Abstract
In this study, we demonstrate that dedifferentiation of round primary chondrocytes into a fibroblast morphology correlates with a profound induction of RhoA protein and stress fibers. Culture of dedifferentiated chondrocytes in alginate gel induces a precipitous loss of RhoA protein and a loss of stress fibers concomitant with the reexpression of the chondrocyte differentiation program. We have found that chondrogenesis in limb bud micromass cultures similarly entails a loss of RhoA protein and that expression of dominant negative RhoA in such cultures can markedly enhance chondrogenesis. Consistent with these results, expression of the Rho antagonist C3 transferase can restore chondrocyte gene expression in dedifferentiated chondrocytes grown on plastic. Transfection of cells with agents that block actin polymerization enhance the ability of either exogenous Sox9 or a Gal4 DBD-Sox9 fusion protein to activate gene expression. Interestingly, the enhancement of Sox9 function by actin depolymerization requires both protein kinase A (PKA) activity and a PKA phosphorylation site in Sox9 (S181) that is known to enhance Sox9 transcriptional activity. Lastly, we demonstrate that RhoA-mediated modulation of actin polymerization regulates the ability of Sox9 to both activate chondrocyte-specific markers and maintain its own expression in chondrocytes via a positive feedback loop.
Vertebrate long bones are formed through a process of endochondral ossification (8, 11). During this process, bone formation begins with the establishment of mesenchymal condensations, which serve as a template for the adult skeletal elements. Chondrocytes then differentiate within the aggregated mesenchyme, generating distinct cartilage primordia that begin longitudinal growth. A very important system to study the events that control the initiation of chondrocyte differentiation is the limb bud micromass culture system. Originally developed by Solursh and colleagues, this culture system employs mesenchymal cells isolated from either chick or mouse embryo limb buds, which undergo chondrogenesis when cultured under conditions of extremely high cell density, termed micromass culture (1, 22). Recently, it was demonstrated that bone morphogenic protein (BMP) signaling is necessary for the initial compaction of limb bud mesenchymal cells into cellular condensations (2). After compaction, the limb bud cells that undergo chondrogenesis assume a round cell shape and coalesce to form a cartilage nodule (1, 2). For many years, it has been appreciated that environmental factors that alter either the cell shape or the actin cytoskeleton can modulate the ability of prechondrogenic cells to undergo chondrogenesis. It was initially noted that in vitro differentiation of limb bud mesenchymal cells into chondrocytes required that these cells be plated at a very high cell density (1, 22), presumably to promote cell-cell interactions. Interestingly, however, chondrogenesis could be induced in low-density cultures of limb bud mesenchymal cells when these cells were plated in either suspension culture (15, 31) or collagen gels (30). Because culture of prechondrogenic cells in either suspension culture or collagen gels induced cell rounding, Solursh and colleagues speculated that a round cell shape might somehow promote chondrogenesis (41). Indeed, transient treatment of subconfluent limb bud mesenchyme cells with the actin binding drug cytochalasin D induced immediate cell rounding and subsequent chondrogenesis in these cultures (41). Consistent with these findings, more recent work has established that culture of “dedifferentiated” primary chondrocytes in either agarose (3) or alginate (4, 25) can induce the redifferentiation of such cultures and reexpression of the chondrocyte phenotype.
Polymerization of actin to generate stress fibers is regulated by the GTPase RhoA (reviewed in reference 5). When bound to GTP, RhoA interacts with and induces the activity of mDia, a member of the formin family, which directly binds to actin and stimulates its polymerization (16, 39). In addition, GTP-bound RhoA activates RhoA kinase (ROCK), which consequently activates LIM kinase, which phosphorylates the actin depolymerizing protein cofilin and thereby stabilizes actin filaments within the cell (reviewed in reference 5). Studies from the Beier lab have established that overexpression of RhoA can block both Sox9 expression and chondrogenic differentiation of the ATDC5 chondrocyte cell line (36). In addition, this group has demonstrated that inhibition of ROCK signaling or pharmacological disruption of the actin cytoskeleton can promote both Sox9 expression and chondrogenic differentiation in some but not all cellular contexts (35, 36).
In the present study, we demonstrate that RhoA signaling and modulation of actin polymerization not only regulates Sox9 expression (35, 36) but in addition directly controls the transcriptional activity of Sox9 protein. We demonstrate that chondrogenesis entails a dramatic loss of RhoA protein levels and that consequent actin depolymerization increases the transcriptional activity of Sox9 protein to both induce its own expression and activate the expression of other downstream targets. Interestingly, enhancement of Sox9 function by actin depolymerization requires both protein kinase A (PKA) activity and a PKA phosphorylation site in Sox9 (S181), suggesting that actin depolymerization and PKA work synergistically to increase Sox9 transcriptional activity in chondrocytes. Together our findings indicate that chondrogenesis entails a dramatic loss of RhoA protein and consequent depolymerization of actin, resulting in a PKA-dependent enhancement of Sox9 transcriptional activity to activate both its own expression and other downstream targets.
MATERIALS AND METHODS
Reagents and antibodies.
H89 and Y27632 were purchased from Calbiochem. Anti-Flag as well as antitubulin, anti-Sox9-p181, and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were purchased from Sigma, Abcam, and Chemicon, respectively.
Isolation and culture of primary chondrocytes.
The caudal third of day 14 chicken embryo sterna was harvested and incubated in Hanks’ balanced salt solution (Gibco) supplemented with 0.6 mg/ml collagenase (Sigma) and 0.04% trypsin (Gibco) for 3 h at 37°C. The single-cell suspension was harvested and plated (as a primary culture) for 5 to 7 days in primary culture medium (Dulbecco's modified Eagle medium [DMEM; Gibco] supplemented with 10% NuSerum [BD Biosciences], 50 U/ml penicillin, and 50 μg/ml streptomycin [Gibco]) to remove any fibroblast contamination. After primary culture, the floating and loosely attached cells were harvested and used as primary chondrocytes. To dedifferentiate cells, the primary chondrocytes were cultured in secondary culture medium (DMEM supplemented with 10% NuSerum, 50 U/ml penicillin, 50 μg/ml streptomycin, 8 μg/ml hyaluronidase [Sigma], and 5 ng/ml basic fibroblast growth factor [Sigma]) for four passages. To induce redifferentiation of the dedifferentiated cells, the dedifferentiated cells were suspended in a 1.2% alginate solution (1.2% alginate, 150 mM sodium chloride, 1 mM calcium chloride, and 20 mM HEPES) at a density of 106 cells/ml and poured as drops into 50 mM of calcium chloride solution using a 22-gauge needle. After 10 min of incubation at room temperature, the beads were washed with DMEM and transferred to differentiation medium (DMEM-F12 [Gibco] supplemented with 10% fetal bovine serum [HyClone], 50 U/ml penicillin, and 50 μg/ml streptomycin) for various time periods. To harvest cells from alginate, the beads were dissociated in dissociation buffer (55 mM sodium citrate and 150 mM sodium chloride) with gentle rocking at 4°C followed by centrifugation at 1,500 rpm for 5 min.
Micromass culture.
Limb bud mesenchymal cells isolated from Hamburger-Hamilton stage 22 to 24 chicken embryos were used for micromass culture. Briefly, limb bud mesenchymal cells suspended at a density of 2 × 107 cells/ml were plated as a 10-μl spot in a tissue culture dish. Following incubation at 37°C for 2 h, differentiation medium was added and cells were incubated for 4 days. For avian retroviral infection, the cells were incubated with virus for 2 h on ice prior to plating as micromass cultures.
Alcian blue staining.
For Alcian blue staining, micromass cultures were fixed with 4% paraformaldehyde for 15 min, rinsed with 0.1 N hydrochloric acid for 15 min, and then stained with 1% Alcian blue 8GX for 2 h. After staining, the cells were washed overnight with 0.1 N hydrochloric acid. To measure the amount of dye taken up by the micromass cultures, the cultures were gently shaken with 4 M guanidine hydrochloride, pH 5.8, for 2 h. Following extraction, Alcian blue levels were assayed by measuring the absorbance at 600 nm.
RT-PCR.
For semiquantitative reverse transcriptase PCR (RT-PCR), RNA was harvested from samples by using the Qiagen RNeasy minikit following the manufacturer's instructions. RT reactions and PCR analysis were carried out as previously described (17, 42). Briefly, for cDNA synthesis, 1 μg of RNA was used in a 30-μl reaction mixture using Moloney murine leukemia virus RT (Invitrogen) as per the manufacturer's instructions. For PCR analysis, GAPDH was used as a control for the amount of cDNA present in each amplification reaction mixture. We normalized all PCR analyses to an arbitrary level of GAPDH in each experiment by phosphorimager quantitation (Bio-Rad). In addition, most PCRs were performed in the linear range and therefore are semiquantitative assessments of gene expression. One microliter of cDNA was used in a 25-μl PCR amplification reaction mixture containing 10 mM Tris-HCl, pH 8.5, 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 200 μM of each deoxynucleoside triphosphate, 0.5 μCi [α-32P]dCTP (specific activity, 3,000 Ci/mmol), 1 μM of the appropriate primers, and 1 U of Taq polymerase (GeneChoice). After an initial denaturation step of 95°C for 4 min, the reaction mixtures were cycled between 95°C (30 s); either 60°C (30 s for GAPDH and aggrecan), 55°C (30 s for endogenous Sox9, viral human Flag-Sox9, and collagen II), or 50°C (30 s for Sox5 and Sox6); and 72°C (1 min) in an MJ Research thermocycler. The reaction products for GAPDH, collagen II, and aggrecan were amplified for 18 cycles. The products of endogenous Sox9, viral Flag-Sox9, and RhoA were amplified for 23 cycles, while the products of Sox5 and Sox6 were amplified for 25 cycles. The primers used for PCR analysis were as follows: GAPDH-F (AGTCATCCCTGAGCTGAATG), GAPDH-R (ACCATCAAGTCCACAACACG), endogenous Sox9-F (CTCCCCCAACGCCATCTTCA), endogenous Sox9-R (AGCTGCTGATGCCGTAGGTA), viral Flag-tagged human Sox9-F (GGATCTGACTACAAAGACGAT), viral Flag-tagged human Sox9-R (CTGAGCTCGGCGTTGTGCAAG), Sox5-F (GCCACCTGATTAGGGTCCAA), Sox5-R (TTCTGATGCAGGCTGATGTC), Sox6-F (GCAGCTGATGGAGAGGAAAC), Sox6-R (CATCTTGTTGGATGGTCGTG), RhoA-F (CCCAACGTGCCTATCATCTT), RhoA-R (GCAGCTCTAGTGGCCATTTC), Collagen II-F (AAAGATGTTGTAGGACCCCG), Collagen II-R (GCAAAGTTTCCACCAAGTCC), Aggrecan-F (CCTGCCTGACCTCTTTGC), and Aggrecan-R (TGGGGAGGAGGGCAACAT).
Immunofluorescence staining.
Micromass cultures or caudal sternal chondrocytes were washed twice with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 15 min. Cells were permeabilized with ice-cold 0.1% Triton X-100-PBS for 15 min and then washed three times with PBS. After blocking with 5% bovine serum albumin in PBS for 30 min, the cells were incubated with either Alexa Fluor 546 phalloidin (diluted 1:200; Invitrogen) or the primary antibodies mouse anti-RhoA immunoglobulin G1 (IgG1; diluted 1:250; Cytoskeleton) and mouse anti-collagen II IgG2a (diluted 1:250; Developmental Studies Hybridoma Bank, Iowa City, IA) in blocking buffer (5% bovine serum albumin in PBS) for 1 h. Cells were washed with wash buffer (0.1% Tween 20-PBS) three times and incubated with the secondary antibodies anti-mouse IgG1 conjugated to Alexa Fluor 594 (Invitrogen) and anti-mouse IgG2a conjugated to Alexa Fluor 488 (Invitrogen) for 1 h. After three washes, the samples were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min and then covered with a coverslip using mounting medium (Vector Laboratories). Images were obtained using either a Zeiss microscope with an oil immersion objective or a confocal microscope (Nikon A1/C1) with a Plan Apochromat VC ×20 objective. Fluorescence intensities were measured using MetaMorph software.
Rho assay and Western blotting.
The amount of active RhoA was measured by using a RhoA assay kit (Cytoskeleton) as per the manufacturer's instructions. Briefly, the cells were washed twice in PBS and lysed, and the solution was clarified by centrifugation at 8,000 rpm for 5 min. The supernatant was then incubated with rhotekin-Rho binding domain beads for 1 h with gentle rocking at 4°C. The beads were then washed once with 10 volumes of lysis buffer and thrice with 10 volumes of washing buffer. The beads were resuspended in 10 μl of Laemmli buffer. The samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotting. For Western blotting, cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 1 mM NaF) supplemented with protease inhibitor cocktail (Roche). The cell lysates were clarified of cellular debris by centrifugation (10,000 × g) for 10 min at 4°C and were then subjected to SDS-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Millipore). After being blocked with 5% nonfat milk in PBS containing 0.05% Tween 20, the membranes were probed with the indicated primary antibodies. After three washes, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Jackson Laboratories). The interactions were detected using enhanced chemiluminescence (Pierce).
Plasmids and reporter assay.
The following plasmids, 4x48p89-collagen II-luciferase, pcDNA3-Sox9, pcDNA3-Sox5, and pcDNA3-Sox6, were generously supplied by Benoit de Crombrugghe. The expression plasmids for wild-type (WT) actin and actin with a mutation (R62D) were generously supplied by Richard Treisman. The expression plasmids for constitutively activated mDia (mDia-CA) and a dominant negative version of mDia (mDia-DN) were generously supplied by Shuh Narumiya. The expression plasmid for RhoA-N14 was generously supplied by Xi He. The pGal4DBD-Sox9 was made by inserting human Sox9 (hSox9) cDNA in frame with the Gal4 DNA binding domain (DBD) at the BamHI and NotI sites in the pCMV-Gal4DBD vector (a kind gift from H. Sasaki). For reporter assays, cells were transfected in triplicate with plasmid DNA using FuGENE 6 (Roche) as per the manufacturer's instructions. Forty-eight hours after transfection, the cells were harvested; firefly and Renilla luciferase expression levels were measured using the dual-luciferase assay kit (Promega) as per the manufacturer's instructions.
Expression and purification of Tat-myc-C3 transferase protein.
Tat-myc-C3 transferase protein was bacterially expressed and purified as follows. Briefly, competent Escherichia coli BL21(DE3) pLysS cells were transformed with pGEXKG Tat-myc-C3 plasmid (generously provided to us by Chris Marshall, Cancer Research Center, United Kingdom). A single colony was picked and incubated at 37°C in 50 ml of LB broth supplemented with 20 μg/ml chloramphenicol and 100 μg/ml ampicillin. The overnight culture was diluted 1:10 in LB broth supplemented with 100 μg/ml ampicillin and grown to a 0.6 to 1.0 optical density at 600 nm before inducing with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Cells were pelleted by centrifugation and resuspended in 10 ml of Tris-buffered saline (TBS) lysis buffer (137 mM NaCl, 5 mM KCl, 25 mM Tris-HCl, pH 7.4, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride). The cells were then disrupted by three 1-min rounds of sonication at 20% intensity using a Branson Sonifier, followed by removal of debris by centrifugation (10,000 × g) for 20 min at 4°C. Clarified supernatants were incubated with a 0.5 ml glutathione-Sepharose (Sigma) bead slurry for 2 h at 4°C to bind the glutathione S-transferase (GST) fusion protein. Beads were washed six times with 50 ml of TBS lysis buffer, followed by two washes with 50 ml of thrombin cleavage buffer (1 mM magnesium chloride, 1 mM calcium chloride, 1 mM dithiothreitol in TBS). The beads were resuspended in 0.5 ml of thrombin cleavage buffer, and Tat-myc-C3 was released from the GST moiety by incubation with 25 U of bovine thrombin (Sigma) overnight at 4°C. The supernatant was collected, beads were washed twice with 0.5-ml thrombin cleavage buffer, and the collected supernatants were incubated with 30 μl of TBS-washed p-aminobenzamide beads (Sigma) for 1 h to remove thrombin before snap freezing aliquots and storing them at −80°C.
RESULTS
Loss of chondrocyte differentiation correlates with increased RhoA signaling.
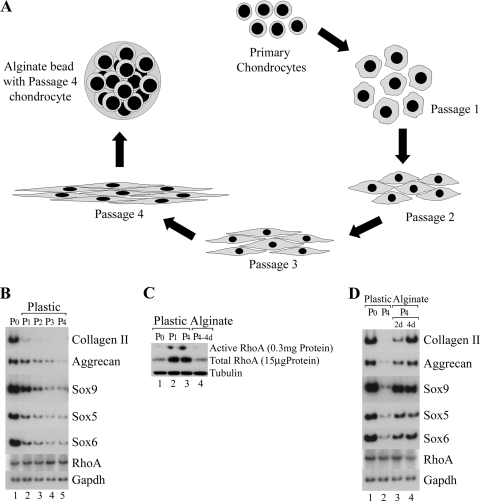
To determine whether RhoA signaling is regulated during chondrocyte differentiation, we employed primary caudal sternal chondrocytes (isolated from day-14 chicken embryos), which display an immature chondrocyte phenotype and appear as very refractile round cells in tissue culture (data not shown). Upon repeated passage, these cells lose their round cell shape and assume a fibroblast morphology (diagrammed in Fig. 1A). We found that both cartilage structural genes (i.e., collagen II and aggrecan) and chondrocyte transcriptional regulators (i.e., Sox9, Sox5, and Sox6) were robustly expressed in the caudal sternal chondrocytes after their initial plating (P0) (Fig. 1B, lane 1) and that expression of all of these chondrocyte markers was lost during continued passage of these cells (P1 to P4) (Fig. 1B, lanes 2 to 5). We assayed RhoA levels in these cultures by using Western blotting and assayed RhoA activity by using a rhotekin-GST pull-down assay (27). Rhotekin is a RhoA effector, which can bind only to the GTP-bound form of RhoA (i.e., “activated” RhoA) (26), and therefore rhotekin-GST can be employed to isolate active RhoA, which is subsequently detected by Western blotting using a RhoA antibody. Interestingly, we found that freshly isolated caudal sternal chondrocytes contained relatively low levels of total RhoA and only trace levels of active (GTP-bound) RhoA (Fig. 1C, lane 1). In striking contrast, we found that passage of these cells on plastic led to a significant increase in the levels of both total RhoA protein and active GTP-bound RhoA (Fig. 1C, compare lane 1 with lanes 2 to 3). Thus, there appears to be an inverse correlation between the levels of active RhoA protein in a cell and the expression of both chondrogenic transcription factors and differentiation markers. Interestingly, the dramatic increase in the total levels of RhoA protein in dedifferentiated cells compared to their chondrogenic progenitors (Fig. 1C) was not accompanied by any alteration in RhoA transcript levels (Fig. 1B).
FIG. 1.
Reactivation of the chondrocyte differentiation program in alginate culture correlates with a loss of both RhoA expression and activity. (A) Protocol to isolate caudal sternal chondrocytes, derive dedifferentiated chondrocytes from these cultures, and restore differentiation of these latter cells in alginate culture conditions. Caudal sternal chondrocytes were isolated from day 14 chicken embryos and either harvested after 5 days in culture following their initial plating (P0) or repeatedly passaged (P1 to P4) in medium containing hyaluronidase (8 μg/ml). P4 dedifferentiated cells were subsequently plated in alginate beads for 2 or 4 days to restore chondrocyte gene expression. (B) Dedifferentiation of primary sternal chondrocytes after continued passage in tissue culture. Caudal sternal chondrocytes were isolated from day 14 chicken embryos and either harvested after 5 days in culture following their initial plating (P0) or repeatedly passaged (P1 to P4) in medium containing hyaluronidase (8 μg/ml). Gene expression was assayed by RT-PCR analysis. (C) RhoA levels and activity increase during chondrocyte dedifferentiation, whereas culture of dedifferentiated chondrocytes in alginate dramatically lowers the levels of both total and active RhoA. Total RhoA levels were assayed by Western blot analysis, and GTP-bound “activated” RhoA protein levels were assayed by rhotekin binding followed by Western blot analysis in extracts derived from P0 chondrocytes or passaged (P1) cells, P4 cells cultured on plastic, or in P4 cells cultured for 4 days in alginate (lanes 1 to 4, respectively). We have obtained similar results in four independent experiments. (D) Culture of dedifferentiated chondrocytes in alginate dramatically results in reexpression of chondrogenic differentiation markers. Caudal sternal chondrocytes were isolated from day 14 chicken embryos and either harvested following their initial plating (P0; lane 1) or passaged four times (P4; lane 2). P4 cells were subsequently cultured in alginate beads for either 2 or 4 days (P4; lanes 3 and 4). Gene expression was assayed by RT-PCR analysis.
Culture of dedifferentiated chondrocytes in alginate gel induces chondrogenic gene expression and dramatically lowers the levels of total and active RhoA protein.
We investigated whether restoration of the chondrogenic phenotype by culture of dedifferentiated cells in an alginate gel would concomitantly affect RhoA protein levels and/or activity. Caudal sternal chondrocytes were passaged four times (P4; during which time they lost expression of chondrocyte-specific markers) and cultured for either 2 or 4 days in alginate beads (Fig. 1D). Culture of the dedifferentiated cells (P4) in alginate for 2 or 4 days led to a dramatic increase in chondrocyte-specific gene expression (Fig. 1D, compare lane 2 with lanes 3 and 4), which was nearly equal to that in freshly plated (P0) chondrocytes (Fig. 1D, lane 1). Interestingly, reexpression of chondrocyte differentiation markers in the cells cultured in alginate correlated with a profound decrease in the levels of both total and active (i.e., GTP-bound) RhoA (Fig. 1C, compare lanes 3 and 4) in the absence of altering RhoA transcript levels (Fig. 1D). These findings indicate that culturing dedifferentiated chondroctyes in alginate gel both restores high levels of chondrocyte gene expression and simultaneously promotes a precipitous loss of RhoA protein.
Chondrogenic gene expression is inversely correlated with RhoA protein accumulation and the appearance of stress fibers.
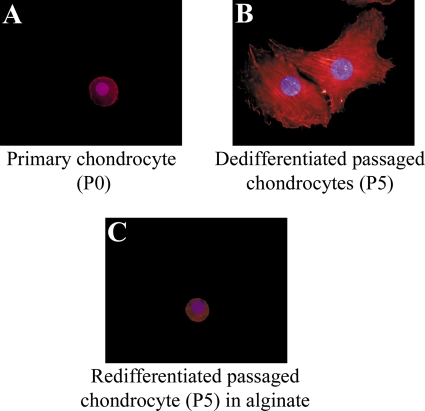
Our results indicate that there is an inverse correlation between chondrocyte differentiation and the level of activated (GTP-bound) RhoA. As activation of RhoA is known to control the polymerization of actin into stress fibers (5), we compared the state of the actin cytoskeleton in both primary chondrocytes and in dedifferentiated chondrocytes cultured either on plastic or in alginate gel. To visualize the polymerization state of actin in both primary chondrocytes and their dedifferentiated progeny, we employed phalloidin, which binds to polymeric actin (38). We observed that while P0 chondrocytes display low levels of diffusely staining cytoplasmic actin (Fig. 2A), dedifferentiated P5 cells show very prominent stress fibers (Fig. 2B). Culture of these dedifferentiated cells in alginate culture not only reinduces chondrogenic gene expression (Fig. 1D) but also restores the actin cytoskeleton to a chondrocyte-like appearance, marked by the absence of detectable stress fibers (Fig. 2C).
FIG. 2.
Chondrocyte-specific gene expression inversely correlates with the appearance of polymerized actin in the form of stress fibers in the cell. Shown are phalloidin staining images of the following cells: caudal sternal chondrocytes isolated from day 14 chicken embryos after 5 days in culture following their initial plating (P0) (A), dedifferentiated progeny of these cells that had been passaged five times (P5) in medium containing hyaluronidase (B), or P5 cells that had been cultured for 4 days in alginate beads and were found to reexpress chondrocyte-specific differentiation markers (C).
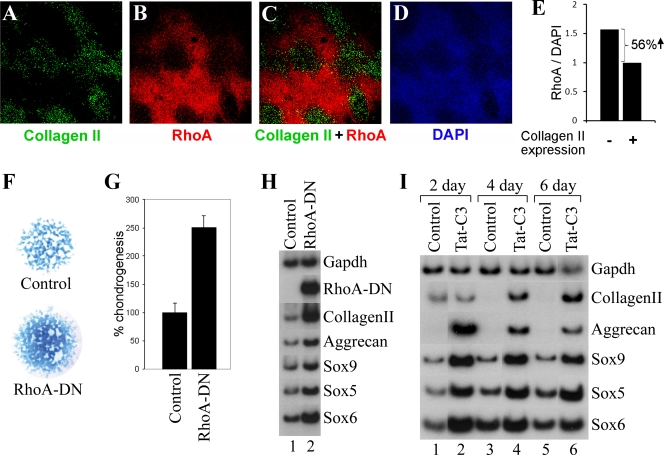
We examined whether expression of collagen II was also inversely correlated with accumulation of RhoA protein in micromass cultures of limb bud mesenchymal cells isolated from HH stage 22 to 24 chicken embryos. We found that the expression of collagen II was specifically confined to areas of the micromass culture displaying the lowest levels of RhoA protein (Fig. 3A to D). We quantitated the RhoA/DAPI staining ratio in differing regions of the micromass culture and found that regions of the culture lacking collagen II displayed a 56% greater RhoA/DAPI staining ratio than regions of the culture expressing collagen II (Fig. 3E). These findings suggest that chondrogenic differentiation in limb bud mesenchymal cells similarly entails a loss of RhoA protein. Thus, in both dedifferentiated chondrocytes cultured either on plastic or in alginate gel and in limb bud micromass cultures, chondrocyte-specific gene expression inversely correlates with the accumulation of both RhoA protein and stress fibers.
FIG. 3.
RhoA signaling represses chondrocyte differentiation in both of the limb bud micromass cultures or in dedifferentiated chondrocytes maintained on plastic. (A to E) Chicken limb bud mesenchymal cells grown in high-density micromass culture were immunostained with anti-collagen II, anti-RhoA, or DAPI as indicated. In panel E, the RhoA/DAPI staining ratio in regions of the micromass culture that either lack or express collagen II is displayed. (F to H) Chicken limb bud mesenchymal cells were infected with either RCAS-GFP (as a control) or RCAS-RhoA-DN (encoding dominant negative RhoA) and plated in high-density micromass culture and stained with Alcian blue (displayed in panel F and quantitated in panel G) or RNA was harvested and gene expression was analyzed by quantitative RT-PCR (H). (I) Administration of the pan-Rho antagonist C3 transferase can promote the redifferentiation of dedifferentiated chondrocytes. Dedifferentiated chondrocytes (P4) were cultured on plastic in either the absence or presence of a Tat-C3 transferase fusion protein for the indicated number of days. Gene expression was assayed by RT-PCR analysis.
RhoA activity represses chondrogenesis in micromass cultures of chicken limb bud mesenchymal cells.
Because RhoA protein was highest in areas of the micromass culture that failed to express collagen II (Fig. 3A to E), we examined whether inhibition of RhoA signaling in such cultures would augment chondrogenesis. We infected limb bud mesenchymal cells with an avian retrovirus encoding either a dominant negative form of RhoA (RhoA-DN, encoded by RhoA mutant RhoA-N19) (10) or green fluorescent protein (GFP) as a control. The infected cells were then plated under micromass conditions for 4 days. We evaluated both proteoglycan expression by using Alcian blue staining (Fig. 3F and G) and the expression of chondrocyte marker genes by semiquantitative RT-PCR (Fig. 3H). Expression of RhoA-DN significantly increased both the extent of Alcian blue staining in these cultures and the expression of chondrogenic marker genes (Fig. 3F to H). Thus, repressing RhoA signaling with a dominant negative form of RhoA enhances chondrogenesis in limb bud micromass cultures, indicating that RhoA signaling limits the amount of chondrocyte differentiation in such cultures.
Inhibition of Rho signaling by the pan-Rho antagonist, C3 transferase, can restore chondrocyte gene expression in dedifferentiated chondrocytes grown on plastic.
Because dedifferentiated chondrocytes grown on plastic show high levels of activated RhoA and extensive stress fibers (Fig. 1 and 2), we evaluated whether administration of the botulinum ADP-ribosyltransferase (C3 transferase) that inhibits RhoA activity (23, 29) would increase the expression of chondrocyte-specific genes in otherwise dedifferentiated chondrocytes grown on a plastic substrate. C3 transferase blocks Rho signaling by catalyzing ADP ribosylation of all Rho family members at amino acid Asn-41 (29). We found that culturing dedifferentiated chondrocytes on plastic in the presence of a fusion protein encoding C3 transferase fused to the leader sequence of the human immunodeficiency virus Tat protein (that promotes the transport of exogenous proteins into the cell) (28) significantly induced the expression of collagen II and aggrecan and increased the expression of endogenous Sox9, Sox5, and Sox6 (Fig. 3I). Induction of chondrocyte-specific gene expression by administration of C3 transferase to dedifferentiated chondrocytes suggests that Rho family signaling represses chondrogenesis in such cells and is consistent with prior findings that inhibition of the RhoA-activated kinase, ROCK, can promote chondrogenesis in some cellular contexts (35).
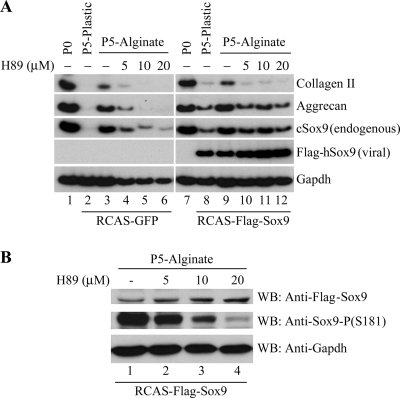
Alginate culture regulates the ability of exogenous Sox9 to induce chondrocyte-specific gene expression in a PKA-dependent manner.
We noticed that culture conditions that induce the loss of RhoA protein in dedifferentiated chondrocytes (such as alginate culture) or agents that inhibit RhoA signaling (such as dominant negative RhoA or the pan-Rho antagonist C3 transferase) robustly induce the expression of both chondrocyte-specific differentiation markers (e.g., collagen II and aggrecan) and the chondrocyte transcriptional regulators Sox9, Sox5, and Sox6. Thus, it seems possible that the loss of RhoA protein or activity promotes chondrogenesis by inducing the expression of these key chondrocyte transcriptional regulators. However, as many cell-type-specific transcriptional regulators maintain their own expression via positive feedback loops (e.g., MyoD) (33), it is possible that increased expression of the chondrogenic Sox transcription factors induced by the loss of RhoA protein/activity reflects an increase in the transcriptional activity of Sox9. To clarify whether alginate culture conditions induce the differentiation of dedifferentiated chondrocytes by solely enhancing the expression of Sox9 or, in addition, by promoting the transcriptional activity of this transcription factor, we infected dedifferentiated chondrocytes (passage 5) with a retrovirus encoding Flag-tagged hSox9 (RCAS-Flag-hSox9). We observed that forced expression of exogenous Flag-tagged hSox9 in dedifferentiated chondrocytes cultured on plastic increased expression of both endogenous chicken Sox9 and aggrecan and induced trace levels of collagen II (Fig. 4A, compare lanes 2 and 8). Interestingly however, the culture of such RCAS-Flag-hSox9-infected cells in alginate gel further increased the expression of all of these chondrocyte markers while not affecting the expression of virus-encoded hSox9 (Fig. 4A, compare lanes 8 and 9). Expression of exogenous RCAS-encoded Flag-tagged hSox9 and endogenous chicken Sox9 were distinguished by employing human and chicken Sox9 primers, respectively, in RT-PCR analysis. These findings indicate that alginate culture and the associated loss of RhoA signaling (Fig. 1) both increase the expression of endogenous Sox9 and enhance the activity of exogenous Sox9 to activate downstream targets.
FIG. 4.
The ability of Sox9 to activate both its own expression and that of other chondrocyte markers is boosted by culture of dedifferentiated cells in alginate gel. (A) Freshly isolated caudal sternal chondrocytes (P0) or dedifferentiated chondrocytes (P5) that have been infected with either RCAS-GFP or RCAS-Flag-hSox9 were cultured either on plastic or in alginate gel as indicated. In some cases, the cultures were exposed to increasing levels of the PKA antagonist H89, as indicated. Gene expression was analyzed by RT-PCR. (B) RCAS-Flag-hSox9 maintains the expression of endogenous chicken Sox9 in the presence of levels of H89 that block phosphorylation of Sox9 S181. Dedifferentiated chondrocytes (P5) infected with RCAS-Flag-hSox9 were cultured in alginate gel and treated with increasing levels of the PKA antagonist H89. Western blot analysis is displayed for Flag-tagged hSox9, Sox9 phospho-S181, and GAPDH.
In addition to BMP signaling and a round cell shape, another factor that seems to play an important role in chondrocyte differentiation is PKA. Treatment of micromass cultures of limb bud mesenchymal cells with the cyclic AMP analogue, dibutyryl cyclic AMP, enhances chondrocyte differentiation in such cultures (12, 32). In addition, PKA activity has been found to rise during chondrogenic differentiation of limb bud micromass cultures (12), and pharmacological inhibition of PKA with the PKA antagonist H89 efficiently blocks chondrogenic differentiation of limb bud micromass cultures (12, 40). Similar to its effect on limb bud cultures, H89 also blocks the induction of endogenous Sox9 and other chondrocyte differentiation markers in dedifferentiated chondrocytes cultured in alginate gel (Fig. 4A, lanes 4 to 6). In striking contrast, forced expression of RCAS-Flag-hSox9 can boost the expression of both endogenous Sox9 and aggrecan in dedifferentiated chondrocytes cultured in alginate gel in the presence of H89 (Fig. 4A, compare lanes 10 to 12). Induction of both endogenous Sox9 and aggrecan by RCAS-Flag-hSox9 occurred in the presence of H89 despite the inhibition of a PKA-dependent activating phosphorylation of Sox9-S181 (9) (Fig. 4B). These findings suggest that a kinase whose activity is blocked by H89 (perhaps PKA) is necessary to promote the expression of endogenous Sox9 in dedifferentiated chondrocytes cultured in alginate gel and that forced expression of RCAS-Flag-hSox9 can activate the expression of endogenous Sox9 even in the presence of this kinase inhibitor.
Most importantly, the induction of both endogenous Sox9 and other chondrocyte markers (i.e., aggrecan and collagen II) by RCAS-Flag-hSox9 was considerably augmented by culture of the dedifferentiated cells in alginate (Fig. 4A, compare lanes 8 and 9). Interestingly however, the ability of alginate culture to enhance the transcriptional activity of exogenous Sox9 was blocked by administration of H89 (Fig. 4A, compare lanes 9 and 12). These findings indicate that alginate culture conditions, which entail a loss of RhoA protein (Fig. 1), increase the transcriptional activity of exogenous Sox9 in a PKA-dependent fashion.
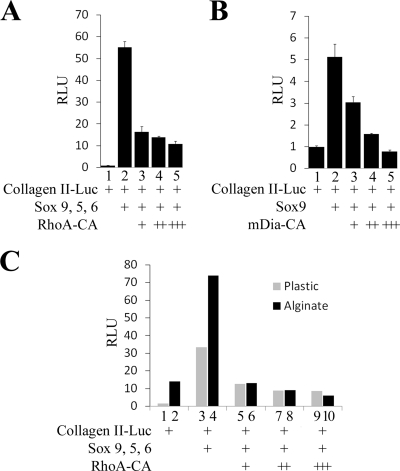
Constitutively active RhoA or mDia can repress the ability of cotransfected chondrogenic Sox factors to activate expression of a collagen II reporter.
Our results indicate that culturing dedifferentiated chondrocytes in alginate gel, with consequent loss of RhoA protein, promotes chondrocyte gene expression by modulating the transcriptional activity of Sox9. To investigate whether RhoA (or its downstream effectors) could similarly modulate the activity of exogenous, transfected Sox9, we evaluated whether activated RhoA or mDia would affect the ability of exogenous Sox9, Sox5, and Sox6 to activate expression of a collagen II-luciferase reporter (4x48p89-Col.II-Luc) (14). Cotransfection of cytomegalovirus-driven Sox9, Sox5, and Sox6 robustly induced expression of the 4x48p89-Col.II-Luc reporter in caudal sternal chondrocytes (Fig. 5A, compare lanes 1 to 2). Addition of an expression vehicle encoding constitutively activated RhoA (RhoA-CA) dramatically blocked the abilities of cotransfected Sox9, Sox5, and Sox6 to induce the expression of this reporter in a dose-dependent manner (Fig. 5A, compare lanes 2 with 3 to 5). Thus, activation of RhoA signaling can block the ability of cotransfected chondrogenic Sox transcription factors to activate expression of a collagen II reporter construct.
FIG. 5.
Activated RhoA or activated mDia can block the abilities of cotransfected Sox9, Sox5, and Sox6 to activate a collagen II reporter and block the prochondrogenic effects of alginate culture. (A) Caudal sternal chondrocytes were cotransfected with a 4x48p89Col.II-firefly luciferase reporter together with expression vectors encoding Sox9, Sox5, Sox6, and RhoA-CA (constitutively activated RhoA) as indicated. In addition, cells were transfected with a simian virus 40 (SV40)-Renilla luciferase construct. The cells were harvested 48 h after transfection followed by lysis of the cells and luciferase assay. 4x48p89Col.II-firefly luciferase activity was normalized to the expression of the cotransfected SV40-Renilla luciferase reporter. (B) Forced expression of a constitutively active form of mDia1 can block the ability of Sox9 to activate a chondrocyte-specific reporter. Caudal sternal chondrocytes were transfected with the 4x48p89Col.II-luciferase reporter and an expression vector encoding Sox9 plus increasing amounts of an expression vector encoding the constitutively active form of mDia (mDia1-CA), which constitutively promotes actin polymerization even in the absence of RhoA signaling (18). In addition, cells were transfected with an SV40-Renilla luciferase construct. The cells were harvested 48 h after transfection followed by lysis of the cells and luciferase assay. 4x48p89Col.II-firefly luciferase activity was normalized to the expression of the cotransfected SV40-Renilla luciferase reporter. (C) Dedifferentiated caudal sternal chondrocytes (which had been passaged five times in plastic) were cotransfected with the 4x48p89Col.II-luciferase reporter, an SV40-Renilla luciferase reporter, and expression vectors for Sox9 and constitutively active RhoA-CA as indicated. After transfection, cells were cultured either on plastic or in alginate beads for 72 h. 4x48p89Col.II-firefly luciferase activity was normalized to the expression of the cotransfected SV40-Renilla luciferase reporter.
While active RhoA has many possible effector molecules, one of its principal activities within the cell is to control the polymerization of actin into stress fibers (5). To determine whether activated RhoA blocks Sox9 transcriptional activity by increasing the levels of polymerized actin within the cell, we employed a constitutively active mutant form of mDia1 (mDia1-CA), encoded by mDiaΔN3 (34), which constitutively promotes actin polymerization even in the absence of RhoA signaling (18). We found that transfection of an expression vehicle encoding mDia1-CA efficiently blocked the ability of cotransfected Sox9 to activate expression of a collagen II-luciferase reporter in a dose-dependent manner (Fig. 5B, compare lane 2 with lanes 3 to 5). Because expression of a constitutively active form of mDia1 blocks Sox9 transcriptional activity, as does expression of a constitutively active form of RhoA, it seems most likely that RhoA signaling blocks chondrogenesis by in part activating the ability of mDia1 to promote actin polymerization.
Culturing dedifferentiated chondrocytes in alginate beads augments the ability of Sox9 to activate gene expression; activated RhoA blocks this effect.
Because alginate culture conditions promote chondrocyte-specific gene expression while promoting the disassembly of stress fibers within the cell (Fig. 1 and 2), we were curious to know if forced expression of activated RhoA (which promotes the formation of stress fibers) (5) would reverse the prochondrogenic effects of alginate culture. To investigate this possibility, we monitored the expression of a transfected collagen II-luciferase reporter in dedifferentiated (P5) chondrocytes that had been cultured either on plastic or in alginate gel. As observed for expression of endogenous chondrocyte-specific genes, expression of the collagen II-luciferase reporter was significantly boosted in P5-dedifferentiated chondrocytes when cultured in alginate gel (Fig. 5C, compare lanes 1 and 2). In addition, we found that the ability of Sox9 to induce expression of the collagen II-luciferase reporter was significantly increased in cells cultured in alginate (Fig. 5C, compare lanes 3 and 4). To evaluate if RhoA signaling could negate the effects of alginate culture conditions, we cotransfected the cells with an expression vehicle encoding activated RhoA (RhoA-CA). While the ability of Sox9 to induce expression of the collagen II reporter was augmented in cells cultured in alginate relative to cells cultured on plastic, cotransfection of constitutively active RhoA blocked this effect (Fig. 5C, compare lanes 3 and 4 with lanes 9 and 10). Thus, constitutively activated RhoA can reverse the ability of alginate culture conditions to promote chondrocyte-specific gene expression.
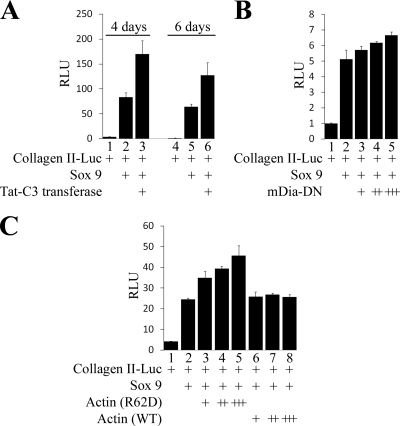
Blockade of actin polymerization enhances the ability of Sox9 to activate chondrocyte-specific gene expression.
Because our findings suggest that active (GTP-bound) RhoA blocks the activity of exogenous Sox9 by promoting actin polymerization, we investigated whether expression of agents that block either RhoA activity or actin polymerization would conversely enhance the transcriptional activity of Sox9. We found that culture of dedifferentiated chondrocytes in medium containing the Tat-C3 transferase fusion protein did in fact augment the ability of exogenous Sox9 to activate a collagen II-luciferase reporter (Fig. 6A), indicating that Rho signaling inhibits the activity of Sox9 in dedifferentiated chondrocytes. To explore whether inhibition of actin polymerization would similarly enhance the transcriptional activity of Sox9, we cotransfected Sox9 with either a dominant negative version of mDia (mDia-DN) (34) or a mutant form of actin [actin (R62D)] (24), both of which reduce actin polymerization. We found that cotransfection of either mDia-DN (Fig. 6B) or actin (R62D) (Fig. 6C) boosted Sox9 induction of a collagen II-luciferase reporter. Thus, agents that block either RhoA function or actin polymerization boost the ability of exogenous Sox9 to induce chondrocyte gene expression.
FIG. 6.
Blockade of actin polymerization with either Tat-C3 transferase, dominant negative mDia, or overexpression of an actin mutant that blocks actin polymerization and augments the ability of Sox9 to activate chondrocyte-specific gene expression. (A) Dedifferentiated chondrocytes (P4) were cotransfected with the 4x48p89Col.II-firefly luciferase reporter, a simian virus 40 (SV40)-Renilla luciferase reporter, plus a cytomegalovirus-driven Sox9 expression vehicle as indicated. The cells were cultured in either the absence or presence of Tat-C3 transferase, as indicated, harvested after either 4 or 6 days in culture, and assayed for relative luciferase units (RLU). (B) Caudal sternal chondrocytes were cotransfected with the 4x48p89Col.II-firefly luciferase reporter and an SV40-Renilla luciferase reporter together with expression vectors encoding Sox9 and a mDia-DN (34) as indicated. 4x48p89Col.II-firefly luciferase activity was normalized to the expression of the cotransfected SV40-Renilla luciferase reporter. (C) Dedifferentiated chondrocytes (P4) were cotransfected with the 4x48p89Col.II-luciferase reporter, an SV40-Renilla luciferase reporter, and an expression vector encoding Sox9 and increasing amounts of expression vectors encoding either a mutant form of actin [actin (R62D)] that reduces actin polymerization (24) or WT actin as indicated. 4x48p89Col.II-firefly luciferase activity was normalized to the expression of the cotransfected SV40-Renilla luciferase reporter.
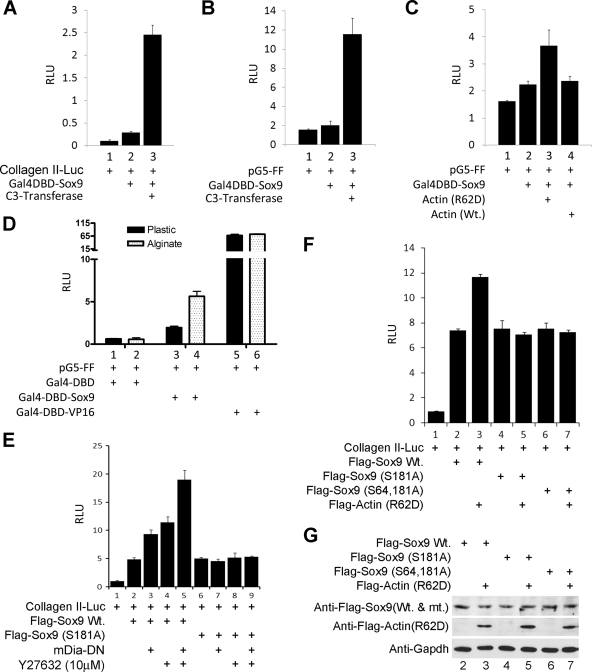
C3 transferase or alginate culture boosts the transcriptional activity of a Gal4-Sox9 fusion protein.
Because the collagen II-luciferase reporter (4x48p89-Col.II-Luc) (14) contains binding sites for Sox5, Sox6, and Sox9 (13), augmented expression of this reporter by agents that block either RhoA function or actin polymerization could reflect the enhancement of either the expression or activity of any of these Sox transcription factors. To evaluate if agents that block actin polymerization directly affect the transcriptional activity of Sox9, we employed a chimeric protein consisting of the DBD of Gal4 fused to Sox9 (Gal4DBD-Sox9). Cotransfection of dedifferentiated chondrocytes with an expression vehicle encoding C3 transferase significantly augmented the ability of Gal4DBD-Sox9 to activate the expression of either the 4x48p89-Col.II-luciferase reporter (Fig. 7A) or the pG5-luciferase reporter (Fig. 7B), whose expression is driven by reiterated Gal4 binding sites. In addition, cotransfection of dedifferentiated chondrocytes with actin (R62D), which blocks actin polymerization, but not with WT actin, significantly augmented the induction of the pG5-luciferase reporter by Gal4DBD-Sox9 (Fig. 7C). Because culture of dedifferentiated chondrocytes in alginate gel represses RhoA activity and induces a precipitous loss of stress fibers (Fig. 1 and 2), we examined whether these culture conditions would augment the transcriptional activity of Gal4DBD-Sox9. Indeed, we found that induction of the pG5-luciferase reporter by Gal4DBD-Sox9 was increased approximately threefold by culturing the cells in alginate gel versus on plastic (Fig. 7D). In contrast, induction of the pG5-luciferase reporter by Gal4DBD-VP16 (which contains the herpes simplex virus VP16 activation domain) was equivalent in cells cultured in alginate gel versus those on plastic (Fig. 7D). Together, these findings indicate that agents that decrease either RhoA function (C3 transferase) or expression (alginate gel culture conditions) or that block actin polymerization [actin (R62D)] all induce the transcriptional activity of a Gal4-Sox9 fusion protein, and by implication induce the transcriptional activity of Sox9.
FIG. 7.
C3 transferase or alginate culture boosts the transcriptional activity of a Gal4-Sox9 fusion protein. (A and B) Dedifferentiated chondrocytes were cotransfected with either the 4x48p89Col.II-firefly (FF) luciferase reporter (A) or the pGalX5-firefly luciferase reporter (pG5-FF) (B) plus the simian virus 40 (SV40)-Renilla luciferase reporter and expression vehicles encoding a Gal4DBD-Sox9 fusion protein or C3 transferase, as indicated. In this and subsequent parts of this figure, the 4x48p89Col.II-FF or pG5-FF luciferase activity was normalized to the expression of the cotransfected SV40-Renilla luciferase reporter. (C) Dedifferentiated chondrocytes were cotransfected with the pGalX5-firefly luciferase reporter (pG5-FF) plus the SV40-Renilla luciferase reporter and expression vehicles encoding a Gal4DBD-Sox9 fusion protein, actin (R62D) or WT actin as indicated. Cells were processed as for panels A and B. (D) Dedifferentiated chondrocytes were cotransfected with the pGalX5-firefly luciferase reporter (pG5-FF) plus the SV40-Renilla luciferase reporter and expression vehicles encoding either Gal4DBD or Gal4DBD-Sox9 fusion protein or Gal4DBD-VP16, as indicated. After transfection, cells were cultured either on plastic or in alginate beads for 72 h. pGalX5-firefly luciferase activity was normalized to the expression of the cotransfected SV40-Renilla luciferase reporter. (E to G) Dedifferentiated chondrocytes were cotransfected with the 4x48p89Col.II-luciferase reporter, an SV40-Renilla luciferase reporter, and an expression vector encoding WT Sox9, Sox9 (S181A), or Sox9 (S64, 181A) and either treated with the ROCK antagonist, Y27632, or cotransfected with an expression vector encoding mDia-DN or actin (R62D) as indicated. The 4x48p89Col.II-FF luciferase activities were normalized to the expression of the cotransfected SV40-Renilla luciferase reporter. (G) Expression levels of WT Sox9, Sox9 (S181A), or Sox9 (S64, 181A) and actin (R62D) were assessed by Western blotting. GAPDH levels are displayed as a loading control for Western blotting.
Sox9 S181 is required for actin depolymerization to boost Sox9 function.
While culture of dedifferentiated chondrocytes in alginate gel boosted the ability of exogenous Sox9 to induce chondrocyte gene expression, we noted that treatment of such cultures with the PKA antagonist H89 blocked the ability of alginate gel to enhance exogenous Sox9 function (Fig. 4). This finding suggests that enhancement of Sox9 activity by actin depolymerization requires PKA activity. To directly explore whether PKA-mediated phosphorylation of Sox9 is necessary for actin depolymerization to augment Sox9 function, we investigated whether administration of the ROCK inhibitor, Y27632, or cotransfection of either mDia-DN or actin (R62D) could enhance the activity of mutant forms of Sox9-containing mutant PKA phosphorylation sites (S181A or S64,181A) (9). WT Sox9, Sox9-S181A, and Sox9-S64,181A each induce comparable levels of expression of a cotransfected collagen II-luciferase reporter in dedifferentiated chondrocytes (Fig. 7E, lanes 2, 4, and 6). Administration of the ROCK inhibitor, Y27632, plus cotransfection of mDia-DN work synergistically to depolymerize actin (5) and increased the induction of the collagen II-luciferase reporter by Sox9-WT by over 300% (Fig. 7E, lanes 2 to 5). In contrast, the combination of both these reagents failed to affect induction of this reporter by Sox9-S181A (Fig. 7E, lanes 6 to 9). Similarly, cotransfection of actin (R62D), which also blocks actin polymerization, increased the ability of WT Sox9 to induce expression of the collagen II-luciferase reporter yet failed to affect induction of this reporter by either Sox9-S181A or Sox9-S64,181A (Fig. 7F). Importantly, each of these forms of Sox9 was equivalently expressed in either the absence or presence of actin (R62D) (Fig. 7G). These findings suggest that phosphorylation of Sox9-S181 is necessary for actin depolymerization to enhance Sox9 function.
DISCUSSION
RhoA signaling and actin polymerization negatively regulate the transcriptional activity of Sox9 during chondrogenesis.
Our findings indicate that culture of dedifferentiated chondrocytes in alginate gel results in a precipitous loss of RhoA protein and a corresponding loss of actin polymerization into stress fibers. Culturing dedifferentiated chondrocytes in alginate gel is sufficient to completely restore expression of both the chondrogenic Sox transcription factors and chondrocyte differentiation markers such as aggrecan and collagen II to levels found in nonpassaged chondrocytes. In contrast, while forced expression of exogenous retrovirus-encoded Sox9 in dedifferentiated chondrocytes maintained on plastic can induce expression of readily detectable levels of endogenous Sox9 and aggrecan (and trace levels of collagen II), activation of all these markers by exogenous Sox9 is significantly augmented by culturing cells in alginate gel. In addition, we have found that alginate gel culture conditions enhance the transcriptional activity of either WT Sox9 or a Gal4 DBD-Sox9 fusion protein (Gal4DBD-Sox9), indicating that the transcriptional activity of Sox9 is modulated by cell shape. The ability of alginate gel culture to enhance Sox9 transcriptional activity was abrogated by cotransfection of a constitutively activated form of RhoA. Thus, the loss of RhoA activity is necessary for alginate gel culture to augment Sox9 transcriptional activity. Conversely, agents that block actin polymerization, such as dominant negative mDia or a mutant form of actin (R62D), both increase the transcriptional activity of either exogenous WT Sox9 or a Gal4-Sox9 fusion protein. Together, these findings indicate that culture of dedifferentiated chondrocytes in alginate gel enhances both the expression and transcriptional activity of Sox9 and that the loss of RhoA activity and consequent depolymerization of actin filaments enhance the transcriptional activity of Sox9.
PKA-dependent phosphorylation of Sox9 is apparently required for actin depolymerization to enhance Sox9 transcriptional activity. We have observed that the PKA antagonist H89 blocks the ability of alginate culture to enhance Sox9 function and that mutation of a PKA phosphorylation site in Sox9 (S181) blocks the ability of agents that induced actin depolymerization to boost Sox9 activity while not affecting the basal activity of Sox9 in dedifferentiated chondrocytes. Together these findings suggest that PKA-mediated phosphorylation of Sox9-S181 is necessary for actin depolymerization to enhance Sox9 function. Future analysis will determine whether actin depolymerization augments Sox9 function either by regulating the extent of Sox9 S181 phosphorylation or by regulating the association of Sox9 phospho-S181 with transcriptional coactivators.
Sox9 activates its own expression in chondrocytes.
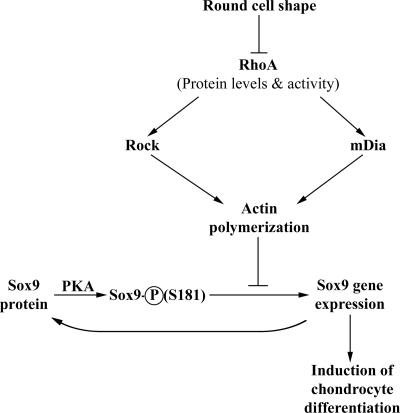
Exogenous retrovirus-encoded Sox9 induced the expression of endogenous Sox9 in dedifferentiated chondrocytes, indicating that Sox9 has the capacity to activate its own synthesis in dedifferentiated chondrocytes. We observed that while administration of the PKA antagonist H89 could efficiently block the expression of endogenous Sox9 in dedifferentiated cells cultured in alginate gel, the expression of retrovirus-encoded Sox9 could maintain the expression of endogenous Sox9 in such H89-treated cells. These findings suggest that Sox9 protein may act to maintain expression of its own locus in differentiated chondrocytes and that signals/culture conditions that inhibit RhoA activity enhance the transcriptional activity of Sox9 and may therefore act to establish this positive autoregulatory loop (summarized in Fig. 8).
FIG. 8.
Model for how RhoA signaling controls chondrogenesis. Our findings indicate that culture of dedifferentiated chondrocytes in alginate gel induces a precipitous loss of RhoA protein, thus resulting in a loss of polymerized actin in the cell. Enhancement of Sox9 function by actin depolymerization requires both PKA activity and a PKA phosphorylation site in Sox9 (S181) that is known to enhance Sox9 transcriptional activity (9). It is possible that actin depolymerization augments Sox9 function by either regulating the extent of Sox9 S181 phosphorylation by PKA or regulating the association of Sox9 phospho-S181 with transcriptional coactivators. Because Sox9 has the capacity to induce its own expression, RhoA signaling may in addition block a Sox9 autoregulatory loop and thereby inhibit Sox9 gene expression.
Chondrogenesis downregulates the amount and activity of RhoA protein but not RhoA RNA.
The induction of Sox9 expression in dedifferentiated chondrocytes cultured in alginate correlates with a loss of RhoA protein but not RhoA RNA. It is possible that the uncoupling of RhoA RNA from RhoA protein levels in either chondrocytes or dedifferentiated chondrocytes that are cultured in alginate gel reflects either a differential stability of RhoA protein in cells cultured in alginate or a differential translation of RhoA RNA in such cells. In either case, the culture of cells in alginate gel induces a precipitous loss of both total and active (GTP-bound) RhoA protein and induces a consequent loss of polymerized actin localized to stress fibers. Consistent with these findings, we observed that the expression of collagen II in limb bud micromass cultures was specifically confined to areas of the micromass culture displaying the lowest levels of RhoA protein, suggesting that chondrogenic differentiation in limb bud mesenchymal cells similarly entails a loss of RhoA protein. The modulation of RhoA protein levels and activity by cell shape change is no doubt relevant to the differentiation status of chondrocytes, as we have observed that treatment of either dedifferentiated chondrocytes with the pan-Rho antagonist, Tat-C3 transferase, or expression of dominant negative RhoA in chick limb bud micromass cultures induces a robust increase in chondrocyte differentiation. How is RhoA signaling controlled during chondrogenesis? Cellular condensation within the prechondrogenic regions correlates with a dramatic increase in the expression of N-cadherin, whose expression subsequently diminishes in the chondrogenic core (20, 21). Blocking N-cadherin function with either antibodies (20, 21) or dominant negative N-cadherin constructs (6, 7) has been found to block both cellular condensation and subsequent chondrogenesis of limb bud mesenchyme in vitro and in vivo. Interestingly, N-cadherin expression in condensing chondrocytes is itself dependent upon Rac1 signaling (37), another GTP binding protein that controls actin dynamics. Because cadherin function is apparently necessary for chondrogenesis to initiate, it may be relevant in this regard that cadherin engagement has been demonstrated to activate a RhoA GAP and thus diminish levels of active RhoA in the cell (19). Future studies will be necessary to address whether cadherin engagement during the condensation stage of chondrogenesis acts similarly to depress RhoA activity and thereby enhance Sox9 transcriptional activity.
Acknowledgments
This work was supported by an NIH grant to A.B.L. (AR048524). D.K. was supported by a postdoctoral fellowship from the Arthritis Foundation.
We thank Benoit de Crombrugghe, Xi He, Chris Marshall, Shuh Narumiya, H. Sasaki, and Richard Treisman for generously providing us with plasmids.
Footnotes
Published ahead of print on 26 May 2009.
REFERENCES
- 1.Ahrens, P. B., M. Solursh, and R. S. Reiter. 1977. Stage-related capacity for limb chondrogenesis in cell culture. Dev. Biol. 6069-82. [DOI] [PubMed] [Google Scholar]
- 2.Barna, M., and L. Niswander. 2007. Visualization of cartilage formation: insight into cellular properties of skeletal progenitors and chondrodysplasia syndromes. Dev. Cell 12931-941. [DOI] [PubMed] [Google Scholar]
- 3.Benya, P. D., and J. D. Shaffer. 1982. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell 30215-224. [DOI] [PubMed] [Google Scholar]
- 4.Bonaventure, J., N. Kadhom, L. Cohen-Solal, K. H. Ng, J. Bourguignon, C. Lasselin, and P. Freisinger. 1994. Reexpression of cartilage-specific genes by dedifferentiated human articular chondrocytes cultured in alginate beads. Exp. Cell Res. 21297-104. [DOI] [PubMed] [Google Scholar]
- 5.Burridge, K., and K. Wennerberg. 2004. Rho and Rac take center stage. Cell 116167-179. [DOI] [PubMed] [Google Scholar]
- 6.DeLise, A. M., and R. S. Tuan. 2002. Alterations in the spatiotemporal expression pattern and function of N-cadherin inhibit cellular condensation and chondrogenesis of limb mesenchymal cells in vitro. J. Cell. Biochem. 87342-359. [DOI] [PubMed] [Google Scholar]
- 7.Delise, A. M., and R. S. Tuan. 2002. Analysis of N-cadherin function in limb mesenchymal chondrogenesis in vitro. Dev. Dyn. 225195-204. [DOI] [PubMed] [Google Scholar]
- 8.Erlebacher, A., E. H. Filvaroff, S. E. Gitelman, and R. Derynck. 1995. Toward a molecular understanding of skeletal development. Cell 80371-378. [DOI] [PubMed] [Google Scholar]
- 9.Huang, W., X. Zhou, V. Lefebvre, and B. de Crombrugghe. 2000. Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9's ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol. Cell. Biol. 204149-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khosravi-Far, R., P. A. Solski, G. J. Clark, M. S. Kinch, and C. J. Der. 1995. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol. Cell. Biol. 156443-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kronenberg, H. M. 2003. Developmental regulation of the growth plate. Nature 423332-336. [DOI] [PubMed] [Google Scholar]
- 12.Lee, Y. S., and C. M. Chuong. 1997. Activation of protein kinase A is a pivotal step involved in both BMP-2- and cyclic AMP-induced chondrogenesis. J. Cell Physiol. 170153-165. [DOI] [PubMed] [Google Scholar]
- 13.Lefebvre, V., W. Huang, V. R. Harley, P. N. Goodfellow, and B. de Crombrugghe. 1997. SOX9 is a potent activator of the chondrocyte-specific enhancer of the proα1(II) collagen gene. Mol. Cell. Biol. 172336-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebvre, V., G. Zhou, K. Mukhopadhyay, C. N. Smith, Z. Zhang, H. Eberspaecher, X. Zhou, S. Sinha, S. N. Maity, and B. de Crombrugghe. 1996. An 18-base-pair sequence in the mouse proα1(II) collagen gene is sufficient for expression in cartilage and binds nuclear proteins that are selectively expressed in chondrocytes. Mol. Cell. Biol. 164512-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levitt, D., and A. Dorfman. 1972. The irreversible inhibition of differentiation of limb-bud mesenchyme by bromodeoxyuridine. Proc. Natl. Acad. Sci. USA 691253-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, F., and H. N. Higgs. 2003. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 131335-1340. [DOI] [PubMed] [Google Scholar]
- 17.Münsterberg, A. E., J. Kitajewski, D. A. Bumcrot, A. P. McMahon, and A. B. Lassar. 1995. Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 92911-2922. [DOI] [PubMed] [Google Scholar]
- 18.Nakano, K., K. Takaishi, A. Kodama, A. Mammoto, H. Shiozaki, M. Monden, and Y. Takai. 1999. Distinct actions and cooperative roles of ROCK and mDia in Rho small G protein-induced reorganization of the actin cytoskeleton in Madin-Darby canine kidney cells. Mol. Biol. Cell 102481-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noren, N. K., C. M. Niessen, B. M. Gumbiner, and K. Burridge. 2001. Cadherin engagement regulates Rho family GTPases. J. Biol. Chem. 27633305-33308. [DOI] [PubMed] [Google Scholar]
- 20.Oberlender, S. A., and R. S. Tuan. 1994. Expression and functional involvement of N-cadherin in embryonic limb chondrogenesis. Development 120177-187. [DOI] [PubMed] [Google Scholar]
- 21.Oberlender, S. A., and R. S. Tuan. 1994. Spatiotemporal profile of N-cadherin expression in the developing limb mesenchyme. Cell Adhes. Commun. 2521-537. [DOI] [PubMed] [Google Scholar]
- 22.Osdoby, P., and A. I. Caplan. 1979. Osteogenesis in cultures of limb mesenchymal cells. Dev. Biol. 7384-102. [DOI] [PubMed] [Google Scholar]
- 23.Paterson, H. F., A. J. Self, M. D. Garrett, I. Just, K. Aktories, and A. Hall. 1990. Microinjection of recombinant p21rho induces rapid changes in cell morphology. J. Cell Biol. 1111001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posern, G., A. Sotiropoulos, and R. Treisman. 2002. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 134167-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reginato, A. M., R. V. Iozzo, and S. A. Jimenez. 1994. Formation of nodular structures resembling mature articular cartilage in long-term primary cultures of human fetal epiphyseal chondrocytes on a hydrogel substrate. Arthritis Rheum. 371338-1349. [DOI] [PubMed] [Google Scholar]
- 26.Reid, T., T. Furuyashiki, T. Ishizaki, G. Watanabe, N. Watanabe, K. Fujisawa, N. Morii, P. Madaule, and S. Narumiya. 1996. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J. Biol. Chem. 27113556-13560. [DOI] [PubMed] [Google Scholar]
- 27.Ren, X. D., W. B. Kiosses, and M. A. Schwartz. 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahai, E., and M. F. Olson. 2006. Purification of TAT-C3 exoenzyme. Methods Enzymol. 406128-140. [DOI] [PubMed] [Google Scholar]
- 29.Sekine, A., M. Fujiwara, and S. Narumiya. 1989. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J. Biol. Chem. 2648602-8605. [PubMed] [Google Scholar]
- 30.Solursh, M., T. F. Linsenmayer, and K. L. Jensen. 1982. Chondrogenesis from single limb mesenchyme cells. Dev. Biol. 94259-264. [DOI] [PubMed] [Google Scholar]
- 31.Solursh, M., and R. S. Reiter. 1975. Determination of limb bud chondrocytes during a transient block of the cell cycle. Cell Differ. 4131-137. [DOI] [PubMed] [Google Scholar]
- 32.Solursh, M., R. S. Reiter, P. B. Ahrens, and B. M. Vertel. 1981. Stage- and position-related changes in chondrogenic response of chick embryonic wing mesenchyme to treatment with dibutyryl cyclic AMP. Dev. Biol. 839-19. [DOI] [PubMed] [Google Scholar]
- 33.Thayer, M. J., S. J. Tapscott, R. L. Davis, W. E. Wright, A. B. Lassar, and H. Weintraub. 1989. Positive autoregulation of the myogenic determination gene MyoD1. Cell 58241-248. [DOI] [PubMed] [Google Scholar]
- 34.Tsuji, T., T. Ishizaki, M. Okamoto, C. Higashida, K. Kimura, T. Furuyashiki, Y. Arakawa, R. B. Birge, T. Nakamoto, H. Hirai, and S. Narumiya. 2002. ROCK and mDia1 antagonize in Rho-dependent Rac activation in Swiss 3T3 fibroblasts. J. Cell Biol. 157819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woods, A., and F. Beier. 2006. RhoA/ROCK signaling regulates chondrogenesis in a context-dependent manner. J. Biol. Chem. 28113134-13140. [DOI] [PubMed] [Google Scholar]
- 36.Woods, A., G. Wang, and F. Beier. 2005. RhoA/ROCK signaling regulates Sox9 expression and actin organization during chondrogenesis. J. Biol. Chem. 28011626-11634. [DOI] [PubMed] [Google Scholar]
- 37.Woods, A., G. Wang, H. Dupuis, Z. Shao, and F. Beier. 2007. Rac1 signaling stimulates N-cadherin expression, mesenchymal condensation, and chondrogenesis. J. Biol. Chem. 28223500-23508. [DOI] [PubMed] [Google Scholar]
- 38.Wulf, E., A. Deboben, F. A. Bautz, H. Faulstich, and T. Wieland. 1979. Fluorescent phallotoxin, a tool for the visualization of cellular actin. Proc. Natl. Acad. Sci. USA 764498-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, Y., J. B. Moseley, I. Sagot, F. Poy, D. Pellman, B. L. Goode, and M. J. Eck. 2004. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell 116711-723. [DOI] [PubMed] [Google Scholar]
- 40.Yoon, Y. M., C. D. Oh, S. S. Kang, and J. S. Chun. 2000. Protein kinase A regulates chondrogenesis of mesenchymal cells at the post-precartilage condensation stage via protein kinase C-alpha signaling. J. Bone Miner Res. 152197-2205. [DOI] [PubMed] [Google Scholar]
- 41.Zanetti, N. C., and M. Solursh. 1984. Induction of chondrogenesis in limb mesenchymal cultures by disruption of the actin cytoskeleton. J. Cell Biol. 99115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng, L., H. Kempf, L. C. Murtaugh, M. E. Sato, and A. B. Lassar. 2002. Shh establishes an Nkx3.2/Sox9 autoregulatory loop that is maintained by BMP signals to induce somitic chondrogenesis. Genes Dev. 161990-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]