-A changes/sequencec | |
|---|---|---|---|---|
| Totala | With hypermutationb | |||
| Donor A | Q23 | 11 | 0 | 0 |
| Q23Δvif | 1 | 1 | 6 | |
| QA039 | 11 | 2 | 8, 13 | |
| QB368 | 13 | 0 | 0 | |
| QA268 | 10 | 0 | 0 | |
| QC440 | 10 | 0 | 0 | |
| Donor B | Q23 | 10 | 0 | 0 |
| Q23Δvif | 2 | 2 | 13, 10 | |
| QA039 | 20 | 4 | 3, 6, 3, 3 | |
| QB368 | 11 | 0 | 0 | |
| Donor C | Q23 | 9 | 0 | 0 |
| Q23Δvif | 1 | 1 | 5 | |
| QA039 | 16 | 2 | 7, 2 | |
The total number of gag proviral 10-copy sequencing reactions.
The number of sequencing reactions with G-to-A substitutions or mixed G-A peaks detected.
The number of G-to-A substitutions or mixed G-A peaks in each hypermutated sequencing reaction, with different hypermutated sequencing reactions from each culture separated by commas.
In order to verify that hypermutation was occurring with the QA039 vif, we repeated the cultures in PBMC from two additional donors. In the second set of cultures, PBMC (from donor B) were infected using only the variants of interest and controls. As shown in Fig. 4c, replication of all viruses was delayed compared to the first set of cultures, potentially reflecting donor-to-donor variability in PBMC cultures. Once again, we did not detect hypermutation in at least 10 gag 10-copy sequencing reactions from Q23/Q23vif or Q23/QB368vif, whereas there was extensive hypermutation in both of the sequencing reactions from Q23Δvif (Table 3). We also observed evidence of hypermutation in 4 of 20 sequencing reactions from Q23/QA039vif (Table 3 and Fig. 4d). In the third set of cultures (donor C), Q23/QA039vif replicated to an intermediate level between Q23/Q23vif and Q23Δvif (Fig. 4e). We found hypermutation in 2 of 16 sequencing reactions from the Q23/QA039vif culture (Table 3 and Fig. 4d).
We also investigated whether there was detectable hypermutation in the env V1-V5 region using the Q23/QA039vif donor C culture sample. We examined a total of 12 10-copy sequences and found no hypermutation in this region (not shown). To assess whether the lack of hypermutation detected in env was due to the specific primers used, we tested two additional sets of env primers using the Q23/QA039vif culture samples. We did not detect any hypermutation in a total of 11 sequencing reactions encompassing the V1-V3 region and four sequencing reactions encompassing the open reading frame encoding gp160 (not shown).
In summary, in all three sets of PBMC cultures, a low level of hypermutation was consistently detected in gag sequences from the Q23/QA039vif cultures but not from the control Q23/Q23vif. In cultures containing other vif variants, including the vif from the second individual with a high level of hypermutation in vivo (QB368), there was also no hypermutation detected. Although hypermutation was consistently detected with the Q23/QA039vif, it was not as extensive as with the Q23Δvif negative control, leading to our assessment of this variant as a “partially defective Vif.”
Longitudinal analysis of hypermutation in QA039.
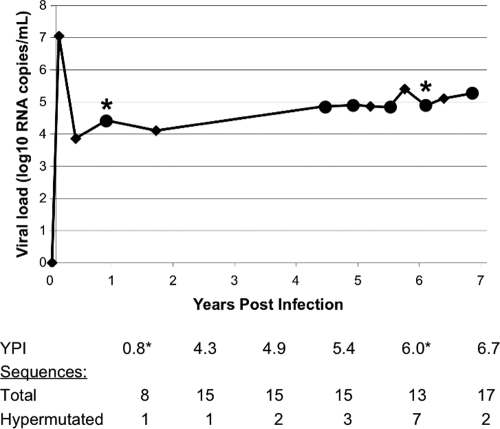
Because the virus from QA039 had a partially defective vif that was present in both early infection and chronic infection, we investigated the level of hypermutation throughout infection in this individual. Figure 5 shows a plot of this individual's plasma viral load through time, with the original early and chronic infection time points marked by asterisks. We also sampled gag single-copy sequences from three intervening time points—4.3, 4.9, and 5.4 ypi—as well as a later time point, 6.7 ypi. We found 8 hypermutated sequences out of 58 total sequences from these additional time points (14%), and at least one hypermutated sequence was present at each time point. It is also evident from this figure that QA039 had a stable and moderately high viral load, on the order of 4 to 5 log10 copies/ml of plasma, through the almost 7 years of available follow-up. This viral load pattern is typical for individuals in this cohort (19, 20) and suggests that hypermutation did not significantly impair HIV-1 replication in this individual.
FIG. 5.
Evaluation of hypermutation throughout infection in QA039. The plot indicates this individual's viral load through time. Diamonds represent follow-up visits, and those at which sequences were examined are marked with large circles. The early infection and chronic infection time points used in the initial screen for hypermutation are marked by asterisks. The table below the graph shows the total number of gag proviral sequences that were examined at each time point, as well as the number of sequences that were found to be hypermutated.
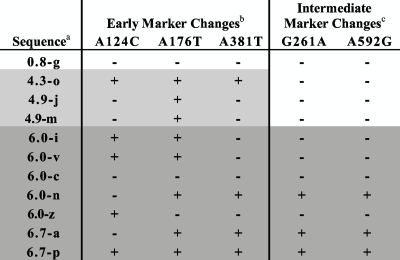
Using the sequences obtained from longitudinal samples, we evaluated whether the hypermutated sequences arose throughout infection or represented archived viruses from early in infection. Based on an alignment of all longitudinal sequences from QA039 (see Fig. S1 in the supplemental material), we identified several non-G→A changes that became fixed at various times in infection. For example, changes at positions 124, 176, and 371 were not present at 0.8 ypi but were present in most sequences after that time point; thus, these serve as “early marker changes” for sequences arising after 0.8 ypi. Similarly, changes at positions 261 and 592 appeared in a few sequences at 4.9 ypi and became predominant at 6.0 ypi; thus, these serve as “intermediate marker changes” for sequences that arose after 4.9-6.0 ypi.
By determining whether each hypermutated sequence contained these marker changes, we estimated approximately when the hypermutated sequences in QA039 were generated. Table 4 indicates whether or not each hypermutated sequence had each of the five marker changes. For example, sequence 0.8-g (sampled at 0.8 ypi) does not contain any marker changes, which is consistent with its early time of sampling. Sample 4.3-o contains all three of the early marker changes, indicating that it became hypermutated between 0.8 and 4.3 ypi. In contrast, sequences 4.9-m and 4.9-j (which are identical) contain only one of the three early marker changes and therefore are likely to represent sequences archived from earlier in infection. Three hypermutated sequences (6.0-n, 6.7a, and 6.7p) contain all or almost all of the marker changes, suggesting that they arose after the intermediate marker changes became fixed, between 4.9 and 6.0 ypi. Overall, there was some evidence for archived hypermutated sequences. However, some sequences continued to become hypermutated throughout infection, suggesting that the process of hypermutation in this individual was ongoing.
TABLE 4.
Presence or absence of marker changes in hypermutated gag single-copy sequences from QA039 longitudinal samples
a The names of hypermutated sequences from QA039 indicate the ypi from which the sequence was obtained, followed by a unique identifier.
b Early marker changes at positions 124, 176, and 371 became fixed after 0.8 ypi. Sequences that were sampled after this time are highlighted in gray boxes. For each sequence, the presence or absence of a marker change is indicated by “+” or “−”, respectively.
c Intermediate marker changes at positions 261 and 592 became fixed between 4.9 and 6.0 ypi. Sequences that were sampled after this time are highlighted in dark gray boxes. For each sequence, the presence or absence of a marker change is indicated by “+” or “−”, respectively.
DISCUSSION
The results of this study demonstrate that HIV-1 hypermutation can be detected in many antiretroviral-naive, HIV-infected individuals, and the level of hypermutation in these individuals is quite variable. Using single-copy sequencing, we found that over half of the individuals in the present study (16/28) had detectable hypermutation in gag. This rate of detection is consistent with prior studies that sequenced many genomes (9, 15, 17) and is substantially higher than studies that used population sequencing (18, 31), as would be expected given the increased sensitivity of single-copy sequencing. We found no association between the level of hypermutation and either viral load or CD4 count. Thus, our results indicate that there is no quantitative relationship between hypermutation and markers of disease progression across the spectrum of hypermutation that is observed in most individuals.
In comparing methods to define hypermutated sequences, we found that using the individual MRCA, which was reconstructed from sequences from early infection, was more sensitive in detecting hypermutation than using a subtype consensus sequence. This comparison highlights the importance of using a common method to define hypermutation. We also compared two methods that have been used to define hypermutated sequences, the LANL Hypermut 2.0 program and a cluster analysis similar to that used by Pace et al. (31), and found good concordance between these methods.
It is puzzling that we found greater hypermutation in gag compared to env, since this is inconsistent with prior studies that have described a gradient of hypermutation increasing from the 5′ to the 3′ end of the genome in in vitro studies (5, 46). Another study of sequences from HIV-infected individuals found no difference between gag and env (32). The difference that we observed is not due to the specific primers used to amplify env, since other primers were tested with no additional hypermutation found. It is also not likely to be due to differences in the number of potential APOBEC3G target sites in each gene. The gag and env regions examined had a similar number of guanine positions (135 and 140, respectively) and GG dinucelotide sites (40 and 30, respectively).
One question that was not fully addressed by our study is whether very high levels of hypermutation are associated with improved clinical outcome, as has been suggested by two prior studies (18, 32). Because individuals with such high levels of hypermutation are rare, we did not detect many in this relatively small study. Four individuals had >20% hypermutated sequences, which roughly corresponds to what would be detected by population sequencing. There was a trend for higher CD4 count among these individuals, which is consistent with a prior study (18). Larger studies are needed to unravel the relationship between hypermutation and disease progression; in particular, larger studies examining multiple sequences per individual may help to identify a threshold level of hypermutation that offers clinical benefit.
However, the results of the present study suggest that a high level of hypermutation does not always confer a clinical benefit. We identified two individuals who had quite high levels of hypermutation (40% or more hypermutated gag sequences) but sustained viral loads that were typical for this cohort (19, 20). Upon evaluating the potential role of Vif in contributing to the high level of hypermutation in these individuals, we found that a virus encoding the QA039 vif consistently generated hypermutated sequences, while a virus with the QB368vif did not have detectable hypermutation. There is a plausible biological explanation for this difference, in that the two-amino-acid insertion found only in QA039 occurred in a domain that is important for Vif dimerization, PPLP (45). Mutation of this domain leads to impaired APOBEC degradation (6). In contrast, the glutamine substitution found in both individuals may not be functionally significant because it occurs in a region of Vif that can be truncated, sometimes without consequences for virus replication (30, 39). It is likely that there is another factor contributing to the extensive hypermutation found in QB368, such as increased APOBEC3G level or activity. Comparison of these two cases highlights the large degree of complexity in the potential determinants of hypermutation.
In the case of QA039, it is intriguing that we observed consistent hypermutation both in vivo and in vitro, but there was no overall impairment of virus replication in either setting. The hypermutated proviruses themselves were almost certainly defective; they frequently contained stop codons, and they did not appear in the pool of replicating viruses in plasma. These observations suggest that the HIV-1 population in this individual was able to sustain a moderate amount of hypermutation (leading to defective proviruses) without affecting the overall level of replication of the virus population. In addition, the persistence of the partially defective vif over 5 years of infection suggests that there was no significant selective pressure against it. Similar to our results, a recent study demonstrated that vif-deficient viruses, which had acquired compensatory mutations, were able to replicate despite extensive G-to-A changes (11).
Overall, the results of the present study indicate that a small to moderate amount of hypermutation is common in HIV-infected individuals and is not associated with improved markers of disease progression in untreated individuals. Our results do not exclude the possibility that a high level of hypermutation may be associated with a clinical benefit in some individuals, as has been suggested by others (18, 31); however, this may not be true for all individuals. This is an important consideration in light of proposals to use Vif/APOBEC as targets for novel antiviral strategies (16, 26, 28, 40, 47). Although there is also evidence for a nonenzymatic effect of APOBEC (2, 3, 22, 29), the relative contribution of nonenzymatic and enzymatic (hypermutation) mechanisms to HIV restriction remains unclear, especially for APOBEC3G (3, 14, 27, 37). Our results emphasize that a potential antiviral strategy based on hypermutation may require a very high level of hypermutation to impair virus replication.
Supplementary Material
Acknowledgments
We thank the women who participated in this study. We also thank the Mombasa clinic and lab staff for technical assistance and Jaisri Lingappa and Michael Emerman for helpful discussions and comments on the manuscript.
This study was supported by NIH R01 AI38518 to J.O. A.P. was supported in part by training grants T32 A107140 and T32 GM07266, and B.C. was supported by D43-00007 through the University of Washington International AIDS Research and Training Program.
Footnotes
Published ahead of print on 3 June 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Benki, S., R. S. McClelland, S. Emery, J. M. Baeten, B. A. Richardson, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2006. Quantification of genital human immunodeficiency virus type 1 (HIV-1) DNA in specimens from women with low plasma HIV-1 RNA levels typical of HIV-1 nontransmitters. J. Clin. Microbiol. 444357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 808450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, K. N., M. Verma, E. Y. Kim, S. M. Wolinsky, and M. H. Malim. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blish, C. A., M. A. Nguyen, and J. Overbaugh. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelico, L., P. Pham, P. Calabrese, and M. F. Goodman. 2006. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13392-399. [DOI] [PubMed] [Google Scholar]
- 6.Donahue, J. P., M. L. Vetter, N. A. Mukhtar, and R. T. D'Aquila. 2008. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology 37749-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emery, S., S. Bodrug, B. A. Richardson, C. Giachetti, M. A. Bott, D. Panteleeff, L. L. Jagodzinski, N. L. Michael, R. Nduati, J. Bwayo, J. K. Kreiss, and J. Overbaugh. 2000. Evaluation of performance of the Gen-Probe human immunodeficiency virus type 1 viral load assay using primary subtype A, C, and D isolates from Kenya. J. Clin. Microbiol. 382688-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 666489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandhi, S. K., J. D. Siliciano, J. R. Bailey, R. F. Siliciano, and J. N. Blankson. 2008. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J. Virol. 823125-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goila-Gaur, R., and K. Strebel. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hache, G., K. Shindo, J. S. Albin, and R. S. Harris. 2008. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 18819-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna, G. J., and R. T. D'Aquila. 2001. Clinical use of genotypic and phenotypic drug resistance testing to monitor antiretroviral chemotherapy. Clin. Infect. Dis. 32774-782. [DOI] [PubMed] [Google Scholar]
- 13.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4868-877. [DOI] [PubMed] [Google Scholar]
- 14.Holmes, R. K., M. H. Malim, and K. N. Bishop. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32118-128. [DOI] [PubMed] [Google Scholar]
- 15.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 757973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeffrey, F. W. 2005. A new approach to an AIDS vaccine: creating antibodies to HIV Vif will enable apobec3G to turn HIV-infection into a benign problem. Med. Hypotheses 64261-263. [DOI] [PubMed] [Google Scholar]
- 17.Kieffer, T. L., P. Kwon, R. E. Nettles, Y. Han, S. C. Ray, and R. F. Siliciano. 2005. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 791975-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Land, A. M., T. B. Ball, M. Luo, R. Pilon, P. Sandstrom, J. E. Embree, C. Wachihi, J. Kimani, and F. A. Plummer. 2008. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. J. Virol. 828172-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavreys, L., J. M. Baeten, V. Chohan, R. S. McClelland, W. M. Hassan, B. A. Richardson, K. Mandaliya, J. O. Ndinya-Achola, and J. Overbaugh. 2006. Higher set point plasma viral load and more-severe acute HIV type 1 (HIV-1) illness predict mortality among high-risk HIV-1-infected African women. Clin. Infect. Dis. 421333-1339. [DOI] [PubMed] [Google Scholar]
- 20.Lavreys, L., J. M. Baeten, J. K. Kreiss, B. A. Richardson, B. H. Chohan, W. Hassan, D. D. Panteleeff, K. Mandaliya, J. O. Ndinya-Achola, and J. Overbaugh. 2004. Injectable contraceptives use and genital ulcer disease during early human immunodeficiency virus (HIV) type 1 infection increase plasma virus load among women. J. Infect. Dis. 189303-311. [DOI] [PubMed] [Google Scholar]
- 21.Long, E. M., S. M. J. Rainwater, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retrovir. 18567-576. [DOI] [PubMed] [Google Scholar]
- 22.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 817238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddison, D., and W. Maddison. 2005. Macclade 4: analysis of phylogeny and character evolution. Sinauer, Sunderland, MA. [DOI] [PubMed]
- 24.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 42499-103. [DOI] [PubMed] [Google Scholar]
- 25.Martin, H. L., P. M. Nyange, B. A. Richardson, L. Lavreys, K. Mandaliya, D. J. Jackson, J. O. Ndinya-Achola, and J. Kreiss. 1998. Hormonal contraception, sexually transmitted diseases, and the risk of heterosexual transmission of HIV-1. J. Infect. Dis. 1781053-1059. [DOI] [PubMed] [Google Scholar]
- 26.Mezei, M., and J. Minarovits. 2006. Reversal of HIV drug resistance and novel strategies to curb HIV infection: the viral infectivity factor Vif as a target and tool of therapy. Curr. Drug Targets 7881-885. [DOI] [PubMed] [Google Scholar]
- 27.Miyagi, E., S. Opi, H. Takeuchi, M. Khan, R. Goila-Gaur, S. Kao, and K. Strebel. 2007. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 8113346-13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathans, R., H. Cao, N. Sharova, A. Ali, M. Sharkey, R. Stranska, M. Stevenson, and T. M. Rana. 2008. Small-molecule inhibition of HIV-1 Vif. Nat. Biotechnol. 261187-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15166-170. [DOI] [PubMed] [Google Scholar]
- 30.Ochsenbauer, C., V. Bosch, I. Oelze, and U. Wieland. 1996. Unimpaired function of a naturally occurring C terminally truncated vif gene product of human immunodeficiency virus type 1. J. Gen. Virol. 77(Pt. 7)1389-1395. [DOI] [PubMed] [Google Scholar]
- 31.Pace, C., J. Keller, D. Nolan, I. James, S. Gaudieri, C. Moore, and S. Mallal. 2006. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J. Virol. 809259-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piantadosi, A., B. Chohan, V. Chohan, R. S. McClelland, and J. Overbaugh. 2007. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 3e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poss, M., and J. Overbaugh. 1999. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 735255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose, P. P., and B. T. Korber. 2000. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics 16400-401. [DOI] [PubMed] [Google Scholar]
- 35.Rousseau, C. M., R. W. Nduati, B. A. Richardson, G. C. John-Stewart, D. A. Mbori-Ngacha, J. K. Kreiss, and J. Overbaugh. 2004. Association of levels of HIV-1-infected breast milk cells and risk of mother-to-child transmission. J. Infect. Dis. 1901880-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakurai, A., A. Jere, A. Yoshida, T. Yamada, A. Iwamoto, A. Adachi, and M. Fujita. 2004. Functional analysis of HIV-1 vif genes derived from Japanese long-term nonprogressors and progressors for AIDS. Microbes Infect. 6799-805. [DOI] [PubMed] [Google Scholar]
- 37.Schumacher, A. J., G. Hache, D. A. Macduff, W. L. Brown, and R. S. Harris. 2008. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 822652-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 91404-1407. [DOI] [PubMed] [Google Scholar]
- 39.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stopak, K., and W. C. Greene. 2005. Protecting APOBEC3G: a potential new target for HIV drug discovery. Curr. Opin. Investig. Drugs 6141-147. [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulenga, N. K., A. D. Sarr, D. Hamel, J. L. Sankale, S. Mboup, and P. J. Kanki. 2008. The level of APOBEC3G (hA3G)-related G-to-A mutations does not correlate with viral load in HIV type 1-infected individuals. AIDS Res. Hum. Retrovir. 241285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vodicka, M. A., W. C. Goh, L. I. Wu, M. E. Rogel, S. R. Bartz, V. L. Schweickart, C. J. Raport, and M. Emerman. 1997. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology 233193-198. [DOI] [PubMed] [Google Scholar]
- 44.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. Hahn, M. S. Saag, and G. M. Shaw. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373117-126. [DOI] [PubMed] [Google Scholar]
- 45.Yang, W., J. P. Bielawski, and Z. Yang. 2003. Widespread adaptive evolution in the human immunodeficiency virus type 1 genome. J. Mol. Evol. 57212-221. [DOI] [PubMed] [Google Scholar]
- 46.Yu, K., F. T. Huang, and M. R. Lieber. 2004. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2796496-6500. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 42494-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwickl, D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas, Austin.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.