Abstract
ADAMTS-7, a metalloproteinase that belongs to ADAMTS family, is important for the degradation of cartilage extracellular matrix proteins in arthritis. Herein we report that ADAMTS-7 is upregulated during chondrocyte differentiation and demonstrates the temporal and spatial expression pattern during skeletal development. ADAMTS-7 potently inhibits chondrocyte differentiation and endochondral bone formation, and this inhibition depends on its proteolytic activity. The cysteine-rich domain of ADAMTS-7 is required for its interaction with the extracellular matrix, and the C-terminal four-thrombospondin motifs are necessary for its full proteolytic activity and inhibition of chondrocyte differentiation. ADAMTS-7 is an important target of canonical PTHrP signaling, since (i) PTHrP induces ADAMTS-7, (ii) ADAMTS-7 is downregulated in PTHrP null mutant (PTHrP−/−) growth plate chondrocytes, and (iii) blockage of ADAMTS-7 almost abolishes PTHrP-mediated inhibition of chondrocyte hypertrophy and endochondral bone growth. ADAMTS-7 associates with granulin-epithelin precursor (GEP), an autocrine growth factor that has been implicated in tissue regeneration, tumorigenesis, and inflammation. In addition, ADAMTS-7 acts as a new GEP convertase and neutralizes GEP-stimulated endochondral bone formation. Collectively, these findings demonstrate that ADAMTS-7, a direct target of PTHrP signaling, negatively regulates endochondral bone formation by associating with and inactivating GEP chondrogenic growth factor.
The ADAMTS family consists of secreted zinc metalloproteinases with a precisely ordered modular organization that includes at least one thrombospondin type I repeat (51, 53). Important functions have been established for several members of the ADAMTS family. ADAMTS-1, ADAMTS-4, ADAMTS-5, ADAMTS-8, ADAMTS-9, ADAMTS-16, and ADAMTS-18 degrade the cartilage aggrecan (1, 19, 36, 61, 84, 88), and ADAMTS-5 plays a primary role in aggrecan loss in arthritis (36, 84). ADAMTS-2, ADAMTS-3, and ADAMTS-14 are procollagen N-propeptidases (18, 30). ADAMTS-2 mutations cause dermatosparaxis, an inherited disorder characterized by severe skin fragility (17). ADAMTS-13 is a von Willebrand factor-cleaving protease, and its mutations lead to heritable life-threatening thrombocytopenic purpura (65). ADAMTS-12 and ADAMTS-7 share the same domain organization and structure and form a subgroup within the ADAMTS family (13, 83). These two enzymes have been found to associate with alpha-2-macroglobulin (13, 70, 83), and ADAMTS-12 also degrades aggrecan (68). Studies from our group demonstrated that ADAMTS-7 and ADAMTS-12 directly associate with and degrade COMP, a prominent noncollagenous component of cartilage (66, 67). COMP is a 524-kDa, pentameric, disulfide-bonded, multidomain glycoprotein composed of approximately equal subunits (∼110 kDa each) (43, 75). Although the function of COMP is not completely understood, it appears to mediate chondrocyte attachment by an integrin receptor (15, 29), and accumulating evidence suggests that COMP may function to stabilize the extracellular matrix (ECM) of articular cartilage by specific cation-dependent interactions with matrix components, including collagen type II (Col II) and Col IX, aggrecan, and fibronectin (14, 26, 72, 80). In addition, mutations in the human COMP gene have been linked to the development of pseudoachondroplasia and multiple epiphyseal dysplasia (10-12, 16, 41, 42, 86), autosomal-dominant forms of short-limb dwarfism (32, 76). COMP was also found to associate with granulin-epithelin precursor (GEP), a novel chondrogenic growth factor, and it affects GEP activity in chondrocytes (95).
GEP, also known as PC-cell-derived growth factor, acrogranin, or GP80, was first purified as a growth factor from conditioned tissue culture medium (94, 98). GEP is a 593-amino acid (aa)-secreted glycoprotein with an apparent molecular mass of 80 kDa (3, 79), which acts as an autocrine growth factor. GEP contains seven and a half repeats of a cysteine-rich motif (CX5-6CX5CCX8CCX6CCXDX2HCCPX4CX5-6C) in the order P-G-F-B-A-C-D-E, where A to G are full repeats and P is the half-motif. Notably, GEP undergoes proteolytic processing with the liberation of small, ∼6-kDa repeat units known as granulins (or epithelins), which retain biological activity (23); peptides are active in cell growth assays (96) and may be related to inflammation (69).
GEP is abundantly expressed in rapidly cycling epithelial cells, in cells of the immune system, in neurons (3, 6, 22, 69), and in several human cancers (9, 23, 38-40, 54, 92, 97). Increasing evidence has implicated GEP in the regulation of differentiation, development, and pathological processes. It has been isolated as a differentially expressed gene from mesothelial differentiation (85), sexual differentiation of the brain (87), macrophage development (8), and synovium in rheumatoid arthritis and osteoarthritis (55). GEP was also shown to be a crucial mediator of wound response and tissue repair (40, 99). GEP has potent antiinflammatory activity, and proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating the antiinflammatory activity of GEP (59). In addition, mutations in GEP cause tau-negative frontotemporal dementia linked to chromosome 17 (7, 21, 34, 81).
In addition to their cartilage-degrading role in arthritis, several metalloproteinases have been shown to play important functions in regulating chondrogenesis (50, 52, 74). Well- orchestrated chondrogenesis is controlled exquisitely by cellular interactions with the growth factors and surrounding matrix proteins, including metalloproteinases, that mediate cellular signaling pathways and transcription of specific genes in a temporal-spatial manner (20, 33, 37). In this study, we report that ADAMTS-7 is a direct target of parathyroid hormone-related peptide (PTHrP) and negatively mediates chondrocyte differentiation and endochondral bone growth through interaction with GEP chondroinductive growth factor.
MATERIALS AND METHODS
Plasmid constructs.
Construction of human ADAMTS-7 and its series of C-terminal domain deletion mutants: full-length human pOTB7-ADAMTS-7 was purchased from Invitrogen. Full-length ADAMTS-7 and its domain deletion mutants were created using the PCR method. PCR analysis was performed with pOTB7-ADAMTS-7 as a template and amplified by PfuTurbo DNA polymerase (Promega) using two primers: the forward primer ADAMTS-7 FW (5′-ATGCAAAGCTTGGTTCCTGCCATGCCCGGCGGCCCCAGTCCCCG), containing a HindIII restriction site (underlined); and the reverse primer ADAMTS-7 RV (5′-ATGCAGAATTCGCGGCGGGCAACCCGCTGAT-3′), ADAMTS-7-M1 RV (5′-ATGCAGAATTCGCCGGCGTTCCTGACAACCA-3′), ADAMTS-7-M2 RV (5′-ATGCAGAATTCCCGACAGAGTGGCAGAGAGC-3′), ADAMTS-7-M3 RV (5′-ATGCAGAATTCGGAGAACACGGGCGGCGGGA-3′), ADAMTS-7-M4 RV (5′-ATGCAGAATTCGGTGGAGCCGTTGCCGTGGC-3′), ADAMTS-7-M5 RV (5′-ATGCAGAATTCGCCACCATCCACGGCCTCG-3′), or ADAMTS-7-M6 RV (5′-ATGCAGAATTCGGGGAAGTCGATAATGTCCT-3′), containing an EcoRI restriction enzyme site (underlined). The PCR was carried out for 35 cycles of denaturation (60 s at 94°C), annealing (60 s at 60°C), and extension (7 min at 72°C). The PCR products were ligated into the pcDNA3.1/myc-HisA vector, using the HindIII and EcoRI restriction enzyme sites. A diagram of the ADAMTS-7 constructs (TS7-TS7-M6) created is shown in Fig. 5.
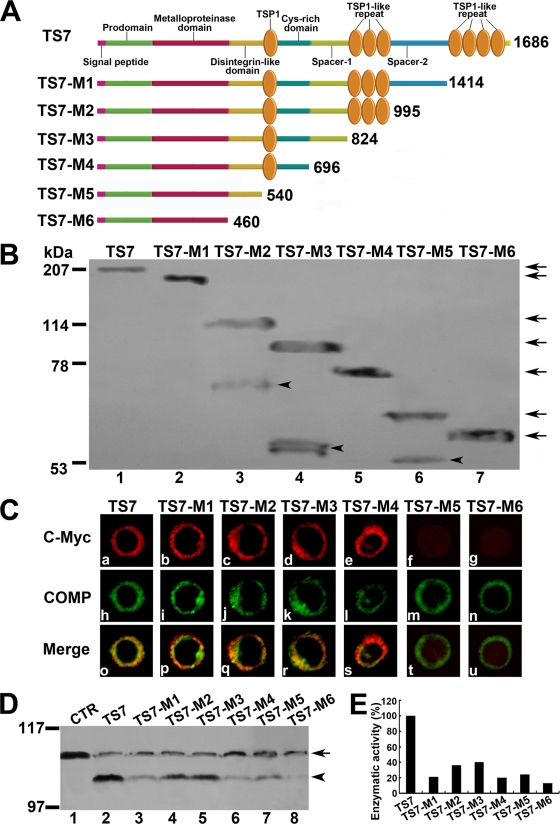
FIG. 5.
Effects of ADAMTS-7 functional domains on its cell surface location and enzymatic activity. (A) Schematic structure of the ADAMTS-7 and its C-terminal deletion mutants. Numbers refer to amino acid residues in ADAMTS-7. The functional domains are indicated. (B) Western blotting analysis of ADAMTS-7 and its C-terminal deletion mutants. The conditioned medium collected from HEK 293-ENBA cells stably transfected by either ADAMTS-7 or its C-terminal deletion mutants was detected by Western blotting analysis with anti-c-Myc antibodies. The arrows indicate the full length of the ADAMTS-7 and its series of mutants, and the arrowheads indicate the fragments resulted from several ADAMTS-7 deletion mutants. (C) The subcellular localization of the ADAMTS-7 and its domain deletion mutants in RCS cells. RCS chondrocytes were transiently transfected with expression constructs encoding the ADAMTS-7 and its C-terminal deletion mutants. The expression of ADAMTS-7 or its C-terminal deletion mutants was visualized with anti-c-Myc antibodies. COMP was visualized by cell staining with polyclonal anti-COMP antibodies. Overlapping signals are indicated with “Merge.” (D) In vitro digestion assays of COMP mediated by ADAMTS-7 and its C-terminal domain deletion mutants. Purified COMP (200 nM) was incubated with the conditioned medium collected from HEK 293-EBNA cells stably transfected by pcDNA3.1 (CTR), ADAMTS-7 (TS7), or its C-terminal domain deletion mutants (TS7-M1 to TS7-M6), as indicated. The cleaved products were subjected to reduced 8% SDS-PAGE and detected with polyclonal anti-COMP antibodies. The intact COMP and its digested fragments are indicated with an arrow and arrowhead, respectively. (E) Qualitative analysis of enzymatic activity of ADAMTS-7 and its C-terminal domain deletion mutants. The values were calibrated against ADAMTS-7 (TS7), here given the value of 100%.
Yeast expression constructs encoding various deletion mutants of ADAMTS-7 or GEP.
Yeast expression vectors pDBleu and pPC86 were obtained from Life Technologies (Gaithersburg, MD). The segment encoding series deletion mutants of ADAMTS-7 was cloned into pPC86 vector and reported previously (66). cDNA inserts encoding an individual granulin unit or a partial unit of human GEP (see Fig. S2 in the supplemental material) were cloned in frame into the SalI/NotI sites of pDBleu vector to generate the plasmids indicated above.
All constructs were verified by nucleic acid sequencing; subsequent analysis was performed using CuraTools (Curagen, New Haven, CT) and BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/).
Site-directed point mutagenesis.
The H388L and H392G mutations in the metalloproteinase domain of ADAMTS-7 were carried out using by the QuikChange XL site-directed mutagenesis kit (Stratagene). An oligonucleotide containing three mutations (5′-GGCCTTCACTGTAGCCCATCGAGCTCGGGGGCAGTTTTGGC; with underlined T, G, and G indicating changes A to T, C to G, and A to G in the original sequence, respectively) and a second oligonucleotide (5′-GCCAAAACTGCCCCCGAGCTCGAGGGCTACAGTGAAGGCC) were used to PCR amplify a mutant under the following conditions: 95°C for 2 min (1 cycle) and 95°C for 1 min, 60°C for 50 s, and 68°C for 20 min (18 cycles). The presence of the mutations in the plasmid (ADAMTS-7-PM) was confirmed by nucleotide sequencing.
Transfection of the cells.
HEK 293-EBNA, COS7, C3H10T1/2, ATDC5, and rat chondrosarcoma cells (RCS) were seeded at 5 × 106 cells/well in a six-well plate in medium containing 10% fetal calf serum (Gibco). The cells were grown overnight and transfected the following day with 6 μg of DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The stable lines were selected with 1 mg/ml of G418 (Sigma) and maintained with 200 μg/ml of G418. To obtain the conditioned medium from various stable lines, the cells were washed once with serum-free Dulbecco's modified Eagle's medium (DMEM) to remove any remaining serum, and 1 ml of serum-free DMEM with or without 100 μg/ml heparin was added to each culture and incubated for an additional 48 h.
RNA preparation and real-time PCR.
Total RNA was extracted from micromass cultures of control cells, the ADAMTS-7 stable line cells, or C3H10T1/2 cells treated with 300 ng/ml of recombinant bone morphogenetic protein 2 (BMP-2) for various time points in the absence or presence of the conditioned medium of ADAMTS-7 and its series of mutants using RNeasy kit (Qiagen). One microgram of total RNA per sample was reverse transcribed using the ImProm-II reverse transcription system (Promega). The following sequence-specific primers were synthesized: 5′-TGGTGGAGCAGCAAGAGCAA-3′ and 5′-CAGTGGACAGTAGACGGAGGAAA-3′ for mouse Col II, 5′-CTGCTGCTAATGTTCTTGAC-3′ and 5′-ACTGGAATCCCT TTACTCTTT-3′ for mouse Col X, 5′-TGATGACACTGCCACCTGTG-3′ and 5′-ACTCTGGCTTTGGGAAGAGC-3′ for mouse core-binding factor α1 (Cbfa1), 5′-CGCTCGCAATACGACTACGC-3′ and 5′-TAGAGCCCTGAGCCCTGTCC-3′ for mouse Sox9, 5′-CAGTGGAGTGTCCTGGTATT-3′ and 5′-GATCTCCGCGATCAGATGGT-3′ for mouse PTHrP, 5′-GCTCGTGCCTCTTGCCTACA-3′ and 5′-CGTGTTCTCCTCGTCCTTGA-3′ for mouse Indian hedgehog (IHH), 5′-GTGAGCCATGATTCGCCTCGG-3′ and 5′-CACCAGGTTCACCAGGATTGCC-3′ for human Col II, and 5′-CCCTTTTTGCTGCTAGTATCC-3′ and 5′-CTGTTGTCCAGGTTTTCCTGGCAC-3′ for human Col X. The following pair of oligonucleotides was used as internal controls: 5′-AGGTCGGTGTGAACGGATTTG-3′ and 5′-TGTAGACCATGTAGTTGAGGTCA-3′ for mouse GAPDH; 5′-ATGACATCAAGAAGGTGGTG-3′ and 5′-CATACCAGGAAATGAGCTTG-3′ for human GAPDH. Reactions were performed in a 50-μl Sybr green PCR volume in a 96-well optical reaction plate formatted in the 7300 sequence detection system (Applied Biosystems) under the following PCR conditions: 40 cycles at 95°C for 15 s and at 60°C for 1 min. The transcript of GAPDH mRNA was employed as an internal control for RNA quality. For each gene, three independent PCRs from the same reverse transcription sample were performed. The presence of a single specific PCR product was verified by using melting curve analysis, confirmed on an agarose gel, and further sequenced by the Applied Biosystems sequencing system (Applied Biosystems).
To examine whether PTHrP induces the expression of ADAMTS-7, C3H10T1/2, and ATDC5 cells were treated with or without 10−7 M of PTHrP for various time points, and the level of ADAMTS-7 was analyzed by real-time PCR with specific primers (5′-AGTCTCCTCCTGATCCTGTG-3′ and 5′-TGGTAGAAAGCAGACGAAGC-3′).
Western blotting.
Total cell extracts, conditioned medium, or in vitro-digested products were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and examined by Western blotting with mouse monoclonal anti-c-Myc antibodies (Santa Cruz Biotechnology), rabbit polyclonal anti-COMP antibodies (25, 27, 28, 66), rabbit polyclonal anti-ADAMTS-7 antibodies (66), or goat polyclonal anti-GEP antibodies (Santa Cruz Biotechnology) and then followed by anti-mouse immunoglobulin G (IgG)-conjugated horseradish peroxidase, anti-rabbit IgG-conjugated horseradish peroxidase, or anti-goat IgG-conjugated horseradish peroxidase at a 1:1,000 dilution. The signals were detected using the enhanced chemiluminescence system (GE Healthcare Life Sciences).
In order to examine ADAMTS-7 expression in the course of chondrogenesis, micromass cultures of C3H10T1/2 cells treated with 300 ng/ml recombinant BMP-2 for various time points were detected by rabbit polyclonal anti- ADAMTS-7 antibodies or antitubulin antibodies (internal control) (Santa Cruz Biotechnology), and the signal was visualized as described above.
Immunohistochemistry.
For histological examination, tissue samples from the embryos were immediately fixed in 4% paraformaldehyde-phosphate-buffered saline after dissection, dehydrated with gradually increasing concentrations of ethanol, and embedded in paraffin. Tissue samples from adult mice were decalcified in 2% EDTA for 2 weeks after fixation and before being embedded in paraffin. Five-micrometer-thick, formalin-fixed paraffin sections of different time point embryos during cartilage development or 18.5-day-old PTHrP null mutant (PTHrP−/−) and wild-type (PTHrP+/+) embryonic murine limbs were immunostained for ADAMTS-7. The sections were pretreated with chondroitinase (Sigma) for 30 min at 37°C and then followed by protein blocking (Dako serum-free protein block) for 10 min at room temperature to reduce nonspecific staining. Affinity-purified polyclonal anti-ADAMTS-7 (66) was diluted at 1:200 and incubated overnight at 4°C. For detection, anti-rabbit link and streptavidin label from the Super Sensitive alkaline phosphatase (BioGenex) was used. A total of 0.5 mg/ml 3,3′-diaminobenzidine (DAB) in 50 mM Tris-Cl substrate (Sigma) was used for visualization, and sections were then counterstained with methyl green.
Immunocytostaining of ADAMTS-7 and its domain deletion mutants.
The RCS cells were transiently transfected with the pcDNA3.1 expression vector containing cDNA encoding ADAMTS-7 or its mutants and then cultured. To localize each recombinant ADAMTS-7 protein, the cells were washed, fixed with 100% methanol in the freezer compartment for 5 min, washed twice in 4°C phosphate-buffered saline for 5 min, and then incubated with 30% goat serum in phosphate-buffered saline for 30 min; the cells were incubated with primary antibodies (i.e., mouse monoclonal anti-c-Myc antibodies and rabbit polyclonal anti-COMP antibodies) at room temperature for 1 h. After being washed with phosphate-buffered saline, the coverslips were incubated with secondary antibodies (i.e., goat anti-mouse IgG conjugated with rhodamine; Santa Cruz Biotechnology; diluted 1:100) and goat anti-rabbit IgG conjugated with fluorescein isothiocyanate (Santa Cruz Biotechnology; diluted 1:100) for 1 h. The specimens were observed under a fluorescence microscope with appropriate optical filters. Microscopic images were captured by using the Image-Pro program (Media Cybernetics) and an Olympus microscope. Images were arranged using Adobe Photoshop.
For visualizing the levels of ADAMTS-7 in the control and ADAMTS-7 stable lines derived form C3H10T1/2 cells, the cultures were processed as described above except that affinity-purified anti-ADAMTS-7 antibodies were used to replace anti-c-Myc antibodies. To reveal the induction of ADAMTS-7 by PTHrP, C3H10T1/2 cells were treated with 10−7 M of PTHrP for 3 days and the expression of ADAMTS-7 was visualized with affinity-purified anti-ADAMTS-7 antibodies.
Alcian blue staining.
To assess the extent of chondrogenesis, micromass cultures of C3H10T1/2 cells maintained in DMEM medium containing 10% fetal calf serum incubated with 300 ng/ml recombinant BMP-2 in the presence of various combinations of conditioned medium as described above for 14 days were fixed in Kahle's fixative, washed with water, and stained overnight with 1% alcian blue 8GX (Sigma) (31).
Culture of fetal mouse bone explants.
Fetal mouse metatarsals were dissected from fetal FVB/N mice (15-day-old embryos) and cultured in DMEM (Gibco) containing 1% heat-inactivated fetal calf serum (Invitrogen) and 100 U penicillin-streptomycin per milliliter in the absence or presence of various of stimuli, as indicated in Fig. 4, 8, and 10, for 5 days in sextuple. PTHrP (10−7 M) was used as a positive control because it was known to strongly inhibit mineralization in this system (24). For alizarin red and alcian blue staining (staining for bone and cartilage), the explants were placed in 4% paraformaldehyde in phosphate-buffered saline for overnight fixation. Subsequently, explants were placed in staining solution (0.05% alizarin red, 0.015% alcian blue, 5% acetic acid in 70% ethanol) for 45 to 60 min. Digital images of stained bones will be analyzed. For safranin O-fast green staining, explants were fixed in 96% alcohol and processed for paraffin embedding. Sections were stained with 0.1% safranin O (orange stain) to evaluate cartilage matrices and with 0.03% fast green to evaluate morphological features as previously described (89).
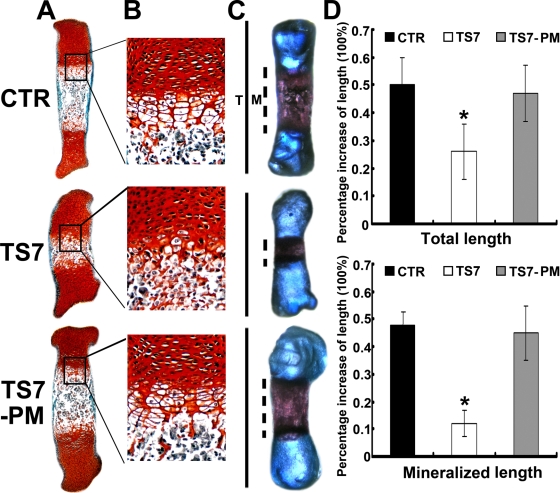
FIG. 4.
ADAMTS-7-mediated chondrocyte hypertrophy, mineralization, and bone length depends on its enzymatic activity. (A and B) Safranin O-fast green staining of metatarsals. Metatarsals were explanted from 15-day-old mouse embryos and cultured in the presence of conditioned medium of control (CTR), ADAMTS-7 (TS7), or its point mutant (TS7-PM). After 5 days of culture, the explants were fixed and safranin O-fast green staining was observed in low-power (A) or high-power (B) microphotography. (C) Alizarin red S and alcian blue staining of metatarsals. The explants were fixed and processed for alizarin red S and alcian blue staining, and a representative photograph of an explanted metatarsal after 5 days of culture is presented. (D) Percent increase in total and mineralization length of metatarsal bones. Metatarsals were cultured as described above, total length or mineralization length was determined, and the percent increase was calculated (percent increase = [length at day 5 − length at day 0]/length at day 0). Asterisks indicate a significant difference from the control (P < 0.05).
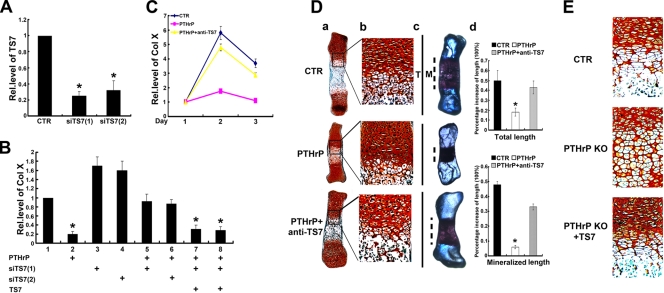
FIG. 8.
ADAMTS-7 is required for the PTHrP-mediated inhibition on chondrocyte differentiation and endochondral bone formation. (A) ADAMTS-7 siRNAs efficiently repress the expression of endogenous ADAMTS-7 in C3H10T1/2 cells. C3H10T1/2 cells were transfected with either pSuper vector (CTR) or one of two ADAMTS-7 siRNAs cloned into pSuper, as indicated, and the level of ADAMTS-7 was measured using real-time PCR. (B) Knockdown of ADAMTS-7 almost abolished PTHrP-mediated inhibition of Col X expression, whereas its reexpression largely restored PTHrP action. Micromass cultures of C3H10T1/2 cells transfected with pSuper, siTS7 [siTS7(1), siTS7(2)], ADAMTS-7 (TS7) expression plasmid, or various combinations as indicated were pretreated with BMP-2 and then cultured in the presence or absence of PTHrP (10−7 M), and the level of Col X was determined by real-time PCR. The units are arbitrary, and the normalized values were calibrated against the control, here given the value of 1. (C) ADAMTS-7 antibody dramatically attenuated PTHrP-mediated inhibition of Col X. ATDC5 cells pretreated with BMP-2 for 3 days were cultured in the absence (CTR) or presence of PTHrP (10−7M) without (PTHrP) or with anti-ADAMTS-7 (1 μg/ml) antibodies (PTHrP + anti-TS7) for various time points as indicated, and Col X was measured and analyzed as described for panel B. (D) ADAMTS-7 blocking antibody completely neutralized PTHrP-mediated chondrocyte hypertrophy, mineralization, and bone length. (a and b) Safranin O-fast green staining of metatarsals. Metatarsals were explanted from 15-day-old mouse embryos and cultured in the absence (CTR) or presence of PTHrP (10−7 M) with or without anti-ADAMTS7 (1 μg/ml) antibodies. The explants were cultured for 5 days, and safranin O-fast green staining was observed by using low-power (a) or high-power (b) microphotography. (c) Alizarin red S and alcian blue staining of metatarsals. Metatarsals were cultured as described above and processed for alizarin red S and alcian blue staining; a representative photograph is presented. (d) Percent increase in total and mineralization length of metatarsal bones. Metatarsals were cultured as described above, total or mineralization length was determined, and the percent increase was calculated (percent increase = [length at day 5 − length at day 0]/length at day 0). Asterisk indicates a significant difference from the control (P < 0.05). (E) ADAMTS-7 corrects the defects in chondrocyte hypertrophy in the metatarsal bones of PTHrP null embryos. Metatarsals were explanted from 14.5-day-old wild-type and PTHrP−/− mouse embryos and cultured in the absence (CTR) or presence of ADAMTS-7-conditioned medium. The explants were cultured for 5 days, and safranin O-fast green staining was observed. Rel.level, relative level.
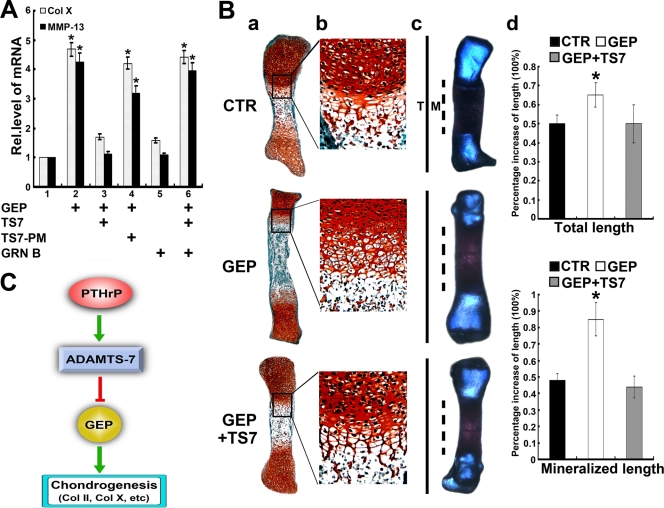
FIG. 10.
ADAMTS-7 inactivates the chondroinductive action of GEP. (A) ADAMTS-7 inhibits GEP-induced Col X and MMP-13 expression, assayed by real-time PCR. ATDC5 cells pretreated with BMP-2 for 5 days were cultured in the absence or presence of GEP, ADAMTS-7 (TS7)-, ADAMTS-7 point mutant (TS7PM)-, or granulin B (GRN B)-conditioned medium or various combinations as indicated for an additional 3 days, and Col X and MMP-13 expression was measured by real-time PCR. The units are arbitrary, and the normalized values were calibrated against controls, here given the value of 1. Asterisks indicate a significant difference from the control (P < 0.05). (B) ADAMTS-7 abolishes GEP-stimulated endochondral ossification. (a and b) Safranin O-fast green staining of metatarsals. Metatarsals were explanted from 15-day-old mouse embryos and cultured in the absence (CTR) or presence of GEP with or without ADAMTS7-conditioned medium. After 5 days of culture, safranin O-fast green staining was observed by using low-power (a) or high-power (b) microphotography. (c) Alizarin red S and alcian blue staining of metatarsals. The explants were fixed and processed for alizarin red S and alcian blue staining; a representative photograph is presented. (d) Percent increase in total and mineralization length of metatarsal bones. Metatarsals were cultured as described above, the total or mineralization length was determined, and the percent increase was calculated (percent increase = [length at day 5 − length at day 0]/length at day 0). Asterisks indicate a significant difference from the control (P < 0.05). (C) Proposed model for explaining the role and regulation of ADAMTS-7 in chondrocyte differentiation. ADAMTS-7 metalloproteinase, a direct target of PTHrP, negatively regulates chondrocyte differentiation by associating with GEP and inactivating chondrogenic activity of GEP growth factor. Rel.level, relative level.
Assay of protein-protein interactions using the yeast two-hybrid system.
Three independent colonies were analyzed for interaction in yeast of two proteins, one of which was fused to the Gal4 DNA binding domain and the other to the VP16 transactivation domain. The procedures of Vojtek et al. (90) and Hollenberg et al. (44) were followed for (i) growing and transforming the yeast strain MAV203 with the selected plasmids and (ii) testing β-galactosidase activity.
Coimmunoprecipitation (Co-IP).
Primary human chondrocytes were extracted with immunoprecipitation buffer (50 mM Tris-HCl, pH 7.4, containing the proteinase inhibitors 1 mM phenylmethylsulfonyl fluoride, 2 mM N-ethylmaleimide, and 0.025 mg/ml leupeptin). Approximately 500 μg of cell extract was incubated with anti-GEP (25 μg/ml), anti-ADAMTS-7 (25 μg/ml), or control rabbit IgG (25 μg/ml) antibodies for 1 h, followed by incubation with 30 μl of protein A-agarose (Invitrogen) at 4°C overnight. After washing five times with immunoprecipitation buffer, bound proteins were released by boiling in 20 μl of 2× SDS loading buffer for 3 min. Released proteins were examined by Western blotting with anti-GEP antibodies, and the signal was detected by using the enhanced chemiluminescence system (Amersham Biosciences).
Assay for the dependence of PTHrP activity on ADAMTS-7.
Two regions of mouse ADAMTS-7 were targeted for small interfering RNA (siRNA) using mammalian expression pSuper vector (OligoEngine) according to the manufacturer's instructions. To generate each siRNA, equimolar amounts of complementary sense and antisense strands were mixed and annealed slowly by cooling to 10°C in a 50-μl reaction buffer (100 mM NaCl and 50 mM HEPES, pH 7.4). The annealed oligonucleotides were inserted into the BglII/HindIII sites of pSuper vector. The resulting plasmids and control vector pSuper were transfected into C3H10T1/2 cells using Lipofectamine 2000 reagent (Invitrogen), and the level of ADAMTS-7 was monitored using real-time PCR and immunofluorescence cell staining. The data demonstrated that the siRNAs 5′-CGAGGAGAAGGACTTAAAG-3′ and 5′-CAGGAGAATGAGAAGGACT-3′ were able to efficiently reduce the expression of mouse ADAMTS-7. Micromass cultures of C3H10T1/2 cells transfected with either pSuper (control [CTR]), pSuper-siTS7 [siTS7(1), siTS7(2)], pcDNA3.1-ADAMTS-7 (TS7), or various combinations as indicated above were pretreated with 300 ng/ml of BMP-2 for 7 days and then cultured with or without PTHrP (10−7 M), and the level of Col X was determined by real-time PCR as described above. In the case of the antibody blocking assay, C3H10T1/2 and ATDC5 cells pretreated with 300 ng/ml of BMP-2 for 5 days were cultured in the absence or presence of 10−7 M of PTHrP without or with 1 μg/ml of anti-ADAMTS-7 antibody for various time points, and Col X was measured as described above.
Statistical test.
One-way analysis of variance was performed using R software to determine the significant differences (F > 3.35; alpha < 0.05) of the activity among different doses. In addition, Tukey's test was also used in conjunction with an analysis of variance to find significant differences (P < 0.05; P < 0.01) of the levels of genes of interest.
RESULTS
Differential expression of ADAMTS-7 in chondrogenesis of a micromass culture of C3H10T1/2 cells.
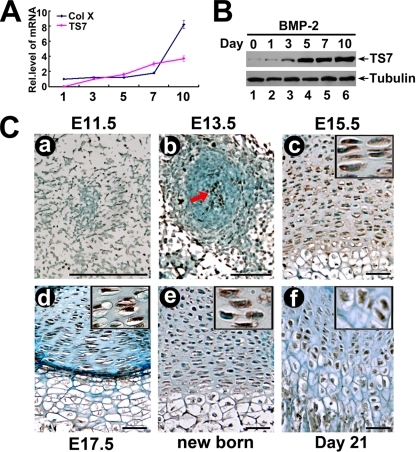
Given that several metalloproteinases play important roles in chondrogenesis and cartilage development (50, 52, 74) and that ADAMTS-7 is highly expressed in the musculoskeletal tissues, including cartilage and bone (66), we examined whether ADAMTS-7 is involved in chondrogenesis by testing its expression profiling during this process. The mouse embryonic mesenchymal stem cell (MSC) line C3H10T1/2 is a pluripotent cell line. It differentiates specifically to the cartilage lineage at high yields when inoculated under high-cell-density micromass cultures as well as when exposed to chondroinductive factors such as BMP-2 (5). Therefore, C3H10T1/2 cells have the potential to become chondrocytes, making them a valuable in vitro correlate for studying the mechanisms of chondrogenesis. To obtain ADAMTS-7 expression profiles during chondrocyte differentiation, micromass cultures of C3H10T1/2 progenitor cells were incubated in the presence of 300 ng/ml of recombinant BMP-2 for induction of chondrocyte differentiation. Cells were harvested at various time points and then followed by real-time PCR for measures of ADAMTS-7 and Col X (a specific marker for hypertrophic chondrocytes). As shown in Fig. 1A, the level of ADAMTS-7 mRNA was relatively low until day 5; at day 7 it tripled and thereafter remained at high levels during the stage representing terminal differentiation marked by the increase in Col X expression. We next examined the level of ADAMT-7 protein. Cells were harvested at various time points, followed by Western blotting (Fig. 1B). ADAMTS-7 protein was markedly elevated at day 5 and thereafter remained at high levels.
FIG. 1.
Expression of ADAMTS-7 in the course of chondrogenesis in vitro and in the growth plate chondrocytes in vivo. (A) Expression of ADAMTS-7 and collagen X was examined in the course of chondrogenesis of a micromass culture of C3H10T1/2 cells. Micromass cultures of C3H10T1/2 cells were stimulated with BMP-2 protein for various time points as indicated, and the mRNA levels of ADAMTS-7 (TS7) and Col X were assayed using real-time PCR. Rel.level, relative level. (B) Differential expression of ADAMTS-7 protein during chondrogenesis of a micromass culture of C3H10T1/2 cells. C3H10T1/2 cells were cultured as described for panel A, and the levels of ADAMTS-7 (TS-7) and tubulin (serving as an internal control) were detected by using immunoblotting. (C) Temporal and spatial expression of ADAMTS-7 during chondrogenesis in vivo, assayed by immunohistochemistry. The sections of long bone from various embryonic and postnatal developmental stages of mice, as indicated, were stained with anti-ADAMTS7 antiserum (brown) and counterstained with methyl green (green). Immunostaining revealed positive cytoplasmic staining in chondrocytes (insets). Bar = 100 μm.
ADAMTS-7 is expressed in the growth plate chondrocytes in vivo.
To characterize the temporal and spatial expression pattern of ADAMTS-7 protein during skeletal development, we performed immunohistochemistry assays at multiple time points, including the following critical developmental stages: embryonic day 11.5 (E11.5) (onset of chondrogenesis that begins with the proliferation and subsequent condensation of mesenchymal cells), E13.5 (right after cartilage formation but before endochondral bone formation), and E17.5 (onset of skeletal growth), as well as postnatal developmental stages (newborn and 21-day-old mice). As revealed in Fig. 1C, ADAMTS-7 expression seems to stain in the proliferating mesenchymal cells at E11.5 (Fig. 1C, panel a) and is clearly detectable in the center of the condensation and around it at E13.5 (Fig. 1C, panel b). ADAMTS-7 demonstrates prominent expression in the proliferating and prehypertrophic chondrocytes at E15.5 (Fig. 1C, panel c) when typical growth plates are formed; a similar expression pattern in growth plates is observed in mice at E17.5 (Fig. 1C, panel d) and in newborn mice (Fig. 1C, panel e). In the case of the 21-day-old mice, ADAMTS-7 is expressed throughout the growth plate chondrocytes (Fig. 1C, panel f). This information suggests that the expression profile of ADAMTS-7 is closely linked to the entire chondrogenesis.
ADAMTS-7 is a potent negative regulator of chondrocyte differentiation.
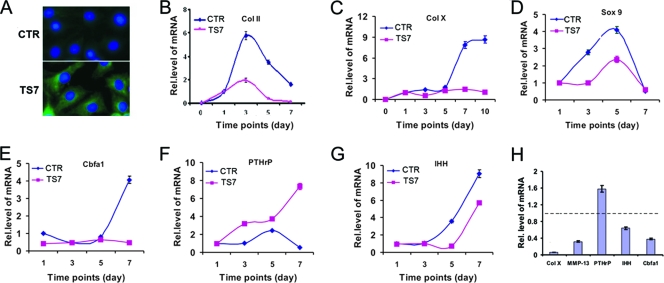
We next sought to determine the role of ADAMTS-7 in chondrogenesis. We generated a control and an ADAMTS-7 stable line based on C3H10T1/2 cells; overexpression of ADAMTS-7 in its stable line is shown in Fig. 2A. Micromass cultures of both pcDNA3.1 vector (CTR) and ADAMTS-7 (TS7) stable cell lines were stimulated with 300 ng/ml BMP-2 protein for different time points, and real-time PCRs were performed using specific primers to mouse Col II, Col X, Sox9, Cbfa1 (also defined as Runx2), PTHrP, and IHH. The data in Fig. 2B to G show that ADAMTS-7 was a potent inhibitor of chondrocyte differentiation, since it suppressed the expression of marker genes for chondrogenesis, including Col II, Col X, Sox9, Cbfa1, and IHH, whereas it enhances the expression of PTHrP. Especially, Col X and Cbfa1 were completely inhibited by the ectopic expression of ADAMTS-7, indicating that ADAMTS-7 is a critical mediator for chondrocyte hypertrophy. Furthermore, this potent inhibition on the hypertrophic chondrocyte differentiation was verified using ADAMTS-7-conditioned medium (collected from HEK 293-EBNA cells stably transfected with an ADAMTS-7 expression plasmid) (Fig. 2H). Briefly, C3H10T1/2 cells were micromass cultured in the presence of either control or ADAMTS-7-conditioned medium supplemented with BMP-2 for 7 days. Compared to that of the control group (the value of each gene in this group was set as 1), the expressions of Col X, matrix metalloproteinase 13 (MMP-13), Cbfa1, and IHH, markers for hypertrophic chondrocytes, were dramatically reduced.
FIG. 2.
ADAMTS-7 is a negative regulator of chondrocyte differentiation. (A) Overexpression of ADAMTS-7 in the ADAMTS-7 stable line. Both control (CTR) (top) and ADAMTS-7 (TS7) (bottom) cell lines based on C3H10T1/2 cells were stained with anti-ADAMTS-7 antibodies. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). (B to G) Overexpression of ADAMTS-7 regulates the expression of genes critical for chondrogenesis. Micromass cultures of both control and ADAMTS-7 stable lines were stimulated with BMP-2 protein for various time points as indicated, and the levels of mRNAs were determined using real-time PCR. (H) Effect of ADAMTS-7-conditioned medium on the expression of molecules controlling chondrocyte maturation. Micromass cultures of C3H10T1/2 cells were cultured in the presence of either control or ADAMTS-7-conditioned medium for 7 days. The units are arbitrary, and the normalized values were calibrated against controls, here given the value of 1. Rel.level, relative level.
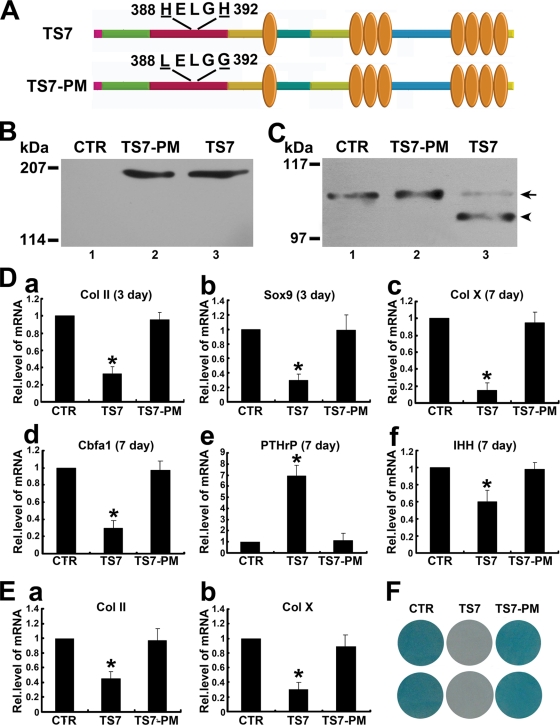
Mutations of conserved histidines in the consensus sequence of ADAMTS-7 catalytic domain inactivate ADAMTS-7 cleavage of COMP.
Similar to other members of the ADAMTS family (53), the catalytic domain of ADAMTS-7 also includes the consensus sequence HEXXHXXGXXHD (at positions 388 to 399) that is involved in the coordination of the catalytic zinc atom at the active site of the metalloproteinase. Substitution of histidines in the consensus sequence of ADAMTS-12, a close homolog of ADAMTS-7, abolished the enzymatic activity (13). To examine whether this is also true for ADAMTS-7, we next prepared an expression vector containing two point mutations in the cDNA for this protein (pcDNA3-ADAMTS7-MUT). These mutations would lead to the production of a protein with changes in two His residues essential for the catalytic activity of metalloproteinases. As shown in Fig. 3A, the amount of ADAMTS-7 point mutant (TS7-PM) this construct expressed was comparable to that of the construct encoding wild-type ADAMTS-7 (TS7) in transfected HEK 293-EBNA cells (Fig. 3B). However, this point mutant of ADAMTS-7 did not produce any COMP-degrading activity (Fig. 3C), demonstrating that the consensus sequence HEXXHXXGXXHD in the catalytic domain is essential for ADAMTS-7-mediated COMP digestion.
FIG. 3.
The proteolytic activity of ADAMTS-7 is required for its regulation of chondrogenesis. (A) Schematic structure of the ADAMTS-7 and its point mutant. HELGH in the catalytic domain was substituted by LELGG, as indicated. (B) Western blot analysis of ADAMTS-7 and its point mutant. The HEK 293-EBNA cells were transfected with either control (CTR) (lane 1) or expression constructs encoding wild-type ADAMTS-7 (TS7) (lane 3) or ADAMTS-7 with mutant catalytic domain (TS7-PM) (lane 2). The conditioned medium was subjected to reduced 8% SDS-PAGE and detected with polyclonal anti-ADAMTS-7 antibodies. (C) In vitro digestion assay of COMP by ADAMTS-7 and its point mutant. COMP was incubated with the conditioned medium collected from HEK 293-EBNA cells stably transfected by pcDNA3.1 vector (CTR) (lane 1) or expression constructs encoding wild-type ADAMTS-7 (TS7) (lane 3) or an ADAMTS-7 point mutant (TS7-PM) (lane 2), as indicated. The cleaved products were subjected to reduced 8% SDS-PAGE and detected with polyclonal anti-COMP antibodies. The intact COMP and its digested fragments are indicated with an arrow and arrowhead, respectively. (D) ADAMTS-7 inhibits chondrocyte differentiation, whereas the point mutant of ADAMTS-7 lacking enzymatic activity loses this inhibition. Micromass cultures of C3H10T1/2 cells stably transfected with a control (CTR), ADAMTS-7 (TS7), and its point mutant (TS7-PM) expression plasmid were incubated with 300 ng/ml of BMP-2, and the mRNA levels of Col II (a), Sox9 (b), Col X (c), Cbfa1 (d), PTHrP (e), and IHH (f) were determined by using real-time PCR. The units are arbitrary, and the normalized values were calibrated against the control (CTR), here given the value of 1. Asterisks indicate a significant increase or decrease compared to the control (P < 0.05). (E) Effect of ADAMTS-7 and its point mutant on chondrocyte differentiation using human MSCs. Human MSCs pellets were incubated with BMP-2 for 14 days in the presence of the conditioned medium obtained from either control (CTR), ADAMTS-7 (TS7), or its point mutant (TS7-PM) stably transfected HEK 293-EBNA cell lines, and Col II (a) and Col X (b) expression was analyzed by real-time PCR. (F) Whole-mount alcian blue histochemistry. Staining was performed on the micromass culture of pcDNA3.1 (CTR), ADAMTS-7 point mutant (TS7-PM), and wild-type ADAMTS-7 (TS7) stable lines based on C3H10T1/2 cells treated with BMP-2 protein for 14 days. Rel.level, relative level.
Enzymatic activity of ADAMTS-7 is required for ADAMTS-7-mediated inhibition of chondrocyte differentiation and endochondral bone growth.
We next sought to determine whether the enzymatic activity of ADAMTS-7 is involved in ADAMTS-7-mediated chondrogenesis; stable lines overexpressing wild-type ADAMTS-7 and its point mutant derived from C3H10T1/2 cells were generated. Micromass cultures of control (CTR), wild-type ADAMTS-7 (TS7), and point mutant (TS7-PM) stable lines were stimulated with 300 ng/ml BMP-2 protein for different time points, and real-time PCRs were performed using specific primers to mouse Col II, Col X, Cbfa1, PTHrP, and IHH. As shown in Fig. 3D, panels a to f, ADAMTS-7 potently inhibited expressions of Col II, Col X, Cbfa1, and IHH whereas it increased the expression of PTHrP; however, its point mutant totally lost these regulatory activities, clearly indicating that ADAMTS-7-mediated chondrogenesis depends on its enzymatic activity. These observations were also repeated with the aggregate culture system of human MSCs (Fig. 3E, panels a and b). Human MSC pellets were incubated with BMP-2 for 14 days in the presence of the conditioned medium obtained from control (CTR), ADAMTS-7 (TS7), or its point mutant (TS7-PM) stably transfected HEK 293-EBNA cell lines, and Col II (Fig. 3E, panel a) and Col X (Fig. 3E, panel b) expression was analyzed by real-time PCR. ADAMTS-7 also potently inhibited expressions of Col II and Col X during in vitro chondrogenesis of human MSC pellet cultures, whereas its point mutant failed to do so. In addition, these findings were further verified using whole-mount alcian blue staining, which was performed on the micromass culture of pcDNA3.1 (CTR), ADAMTS-7 point mutant (TS7-PM), and wild-type ADAMTS-7 (TS7) stable lines based on C3H10T1/2 cells treated with BMP-2 protein for 14 days (Fig. 3F).
The effect of ADAMTS-7 on endochondral bone formation as well as the dependence on its proteolytic activity were then studied in an ex vivo model of bone formation using cultures of 15-day-old fetal mouse metatarsals. The metatarsals were cultured for 5 days in the presence of the conditioned medium obtained from control (CTR), ADAMTS-7 (TS7), or its point mutant (TS7-PM) stably transfected HEK 293-EBNA cell lines. At the time of explantation, these explants consisted of undifferentiated cartilage. In a 5-day culture period, these explants underwent all sequential stages of endochondral bone formation. As shown in Fig. 4, ADAMTS-7 strongly inhibited chondrocyte hypertrophy, mineralization, and bone length whereas its point mutant lacking proteolytic activity totally lost these regulatory activities.
Effects of ADAMTS-7 noncatalytic ancillary domains on its biochemical activities and regulation of chondrocyte differentiation.
ADAMTS-7 contains a zinc catalytic domain followed by noncatalytic ancillary domains, including a disintegrin domain, a thrombospondin domain, a cysteine-rich domain (CRD), a spacer 1 domain, three-thrombospondin motifs, a spacer 2 domain, and C-terminal four-thrombospondin motifs (Fig. 5A). As noncatalytic ancillary domains have been shown to play important roles in regulating the subcellular localizations and enzymatic activities of ADAMTS (35, 58), we thus generated various C-terminal domain deletion mutants of ADAMTS-7 in order to dissect their effects on ADAMTS biochemical activities. Constructs encoding either c-Myc-tagged full-length ADAMTS-7 or its C-terminal domain-deleted mutants were transfected into HEK 293-EBNA cells, and the expressions of ADAMTS-7 and its mutants were examined by Western blotting analysis with anti-c-Myc antibody (Fig. 5B). The molecular masses of the recombinant enzymes estimated by SDS-PAGE were slightly higher than those predicted from their amino acid composition, probably due to the glycosylation of the protein that contains 10 potential N glycosylation sites and 32 potential O glycosylation sites (83). Note that some deletion mutants exhibit the smaller fragments in addition to the predicted protein bands, including mutants TS7-M2, TS7-M3, and TS7-5, which likely resulted from the intracellular processing or degradation of the protein.
Immunolocalization of ADAMTS-7 (TS7) in RCS chondrocytes with the anti-c-Myc monoclonal antibody indicated that full-length ADAMTS-7 was associated with the cell surface and ECM and colocalized with its binding partner COMP (Fig. 5C). Removal of C-terminal four-thrombospondin motifs (TS7-M1) that are known to bind COMP (66), further removal of a spacer 2 domain (TS7-M2), further deletion of three-thrombospondin motifs (TS7-M3), and further deletion of a spacer 1 domain (TS7-M4), did not change the cell surface localization of the protein; however, further deletion of the CRD (TS7-M5) released the enzyme from the cell surface into the medium. These data indicated that the CRD of ADAMTS-7 is required for its binding to ECM and cell surface appearance.
In order to define the effects of functional domains of ADAMTS-7 on its enzymatic activities, the conditioned medium obtained from HEK 293 cells stably transfected with ADAMTS-7 and its series of C-terminal deletion mutants, which contains comparable amounts of proteins (Fig. 5B), was incubated with recombinant COMP (Fig. 5D). In accordance with previous report showing the cleavage of COMP by ADAMTS-7 (66), full-length ADAMTS-7 (TS7) efficiently digested COMP, with the release of an approximately 100-kDa fragment; deletion of C-terminal four-thrombospondin motifs (TS7-M1) required for binding COMP substrate (66) dramatically reduced enzymatic activity. Surprisingly, further removal of a spacer 2 domain (TS7-M2) actually enhanced the COMP degradation, indicating that this domain is likely an inhibitory domain for its enzymatic activity; further deletion of three-thrombospondin motifs (TS7-M3) did not affect enzyme activity, but further deletion of a spacer 1 domain (TS7-M4) markedly diminished the COMP cleavage, and deletion of the CRD (TS5-M5) slightly enhanced this activity. In addition, the catalytic domain of ADAMTS-7 alone poorly demonstrated COMP degradation activity. Collectively, this set of experiments clearly indicates that the noncatalytic ancillary domains of ADAMTS-7 are involved in precise regulation of its enzymatic activity. Qualitative analysis of enzymatic activity of ADAMTS-7 (TS7) and various mutants (TS7-M1 to TS7-M6) on the cleavage of COMP is summarized in Fig. 5E.
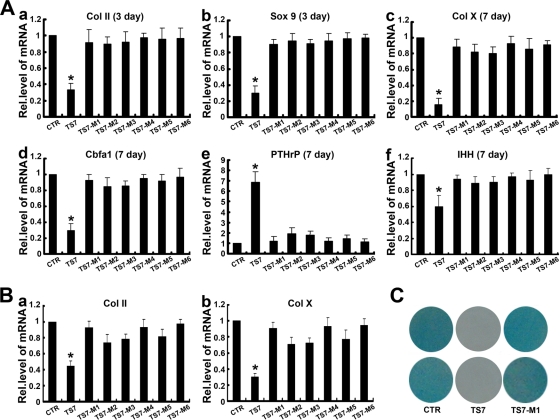
To next examine whether an individual domain of ADAMTS-7 affects its activity in regulating chondrocyte differentiation, micromass cultures of C3H10T1/2 cells were stimulated with 300 ng/ml BMP-2 protein in the presence of various conditioned medium obtained from HEK 293-ENBA cells stably transfected with control (CTR), full-length ADAMTS-7 (TS-7), and its domain deletion mutants (TS7-M1 to TS7-M6) for 3 days (for measuring Col II and Sox9) and 7 days (for determining Col X, Cbfa1, PTHrP, and IHH), and real-time PCRs were performed using specific primers. As shown in Fig. 6A, ADAMTS-7-mediated regulation of chondrogenesis was completely lost when C-terminal four-thrombospondin motifs (TS7-M1) were deleted; similar to the TS7-M1 mutant, none of the other mutations, including further removal of a spacer 2 domain (TS7-M2), further deletion of three-thrombospondin motifs (TS7-M3), and further deletion of a spacer 1 domain (TS7-M4), the CRD (TS7-M5), and the catalytic domain of ADAMTS-7 alone (TS7-M6), exhibited the regulatory activity of chondrogenesis. These findings were also observed with human MSC pellet cultures (Fig. 6B, panels a and b). These findings were further verified using alcian blue staining of corresponding cultures (Fig. 6C). Taken together, C-terminal four-thrombospondin motifs of ADAMTS-7 are critical for its inhibition of chondrocyte differentiation, especially late-stage differentiation.
FIG. 6.
Substrate-binding domain of ADAMTS-7 is important for its regulation of chondrogenesis. (A) Effect of ADAMTS-7 and its C-terminal deletion mutants on chondrocyte differentiation of murine C3H10T1/2 cells. Micromass cultures of control (CTR), wild-type ADAMTS-7 (TS7), or its C-terminal mutants (TS7-M1 to TS7-M6) C3H10T1/2 stable cell lines were stimulated with 300 ng/ml BMP-2 protein for 3 or 7 days, and the mRNA levels of Col II (a), Sox9 (b), Col X (c), Cbfa1 (d), PTHrP (e), and IHH (f) were determined by using real-time PCR. The units are arbitrary, and the normalized values were calibrated against the control (CTR), here given the value of 1. Asterisks indicate a significant increase or decrease from the control (P < 0.05). (B) Effect of ADAMTS-7 and its C-terminal deletion mutants on the expressions of Col II and Col X during chondrogenesis of human MSCs. Human MSCs pellets were cultured for 14 days in the presence of the conditioned medium obtained from the control (CTR), ADAMTS-7 (TS7), or its C-terminal mutants (TS7-M1 to TS7-M6) stably transfected HEK 293-EBNA cell lines, and Col II (a) and Col X (b) expression was analyzed by real-time PCR as described for panel A. (C) Whole-mount alcian blue histochemistry. Staining was performed on the micromass culture of pcDNA3.1 (CTR), wild-type ADAMTS-7 (TS7), and C-terminal four-thrombospondin motifs deletion mutant (TS7-M1) stable lines based on C3H10T1/2 cells treated with BMP-2 protein for 14 days. Rel.level, relative level.
PTHrP induces ADAMTS-7 expressions in C3H10T1/2 and ATDC5 cells.
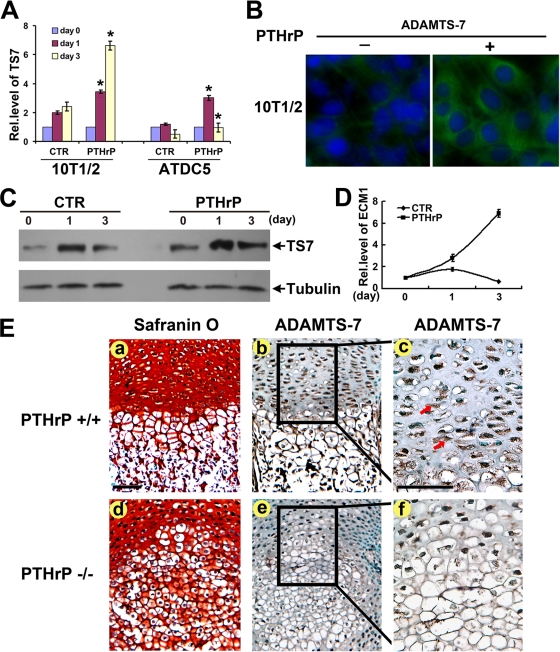
Because ADAMTS-7 is highly expressed in proliferating and prehypertrophic chondrocytes (Fig. 1C) and negatively regulates chondrocyte hypertrophy (Fig. 2), we next used C3H10T1/2 cells and chondroprogenitor ATDC5 cells to examine whether ADAMTS-7 is the downstream molecule of PTHrP, a critical negative signal in this process. Micromass cultures of both C3H10T1/2 and ATDC5 cells pretreated with 300 ng/ml of BMP-2 for 1 week were cultured with or without 10−7 M of PTHrP for various time points, and the level of ADAMTS-7 mRNA was measured by using real-time PCR (Fig. 7A). ADAMTS-7 mRNA was increased to 1.9-fold at day 1 and to 2.3-fold by day 3 in the PTHrP-untreated control C3H10T1/2 cells. PTHrP markedly enhanced the level of ADAMTS-7 mRNA to 3.9-fold at day 1 and to 6.5-fold by day 3. In the case of ATDC5 cells, ADAMTS-7 mRNA was slightly increased by day 1 but was dramatically reduced by day 3 in the PTHrP-untreated cells; PTHrP significantly induced ADAMTS-7 to 2.8-fold by day 1. Although ADAMTS-7 mRNA returned to baseline by day 3, it was still approximately twofold higher than the mRNA in the control at the same time point. Collectively, these findings demonstrate that ADAMTS-7 is a PTHrP-inducible gene in the late stage of chondrogenesis. In addition, induction of the ADAMTS-7 protein level by PTHrP was also visualized by both immunofluorescent cell staining in C3H10T1/2 cells (Fig. 7B) and immunoblotting in ATDC5 cells (Fig. 7C). Furthermore, ECM1, a known late-responsive gene of PTHrP in chondrocytes (45), was employed as a positive control (Fig. 7D).
FIG. 7.
ADAMTS-7 expression depends on PTHrP signaling. (A) PTHrP induces the expression of ADAMTS-7 mRNA, assayed by real-time PCR. 10T1/2 and ATDC5 cells pretreated with recombinant 300 ng/ml of BMP-2 for 1 week were cultured without PTHrP (CTR) or with PTHrP (10−7 M) for various time periods, as indicated. The normalized values against GAPDH were calibrated against controls (day 0), given the value of 1. Asterisk indicates a significant difference from the control at corresponding time points (P < 0.05). (B) PTHrP increases the level of ADAMTS-7 protein, assayed by immunofluorescent cell staining. Micromass cultures of C3H10T1/2 cells treated with or without PTHrP (10−7 M) for 3 days were stained with anti-ADAMTS-7 antibodies (green). The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). (C) PTHrP induces the expression of ADAMTS-7 protein, assayed by Western blotting. ATDC5 cells pretreated with recombinant 300 ng/ml of BMP-2 for 1 week were cultured without PTHrP (CTR) or with PTHrP (10−7 M) for various time periods, as indicated. The cell lysates were detected with either anti-ADAMTS-7 or antitubulin (serving as an internal control) antibodies. (D) PTHrP induces the expression of ECM1 mRNA, assayed by real-time PCR. 10T1/2 cells pretreated with recombinant 300 ng/ml of BMP-2 for 1 week were cultured without PTHrP (CTR) or with PTHrP (10−7 M) for various time periods, as indicated. The normalized values against GAPDH were calibrated against controls (day 0), given the value of 1. (E) ADAMTS-7 is markedly reduced in the growth plate chondrocytes of PTHrP null mice, revealed by immunohistochemistry. Safranin O staining was performed using sections of long bone from 18.5-day-old wild-type (a) and PTHrP null mutant (d) mouse embryos. (b and e) Low-power microphotograph of section stained with anti-ADAMTS-7 antibodies (brown, indicated with arrows) and counterstained with methyl green (green). (c and f) High-power microphotograph of the section shown in panels b and e. Rel.level, relative level. Bar = 100 μm.
In vivo expression of ADAMTS-7 in growth plate chondrocytes depends on PTHrP signaling.
We next determined whether ADAMTS-7 expression depends on PTHrP signaling in vivo by performing immunohistochemistry with sections of long bone from 18.5-day-old wild-type (PTHrP+/+) and PTHrP null mutant (PTHrP−/−) mouse embryos. As expected, ADAMTS-7 demonstrates a prominent expression in the proliferating and prehypertrophic chondrocytes in the embryonic growth plates of control mice (Fig. 7E, panel b and c). Since PTHrP inhibits the hypertrophic chondrocyte differentiation, PTHrP−/− mice display reduced zones of proliferating chondrocytes, which indicates the accelerated onset of hypertrophic differentiation (Fig. 7E, panel d). In the disordered growth plate of PTHrP knockout mice, ADAMTS-7 is downregulated (Fig. 7E, panels e and f), indicating that in vivo expression of ADAMTS-7 is controlled by PTHrP signaling.
ADAMTS-7 is a crucial downstream target for PTHrP action.
We next investigated whether endogenous ADAMTS-7 is required for PTHrP-mediated inhibition of Col X expression during chondrocyte hypertrophy using the siRNA approach. Two siRNAs that specially recognize two distant regions of murine ADAMTS-7 were selected (see Materials and Methods for details). As shown in Fig. 8A, these siRNAs led to approximately 65 to 75% inhibition of endogenous ADAMTS-7 in C3H10T1/2 cells. Micromass cultures of pSuper vector (CTR) and pSuper-TS7 [siTS7(1) or siTS7(2)] stably transfected C3H10T1/2 cells were cultured in the presence of BMP-2 (300 ng/ml) for 5 days and then treated with 10−7 M PTHrP for an additional 3 days. The level of Col X mRNA was determined by real-time PCR. As shown in Fig. 8B, PTHrP strongly inhibited Col X expression in the control cells, but this PTHrP-mediated inhibition was largely lost when ADAMTS-7 was knocked down by siRNA (Fig. 8B, compare bars 2, 5, and 6). In addition, the inhibition was partially restored when ADAMTS-7 was reexpressed via cotransfection of an ADAMTS-7 expression plasmid (Fig. 8B, compare bars 2, 7, and 8). Note that the siRNA-mediated loss of ADAMTS-7 without PTHrP exhibited higher Col X expression than that of control cells (Fig. 8B, compare bars 1, 3, and 4). The requirement of ADAMTS-7 for PTHrP-mediated inhibition on Col X expression was verified by using the antibody blocking assay (Fig. 8C). The dependence on ADAMTS-7 of PTHrP-mediated endochondral bone formation was revealed by using cultures of fetal mouse metatarsals (Fig. 8D). In line with a previous report (24), PTHrP potently inhibited chondrocyte hypertrophy, mineralization, and bone length; however, the addition of anti-ADAMTS-7 antibody largely abolished the PTHrP-mediated inhibition of these activities. Taken together, these results indicated that PTHrP-mediated chondrocyte differentiation and endochondral bone growth depends, at least in part, on ADAMTS-7 metalloproteinase. We also sought to determine whether ADAMTS-7 was able to rescue the defects seen in growth plates of PTHrP null embryos using fetal metatarsals followed by safranin O-fast green staining. As shown in Fig. 8E, disorganized PTHrP null growth plates, including accelerated chondrocyte hypertrophy, were largely corrected in the presence of recombinant ADAMTS-7.
ADAMTS-7 associates with GEP growth factor in yeast and in chondrocytes.
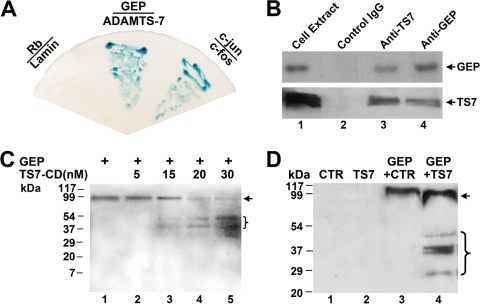
We next sought to elucidate the molecular mechanism by which ADAMTS-7 adversely regulates chondrocyte differentiation and endochondral bone growth. In addition to ADAMTS-7, GEP growth factor was also identified as a COMP binding partner (95). The facts that COMP associates with ADAMTS-7 (66) and that COMP also interacts with GEP (95) promoted us to determine whether GEP also binds to ADAMTS-7. The yeast two-hybrid assay was performed to examine the interaction between ADAMTS-7 and GEP. The plasmid encoding GEP linked to the Gal4 DNA binding domain (Fig. 9A) and the plasmid encoding ADAMTS-7 fused to the VP16AD (Fig. 9A) were used to cotransform the yeast. Like the c-Jun/c-Fos pair, which is known to interact and is used as a positive control, our assays indicated that ADAMTS-7 interacts with GEP in yeast, based on the activation of the LacZ reporter gene.
FIG. 9.
ADAMTS-7 associates with and digests GEP. (A) ADAMTS-7 binds to GEP in yeast. Yeast two-hybrid assay to test the interaction of proteins fused to the VP16 AD and proteins fused to the Gal4 DNA binding domain. Each pair of plasmids, as indicated, encoding proteins fused to VP16 (below the line) in the vector pPC86 (i.e., pPC86-c-jun, pPC86-GEP, and pPC86-Rb) and those encoding proteins fused to Gal4 (above the line) in the vector pDBleu (i.e., pDB-c-fos, pDB-ADAMTS-7, and pDB-lamin) were cotransfected into yeast strain MAV203. Yeast transformants were selected on SD-Leu-Trp plates and tested for β-galactosidase activity. The known interaction between c-Jun and c-Fos was used as a positive control, whereas the lack of interaction between Rb and lamin was used as a negative control. (B) ADAMTS-7 binds to GEP in chondrocytes (Co-IP assay). Cell extracts prepared from human chondrocytes were incubated with anti-GEP antibodies, anti-ADAMTS-7 antibodies, or control IgG. The immunoprecipitated protein complex and cell extracts (lane 1, which provides a positive control) were examined by Western blotting with either anti-GEP (top) or anti-ADAMTS-7 (bottom) antibodies. (C) The catalytic domain of ADAMTS-7 digests GEP in a dose-dependent manner. GEP was incubated with various amounts of catalytic domain of ADAMTS-7 (TS7-CD), as indicated; the cleaved products were separated by nonreduced SDS-PAGE, and intact GEP and fragments were detected with polyclonal anti-GEP antibodies. The intact GEP (arrow) and processed fragments are indicated. (D) Intact ADAMTS-7 cleaves GEP. GEP was incubated with either the control (CTR) (lane 3) or ADAMTS-7-conditioned medium (lane 4) for 4 h. The processed products as well as the control and ADAMTS-7 media were detected with anti-GEP antibodies. The intact GEP (arrow) and processed fragments are indicated.
The in vivo interaction between ADAMTS-7 and GEP was verified using a Co-IP assay in order to determine whether these two proteins are bound in native human chondrocytes. For the Co-IP assays, the cell extracts were incubated with either anti-GEP antibodies (Fig. 9B, lane 3), anti-ADAMTS-7 antibodies (Fig. 9B, lane 4), or control IgG (Fig. 9B, lane 2), and the immunoprecipitated complexes were subjected to a reducing SDS-PAGE and detected with either anti-GEP or anti-ADAMTS-7 antibodies. A specific GEP (Fig. 9B, top) or ADAMTS-7 (Fig. 9B, bottom) band was present in the immunoprecipitated complexes brought down by anti-GEP antibodies (Fig. 9B, lane 3) and anti-ADAMTS-7 antibodies (Fig. 9B, lane 4), but not control IgG antibodies (Fig. 9B, lane 2), demonstrating that GEP specifically binds to ADAMTS-7 in vivo.
In order to identify the GEP-binding motif in ADAMTS-7, we generated various constructs that expressed various ADAMTS-7 deletion mutants in yeast. Results from filter-based β-galactosidase assays (see Fig. S1B in the supplemental material) of all these mutants are summarized (see Fig. S1A in the supplemental material). The prodomain (aa 26 to 246), the metalloproteinase domain plus the disintegrin-like domain and the CRD (aa 238 to 711), the spacer 1 plus a three-thrombospondin-like repeat (aa 703 to 1007), as well as spacer 2 (aa 999 to 1324) did not interact with GEP; whereas the C-terminal region (aa 1324 to 1595) did interact with GEP. Deletion of two-thrombospondin motifs (aa 1433 to 1595) abolished the interaction, indicating that the thrombospondin motifs are required for GEP binding. These two-thrombospondin motifs alone (aa 1324 to 1433) did not bind to GEP, but the four-thrombospondin motif (aa 1324 to 1533) did. Our conclusion from this set of experiments is that that four C-terminal thrombospondin repeats of ADAMTS-7 are required and sufficient for its interaction with GEP.
We also generated various constructs that expressed individual granulin units (i.e., A, B, C, D, E, F, G) or the partial repeat (P) in yeasts for dissecting the GEP segment required for binding ADAMTS-7. Results from filter-based β-galactosidase assays (see Fig. S2B in the supplemental material) of all these mutants are summarized (see Fig. S2A in the supplemental material). This set of experiments clearly indicates that each granulin unit, but not the partial repeat, is sufficient for binding to ADAMTS-7.
ADAMTS-7 is a novel GEP convertase.
GEP undergoes proteolytic processing with the liberation of smaller fragments which retain biological activity (99). The finding that ADAMTS-7 metalloproteinase associates with GEP led us to determine whether ADAMTS-7 metalloproteinase is a novel GEP convertase. We first examined the GEP-converting activities of ADAMTS-7 in an in vitro digestion assay with a recombinant ADAMTS-7 catalytic domain and GEP (Fig. 9C). The ADAMTS-7 catalytic domain did cleave GEP with the liberation of smaller fragments. This GEP-converting action of ADAMTS-7 was further verified with ADAMTS-7-conditioned medium. As indicated in Fig. 9D, ADAMTS-7-conditioned medium also resulted in the proteolytic processing of GEP. Note that more fragments were observed using intact ADAMTS-7 than using the catalytic domain alone.
ADAMTS-7 eliminates the chondroinductive activity of GEP.
The finding that ADAMTS-7 associates with and converts GEP promoted us to examined whether ADAMTS-7-mediated inhibition of chondrocyte hypertrophy and endochondral bone growth is exerted by inactivating GEP's chondroinductive activity. For this purpose, we first examined whether ADAMTS-7 was able to inactivate GEP-stimulated chondrocyte hypertrophy using chondroprogenitor ATDC5. Briefly, ATDC5 cells pretreated with BMP-2 for 1 week were cultured without or with GEP (100 ng/ml), conditioned medium of ADAMTS-7 or its point mutation, or various combinations as indicated in Fig. 10A. GEP-stimulated Col X and MMP-13 expressions were completely abolished by the addition of ADAMTS-7; in addition, this inhibition of GEP action depends on its enzymatic activity, since the ADAMTS-7 point mutation failed to do so. Furthermore, ADAMTS-7-GEP interaction is also needed to counteract the GEP activity in chondrocyte differentiation, and ADAMTS-7 failed to attenuate GEP-mediated chondrocyte hypertrophy in the presence of a recombinant granulin B unit that is sufficient to compete with GEP for binding to ADAMTS-7 (Fig. 10A; also see Fig. S2 in the supplemental material). We next determined whether ADAMTS-7 was also able to counteract the GEP activity in regulating endochondral bone growth (Fig. 10B). As expected, GEP growth factor stimulated chondrocyte maturation, mineralization, and bone growth; however, GEP-mediated endochondral bone growth was completely abolished by the addition of ADAMTS-7-conditioned medium. These observations, together with the finding that ADAMTS-7 converted GEP into small fragments, suggested that ADAMTS-7 negatively regulates chondrocyte hypertrophy and endochondral bone growth by associating with GEP and inactivating its chondrogenic activity.
DISCUSSION
Through a functional genetic screen, ADAMTS-7 was isolated as the first metalloproteinase found to directly associate with and degrade COMP (66). The current study focused on the role of ADAMTS-7 in chondrogenesis as well as the molecular mechanism involved. ADAMTS-7 protein was highly induced in the course of chondrogenesis in vitro and also demonstrated prominent expression in the growth plate chondrocytes in vivo (Fig. 1B and 1C). Real-time PCR for measurements of ADAMTS-7 showed that the level of ADAMTS-7 mRNA was relatively low until day 5, and at day 7 it tripled and thereafter remained at high levels during the late-differential stage (Fig. 1A). The discrepancy between the protein and mRNA of ADAMTS-7 during chondrogenesis suggests that posttranscription regulations, such as translation, mRNA stability, and protein degradation, might be also important in the control of ADAMTS-7 expression during chondrogenesis.
ADAMTS-7 appears to be a potent negative regulator of chondrocyte differentiation and endochondral bone growth, and its inhibitory activities strictly depend on its enzymatic activities, since its point mutant lacking enzymatic activity lost these inhibitions (Fig. 2 and 3). ADAMTS-7 is composed of multiple functional domains, including a prodomain, a catalytic domain, a disintegrin domain, a thrombospondin motif, a CRD, a spacer 1 domain, three-thrombospondin motifs, a spacer 2 domain, and C-terminal four-thrombospondin motifs (Fig. 5A). In addition to its C-terminal four-thrombospondin motifs known to bind to substrates, including COMP (66), the role of an individual domain in regulating the biochemical properties of ADAMTS-7 remains unknown. To address this issue, we generated a series of domain deletion mutants of ADAMTS-7 and found that the CRD is required for ADAMTS-7 binding to the cell surface and ECM in chondrocytes (Fig. 5C). The addition of heparin to the culture medium released the majority of ADAMTS-7 from the cell surface (data not shown). This suggests that ADAMTS-7 binds to negatively charged glycosaminoglycans on the cell surface and in the ECM. Interestingly, this CRD-dependent localization appears to be cell type specific, since ADAMTS-7 and its deletion mutants are predominately localized in the cytoplasm of Cos-7 cells (see Fig. S3 in the supplemental material). In addition, the enzymatic activities of ADAMTS-7 are also precisely regulated by its noncatalytic domains, especially its substrate-capturing C-terminal four-thrombospondin motifs and an inhibitory spacer 2 domain (Fig. 5).
Multiple signaling pathways are involved in endochondral ossification in epiphyseal growth plate (78). Among them, PTHrP and IHH coordinately regulate the rate of chondrocyte differentiation through a negative feedback loop (48, 57, 91). Several lines of evidence indicated that PTHrP negatively regulated the endochondral bone formation. PTHrP prevents chondrocyte hypertrophy in the growth plate and maintains a pool of cells above the hypertrophic zone in a proliferative condition (57). IHH is expressed at the prehypertrophic- hypertrophic boundary so that cells that escape the inhibitory action of PTHrP signaling in the growth plate express IHH, which in turn will stimulate PTHrP expression (49, 60). In embryonic mice lacking PTHrP, chondrocytes stop proliferating prematurely, with accelerated differentiation (71). Mice lacking either PTHrP (56, 64) or the parathyroid hormone (PTH)-PTHrP receptor (2, 63) also have accelerated chondrocyte differentiation and impaired skeletal growth, exhibiting shortened zones of proliferative chondrocytes and premature hypertrophic differentiation. However, animals overexpressing PTHrP in chondrocytes show delayed hypertrophic differentiation (93). Chondrocyte-specific expression of a constitutively active PTH-PTHrP receptor also delays the conversion of proliferative chondrocytes to hypertrophic chondrocytes (82). In mouse models, both PTHrP and PTH-PTHrP receptor knockout mice display advanced endochondral bone formation (2, 56, 63). Our studies demonstrating that (i) overexpressing ADAMTS-7 enhanced the expression of PTHrP whereas it inhibited IHH (Fig. 2F and G), (ii) PTHrP induced ADAMTS-7 expression in the course of chondrogenesis in vitro (Fig. 7A to C), and (iii) ADAMTS-7 expression strictly depends on PTHrP in growth plate chondrocytes in mice (Fig. 7E). These findings suggest that there exists a positive feedback loop between ADAMTS-7 and PTHrP signaling in the course of chondrogenesis. In addition, ADAMTS-7 appears to be a crucial downstream molecule of PTHrP based on the observations that (i) repression of ADAMTS-7 via an siRNA approach or blocking ADAMTS-7 activity using its blocking antibodies almost abolished PTHrP-mediated inhibition of Col X expression, but reexpression of ADAMTS-7 restored PTHrP action (Fig. 8B); (ii) an ADAMTS-7 blocking antibody totally neutralized the inhibition of chondrocyte hypertrophy, mineralization, and bone growth by PTHrP (Fig. 8C and D); and (iii) ADAMTS-7 largely corrected the defects of PTHrP null growth plates (Fig. 8E). Furthermore, similar to PTHrP that is known to stimulate chondrocyte proliferation (73), ADAMTS-7 was also found to increase chondrocyte proliferation, and its stimulation on cell proliferation largely depends on its substrate-binding C-terminal four-thrombospondin motifs (see Fig. S4 in the supplemental material). Because ADAMTS-7-mediated chondrogenesis and endochondral ossification depend on its proteolytic activity (Fig. 3 and 4), and it was known to associate with and degrade COMP (66), whether the effects on PTHrP are dependent on ADAMTS-7 metalloproteinase activity or its interaction with COMP remains to be delineated.
GEP is a growth factor implicated in tissue regeneration, tumorigenesis, and inflammation (39, 40, 59, 99). Several GEP-associated partners have been reported and found to affect GEP action in various processes. One example is the secretory leukocyte protease inhibitor (SLPI). Elastase digests GEP exclusively in the intergranulin linkers, resulting in the generation of granulin peptides, suggesting that this protease may be an important GEP convertase. SLPI blocks this proteolysis either by directly binding to elastase or by sequestering granulin peptides from the enzyme (99). It was found that GEP can modulate transcriptional activities by interacting with human cyclin T1 (47) and Tat-P-TEFb (46). GEP was also found to interact with perlecan, a heparan sulfate proteoglycan; perlecan null mice exhibit severe skeletal defects (4, 38, 62, 77). A recent report reveled that proteinase 3 associated with and processed GEP, and proteinase 3 and elastase enhance neutrophil-dependent inflammation by eliminating the antiinflammatory activity of GEP (59). Here, we present evidence showing that GEP associates with ADAMTS-7 metalloproteinase in chondrocytes and that ADAMTS-7 is able to convert GEP into its processed fragments (Fig. 9); in addition, ADAMTS-7-inhibits chondrocyte differentiation and endochondral bone formation, probably by inactivating the chondrogenic activity of GEP. Our previous report showing that (i) ADAMTS-7 binds to and degrades COMP (66) and (ii) COMP interacts with GEP and potentiates GEP-stimulated chondrocyte functions (95), together with the present study revealing the interaction between ADAMTS-7 and GEP, indicate that ADAMTS-7, GEP, and COMP form an interaction and interplay network in regulating chondrocyte functions (66, 95). It remains to be determined how the interaction network among ADAMTS-7 metalloproteinase, GEP growth factor, and the COMP ECM molecule acts in concert in regulating chondrocyte differentiation and endochondral ossification.
It is interesting to note that the subcellular localization of ADAMTS-7 in chondrocytes appears to depend on the developmental stage and location, since it is primarily intracellular in the early stages of embryonic murine limbs (Fig. 6), whereas it is primarily in the pericellular matrix in chondrocytes isolated from adult human articular cartilage (F. J. Guo and C. J. Liu, unpublished data). In addition, COMP ECM protein and GEP growth factor, two ADAMTS-7 binding partners, were also found to be primarily intracellular in the growth plate chondrocytes (95). Distinct localization of ADAMTS-7 and GEP in various chondrocytes suggests that they may play important roles at various stages in skeletogenesis.
As summarized in Fig. 10C, this study provides novel insights into the role of ADAMTS-7, a novel mediator in PTHrP pathway, in regulating chondrocyte differentiation and endochondral bone formation and sheds light on the molecular mechanism by which ADAMTS-7 adversely regulates chondrogenesis; i.e., ADAMTS-7 metalloproteinase associates with the GEP chondrogenic growth factor and eliminates GEP-stimulated chondrogenesis via conversion of GEP into its processed fragments. In addition, this study also provides us with potential molecule targets for the treatment of cartilage disorders and arthritic conditions.
Supplementary Material
Acknowledgments
This work was aided by NIH research grants AR053210, AR050620, and AG029388 and a grant from the Arthritis National Research Foundation.
Footnotes
Published ahead of print on 1 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abbaszade, I., R. Q. Liu, F. Yang, S. A. Rosenfeld, O. H. Ross, J. R. Link, D. M. Ellis, M. D. Tortorella, M. A. Pratta, J. M. Hollis, R. Wynn, J. L. Duke, H. J. George, M. C. Hillman, Jr., K. Murphy, B. H. Wiswall, R. A. Copeland, C. P. Decicco, R. Bruckner, H. Nagase, Y. Itoh, R. C. Newton, R. L. Magolda, J. M. Trzaskos, T. C. Burn, et al. 1999. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J. Biol. Chem. 27423443-23450. [DOI] [PubMed] [Google Scholar]
- 2.Amizuka, N., H. Warshawsky, J. E. Henderson, D. Goltzman, and A. C. Karaplis. 1994. Parathyroid hormone-related peptide-depleted mice show abnormal epiphyseal cartilage development and altered endochondral bone formation. J. Cell Biol. 1261611-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anakwe, O. O., and G. L. Gerton. 1990. Acrosome biogenesis begins during meiosis: evidence from the synthesis and distribution of an acrosomal glycoprotein, acrogranin, during guinea pig spermatogenesis. Biol. Reprod. 42317-328. [DOI] [PubMed] [Google Scholar]
- 4.Arikawa-Hirasawa, E., H. Watanabe, H. Takami, J. R. Hassell, and Y. Yamada. 1999. Perlecan is essential for cartilage and cephalic development. Nat. Genet. 23354-358. [DOI] [PubMed] [Google Scholar]
- 5.Atkinson, B. L., K. S. Fantle, J. J. Benedict, W. E. Huffer, and A. Gutierrez-Hartmann. 1997. Combination of osteoinductive bone proteins differentiates mesenchymal C3H/10T1/2 cells specifically to the cartilage lineage. J. Cell. Biochem. 65325-339. [PubMed] [Google Scholar]
- 6.Baba, T., H. B. Hoff III, H. Nemoto, H. Lee, J. Orth, Y. Arai, and G. L. Gerton. 1993. Acrogranin, an acrosomal cysteine-rich glycoprotein, is the precursor of the growth-modulating peptides, granulins, and epithelins, and is expressed in somatic as well as male germ cells. Mol. Reprod. Dev. 34233-243. [DOI] [PubMed] [Google Scholar]
- 7.Baker, M., I. R. Mackenzie, S. M. Pickering-Brown, J. Gass, R. Rademakers, C. Lindholm, J. Snowden, J. Adamson, A. D. Sadovnick, S. Rollinson, A. Cannon, E. Dwosh, D. Neary, S. Melquist, A. Richardson, D. Dickson, Z. Berger, J. Eriksen, T. Robinson, C. Zehr, C. A. Dickey, R. Crook, E. McGowan, D. Mann, B. Boeve, H. Feldman, and M. Hutton. 2006. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442916-919. [DOI] [PubMed] [Google Scholar]
- 8.Barreda, D. R., P. C. Hanington, C. K. Walsh, P. Wong, and M. Belosevic. 2004. Differentially expressed genes that encode potential markers of goldfish macrophage development in vitro. Dev. Comp. Immunol. 28727-746. [DOI] [PubMed] [Google Scholar]
- 9.Bateman, A., D. Belcourt, H. Bennett, C. Lazure, and S. Solomon. 1990. Granulins, a novel class of peptide from leukocytes. Biochem. Biophys. Res. Commun. 1731161-1168. [DOI] [PubMed] [Google Scholar]
- 10.Briggs, M. D., S. M. Hoffman, L. M. King, A. S. Olsen, H. Mohrenweiser, J. G. Leroy, G. R. Mortier, D. L. Rimoin, R. S. Lachman, and E. S. Gaines. 1995. Pseudoachondroplasia and multiple epiphyseal dysplasia due to mutations in the cartilage oligomeric matrix protein gene. Nature 10330-336. [DOI] [PubMed] [Google Scholar]
- 11.Briggs, M. D., G. R. Mortier, W. G. Cole, L. M. King, S. S. Golik, J. Bonaventure, L. Nuytinck, A. De Paepe, J. G. Leroy, L. Biesecker, M. Lipson, W. R. Wilcox, R. S. Lachman, D. L. Rimoin, R. G. Knowlton, and D. H. Cohn. 1998. Diverse mutations in the gene for cartilage oligomeric matrix protein in the pseudoachondroplasia-multiple epiphyseal dysplasia disease spectrum. Am. J. Hum. Genet. 62311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briggs, M. D., I. M. Rasmussen, J. L. Weber, J. Yuen, K. Reinker, A. P. Garber, D. L. Rimoin, and D. H. Cohn. 1993. Genetic linkage of mild pseudoachondroplasia (PSACH) to markers in the pericentromeric region of chromosome 19. Genomics 18656-660. [DOI] [PubMed] [Google Scholar]
- 13.Cal, S., J. M. Arguelles, P. L. Fernandez, and C. Lopez-Otin. 2001. Identification, characterization, and intracellular processing of ADAM-TS12, a novel human disintegrin with a complex structural organization involving multiple thrombospondin-1 repeats. J. Biol. Chem. 27617932-17940. [DOI] [PubMed] [Google Scholar]
- 14.Chen, F. H., M. E. Herndon, N. Patel, J. T. Hecht, R. S. Tuan, and J. Lawler. 2007. Interaction of cartilage oligomeric matrix protein/thrombospondin 5 with aggrecan. J. Biol. Chem. 28224591-24598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, F. H., A. O. Thomas, J. T. Hecht, M. B. Goldring, and J. Lawler. 2005. Cartilage oligomeric matrix protein/thrombospondin 5 supports chondrocyte attachment through interaction with integrins. J. Biol. Chem. 28032655-32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohn, D. H., M. D. Briggs, L. M. King, D. L. Rimoin, W. R. Wilcox, R. S. Lachman, and R. G. Knowlton. 1996. Mutations in the cartilage oligomeric matrix protein (COMP) gene in pseudoachondroplasia and multiple epiphyseal dysplasia. Ann. N. Y. Acad. Sci. 785188-194. [DOI] [PubMed] [Google Scholar]
- 17.Colige, A., A. L. Sieron, S. W. Li, U. Schwarze, E. Petty, W. Wertelecki, W. Wilcox, D. Krakow, D. H. Cohn, W. Reardon, P. H. Byers, C. M. Lapiere, D. J. Prockop, and B. V. Nusgens. 1999. Human Ehlers-Danlos syndrome type VII C and bovine dermatosparaxis are caused by mutations in the procollagen I N-proteinase gene. Am. J. Hum. Genet. 65308-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colige, A., I. Vandenberghe, M. Thiry, C. A. Lambert, J. Van Beeumen, S. W. Li, D. J. Prockop, C. M. Lapiere, and B. V. Nusgens. 2002. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J. Biol. Chem. 2775756-5766. [DOI] [PubMed] [Google Scholar]
- 19.Collins-Racie, L. A., C. R. Flannery, W. Zeng, C. Corcoran, B. Annis-Freeman, M. J. Agostino, M. Arai, E. DiBlasio-Smith, A. J. Dorner, K. E. Georgiadis, M. Jin, X. Y. Tan, E. A. Morris, and E. R. LaVallie. 2004. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol. 23219-230. [DOI] [PubMed] [Google Scholar]
- 20.Colnot, C. 2005. Cellular and molecular interactions regulating skeletogenesis. J. Cell. Biochem. 95688-697. [DOI] [PubMed] [Google Scholar]
- 21.Cruts, M., I. Gijselinck, J. van der Zee, S. Engelborghs, H. Wils, D. Pirici, R. Rademakers, R. Vandenberghe, B. Dermaut, J. J. Martin, C. van Duijn, K. Peeters, R. Sciot, P. Santens, T. De Pooter, M. Mattheijssens, M. Van den Broeck, I. Cuijt, K. Vennekens, P. P. De Deyn, S. Kumar-Singh, and C. Van Broeckhoven. 2006. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 442920-924. [DOI] [PubMed] [Google Scholar]
- 22.Daniel, R., Z. He, K. P. Carmichael, J. Halper, and A. Bateman. 2000. Cellular localization of gene expression for progranulin. J. Histochem. Cytochem. 48999-1009. [DOI] [PubMed] [Google Scholar]
- 23.Davidson, B., E. Alejandro, V. A. Florenes, J. M. Goderstad, B. Risberg, G. B. Kristensen, C. G. Trope, and E. C. Kohn. 2004. Granulin-epithelin precursor is a novel prognostic marker in epithelial ovarian carcinoma. Cancer 1002139-2147. [DOI] [PubMed] [Google Scholar]
- 24.Deckers, M. M., P. Smits, M. Karperien, J. Ni, P. Tylzanowski, P. Feng, D. Parmelee, J. Zhang, E. Bouffard, R. Gentz, C. W. Lowik, and J. Merregaert. 2001. Recombinant human extracellular matrix protein 1 inhibits alkaline phosphatase activity and mineralization of mouse embryonic metatarsals in vitro. Bone 2814-20. [DOI] [PubMed] [Google Scholar]
- 25.Di Cesare, P. E., C. S. Carlson, E. S. Stolerman, N. Hauser, H. Tulli, and M. Paulsson. 1996. Increased degradation and altered tissue distribution of cartilage oligomeric matrix protein in human rheumatoid and osteoarthritic cartilage. J. Orthop. Res. 14946-955. [DOI] [PubMed] [Google Scholar]
- 26.Di Cesare, P. E., F. S. Chen, M. Moergelin, C. S. Carlson, M. P. Leslie, R. Perris, and C. Fang. 2002. Matrix-matrix interaction of cartilage oligomeric matrix protein and fibronectin. Matrix Biol. 21461-470. [DOI] [PubMed] [Google Scholar]
- 27.Di Cesare, P. E., C. Fang, M. P. Leslie, C. J. Della Valle, J. M. Gold, H. Tulli, R. Perris, and C. S. Carlson. 1999. Localization and expression of cartilage oligomeric matrix protein by human rheumatoid and osteoarthritic synovium and cartilage. J. Orthop. Res. 17437-445. [DOI] [PubMed] [Google Scholar]
- 28.Di Cesare, P. E., C. Fang, M. P. Leslie, H. Tulli, R. Perris, and C. S. Carlson. 2000. Expression of cartilage oligomeric matrix protein (COMP) by embryonic and adult osteoblasts. J. Orthop. Res. 18713-720. [DOI] [PubMed] [Google Scholar]
- 29.DiCesare, P. E., M. Mörgelin, K. Mann, and M. Paulsson. 1994. Cartilage oligomeric matrix protein and thrombospondin 1. Purification from articular cartilage, electron microscopic structure, and chondrocyte binding. Eur. J. Biochem. 223927-937. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes, R. J., S. Hirohata, J. M. Engle, A. Colige, D. H. Cohn, D. R. Eyre, and S. S. Apte. 2001. Procollagen II amino propeptide processing by ADAMTS-3. Insights on dermatosparaxis. J. Biol. Chem. 27631502-31509. [DOI] [PubMed] [Google Scholar]
- 31.Fischer, L., G. Boland, and R. S. Tuan. 2002. Wnt-3A enhances bone morphogenetic protein-2-mediated chondrogenesis of murine C3H10T1/2 mesenchymal cells. J. Biol. Chem. 27730870-30878. [DOI] [PubMed] [Google Scholar]
- 32.Francomano, C. A., I. McIntosh, and D. J. Wilkin. 1996. Bone dysplasias in man: molecular insights. Curr. Opin. Genet. Dev. 6301-308. [DOI] [PubMed] [Google Scholar]
- 33.Franz-Odendaal, T. A., and M. K. Vickaryous. 2006. Skeletal elements in the vertebrate eye and adnexa: morphological and developmental perspectives. Dev. Dyn. 2351244-1255. [DOI] [PubMed] [Google Scholar]
- 34.Gass, J., A. Cannon, I. R. Mackenzie, B. Boeve, M. Baker, J. Adamson, R. Crook, S. Melquist, K. Kuntz, R. Petersen, K. Josephs, S. M. Pickering-Brown, N. Graff-Radford, R. Uitti, D. Dickson, Z. Wszolek, J. Gonzalez, T. G. Beach, E. Bigio, N. Johnson, S. Weintraub, M. Mesulam, C. L. White III, B. Woodruff, R. Caselli, G. Y. Hsiung, H. Feldman, D. Knopman, M. Hutton, and R. Rademakers. 2006. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum. Mol. Genet. 152988-3001. [DOI] [PubMed] [Google Scholar]
- 35.Gendron, C., M. Kashiwagi, N. H. Lim, J. J. Enghild, I. B. Thogersen, C. Hughes, B. Caterson, and H. Nagase. 2007. Proteolytic activities of human ADAMTS-5: comparative studies with ADAMTS-4. J. Biol. Chem. 28218294-18306. [DOI] [PubMed] [Google Scholar]
- 36.Glasson, S. S., R. Askew, B. Sheppard, B. Carito, T. Blanchet, H. L. Ma, C. R. Flannery, D. Peluso, K. Kanki, Z. Yang, M. K. Majumdar, and E. A. Morris. 2005. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature 434644-648. [DOI] [PubMed] [Google Scholar]
- 37.Goldring, M. B., K. Tsuchimochi, and K. Ijiri. 2006. The control of chondrogenesis. J. Cell. Biochem. 9733-44. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez, E. M., M. Mongiat, S. J. Slater, R. Baffa, and R. V. Iozzo. 2003. A novel interaction between perlecan protein core and progranulin: potential effects on tumor growth. J. Biol. Chem. 27838113-38116. [DOI] [PubMed] [Google Scholar]
- 39.He, Z., and A. Bateman. 2003. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J. Mol. Med. 81600-612. [DOI] [PubMed] [Google Scholar]
- 40.He, Z., C. H. Ong, J. Halper, and A. Bateman. 2003. Progranulin is a mediator of the wound response. Nat. Med. 9225-229. [DOI] [PubMed] [Google Scholar]
- 41.Hecht, J. T., C. A. Francomano, M. D. Briggs, M. Deere, B. Conner, W. A. Horton, M. Warman, D. H. Cohn, and S. H. Blanton. 1993. Linkage of typical pseudoachondroplasia to chromosome 19. Genomics 18661-666. [DOI] [PubMed] [Google Scholar]
- 42.Hecht, J. T., L. D. Nelson, E. Crowder, Y. Wang, F. F. Elder, W. R. Harrison, C. A. Francomano, C. K. Prange, G. G. Lennon, and M. Deere. 1995. Mutations in exon 17B of cartilage oligomeric matrix protein (COMP) cause pseudoachondroplasia. Nat. Genet. 10325-329. [DOI] [PubMed] [Google Scholar]
- 43.Hedbom, E., P. Antonsson, A. Hjerpe, D. Aeschlimann, M. Paulsson, E. Rosa-Pimentel, Y. Sommarin, M. Wendel, A. Oldberg, and D. Heinegard. 1992. Cartilage matrix proteins. An acidic oligomeric protein (COMP) detected only in cartilage. J. Biol. Chem. 2676132-6136. [PubMed] [Google Scholar]
- 44.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 153813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoogendam, J., H. Farih-Sips, E. van Beek, C. W. Lowik, J. M. Wit, and M. Karperien. 2007. Novel late response genes of PTHrP in chondrocytes. Horm. Res. 67159-170. [DOI] [PubMed] [Google Scholar]
- 46.Hoque, M., B. Tian, M. B. Mathews, and T. Pe'ery. 2005. Granulin and granulin repeats interact with the Tat. P-TEFb complex and inhibit Tat transactivation. J. Biol. Chem. 28013648-13657. [DOI] [PubMed] [Google Scholar]
- 47.Hoque, M., T. M. Young, C. G. Lee, G. Serrero, M. B. Mathews, and T. Pe'ery. 2003. The growth factor granulin interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol. Cell. Biol. 231688-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu, Z., M. Yu, and G. Hu. 2007. NDST-1 modulates BMPR and PTHrP signaling during endochondral bone formation in a gene knockout model. Bone 401462-1474. [DOI] [PubMed] [Google Scholar]
- 49.Huang, W., U. I. Chung, H. M. Kronenberg, and B. de Crombrugghe. 2001. The chondrogenic transcription factor Sox9 is a target of signaling by the parathyroid hormone-related peptide in the growth plate of endochondral bones. Proc. Natl. Acad. Sci. USA 98160-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang, W., X. Zhou, V. Lefebvre, and B. de Crombrugghe. 2000. Phosphorylation of SOX9 by cyclic AMP-dependent protein kinase A enhances SOX9's ability to transactivate a Col2a1 chondrocyte-specific enhancer. Mol. Cell. Biol. 204149-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hurskainen, T. L., S. Hirohata, M. F. Seldin, and S. S. Apte. 1999. ADAM-TS5, ADAM-TS6, and ADAM-TS7, novel members of a new family of zinc metalloproteases. General features and genomic distribution of the ADAM-TS family. J. Biol. Chem. 27425555-25563. [DOI] [PubMed] [Google Scholar]
- 52.Jin, E. J., K. S. Park, O. S. Bang, and S. S. Kang. 2007. Akt signaling regulates actin organization via modulation of MMP-2 activity during chondrogenesis of chick wing limb bud mesenchymal cells. J. Cell. Biochem. 102252-261. [DOI] [PubMed] [Google Scholar]
- 53.Jones, G. C., and G. P. Riley. 2005. ADAMTS proteinases: a multi-domain, multi-functional family with roles in extracellular matrix turnover and arthritis. Arthritis Res. Ther. 7160-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones, M. B., M. Spooner, and E. C. Kohn. 2003. The granulin-epithelin precursor: a putative new growth factor for ovarian cancer. Gynecol. Oncol. 88S136-S139. [DOI] [PubMed] [Google Scholar]
- 55.Jüsten, H. P., E. Grunewald, G. Totzke, I. Gouni-Berthold, A. Sachinidis, D. Wessinghage, H. Vetter, K. Schulze-Osthoff, and Y. Ko. 2000. Differential gene expression in synovium of rheumatoid arthritis and osteoarthritis. Mol. Cell. Biol. Res. Commun. 3165-172. [DOI] [PubMed] [Google Scholar]
- 56.Karaplis, A. C., A. Luz, J. Glowacki, R. T. Bronson, V. L. Tybulewicz, H. M. Kronenberg, and R. C. Mulligan. 1994. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 8277-289. [DOI] [PubMed] [Google Scholar]
- 57.Karp, S. J., E. Schipani, B. St-Jacques, J. Hunzelman, H. Kronenberg, and A. P. McMahon. 2000. Indian hedgehog coordinates endochondral bone growth and morphogenesis via parathyroid hormone related-protein-dependent and -independent pathways. Development 127543-548. [DOI] [PubMed] [Google Scholar]
- 58.Kashiwagi, M., J. J. Enghild, C. Gendron, C. Hughes, B. Caterson, Y. Itoh, and H. Nagase. 2004. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J. Biol. Chem. 27910109-10119. [DOI] [PubMed] [Google Scholar]
- 59.Kessenbrock, K., L. Frohlich, M. Sixt, T. Lammermann, H. Pfister, A. Bateman, A. Belaaouaj, J. Ring, M. Ollert, R. Fassler, and D. E. Jenne. 2008. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J. Clin. Investig. 1182438-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kronenberg, H. M. 2006. PTHrP and skeletal development. Ann. N. Y. Acad. Sci. 10681-13. [DOI] [PubMed] [Google Scholar]
- 61.Kuno, K., Y. Okada, H. Kawashima, H. Nakamura, M. Miyasaka, H. Ohno, and K. Matsushima. 2000. ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett. 478241-245. [DOI] [PubMed] [Google Scholar]
- 62.Kvist, A. J., A. E. Johnson, M. Morgelin, E. Gustafsson, E. Bengtsson, K. Lindblom, A. Aszodi, R. Fassler, T. Sasaki, R. Timpl, and A. Aspberg. 2006. Chondroitin sulfate perlecan enhances collagen fibril formation. Implications for perlecan chondrodysplasias. J. Biol. Chem. 28133127-33139. [DOI] [PubMed] [Google Scholar]
- 63.Lanske, B., A. C. Karaplis, K. Lee, A. Luz, A. Vortkamp, A. Pirro, M. Karperien, L. H. Defize, C. Ho, R. C. Mulligan, A. B. Abou-Samra, H. Juppner, G. V. Segre, and H. M. Kronenberg. 1996. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science 273663-666. [DOI] [PubMed] [Google Scholar]
- 64.Lee, K., B. Lanske, A. C. Karaplis, J. D. Deeds, H. Kohno, R. A. Nissenson, H. M. Kronenberg, and G. V. Segre. 1996. Parathyroid hormone-related peptide delays terminal differentiation of chondrocytes during endochondral bone development. Endocrinology 1375109-5118. [DOI] [PubMed] [Google Scholar]
- 65.Levy, G. G., W. C. Nichols, E. C. Lian, T. Foroud, J. N. McClintick, B. M. McGee, A. Y. Yang, D. R. Siemieniak, K. R. Stark, R. Gruppo, R. Sarode, S. B. Shurin, V. Chandrasekaran, S. P. Stabler, H. Sabio, E. E. Bouhassira, J. D. Upshaw, Jr., D. Ginsburg, and H. M. Tsai. 2001. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature 413488-494. [DOI] [PubMed] [Google Scholar]
- 66.Liu, C. J., W. Kong, K. Ilalov, S. Yu, K. Xu, L. Prazak, M. Fajardo, B. Sehgal, and P. E. Di Cesare. 2006. ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 20988-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu, C. J., W. Kong, K. Xu, Y. Luan, K. Ilalov, B. Sehgal, S. Yu, R. D. Howell, and P. E. Di Cesare. 2006. ADAMTS-12 associates with and degrades cartilage oligomeric matrix protein. J. Biol. Chem. 28115800-15808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Llamazares, M., A. J. Obaya, A. Moncada-Pazos, R. Heljasvaara, J. Espada, C. Lopez-Otin, and S. Cal. 2007. The ADAMTS12 metalloproteinase exhibits anti-tumorigenic properties through modulation of the Ras-dependent ERK signalling pathway. J. Cell Sci. 1203544-3552. [DOI] [PubMed] [Google Scholar]
- 69.Lu, R., and G. Serrero. 2000. Inhibition of PC cell-derived growth factor (PCDGF, epithelin/granulin precursor) expression by antisense PCDGF cDNA transfection inhibits tumorigenicity of the human breast carcinoma cell line MDA-MB-468. Proc. Natl. Acad. Sci. USA 973993-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luan, Y., L. Kong, D. R. Howell, K. Ilalov, M. Fajardo, X. H. Bai, P. E. Di Cesare, M. B. Goldring, S. B. Abramson, and C. J. Liu. 2008. Inhibition of ADAMTS-7 and ADAMTS-12 degradation of cartilage oligomeric matrix protein by alpha-2-macroglobulin. Osteoarthritis Cartilage 161413-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacLean, H. E., J. Guo, M. C. Knight, P. Zhang, D. Cobrinik, and H. M. Kronenberg. 2004. The cyclin-dependent kinase inhibitor p57(Kip2) mediates proliferative actions of PTHrP in chondrocytes. J. Clin. Investig. 1131334-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mansson, B., D. Carey, M. Alini, M. Ionescu, L. C. Rosenberg, A. R. Poole, D. Heinegard, and T. Saxne. 1995. Cartilage and bone metabolism in rheumatoid arthritis. Differences between rapid and slow progression of disease identified by serum markers of cartilage metabolism. J. Clin. Investig. 951071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mau, E., H. Whetstone, C. Yu, S. Hopyan, J. S. Wunder, and B. A. Alman. 2007. PTHrP regulates growth plate chondrocyte differentiation and proliferation in a Gli3 dependent manner utilizing hedgehog ligand dependent and independent mechanisms. Dev. Biol. 30528-39. [DOI] [PubMed] [Google Scholar]
- 74.Melton, J. T., N. M. Clarke, and H. I. Roach. 2006. Matrix metalloproteinase-9 induces the formation of cartilage canals in the chondroepiphysis of the neonatal rabbit. J. Bone Joint Surg. Am. 88(Suppl. 3)155-161. [DOI] [PubMed] [Google Scholar]
- 75.Morgelin, M., J. Engel, D. Heinegard, and M. Paulsson. 1992. Proteoglycans from the swarm rat chondrosarcoma. Structure of the aggregates extracted with associative and dissociative solvents as revealed by electron microscopy. J. Biol. Chem. 26714275-14284. [PubMed] [Google Scholar]
- 76.Mundlos, S., and B. R. Olsen. 1997. Heritable diseases of the skeleton. Part II: molecular insights into skeletal development-matrix components and their homeostasis. FASEB J. 11227-233. [PubMed] [Google Scholar]
- 77.Nicole, S., C. S. Davoine, H. Topaloglu, L. Cattolico, D. Barral, P. Beighton, C. B. Hamida, H. Hammouda, C. Cruaud, P. S. White, D. Samson, J. A. Urtizberea, F. Lehmann-Horn, J. Weissenbach, F. Hentati, and B. Fontaine. 2000. Perlecan, the major proteoglycan of basement membranes, is altered in patients with Schwartz-Jampel syndrome (chondrodystrophic myotonia). Nat. Genet. 26480-483. [DOI] [PubMed] [Google Scholar]
- 78.Olsen, B. R., A. M. Reginato, and W. Wang. 2000. Bone development. Annu. Rev. Cell Dev. Biol. 16191-220. [DOI] [PubMed] [Google Scholar]
- 79.Ong, C. H., and A. Bateman. 2003. Progranulin (granulin-epithelin precursor, PC-cell derived growth factor, acrogranin) in proliferation and tumorigenesis. Histol. Histopathol. 181275-1288. [DOI] [PubMed] [Google Scholar]
- 80.Rosenberg, K., H. Olsson, M. Morgelin, and D. Heinegard. 1998. Cartilage oligomeric matrix protein shows high affinity zinc-dependent interaction with triple helical collagen. J. Biol. Chem. 27320397-20403. [DOI] [PubMed] [Google Scholar]
- 81.Rowland, L. P. 2006. Frontotemporal dementia, chromosome 17, and progranulin. Ann. Neurol. 60275-277. [DOI] [PubMed] [Google Scholar]
- 82.Schipani, E., B. Lanske, J. Hunzelman, A. Luz, C. S. Kovacs, K. Lee, A. Pirro, H. M. Kronenberg, and H. Juppner. 1997. Targeted expression of constitutively active receptors for parathyroid hormone and parathyroid hormone-related peptide delays endochondral bone formation and rescues mice that lack parathyroid hormone-related peptide. Proc. Natl. Acad. Sci. USA 9413689-13694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Somerville, R. P., J. M. Longpre, E. D. Apel, R. M. Lewis, L. W. Wang, J. R. Sanes, R. Leduc, and S. S. Apte. 2004. ADAMTS7B, the full-length product of the ADAMTS7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. J. Biol. Chem. 27935159-35175. [DOI] [PubMed] [Google Scholar]
- 84.Stanton, H., F. M. Rogerson, C. J. East, S. B. Golub, K. E. Lawlor, C. T. Meeker, C. B. Little, K. Last, P. J. Farmer, I. K. Campbell, A. M. Fourie, and A. J. Fosang. 2005. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature 434648-652. [DOI] [PubMed] [Google Scholar]
- 85.Sun, X., M. Gulyas, and A. Hjerpe. 2004. Mesothelial differentiation as reflected by differential gene expression. Am. J. Respir. Cell Mol. Biol. 30510-518. [DOI] [PubMed] [Google Scholar]
- 86.Susic, S., J. McGrory, J. Ahier, and W. G. Cole. 1997. Multiple epiphyseal dysplasia and pseudoachondroplasia due to novel mutations in the calmodulin-like repeats of cartilage oligomeric matrix protein. Clin. Genet. 51219-224. [DOI] [PubMed] [Google Scholar]
- 87.Suzuki, M., and M. Nishiahara. 2002. Granulin precursor gene: a sex steroid-inducible gene involved in sexual differentiation of the rat brain. Mol. Genet. Metab. 7531-37. [DOI] [PubMed] [Google Scholar]
- 88.Tortorella, M. D., T. C. Burn, M. A. Pratta, I. Abbaszade, J. M. Hollis, R. Liu, S. A. Rosenfeld, R. A. Copeland, C. P. Decicco, R. Wynn, A. Rockwell, F. Yang, J. L. Duke, K. Solomon, H. George, R. Bruckner, H. Nagase, Y. Itoh, D. M. Ellis, H. Ross, B. H. Wiswall, K. Murphy, M. C. Hillman, Jr., G. F. Hollis, E. C. Arner, et al. 1999. Purification and cloning of aggrecanase-1: a member of the ADAMTS family of proteins. Science 2841664-1666. [DOI] [PubMed] [Google Scholar]
- 89.van der Kraan, P. M., E. L. Vitters, H. M. van Beuningen, L. B. van de Putte, and W. B. van den Berg. 1990. Degenerative knee joint lesions in mice after a single intra-articular collagenase injection. A new model of osteoarthritis. J. Exp. Pathol. (Oxford) 7119-31. [PMC free article] [PubMed] [Google Scholar]
- 90.Vojtek, A. B., S. M. Hollenberg, and J. A. Cooper. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74205-214. [DOI] [PubMed] [Google Scholar]
- 91.Vortkamp, A., K. Lee, B. Lanske, G. V. Segre, H. M. Kronenberg, and C. J. Tabin. 1996. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273613-622. [DOI] [PubMed] [Google Scholar]
- 92.Wang, W., J. Hayashi, W. E. Kim, and G. Serrero. 2003. PC cell-derived growth factor (granulin precursor) expression and action in human multiple myeloma. Clin. Cancer Res. 92221-2228. [PubMed] [Google Scholar]
- 93.Weir, E. C., W. M. Philbrick, M. Amling, L. A. Neff, R. Baron, and A. E. Broadus. 1996. Targeted overexpression of parathyroid hormone-related peptide in chondrocytes causes chondrodysplasia and delayed endochondral bone formation. Proc. Natl. Acad. Sci. USA 9310240-10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wright, W. E., D. A. Sassoon, and V. K. Lin. 1989. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell 56607-617. [DOI] [PubMed] [Google Scholar]
- 95.Xu, K., Y. Zhang, K. Ilalov, C. S. Carlson, J. Q. Feng, P. E. Di Cesare, and C. J. Liu. 2007. Cartilage oligomeric matrix protein associates with granulin-epithelin precursor (GEP) and potentiates GEP-stimulated chondrocyte proliferation. J. Biol. Chem. 28211347-11355. [DOI] [PubMed] [Google Scholar]
- 96.Zanocco-Marani, T., A. Bateman, G. Romano, B. Valentinis, Z. H. He, and R. Baserga. 1999. Biological activities and signaling pathways of the granulin/epithelin precursor. Cancer Res. 595331-5340. [PubMed] [Google Scholar]
- 97.Zhang, H., and G. Serrero. 1998. Inhibition of tumorigenicity of the teratoma PC cell line by transfection with antisense cDNA for PC cell-derived growth factor (PCDGF, epithelin/granulin precursor). Proc. Natl. Acad. Sci. USA 9514202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou, J., G. Gao, J. W. Crabb, and G. Serrero. 1993. Purification of an autocrine growth factor homologous with mouse epithelin precursor from a highly tumorigenic cell line. J. Biol. Chem. 26810863-10869. [PubMed] [Google Scholar]
- 99.Zhu, J., C. Nathan, W. Jin, D. Sim, G. S. Ashcroft, S. M. Wahl, L. Lacomis, H. Erdjument-Bromage, P. Tempst, C. D. Wright, and A. Ding. 2002. Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell 111867-878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.