Abstract
Viral infection triggers host innate immune responses through cellular sensor molecules which activate multiple signaling cascades that induce the production of interferons (IFN) and other cytokines. The recent identification of mammalian cytoplasmic viral RNA sensors, such as retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) and their mitochondrial adaptor, the mitochondrial antiviral signaling protein (MAVS), also called IPS-1, VISA, and Cardif, highlights the significance of these molecules in the induction of IFN. Teleost fish also possess a strong IFN system, but nothing is known concerning the RLRs and their downstream adaptor. In this study, we cloned MAVS cDNAs from several fish species (including salmon and zebrafish) and showed that they were orthologs of mammalian MAVS. We demonstrated that overexpression of these mitochondrial proteins in fish cells led to a constitutive induction of IFN and IFN-stimulated genes (ISGs). MAVS-overexpressing cells were almost fully protected against RNA virus infection, with a strong inhibition of both DNA and RNA virus replication (1,000- and 10,000-fold decreases, respectively). Analyses of MAVS deletion mutants showed that both the N-terminal CARD-like and C-terminal transmembrane domains, but not the central proline-rich region, were indispensable for MAVS signaling function. In addition, we cloned the cDNAs encoding a RIG-I-like molecule from salmonid and cyprinid cell lines. Like the case with MAVS, overexpression of RIG-I CARDs in fish cells led to a strong induction of both IFN and ISGs, conferring on fish cells full protection against RNA virus infection. This report provides the first demonstration that teleost fish possess a functional RLR pathway in which MAVS may play a central role in the induction of the innate immune response.
Host mammalian cells recognize viral components through pattern recognition receptors (PRRs) activating multiple signaling cascades that induce the production of interferons (IFN) and other cytokines (2, 25, 26). This innate immune response plays a major role in the early control of infection and is also involved in the induction of the specific adaptive immune response. To date, the following three classes of PRRs have clearly been shown to be involved in virus-specific component detection: Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and nucleotide oligomerization domain-like receptors (NLRs). These PRRs detect several types of pathogen-associated molecular patterns, such as genomic DNA, single-stranded RNA (ssRNA), double-stranded RNA (dsRNA), 5′-end triphosphate ssRNA (5′pppRNA), and viral proteins. RLRs and TLRs are essential in the induction of type I IFN and proinflammatory cytokines in a cell type-specific manner, whereas NLRs participate in multiprotein complexes, called “inflammasomes,” and play a role in the production of mature interleukin-1β (24). While RLRs and NLRs sense the presence of cytosolic viral components, TLRs are present on the cell surface or in endosomal membranes. Recently, the DNA-dependent activator of IFN regulatory factors (DAI), also called DLM-1/ZBP1, was identified as a candidate cytosolic DNA sensor and should represent another class of PRRs (36).
RLRs known to play a key role in sensing RNA virus invasion comprise three helicases, including RIG-I (also known as DDX58) (44), melanoma differentiation-associated gene 5 (MDA5) (10, 12, 13), and laboratory of genetics and physiology 2 (LGP2) (28, 43). RIG-I and MDA5 consist of a DEXD/H box RNA helicase domain and two N-terminal caspase recruitment domains (CARDs) involved in signaling. LGP2 contains the helicase domain but lacks the CARDs and is thought to be a negative regulator (15, 28, 37). RIG-I binds preferentially, but not exclusively, to ssRNAs phosphorylated at the 5′ end, whereas MDA5 recognizes long dsRNAs that do not require 5′ phosphorylation (31). This difference in ligand preference results in specificity for the recognition of individual viruses (42).
The downstream adaptor protein located on the outer mitochondrial membrane is the mitochondrial antiviral signaling protein (MAVS), also known as IFN-β promoter stimulator 1 (IPS-1), virus-induced signaling adaptor (VISA), and CARD adaptor inducing IFN-β (Cardif) (14, 22, 32, 41). After viral RNA binding, RIG-I undergoes an ATP-dependent conformational change that exposes the two N-terminal CARDs and induces oligomerization, which allows the CARDs to interact with MAVS. Human MAVS contains an N-terminal CARD, a proline-rich (Pro) region, and a C-terminal mitochondrial transmembrane (TM) sequence. Overexpression of MAVS significantly induces type I IFNs and IFN-stimulated genes (ISGs) (14, 22, 32, 41), and conversely, deficiency in MAVS expression impairs the antiviral response and increases susceptibility to RNA virus infection in vitro and in vivo (16, 35), indicating that MAVS plays a central role in the innate antiviral response. Activation of MAVS induces the recruitment of downstream signaling molecules, leading to the activation of the transcription factors interferon regulatory factor 3/7 (IRF-3/7) and NF-κB, allowing their translocation into the nucleus and the expression of the type I IFNs and inflammatory cytokines (42).
In recent years, intensive commercial farming of different fish species and the emergence of zebrafish as a powerful new vertebrate model of human disease have generated increasing interest in fish immune responses (21, 34, 40). Different IFN genes and many ISGs were recently cloned from different fish species (1, 19, 27). Both fish and mammals possess IFN-γ molecules that constitute a distinct lineage (47). In fish cells, viruses induce IFNs that have amino acid sequences most similar to that of mammalian IFN-α (type I) (49), but the sequences of their receptors and the fish IFN gene organization suggest that they may be more closely related to mammalian IFN-λ (type III) (17). The nomenclature of fish IFNs is still a matter of debate, but it is clear that virus-induced fish IFNs and mammalian type I and type III IFNs all derive from intron-bearing genes with likely antiviral activity present in early vertebrates. The IFN-stimulated response elements are similar in fish and mammals (5), and a number of IFN-induced genes are also retrieved from both classes, confirming that the antiviral IFN system has an ancient common origin.
While the control of IFN expression in mammals has been studied extensively, the specific characteristics of the fish IFN system raise the issue of induction pathways. Fish possess TLRs that include orthologs of human TLRs and other TLRs that are not retrieved from mammals, among which several receptors seem to be involved in IFN induction (20, 33). Concerning the RLRs, sequence data present in GenBank suggest that fish also possess RIG-I-like molecules and several of the downstream signaling molecules found in mammals. A recent extensive study of RLR evolution concluded that orthologs of MDA5, but not RIG-I, were found in fish, suggesting that MDA5 appeared before RIG-I in vertebrate evolution (30). In the present study, we cloned MAVS-like cDNAs from several fish species (including salmon and zebrafish) and showed that they were true orthologs of mammalian MAVS. We demonstrated that overexpression of this mitochondrial protein in fish cells led to a constitutive induction of IFN and ISGs and conferred a strong antiviral state against both RNA and DNA viruses. As previously shown for human MAVS, deletion of the N-terminal CARD-like domain and the C-terminal TM domain of fish MAVS, but not its central Pro region, abolished its signaling function. In addition, we identified sequences encoding a RIG-I-like molecule in salmonid and cyprinid cell lines that led to a strong cellular antiviral response when overexpressed in fish cells. These data strongly support the hypothesis that a functional RLR-based induction pathway of IFN is conserved in vertebrates.
MATERIALS AND METHODS
Cells and viruses.
EPC (epithelioma papulosum cyprini) and CHSE 214 cells were maintained in Glasgow's modified Eagle's medium-HEPES 25 mM medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine. The EPC cells (ATCC CRL-2872) were originally described as isolated from common carp (Cyprinus carpio) and were subsequently found to be from fathead minnow (Pimephales promelas) through cytochrome c oxidase subunit I sequencing (J. Winton, personal communication). Genes cloned from this cell line are referred to as EPC genes hereafter. Infectious hematopoietic necrosis virus 32/87 (IHNV), viral hemorrhagic septicemia virus 23/75 (VHSV), spring viremia of carp virus Fijan (SVCV), and epizootic hematopoietic necrosis virus (EHNV) strains were propagated in monolayer cultures of EPC cells at either 14°C (IHNV and VHSV) or 25°C (SVCV and EHNV) in the presence of 2% fetal bovine serum. Virus titers were determined by plaque assay on EPC cells under an agarose overlay (0.35% in Glasgow's modified Eagle's medium-HEPES medium). At 2 to 5 days postinfection, cell monolayers were fixed with 10% Formol and stained with crystal violet.
Molecular cloning and sequencing of MAVS, RIG-I, and IFN.
Partial MAVS and RIG-I sequences were obtained using BLAST analysis of salmon (Salmo salar), zebrafish (Danio rerio), and carp (Cyprinus carpio) expressed sequence tags (ESTs) with homology to mammalian MAVS and RIG-I gene sequences. Total RNAs from TO (salmon) and EPC cells or from the pronephroi of zebrafish were extracted using an RNeasy kit (Qiagen) according to the manufacturer's instructions. The RNAs were used to generate full-length cDNAs, using a Smart RACE cDNA amplification kit (BD Clontech) with universal primers provided by the manufacturer and gene-specific primers designed from the EST sequences. Reverse transcription-PCR (RT-PCR) products were purified with a QIAquick PCR purification kit (Qiagen) and fully sequenced by primer walking. Specific primers were then designed and used to amplify full-length or truncated open reading frames (ORFs) of MAVS and RIG-I (see Table S1 in the supplemental material). The MAVS ΔTM mutant was engineered by introducing a stop codon into the MAVS ORF by site-directed mutagenesis, using a QuikChange kit (Stratagene). The specific primers used to amplify the IFN gene expressed by EPC cells by RT-PCR were designed from the sequence of the crucian carp (Carassius auratus) IFN gene (GenBank accession number AY452069). cDNAs were cloned into the eukaryotic expression vectors pcDNA1.1/Amp (Invitrogen), peGFP-C1, and peGFP-N1 (Clontech), using unique restriction sites (see Table S1 in the supplemental material). The sequence of each cloned gene was confirmed by nucleotide sequencing.
Transfection, infection, and fluorescence microscopy.
CHSE 214 and EPC cells were passaged in six-well plates at a concentration of 5 × 106 cells per well 24 h prior to transfection by electroporation (Amaxa Biosystems). Cells were trypsinized and resuspended in 100 μl of solution T. Cells were then mixed with 2 μg of plasmid DNA and electroporated using the program T-020. Finally, cells were split equally into two wells of 12-well plates. Cell monolayers were washed at 24 h posttransfection, and the cells in one well per transfection were infected with 2 to 5 PFU/cell of IHNV, VHSV, SVCV, or EHNV at 24 h, 48 h, 72 h, or 6 days posttransfection. After 1 hour of adsorption, the inoculum was removed, the cell monolayer was washed twice, and medium samples (0.5 ml of the 2-ml overlay) were taken (zero time point) and replaced by an equivalent volume of fresh medium. At 24 h, 48 h, 72 h, or 4 days postinfection, supernatant aliquots were harvested, and they were analyzed later by plaque assay. Cell monolayers were either stained with crystal violet or subjected to total RNA extraction using an RNeasy kit (Qiagen). For immunofluorescence microscopy, specific subcellular compartments of enhanced green fluorescent protein (EGFP)- and EGFP-MAVS-transfected cells were stained in vivo, using 1 μg/ml of Hoechst 33342 (Sigma) for the nuclei and 400 nM of MitoTracker Red 580 FM (Invitrogen) for the mitochondria, for 2 h at 20°C. Cell monolayers were then visualized with a UV-visible light microscope (Carl Zeiss).
IFN titration.
The protective effects of IFNs secreted after overexpression of MAVS, CARDs of RIG-I (RIG-I Nter), and IFN itself were measured by monitoring the inhibition of the cytopathic effect (CPE) caused by VHSV. Briefly, EPC cells were transfected with eukaryotic expression vectors encoding MAVS, RIG-I Nter, or IFN or with an empty vector as a control. After 4 days of incubation at 20°C, the supernatants were harvested. Fresh EPC cells seeded in 96-well plates (cell density of 2.5 × 105 per well) were then incubated with 1/3 serial dilutions of transfected cell supernatant. After overnight incubation at 20°C to allow the establishment of the antiviral state, the transfected cell supernatants were removed, and the cell monolayers were washed twice and infected with 600 PFU of VHSV per well and incubated for 48 h at 15°C. Finally, the medium was removed and the monolayers were fixed and stained with crystal violet. The IFN antiviral titer was expressed as the supernatant dilution that reduced the CPE by 50%.
RNA isolation and quantitative RT-PCR analysis.
Total RNA was extracted by use of an RNeasy kit (Qiagen) from EPC cells transfected with eukaryotic expression vectors encoding MAVS or RIG-I Nter or with an empty vector as a control and infected or not with 5 PFU/cell of VHSV at 12 and 24 h postinfection. First-strand cDNAs were synthesized using an oligo(dT) primer, Superscript reverse transcriptase (Invitrogen Life Technologies), and RNase-free DNase I-treated RNA.
Real-time RT-PCR was performed using SYBR green reagent (Applied Biosystems) and a MasterCycler Realplex thermal cycler (Eppendorf) according to the manufacturers’ instructions. All reactions were performed in triplicate. Data analysis was performed as described in ABI Prism 7700 sequence detection bulletin no. 2 from Applied Biosystems. Oligonucleotides used for real-time RT-PCR were designed from β-actin, ISG15-1, ISG15-2, and VIG-1 sequences from crucian carp (Carassius auratus), published by Zhang and colleagues (45). The oligonucleotides targeting IFN, RIG-I, and MAVS were designed from the sequence determined in this study (see Table S1 in the supplemental material). The software Primer Express was used for the design of oligonucleotides.
RESULTS
The antiviral protein MAVS has a counterpart in teleost fish.
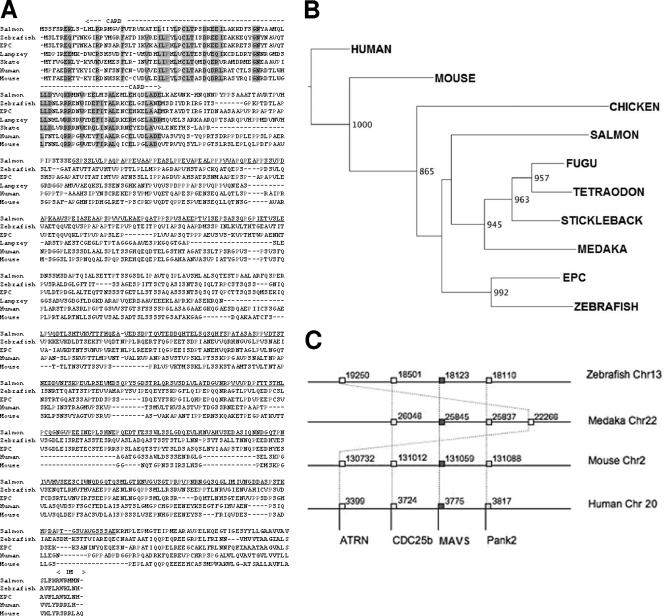
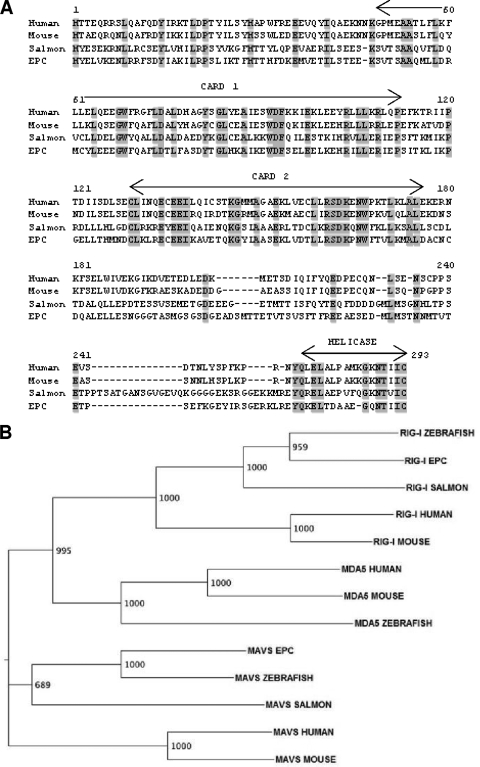
To gain insight into the IFN-inducing pathway in fish, we cloned the full-length MAVS-related cDNAs of Atlantic salmon, zebrafish, and EPC cells. Sequences similar to the human MAVS CARD sequence were identified in fish EST and genome sequence databases. Relevant specific primers were designed from the best hits and were used for 5′ and 3′ rapid amplification of cDNA ends (RACE) experiments with cDNAs from the fish cell lines TO and EPC and from zebrafish pronephroi. The RACE experiments led to the cloning of salmon, zebrafish, and EPC cDNAs of 1,821, 1,758, and 1,758 nucleotides, respectively. They carried typical MAVS ORFs encoding 606 amino acids (aa) (salmon) and 585 aa (zebrafish and EPC), with a CARD motif, a proline-rich region, a TM stretch, and a short intramembrane domain (Fig. 1A). The RACE analysis also revealed that a shorter cDNA variant was expressed, most probably corresponding to an alternative splicing product(s). These sequences were subjected to multiple alignments with the mouse and human MAVS sequences, showing that apart from the conserved CARD, the sequences from fish and humans display only weak similarity (25 to 30%). In contrast, fish and human CARDs showed about 40 to 48% similarity, and different fish CARDs were 50 to 79% similar.
FIG. 1.
MAVS sequences through vertebrate evolution. (A) Multiple alignment of MAVS sequences from fish and other vertebrates. The CARD and the intramembrane (IM) domain are indicated above the alignment. Relevant positions conserved in the CARD are boxed in gray. The residues of the TM region, predicted by the TMpred server, are shown in bold. The region missing in a salmon transcript, probably due to alternate splicing, is underlined. This salmon MAVS splicing variant is called mini-MAVS. The little skate (Leucoraja erinacea) sequence is from GenBank accession no. DT726507, and the lamprey (Petromyzon marinus) sequence is from GenBank accession no. FD727562. (B) Maximum likelihood phylogenetic tree of vertebrate MAVS. Neighbor-joining analysis led to a similar tree (not shown). Sequences from the following organisms (GenBank accession number, unless indicated otherwise) were included: Atlantic salmon (FN178458; this study), Salmo salar; zebrafish (FN178460; this study), Danio rerio; EPC cell line (FN178455; this study); medaka fish (AM299285 and Ensembl no. Chr22_25852569), Oryzia latipes; fugu (Ensembl release 52 no. scaffold338_161560), Fugu rubripes; pufferfish (Ensembl release 52 no. Chr10_12331965), Tetraodon nigroviridis; stickleback (Ensembl no. LG II_11620643), Gasterosteus aculeatus; chicken (NP_001012911), Gallus gallus; mouse (NP_659137), Mus musculus; and human (NP_065797), Homo sapiens. (C) Conserved synteny around the MAVS gene in zebrafish, medakas, mice, and humans. The locations of the different markers and the chromosomes involved are indicated for the different species (data from the assemblies are available at the Ensembl website [http://www.ensembl.org/]).
To gain insight into MAVS evolution, we searched for MAVS genes in different fish genomes as well as in shark and lamprey genomes. We could find only one mavs gene in the zebrafish genome, on chromosome 13. A typical mavs-like gene was also present in the genomes of stickleback, fugu, pufferfish, and medaka, indicating that it is generally conserved in fishes. A phylogenetic analysis suggested that all of these sequences constitute true orthologs (Fig. 1B). Interestingly, we also identified partial MAVS-related sequences in the little skate, a cartilaginous fish (GenBank accession no. DT726507), and in the lamprey, an agnathan (GenBank accession no. FD727562), suggesting that a MAVS-related molecule was already present in the first vertebrates. To confirm that fish sequences correspond to orthologs of the mammalian antiviral mavs gene, we searched for conserved synteny involving mavs and other markers in the neighborhood. We identified three markers that are conservatively found in the genomic region where mavs is located in fishes as well as in mammals (Fig. 1C), reinforcing the idea that they are all true orthologs. From the protein structure, CARD similarity, phylogenetic analysis, and conserved synteny, we therefore concluded that teleost fish possess a mavs gene that is orthologous to the molecule involved in the regulation of IFN expression in humans and mice.
Salmon MAVS is a mitochondrial protein.
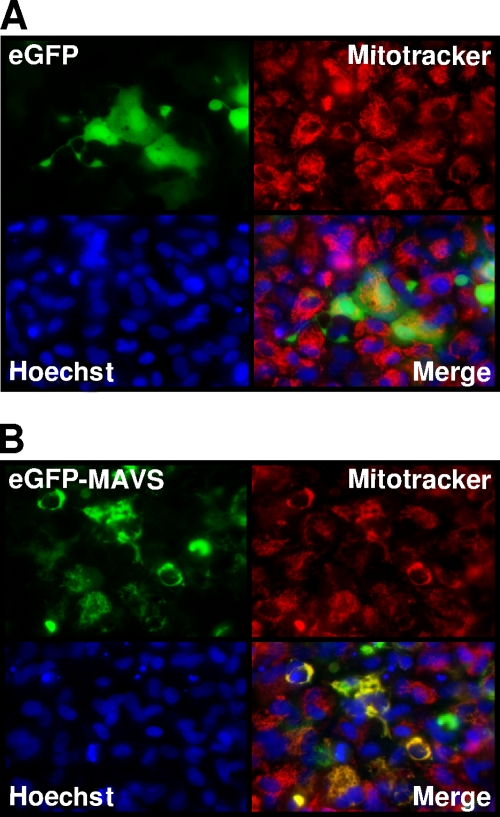
Human MAVS contains a hydrophobic TM domain at the C terminus, targeting the outer mitochondrial membrane (32). A TM domain was also predicted for the C terminus of the salmon, zebrafish, and EPC MAVS (Fig. 1A), suggesting that these proteins may also be targeted to the mitochondria. To determine the subcellular localization of salmon MAVS, EPC cells were transfected with an expression vector, peGFP-MAVS, encoding the fusion protein EGFP-MAVS. Fluorescence microscopy of living cells showed a pattern of salmon MAVS localization that was superposed with the staining pattern of MitoTracker, a fluorescent marker that accumulates specifically in the mitochondria (Fig. 2B). In contrast, cells transfected with the peGFP vector showed a cytosolic protein that did not overlap with MitoTracker staining (Fig. 2A). Thus, salmon MAVS seems to be localized to the mitochondria, like its human ortholog.
FIG. 2.
Localization of salmon MAVS to mitochondria. peGFP-MAVS or peGFP, as a control, was transfected into EPC cells. The mitochondria and the nuclei were stained in vivo with MitoTracker (red) and Hoechst (blue), respectively, and the cells were imaged by microscopy. The yellow staining in the overlay image indicates colocalization of MAVS and MitoTracker.
Overexpression of fish MAVS induces a strong antiviral state in fish cells.
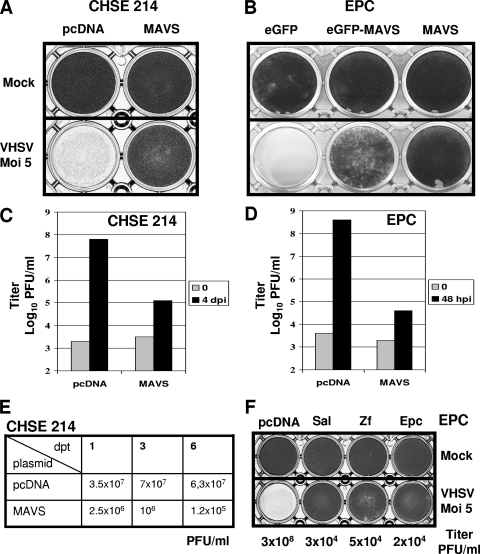
Since human MAVS overexpression is sufficient to delay the replication of vesicular stomatitis virus, an RNA virus of the Rhabdoviridae family, we examined whether salmon MAVS could mediate a similar effect in fish cells. Salmon cells of the CHSE 214 cell line were transfected with 2 μg of pcDNA-MAVS or an empty vector (pcDNA) as a control. At 6 days posttransfection, the cells were infected with a fish rhabdovirus, VHSV, at a multiplicity of infection (MOI) of 5 and incubated at 15°C. Infection of CHSE 214 cells transfected with empty vector by VHSV led to a complete CPE in 5 days at 15°C (Fig. 3A). In contrast, transfection of these cells with a vector encoding the salmon MAVS fully protected them against VHSV infection (Fig. 3A). Measurement of the viral titer showed that overexpression of salmon MAVS decreased the viral titer 525-fold compared to that in control cells (from 6.3 × 107 to 1.3 × 105 PFU/ml at 4 days postinfection) (Fig. 3C). Moreover, the viral titer increased only 40-fold in 4 days in MAVS-expressing cells, compared to 32,000-fold for the control cells. For CHSE cells, it took several days to reach the full antiviral state. As summarized in Fig. 3E, if the cells were challenged with VHSV (MOI of 5) 1 day after transfection of pcDNA-MAVS, the viral titer was decreased only 10-fold compared to that in control cells transfected with the empty vector (pcDNA). The difference in viral titers increased to 70-fold at 3 days posttransfection and reached 525-fold at 6 days posttransfection.
FIG. 3.
Fish MAVS are strong antiviral proteins. CHSE 214 (A) and EPC (B) cells were transfected with 2 μg of pcDNA-MAVS, peGFP-MAVS, or an empty vector (pcDNA) as a control. At 24 h and 6 days posttransfection, EPC and CHSE 214 cells, respectively, were infected with VHSV at an MOI of 5 for 5 days at 15°C. Cell monolayers were then stained with crystal violet. The culture supernatants from cells infected with VHSV were collected at 0 and 4 days postinfection (dpi) for CHSE 214 cells (C) and at 48 h postinfection (hpi) for EPC cells (D), and the viral titer was determined by plaque assay on EPC cells. Each time point was represented by two independent experiments, and each virus titration was done in duplicate. Means are shown. The standard errors were calculated, but the bars are not shown because the errors were very small. (E) CHSE 214 cells were transfected with 2 μg of pcDNA-MAVS or an empty vector (pcDNA) as a control. At 1, 3, or 6 days posttransfection (dpt), CHSE 214 cells were infected with VHSV at an MOI of 5. The culture supernatants were collected at 4 days postinfection, and the viral titer was determined by plaque assay on EPC cells. (F) EPC cells were transfected with 2 μg of pcDNA vector encoding MAVS from salmon (Sal), zebrafish (Zf), or EPC cells or an empty vector (pcDNA) as a control. At 48 h posttransfection, EPC cells were infected with VHSV at an MOI of 5 for 6 days at 15°C. Cell monolayers were then stained with crystal violet, and the viral titer was determined for each culture supernatant by plaque assay.
For more convenience in terms of experiment duration, these results were confirmed in a heterologous system, the EPC cell line, which is more efficient for transfection and more sensitive to challenge by VHSV. Infection of EPC cells transfected with empty vector by VHSV at an MOI of 5 led to a complete CPE in 2 to 3 days at 15°C (Fig. 3B). In contrast, transfection of these cells with a vector encoding the salmon MAVS or the fusion protein EGFP-MAVS (used above to determine the subcellular localization of MAVS) conferred full or high protection, respectively, against VHSV at this high MOI (Fig. 3B). These cells could be kept at 15°C for more than 10 days after challenge without developing any virus-induced CPE. Moreover, the establishment of this strong antiviral effect of MAVS required only 1 day of incubation posttransfection for EPC cells, compared to 6 days for CHSE cells, considerably shortening the experiments. Measurement of the viral titer showed that overexpression of salmon MAVS decreased the viral titer 10,000-fold compared to that in control cells (from 4 × 108 to 4 × 104 PFU/ml at 48 h postinfection) (Fig. 3D). Moreover, the viral titer increased only 20-fold in 48 h in MAVS-expressing cells, compared to 100,000-fold for the control cells. Similar antiviral effects were observed using expression vectors encoding MAVS proteins from three fish species, namely, salmon, zebrafish, and EPC cells (Fig. 3F). For each MAVS protein, the EPC cell monolayers were fully protected against VHSV challenge and the final titer was reduced 10,000-fold compared to that in control cells. These results show that MAVS is a pivotal cellular antiviral protein whose overexpression induces a strong antiviral immunity in fish.
MAVS protein induces strong antiviral immunity against several viruses.
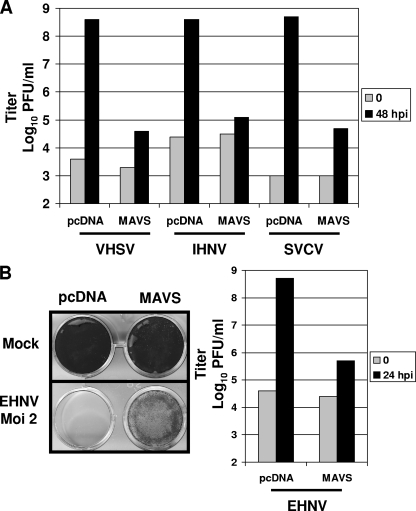
Salmon MAVS overexpression is sufficient to protect fish cells against VHSV. To see whether this protection was also effective against other viruses, we tested the effect of MAVS overexpression on three other fish viruses, namely, IHNV, another novirhabdovirus; SVCV, a vesiculovirus; and EHNV, a dsDNA virus belonging to the Iridoviridae family.
Infection of EPC cells transfected with empty vector by VHSV, IHNV, or SVCV at an MOI of 5 led to a complete CPE in 2 to 4 days. In contrast, transfection of the cells with a vector encoding the salmon MAVS fully protected them against these three viruses (data not shown). Measurement of the viral titer showed that overexpression of salmon MAVS decreased the viral titer 3,000- to 10,000-fold compared to that in control cells (from 4 × 108 to 5 × 108 PFU/ml to 5 × 104 to 13 × 104 PFU/ml at 48 h postinfection) (Fig. 4A). Moreover, viral production in MAVS-expressing cells was very low for each virus at 48 h postinfection, with 4- to 50-fold increases from the initial titer, compared to 16,000- to 500,000-fold increases for the control cells. Thus, MAVS seems to be an important player in antirhabdovirus immunity in fish.
FIG. 4.
Overexpression of MAVS induces antiviral immunity against several viruses. EPC cells were transfected with 2 μg of pcDNA-MAVS or an empty vector (pcDNA) as a control. At 24 h posttransfection, EPC cells were infected with different RNA viruses of the Rhabdoviridae family, i.e., VHSV, IHNV, and SVCV, at an MOI of 5 (A) or with a DNA virus of the Iridoviridae family, i.e., EHNV, at an MOI of 2 (B). The culture supernatants were collected at 48 h or 24 h postinfection, respectively, and the viral titer was determined by plaque assay on EPC cells. For panel B, cell monolayers were then stained with crystal violet. Each time point was represented by two independent experiments, and each virus titration was done in duplicate. Means are shown.
We further examined the effect of MAVS overexpression on the replication of the iridovirus EHNV. Infection of EPC cells transfected with empty vector by EHNV at an MOI of 2 led to a complete CPE in 24 h (Fig. 4B). In contrast, the cells overexpressing salmon MAVS were able to delay the appearance of total CPE. Although the CPE was important, measurement of the viral titer showed that overexpression of salmon MAVS decreased the viral titer 1,000-fold compared to that in control cells (from 5 × 108 to 5 × 105 PFU/ml at 24 h postinfection) (Fig. 4B). Moreover, viral production in MAVS-expressing cells was very low, with a 20-fold increase from the initial titer, compared to a 12,500-fold increase for the control cells. Therefore, MAVS seems to induce strong antiviral immunity against both RNA and DNA viruses.
CARD-like and C-terminal TM domains are essential for MAVS function.
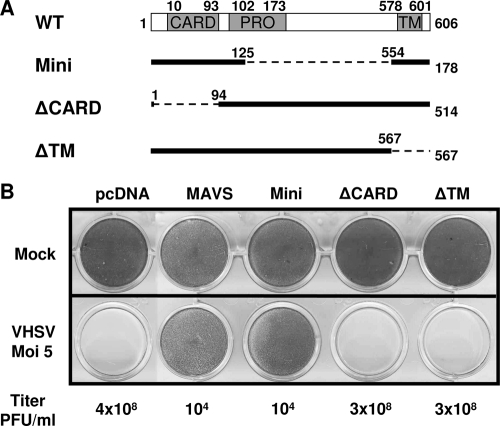
As shown in Fig. 1A, the N-terminal CARD-like domain of MAVS was relatively well conserved during evolution (40 to 48% similarity between the human and fish proteins). In addition, MAVS also contain a Pro region and a C-terminal TM domain, which are conserved (Fig. 5A). However, a potential splicing variant of salmon MAVS was also amplified. This transcript encodes a MAVS with a large deletion of 428 aa, including a large part of the Pro region. This small protein, of 178 aa, compared to the 606 aa of full-length MAVS, was called mini-MAVS (Fig. 5A). In parallel, we engineered expression vectors encoding MAVS deletion mutant proteins lacking either the CARD-like domain (ΔCARD) or the TM domain (ΔTM) (Fig. 5A).
FIG. 5.
The CARD-like domain and the mitochondrial membrane targeting sequence are essential for MAVS signaling. (A) Diagram illustrating the MAVS splicing variant and mutants. Mini, salmon MAVS splicing variant lacking residues 126 to 553; ΔCARD, salmon MAVS mutant lacking the CARD-like domain (residues 2 to 93); ΔTM, mutant lacking the mitochondrial membrane targeting sequence (residues 568 to 606). On the right is the size of each protein, in amino acids. (B) EPC cells were transfected with 2 μg of pcDNA vector encoding MAVS, mini-MAVS, and its mutants (ΔCARD and ΔTM). At 24 h posttransfection, the cells were infected with VHSV at an MOI of 5. The culture supernatants were collected at 48 h postinfection, and the viral titer was determined by plaque assay. The cell monolayers were stained with crystal violet at 5 days postinfection.
EPC cells were transfected with each construct and were then infected by VHSV at an MOI of 5 at 24 h posttransfection. As shown in Fig. 5B, deletions of either the CARD-like or TM domain abolished the ability of MAVS to fully protect EPC cells against VHSV challenge. Indeed, the EPC cells expressing MAVS mutants showed full CPE at 48 h postinfection, and the virus titers were roughly similar to that measured for the control transfected with an empty vector (>108 PFU/ml). In contrast, mini-MAVS overexpression was able to fully protect EPC cells against VHSV challenge as efficiently as that of full-length MAVS. Moreover, measurement of the viral titer showed that overexpression of mini-MAVS, as well as full-length MAVS, decreased the viral titer 40,000-fold compared to that in control cells (from 4 × 108 to 104 PFU/ml at 48 h postinfection) (Fig. 5B). Thus, the CARD-like and TM domains are both necessary and sufficient for MAVS signaling.
Fish express a counterpart of the cytosolic RNA helicase RIG-I.
MAVS was described as an adaptor that provides a link between the cytoplasmic RNA helicase RIG-I sensing of intracellular viral infection and downstream activation events leading to the production of type I IFNs. Therefore, we investigated the presence of such a protein upstream of MAVS in fish. Sequences with significant similarity to mammalian RIG-I sequences were identified using BLAST analysis of salmon (Salmo salar) and carp (Cyprinus carpio) ESTs. Relevant specific primers were designed from sequences encoding the CARD and helicase domains most similar to those of human RIG-I and were used for 5′- and 3′-RACE reactions, using total RNAs prepared from TO and EPC cells. The RACE experiments led to the cloning of salmon and EPC cell cDNAs, of 2,889 and 2,823 nucleotides, respectively, encoding a RIG-I-like protein (GenBank accession no. FN178459 and FN394062, respectively). They carried ORFs encoding proteins of 962 aa (salmon) and 940 aa (EPC cells), which is slightly longer than their human and mouse counterparts (925 and 926 aa, respectively), with two N-terminal CARD-like domains and a DEXD/H box RNA helicase domain at the C terminus. Fish RIG-I-like proteins are relatively well conserved between salmonids and cyprinids, with 71% similarity, as well as with their mouse and human counterparts, ranging from 55 to 58% similarity.
The CARD-like domains of these RIG-I-like proteins were subjected to multiple alignments with the CARDs of human and mouse RIG-I (Fig. 6A). The N-terminal regions of these RIG-I and RIG-I-like proteins containing two CARD-like domains present 44 to 50% similarity between fish and mammals. A phylogenetic analysis based on a multiple alignment of the CARD-like domains from RIG-I and related proteins, i.e., MDA5 and MAVS of several vertebrates (see Fig. S1 in the supplemental material), was also performed (Fig. 6B). The phylogenetic tree shows specific branches for RIG-I, MDA5, and MAVS, which are supported by high bootstrap values. While a fish MDA5 gene was recently reported (30), these observations clearly indicate that fish also have a CARD-containing protein more closely related to RIG-I than to MDA5, which we call RIG-I hereafter.
FIG. 6.
CARD-like domains of fish RIG-I and MAVS. (A) Multiple alignment of RIG-I CARDs from fish and other vertebrates. The conserved residues are boxed in gray. The CARDs and the beginning of the helicase domain are mapped. Sequences for the following (GenBank accession numbers) were used: salmon RIG-I (FN178459; this study), from Salmo salar; EPC RIG-I (FN178456; this study); mouse RIG-I (NP_766277), from Mus musculus; and human RIG-I (NP_055129), from Homo sapiens. (B) Neighbor-joining phylogenetic tree of vertebrate CARDs from RIG-I, MDA5, and MAVS. The tree was calculated by ClustalW, based on a multiple alignment of RIG-I, MDA5, and MAVS CARD-like domains from fish and other vertebrates (see Fig. S1 in the supplemental material), and a 1,000-bootstrap analysis was performed. High bootstrap values (>800) are indicated. The following sequences (GenBank accession number, unless indicated otherwise) were used: salmon RIG-I (FN178459; this study) and MAVS (FN178458; this study), from Salmo salar; zebrafish RIG-I (Ensembl protein no. ENSDARP00000058175; gene located at Chr23 positions 46,283,361 to 46,298,826), MDA5 (NC_007120), and MAVS (FN178460; this study), from Danio rerio; EPC RIG-I (FN178456; this study) and MAVS (FN178455; this study); mouse RIG-I (NP_766277), MDA5 (AAM21359), and MAVS (NP_659137), from Mus musculus; and human RIG-I (NP_055129), MDA5 (AAG34368), and MAVS (NP_065797), from Homo sapiens.
Overexpression of MAVS or the CARD-like domains of RIG-I induces powerful antiviral immunity.
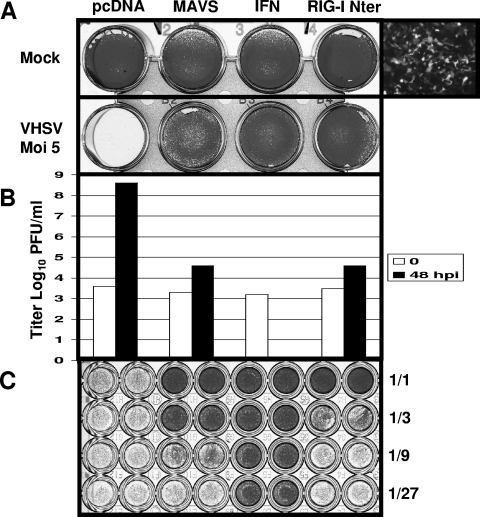
It was previously shown that the CARD-like domains of RIG-I interact with the N-terminal CARD-like structure of MAVS, leading to subsequent activation of important transcription factors, such as IRFs and NF-κB, and induction of type I IFN and IFN-inducible genes (14). Moreover, overexpression of the N-terminal CARD of RIG-I was sufficient to constitutively activate the downstream adaptor MAVS and then the expression of endogenous IFN (44). Therefore, the N-terminal sequence of fish RIG-I (first 275 aa) from EPC cells was cloned into an expression vector in fusion, at its C terminus, with EGFP (RIG-I Nter). EPC cells transfected with this plasmid construct showed cytosolic protein expression (Fig. 7A). As a control, EPC cells were transfected with an empty vector or an expression vector encoding either the MAVS or the IFN from EPC cells. The IFN was amplified from virus-infected EPC cells by use of primers derived from the IFN sequence of the crucian carp (Carassius auratus; GenBank accession no. AY452069). At 24 hours posttransfection, cell monolayers were infected with VHSV at a high MOI of 5 and incubated at 15°C. Infection of EPC cells transfected with empty vector by VHSV led to complete CPE in 2 days at 15°C (Fig. 7A). In contrast, transfection of these cells with a vector encoding RIG-I Nter, as well as MAVS and IFN, fully protected them against VHSV at this high MOI (Fig. 7A). These transfected cells could be kept for over 10 days at 15°C without developing any CPE. Measurement of the viral titer showed that overexpression of RIG-I Nter, as previously observed with MAVS, decreased the viral titer 10,000-fold compared to that in control cells (from 4 × 108 to 4 × 104 PFU/ml at 48 h postinfection) (Fig. 7B). For IFN-transfected cells, no virus could be detected by plaque assay, probably due to IFN saturation of the supernatant. Overexpression of CARD-like domains of RIG-I, as previously observed with MAVS, fully protected cells against RNA virus infection. A high level of protection was also observed following overexpression of the full-length RIG-I molecule, with the viral titer decreased 3,000-fold compared to that in control cells (from 3 × 108 to 105 PFU/ml at 4 days postinfection) (data not shown).
FIG. 7.
Overexpression of MAVS or the N-terminal region of RIG-I induces powerful antiviral immunity. (A) EPC cells were transfected with 2 μg of pcDNA vector encoding MAVS, RIG-I Nter (first 275 aa), or the IFN from EPC cells (FN178457; this study) or an empty vector (pcDNA) as a control. At 24 h posttransfection, EPC cells were infected with VHSV at an MOI of 5 for 6 days at 15°C. Cell monolayers were then stained with crystal violet. The UV-visible light microscope picture on the right shows the level of RIG-I Nter-GFP fusion protein expression achieved in EPC cells at 24 h posttransfection. (B) The viral titer for each culture supernatant was determined by plaque assay at 48 h postinfection. Each time point is represented by two independent experiments, and each virus titration was done in duplicate. Means are shown. (C) The culture supernatants of mock-infected wells were harvested at 4 days posttransfection, and the protective effects of the IFNs secreted after overexpression of MAVS, CARDs of RIG-I (RIG-I Nter), and IFN itself were measured by monitoring the inhibition of CPE caused by VHSV.
Overexpression of MAVS, as well as the CARD-like domains of RIG-I, constitutively induces expression of both IFN and ISGs.
To detect potential IFN induction by RIG-I Nter and MAVS overexpression, the supernatant of mock-infected, RIG-I Nter- or MAVS-transfected cells was harvested at 4 days posttransfection and subjected to biological titration of the secreted IFN. As expected, supernatant from IFN-transfected cells was positive for IFN and fully protected fresh EPC cells against VHSV infection at a dilution of >1/243 (Fig. 7C and data not shown). To a lesser extent, MAVS and RIG-I Nter overexpression also led to the secretion of detectable amounts of IFN in the supernatant that were able to protect fresh EPC cells against VHSV, up to dilutions of 1/9 and 1/3, respectively (Fig. 7C). In contrast, the supernatant from EPC cells transfected with empty vector failed to protect fresh cells against VHSV. Overexpression of the N-terminal CARDs of RIG-I, as well as MAVS, was sufficient to fully protect cells against virus infection by constitutively inducing endogenous IFN expression.
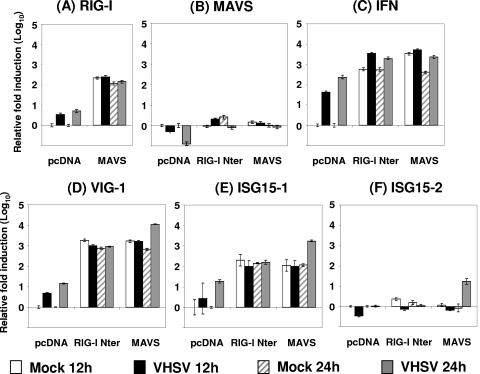
Using real-time RT-PCR, we further analyzed the induced mRNA expression of IFN and ISGs after overexpression of salmon MAVS or EPC RIG-I Nter in EPC cells. These cells were then infected or not by VHSV at an MOI of 5, and total RNAs were extracted at 12 and 24 h postinfection. First, variations in MAVS and RIG-I expression were examined for each condition. These experiments showed that RIG-I was constitutively expressed in resting EPC cells (see Table S2 in the supplemental material) and that VHSV infection and MAVS overexpression significantly upregulated the expression of RIG-I (Fig. 8A). MAVS overexpression was particularly efficient in RIG-I mRNA upregulation, with a 218-fold increase, compared to a 3.4-fold increase induced by VHSV infection. No cumulative effect on RIG-I mRNA expression could be observed after MAVS transfection followed by VHSV infection. In contrast, although MAVS was also constitutively expressed in resting EPC cells (see Table S2 in the supplemental material), neither VHSV infection nor RIG-I Nter overexpression upregulated its expression. Moreover, the overexpression of a heterologous MAVS—from Atlantic salmon—did not affect the autologous expression of MAVS either (Fig. 8B). Similar observations were made after poly(I-C) stimulation for 24 h (data not shown).
FIG. 8.
Overexpression of MAVS and RIG-I induces IFN and ISG expression. EPC cells were transfected with 2 μg of pcDNA vector encoding salmon MAVS or RIG-I Nter (first 275 aa) or an empty vector (pcDNA) as a control. At 24 h posttransfection, EPC cells were infected or not with VHSV at an MOI of 5 and incubated at 15°C for 12 and 24 h before total RNA extraction and RT. Quantitative real-time PCR was conducted using primers targeting RIG-I (A), MAVS (B), IFN (C), VIG-1 (D), ISG15-1 (E), and ISG15-2 (F) (see Table S1 in the supplemental material). The β-actin gene was used as an internal control to normalize the cDNA template and for real-time PCR calculations. The standard deviations for triplicate experiments are shown.
These experiments confirmed that both MAVS and RIG-I overexpression has a strong effect on IFN induction, with 3,400- and 590-fold mRNA increases, respectively, compared to the empty vector-transfected control (Fig. 8C). These results are in accordance with the previous experiment showing an IFN protective effect in the supernatants of MAVS- and RIG-I Nter-overexpressing cells. VHSV infection was also followed by induction of IFN mRNA synthesis, and a clear but limited cumulative effect could be observed in cells overexpressing MAVS or RIG-I Nter and infected with VHSV at any time point (Fig. 8C). We then examined the induction of two previously described ISGs, namely, vig-1 (virus-induced gene 1), the fish ortholog of the mammalian viperin gene (3, 4, 7), and the fish ortholog of isg15 (isg15-1 or vig-3) (18, 23). As expected, these two genes were induced by VHSV infection as soon as 12 h postinfection (Fig. 8D and E). In cells overexpressing MAVS or RIG-I Nter, both genes were strongly induced. Vig-1 mRNA expression increased 1,650- and 1,800-fold, respectively, compared to its expression in an empty vector-transfected control (Fig. 8D). Similarly, Isg15-1 mRNA expression increased 110- and 200-fold, respectively (Fig. 8E). Interestingly, in the case of MAVS-overexpressing cells, the induction of vig-1 and isg15-1 was significantly increased by subsequent VHSV infection, by 17- and 15-fold, respectively, compared to the mock-infected condition at 24 h postinfection. Finally, we were interested in another variant of crucian carp isg15, the isg15-2 gene, which encodes a protein 78.9% similar to ISG15-1 but for which no function is described to date (46). The igs15-2 gene was described as moderately induced by virus and IFN stimulation in crucian carp (Carassius auratus) blastula embryonic cells (CAB). Our results for EPC cells are generally consistent with this pattern, since isg15-2 was not induced by either RIG-I Nter or MAVS expression or the virus infection alone (Fig. 8F). Interestingly, the VHSV infection significantly increased ISG15-2 mRNA expression, by 17-fold, in cells overexpressing MAVS only. Thus, overexpression of MAVS and RIG-I Nter led to a strong induction of both IFN and ISGs.
DISCUSSION
In this article, we report the identification and functional characterization of MAVS from different fish species, including salmonids and cyprinids, as a potential key cellular factor in the innate immune defense against viruses. We also identify a CARD-bearing protein similar to RIG-I as an efficient IFN inducer, shedding light on the origin of the RLR pathway of IFN production.
The fish mavs genes described here encode proteins with the typical domain organization (CARD-Pro region-mitochondrial TM domain) of the mammalian adaptor encoded by mavs/ips-1/cardif/visa (14, 22, 32, 41). A putative tumor necrosis factor receptor-associated factor (TRAF3)-interacting motif is also found in fish MAVS, with a consensus sequence (PVQ/RD/ET) that is relatively well conserved compared to that found in mammalian MAVS (PVQD/ET), although it is not located in exactly the same region of the protein. TRAF3 is required for the activation of IRF3, and the specific interaction between the TRAF domain of TRAF3 and the TRAF3-interacting motif of MAVS is essential for an optimal MAVS-mediated antiviral response in mammals (29). In mammals, the C-terminal TM domain is essential for both localization of MAVS to the mitochondrial outer membrane and IFN induction. The fish MAVS is also found on the mitochondria, confirming that it shares this essential location with its mammalian counterpart. In addition, the experiments with a truncated MAVS lacking the TM domain demonstrated that it is also required for the function of the fish protein. Finally, a phylogenetic analysis of the fish sequences indicated that fish MAVS is likely the ortholog of mammalian MAVS, which is further confirmed by the conserved synteny linking the genomic regions where human and zebrafish mavs genes are located. These indications firmly establish that a mavs gene encoding a mitochondrial protein was already expressed by the common ancestor of fish and mammals.
Although the induction of a mavs transcript by poly(I-C) was previously reported for zebrafish (9), its implication in IFN production remained unclear. As previously shown for mammalian MAVS, this work demonstrates that overexpression of fish MAVS is sufficient to induce a high level of expression of both ifn and isg genes, such as the fish orthologs of mammalian isg15 and viperin. More interestingly, overexpression of these MAVS in fish cells (salmon and EPC cell lines) has a deleterious effect on infection by RNA viruses, as previously observed in mammalian models (14, 32). Indeed, a delay in the appearance of total CPE is generally observed in mammalian cells overexpressing MAVS after a VSV challenge at a high MOI, with a 20- to 100-fold decrease of the viral titer at 24 h postinfection. In contrast, fish rhabdovirus replication was almost fully inhibited in fish cells, with only a 30-fold increase in viral titer at 48 h postinfection, compared to a 200,000-fold increase in the control (averages observed with the three different rhabdoviruses used in this report). Moreover, fish cell monolayers survived these high-MOI virus challenges and could be kept at 15°C over a period of 10 days postinfection without developing any CPE. Infection of MAVS-overexpressing cells with a recombinant IHNV expressing a fluorescent protein allowed us to detect viral replication in only a few cells, which confirmed the almost full inhibition of virus replication (data not shown).
The MAVS-overexpressing cells were also able to delay the appearance of total CPE after infection by a dsDNA virus (iridovirus) and to reduce the final virus titer 1,000-fold. Moreover, viral production was reduced 625-fold in MAVS-overexpressing cells compared to control cells at 24 h postinfection. Recent reports have shown that maximal activation of the IFN-β promoter by DNA virus infection in mammalian cells requires functional RIG-I and MAVS (6, 8, 11, 16, 38). Although a cytoplasmic DNA sensor, DAI, was recently characterized and shown to mediate type I IFN induction following DNA virus infection (36), evidence for redundancy in the cytosolic DNA sensing system was provided, depending on the cell type (39). Since RIG-I binds and responds to either short dsRNA or 5′pppRNA (31), it is most likely that its cytosolic targets are RNA intermediates synthesized during DNA virus gene expression. Altogether, these findings underline a potential role of the MAVS-dependent RNA sensing pathway in triggering the innate immune response against DNA viruses, at least in mammalian cells.
Since fish express a typical MAVS involved in IFN production, they should also possess specific sensors of the RLR pathways that would be counterparts of mammalian RIG-I and MDA5. A recent survey of sequences sharing the MDA-5 and RIG-I domain arrangement (CARD1-CARD2-helicase-DEAD/DEAH) revealed that fish possess likely counterparts of MDA5 but not RIG-I, suggesting that the complete RLR pathway may have originated recently in the tetrapod or even the mammalian lineage (30). Combining in silico analysis and RACE, we identified a fish transcript encoding a protein most similar to RIG-I, with two CARDs and a helicase motif. (While the manuscript was in revision, Zou et al. also described fish RIG-I-like sequences [48].) The phylogenetic analysis of the first CARD showed that the gene contains a CARD sequence most similar to that of mammalian RIG-I and clearly distinct from the MDA5 CARD1 sequence. The similarities of the full-length fish protein to the mammalian RIG-I and MDA5 proteins (55 to 58% and 36 to 39%, respectively) also confirmed the closest relationship between the fish RIG-I-like protein and mammalian RIG-I. Since the regions encoding the fish RIG-I-like protein include many genes encoding proteins with CARDs, the orthology relationships of the gene are unclear and will await an extensive genomic and expressional analysis of the family to be clarified. In any case, cells overexpressing RIG-I Nter or the full-length RIG-I molecule are efficiently protected against virus infection and secrete large amounts of IFN. Although we did not formally demonstrate by knockout and complementation experiments that the protection afforded by RIG-I Nter involves MAVS-dependent signaling, our observations strongly suggest that the RIG-I-like receptor is a functional counterpart of mammalian RIG-I. In this perspective, the common ancestor of fish and mammals would have possessed a system similar to the canonical RLR antiviral pathway, with RNA sensors related to both MDA5 and RIG-I. Since IFN induction through several TLRs was recently demonstrated for fish (20), as well as the expression of the key adaptor TRIF (9), it seems that old vertebrates were already equipped with two main pathways of RNA virus sensing, leading to the induction of primordial ifn genes with four introns.
Many of the main components of the IFN system were recently identified in teleost fish, including a typical IFN-stimulated response element as well as many IFN-induced genes, most of which are well conserved in vertebrates. Thus, fish counterparts of genes encoding antiviral proteins, such as PKR, Mx, ISG15, and Viperin, have been characterized, indicating that the components of the IFN cascade downstream of IFN were also part of the primordial antiviral system. The coupling between the fish RIG-I-like MAVS and the effectors of the IFN system was further analyzed by the quantification of mavs, rig-I-like, vig-1/viperin, and isg15 transcripts in cells overexpressing fish MAVS or RIG-I Nter. The effects of MAVS and RIG-I Nter are remarkably consistent with the predictions made from our knowledge of the mammalian pathway. The overexpression of RIG-I and MAVS leads to upregulation of IFN and IFN-induced transcripts (one each of ifn, vig-1, and isg15-1) through higher activation of the adaptor MAVS, which occurs by TM domain-dependent dimerization. As in mammals, the level of mavs transcript is not subjected to significant variations by virus or viral compound activation, but the overexpression of MAVS leads to upregulation of the RIG-I transcript in a positive feedback loop. The effects of MAVS overexpression are so massive that the additive effect of viral infection on the level of IFN transcript is very limited, although it is still detectable. Interestingly, isg15-2 gene induction requires both MAVS overexpression and viral infection. This gene was described for crucian carp as a second isg15-like gene. This gene showed very slight induction by viral infection in crucian carp CAB cells but was more upregulated by IFN-containing supernatant and inactivated virus, suggesting a particular induction pathway. The differential upregulation of isg15-2 transcripts in EPC cells at 24 postinfection may not be explained by a higher level of the ifn transcript at this time point but rather may suggest the coupling of MAVS with an alternative pathway that does not involve RIG-I Nter and the ifn gene we measured. Further characterization of TLR and RLR pathways as well as of the ifn repertoire and functional diversity will be needed to clarify this issue.
In conclusion, we have demonstrated that an ortholog of mavs and a rig-I-like gene constitute potential key components of an RLR pathway of ifn induction and antiviral immunity in fish. These observations put forward the RLR pathway as part of the primordial IFN system of vertebrates, in addition to the well-conserved TLR pathways of virus sensing.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the Animal Health Department, INRA, and by The European Community through the EADGENE Network of Excellence.
We thank Alpharma Inc. (Oslo, Norway) for providing the TO cells. We also thank Julie Bernard and Emilie Merour for their technical assistance and Human Rezaei for his protein analysis expertise.
Footnotes
Published ahead of print on 27 May 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altmann, S. M., M. T. Mellon, D. L. Distel, and C. H. Kim. 2003. Molecular and functional analysis of an interferon gene from the zebrafish, Danio rerio. J. Virol. 771992-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beutler, B., C. Eidenschenk, K. Crozat, J. L. Imler, O. Takeuchi, J. A. Hoffmann, and S. Akira. 2007. Genetic analysis of resistance to viral infection. Nat. Rev. Immunol. 7753-766. [DOI] [PubMed] [Google Scholar]
- 3.Boudinot, P., P. Massin, M. Blanco, S. Riffault, and A. Benmansour. 1999. vig-1, a new fish gene induced by the rhabdovirus glycoprotein, has a virus-induced homologue in humans and shares conserved motifs with the MoaA family. J. Virol. 731846-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boudinot, P., S. Riffault, S. Salhi, C. Carrat, C. Sedlik, N. Mahmoudi, B. Charley, and A. Benmansour. 2000. Vesicular stomatitis virus and pseudorabies virus induce a vig1/cig5 homologue in mouse dendritic cells via different pathways. J. Gen. Virol. 812675-2682. [DOI] [PubMed] [Google Scholar]
- 5.Boudinot, P., S. Salhi, M. Blanco, and A. Benmansour. 2001. Viral haemorrhagic septicaemia virus induces vig-2, a new interferon-responsive gene in rainbow trout. Fish Shellfish Immunol. 11383-397. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, G., J. Zhong, J. Chung, and F. V. Chisari. 2007. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc. Natl. Acad. Sci. USA 1049035-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin, K. C., and P. Cresswell. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 9815125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, L., P. Dai, T. Parikh, H. Cao, V. Bhoj, Q. Sun, Z. Chen, T. Merghoub, A. Houghton, and S. Shuman. 2008. Vaccinia virus subverts a mitochondrial antiviral signaling protein-dependent innate immune response in keratinocytes through its double-stranded RNA binding protein, E3. J. Virol. 8210735-10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, S., S. Chen, Y. Liu, Y. Lin, H. Liu, L. Guo, B. Lin, S. Huang, and A. Xu. 2008. Zebrafish TRIF, a Golgi-localized protein, participates in IFN induction and NF-kappaB activation. J. Immunol. 1805373-5383. [DOI] [PubMed] [Google Scholar]
- 10.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 1038459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii, K. J., C. Coban, H. Kato, K. Takahashi, Y. Torii, F. Takeshita, H. Ludwig, G. Sutter, K. Suzuki, H. Hemmi, S. Sato, M. Yamamoto, S. Uematsu, T. Kawai, O. Takeuchi, and S. Akira. 2006. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 740-48. [DOI] [PubMed] [Google Scholar]
- 12.Kang, D. C., R. V. Gopalkrishnan, Q. Wu, E. Jankowsky, A. M. Pyle, and P. B. Fisher. 2002. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA 99637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 14.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 15.Komuro, A., and C. M. Horvath. 2006. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 8012332-12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, H., T. Kawai, H. Kato, S. Sato, K. Takahashi, C. Coban, M. Yamamoto, S. Uematsu, K. J. Ishii, O. Takeuchi, and S. Akira. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 2031795-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levraud, J. P., P. Boudinot, I. Colin, A. Benmansour, N. Peyrieras, P. Herbomel, and G. Lutfalla. 2007. Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J. Immunol. 1784385-4394. [DOI] [PubMed] [Google Scholar]
- 18.Liu, M., R. Reimschuessel, and B. A. Hassel. 2002. Molecular cloning of the fish interferon stimulated gene, 15 kDa (ISG15) orthologue: a ubiquitin-like gene induced by nephrotoxic damage. Gene 298129-139. [DOI] [PubMed] [Google Scholar]
- 19.Lutfalla, G., H. Roest Crollius, N. Stange-Thomann, O. Jaillon, K. Mogensen, and D. Monneron. 2003. Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: the class II cytokine receptors and their ligands in mammals and fish. BMC Genomics 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuo, A., H. Oshiumi, T. Tsujita, H. Mitani, H. Kasai, M. Yoshimizu, M. Matsumoto, and T. Seya. 2008. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J. Immunol. 1813474-3485. [DOI] [PubMed] [Google Scholar]
- 21.Meeker, N. D., and N. S. Trede. 2008. Immunology and zebrafish: spawning new models of human disease. Dev. Comp. Immunol. 32745-757. [DOI] [PubMed] [Google Scholar]
- 22.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 23.O'Farrell, C., N. Vaghefi, M. Cantonnet, B. Buteau, P. Boudinot, and A. Benmansour. 2002. Survey of transcript expression in rainbow trout leukocytes reveals a major contribution of interferon-responsive genes in the early response to a rhabdovirus infection. J. Virol. 768040-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrilli, V., C. Dostert, D. A. Muruve, and J. Tschopp. 2007. The inflammasome: a danger sensing complex triggering innate immunity. Curr. Opin. Immunol. 19615-622. [DOI] [PubMed] [Google Scholar]
- 25.Pichlmair, A., and C. Reis e Sousa. 2007. Innate recognition of viruses. Immunity 27370-383. [DOI] [PubMed] [Google Scholar]
- 26.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signaling, antiviral responses and virus countermeasures. J. Gen. Virol. 891-47. [DOI] [PubMed] [Google Scholar]
- 27.Robertsen, B. 2006. The interferon system of teleost fish. Fish Shellfish Immunol. 20172-191. [DOI] [PubMed] [Google Scholar]
- 28.Rothenfusser, S., N. Goutagny, G. DiPerna, M. Gong, B. G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K. A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 1755260-5268. [DOI] [PubMed] [Google Scholar]
- 29.Saha, S. K., E. M. Pietras, J. Q. He, J. R. Kang, S. Y. Liu, G. Oganesyan, A. Shahangian, B. Zarnegar, T. L. Shiba, Y. Wang, and G. Cheng. 2006. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 253257-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarkar, D., R. Desalle, and P. B. Fisher. 2008. Evolution of MDA-5/RIG-I-dependent innate immunity: independent evolution by domain grafting. Proc. Natl. Acad. Sci. USA 10517040-17045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlee, M., E. Hartmann, C. Coch, V. Wimmenauer, M. Janke, W. Barchet, and G. Hartmann. 2009. Approaching the RNA ligand for RIG-I? Immunol. Rev. 22766-74. [DOI] [PubMed] [Google Scholar]
- 32.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122669-682. [DOI] [PubMed] [Google Scholar]
- 33.Seya, T., M. Matsumoto, T. Ebihara, and H. Oshiumi. 2009. Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunol. Rev. 22744-53. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan, C., and C. H. Kim. 2008. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 25341-350. [DOI] [PubMed] [Google Scholar]
- 35.Sun, Q., L. Sun, H. H. Liu, X. Chen, R. B. Seth, J. Forman, and Z. J. Chen. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24633-642. [DOI] [PubMed] [Google Scholar]
- 36.Takaoka, A., and T. Taniguchi. 2008. Cytosolic DNA recognition for triggering innate immune responses. Adv. Drug Deliv. Rev. 60847-857. [DOI] [PubMed] [Google Scholar]
- 37.Venkataraman, T., M. Valdes, R. Elsby, S. Kakuta, G. Caceres, S. Saijo, Y. Iwakura, and G. N. Barber. 2007. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 1786444-6455. [DOI] [PubMed] [Google Scholar]
- 38.Wang, F., X. Gao, J. W. Barrett, Q. Shao, E. Bartee, M. R. Mohamed, M. Rahman, S. Werden, T. Irvine, J. Cao, G. A. Dekaban, and G. McFadden. 2008. RIG-I mediates the co-induction of tumor necrosis factor and type I interferon elicited by myxoma virus in primary human macrophages. PLoS Pathog. 4e1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Z., M. K. Choi, T. Ban, H. Yanai, H. Negishi, Y. Lu, T. Tamura, A. Takaoka, K. Nishikura, and T. Taniguchi. 2008. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. USA 1055477-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whyte, S. K. 2007. The innate immune response of finfish—a review of current knowledge. Fish Shellfish Immunol. 231127-1151. [DOI] [PubMed] [Google Scholar]
- 41.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19727-740. [DOI] [PubMed] [Google Scholar]
- 42.Yoneyama, M., and T. Fujita. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 22754-65. [DOI] [PubMed] [Google Scholar]
- 43.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 1752851-2858. [DOI] [PubMed] [Google Scholar]
- 44.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y. B., J. Jiang, Y. D. Chen, R. Zhu, Y. Shi, Q. Y. Zhang, and J. F. Gui. 2007. The innate immune response to grass carp hemorrhagic virus (GCHV) in cultured Carassius auratus blastulae (CAB) cells. Dev. Comp. Immunol. 31232-243. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y. B., Y. L. Wang, and J. F. Gui. 2007. Identification and characterization of two homologues of interferon-stimulated gene ISG15 in crucian carp. Fish Shellfish Immunol. 2352-61. [DOI] [PubMed] [Google Scholar]
- 47.Zou, J., A. Carrington, B. Collet, J. M. Dijkstra, Y. Yoshiura, N. Bols, and C. Secombes. 2005. Identification and bioactivities of IFN-gamma in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J. Immunol. 1752484-2494. [DOI] [PubMed] [Google Scholar]
- 48.Zou, J., M. Chang, P. Nie, and C. J. Secombes. 2009. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou, J., C. Tafalla, J. Truckle, and C. J. Secombes. 2007. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J. Immunol. 1793859-3871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.