Abstract
Sister chromatid recombination (SCR) is a potentially error-free pathway for the repair of DNA lesions associated with replication and is thought to be important for suppressing genomic instability. The mechanisms regulating the initiation and termination of SCR in mammalian cells are poorly understood. Previous work has implicated all the Rad51 paralogs in the initiation of gene conversion and the Rad51C/XRCC3 complex in its termination. Here, we show that hamster cells deficient in the Rad51 paralog XRCC2, a component of the Rad51B/Rad51C/Rad51D/XRCC2 complex, reveal a bias in favor of long-tract gene conversion (LTGC) during SCR. This defect is corrected by expression of wild-type XRCC2 and also by XRCC2 mutants defective in ATP binding and hydrolysis. In contrast, XRCC3-mediated homologous recombination and suppression of LTGC are dependent on ATP binding and hydrolysis. These results reveal an unexpectedly general role for Rad51 paralogs in the control of the termination of gene conversion between sister chromatids.
DNA double-strand breaks (DSBs) are potentially dangerous lesions, since their misrepair may cause chromosomal translocations, gene amplifications, loss of heterozygosity (LOH), and other types of genomic instability characteristic of human cancers (7, 9, 21, 40, 76, 79). DSBs are repaired predominantly by nonhomologous end joining or homologous recombination (HR), two evolutionarily conserved DSB repair mechanisms (8, 12, 16, 33, 48, 60, 71). DSBs generated during the S or G2 phase of the cell cycle may be repaired preferentially by HR, using the intact sister chromatid as a template for repair (12, 26, 29, 32, 71). Sister chromatid recombination (SCR) is a potentially error-free pathway for the repair of DSBs, which has led to the proposal that SCR protects against genomic instability, cancer, and aging. Indeed, a number of human cancer predisposition genes are implicated in SCR control (10, 24, 45, 57, 75).
HR entails an initial processing of the DSB to generate a free 3′ single-stranded DNA (ssDNA) overhang (25, 48, 56). This is coupled to the loading of Rad51, the eukaryotic homolog of Escherichia coli RecA, which polymerizes to form an ssDNA-Rad51 “presynaptic” nucleoprotein filament. Formation of the presynaptic filament is tightly regulated and requires the concerted action of a large number of gene products (55, 66, 68). Rad51-coated ssDNA engages in a homology search by invading homologous duplex DNA. If sufficient homology exists between the invading and invaded strands, a triple-stranded synapse (D-loop) forms, and the 3′ end of the invading (nascent) strand is extended, using the donor as a template for gene conversion. This recombination intermediate is thought to be channeled into one of the following two major subpathways: classical gap repair or synthesis-dependent strand annealing (SDSA) (48). Gap repair entails the formation of a double Holliday junction, which may resolve into either crossover or noncrossover products. Although this is a major pathway in meiotic recombination, crossing-over is highly suppressed in somatic eukaryotic cells (26, 44, 48). Indeed, the donor DNA molecule is seldom rearranged during somatic HR, suggesting that SDSA is the major pathway for the repair of somatic DSBs (26, 44, 49, 69). SDSA terminates when the nascent strand is displaced from the D-loop and pairs with the second end of the DSB to form a noncrossover product. The mechanisms underlying displacement of the nascent strand are not well understood. However, failure to displace the nascent strand might be expected to result in the production of longer gene conversion tracts during HR (36, 44, 48, 63).
Gene conversion triggered in response to a Saccharomyces cerevisiae or mammalian chromosomal DSB generally results in the copying of a short (50- to 300-bp) stretch of information from the donor (short-tract gene conversion [STGC]) (14, 47, 48, 67, 69). A minority of gene conversions in mammalian cells entail more-extensive copying, generating gene conversion tracts that are up to several kilobases in length (long-tract gene conversion [LTGC]) (26, 44, 51, 54, 64). In yeast, very long gene conversions can result from break-induced replication (BIR), a highly processive form of gene conversion in which a bona fide replication fork is thought to be established at the recombination synapse (11, 36, 37, 39, 61, 63). In contrast, SDSA does not require lagging-strand polymerases and appears to be much less processive than a conventional replication fork (37, 42, 78). BIR in yeast has been proposed to play a role in LOH in aging yeast, telomere maintenance, and palindromic gene amplification (5, 41, 52). It is unclear to what extent a BIR-like mechanism operates in mammalian cells, although BIR has been invoked to explain telomere elongation in tumors lacking telomerase (13). It is currently unknown whether LTGC and STGC in somatic mammalian cells are products of mechanistically distinct pathways or whether they represent alternative outcomes of a common SDSA pathway.
Vertebrate cells contain five Rad51 paralogs—polypeptides with limited sequence homology to Rad51—Rad51B, Rad51C, Rad51D, XRCC2, and XRCC3 (74). The Rad51 paralogs form the following two major complexes: Rad51B/Rad51C/Rad51D/XRCC2 (BCDX2) and Rad51C/XRCC3 (CX3) (38, 73). Genetic deletion of any one of the rad51 paralogs in the mouse germ line produces early embryonic lethality, and mouse or chicken cells lacking any of the rad51 paralogs reveal hypersensitivity to DNA-damaging agents, reduced frequencies of HR and of sister chromatid exchanges, increased chromatid-type errors, and defective sister chromatid cohesion (18, 72, 73, 82). Collectively, these data implicate the Rad51 paralogs in SCR regulation. The purified Rad51B/Rad51C complex has been shown to assist Rad51-mediated strand exchange (62). XRCC3 null or Rad51C null hamster cells reveal a bias toward production of longer gene conversion tracts, suggesting a role for the CX3 complex in late stages of SDSA (6, 44). Rad51C copurifies with branch migration and Holliday junction resolution activities in mammalian cell extracts (35), and XRCC3, but not XRCC2, facilitates telomere shortening by reciprocal crossing-over in telomeric T loops (77). These data, taken together with the meiotic defects observed in Rad51C hypomorphic mice, suggest a specialized role for CX3, but not for BCDX2, in resolving Holliday junction structures (31, 58).
To further address the roles of Rad51 paralogs in late stages of recombination, we have studied the balance between long-tract (>1-kb) and short-tract (<1-kb) SCR in XRCC2 mutant hamster cells. We found that DSB-induced gene conversion in both XRCC2 and XRCC3 mutant cells is biased in favor of LTGC. These defects were suppressed by expression of wild-type (wt) XRCC2 or XRCC3, respectively, although the dependence upon ATP binding and hydrolysis differed between the two Rad51 paralogs. These results indicate that Rad51 paralogs play a more general role in determining the balance between STGC and LTGC than was previously appreciated and suggest roles for both the BCDX2 and CX3 complexes in influencing the termination of gene conversion in mammals.
MATERIALS AND METHODS
Plasmids.
The HR/SCR reporter and I-SceI expression vector design and construction were reported previously (51). wt human XRCC2 (hXRCC2) and its mutants, as well as Rad51B, Rad51C, Rad51D, and Rad51 expression plasmids, have been described previously (17, 20, 70). The wt hXRCC3 was PCR amplified from the C-terminal His-tagged hXRCC3 expression plasmid pDS158 (80) and cloned in native form into the modified pcDNA3β vector (51) by standard cloning methods. XRCC3 point mutants were generated by PCR and confirmed by sequencing.
Antibodies and Western blotting.
Stably transfected cells were lysed in radioimmunoprecipitation assay buffer, and cell extracts were resolved with 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting using monoclonal antibody against XRCC2 (Abcam) or polyclonal antibodies against XRCC3 (kindly provided by Patrick Sung). Beta-actin was analyzed as a loading control using a specific monoclonal antibody (Abcam).
Cell lines and cell culture.
The cell lines irs1 (XRCC2−/−), V79 (wt parental cell line from which irs1 was derived), irs1SF (XRCC3−/−), and CHO-AA8 (wt parental cell line from which irs1SF was derived) were cultured as described previously (20). Single-copy HR/SCR reporters were established in these cells as described previously (44, 51). Briefly, 1 μg of AflIII-linearized SCR reporter was electroporated (Amaxa Biosystems) into 3 × 106 cells and selected in puromycin (2 μg/ml). Individual puromycin-resistant clones were expanded for genomic DNA preparation, which was analyzed by Southern blotting using multiple parallel restriction digests to identify clones that contained only one intact copy of the randomly integrated HR/SCR reporter. Stable expression of wt and mutant XRCC2 and XRCC3 clones was achieved by transfection using Lipofectamine 2000 (Invitrogen), followed by selection in G418. To avoid experimental error caused by clone-to-clone variation, these stable populations were studied as pools, without recloning, and were expanded for analysis, as described previously (44).
Recombination assays.
Recombination assays were carried out as described previously (44). Briefly, 5 × 106 cells were electroporated with 14 μg of pcDNA3β-myc-NLS-I-SceI or control pcDNA3β vector. Transfection efficiency (typically ∼90%) was measured by cotransfection in a ratio of 1:1 of a GFP expression plasmid to total DNA, as described previously (44). Green fluorescent protein-positive (GFP+) cells were analyzed by fluorescence-activated cell sorting 72 h after transfection, as described previously (51), and the data were obtained from multiple experiments. In all experiments, the background frequency of GFP+ cells was subtracted from the I-SceI-induced frequency, and I-SceI-induced values were corrected for I-SceI transfection efficiency. Long-tract SCR was measured by colony formation in blasticidin (BSD) in parallel in I-SceI-transfected or control vector-transfected cells, with correction for background BsdR+ frequency, transfection efficiency, and plating efficiency, as described above and previously (44, 51). The plating efficiency for the cell lines examined varied from 20 to 70% and was increased by 10 to 20% by expression of the wt Rad51 paralog gene product. The background frequencies of BsdR+ colonies were consistently less than 0.003% of I-SceI-induced frequencies. Trypsinized cells were counted and replated at a density of 1 × 106 to 3 × 106 cells per 10-cm plate and cultivated in 5 μg/ml BSD 24 h after plating. Colonies generated after 2 to 3 weeks of selection were stained and counted for BsdR+ frequency. In parallel experiments, I-SceI-induced BsdR+ colonies were expanded for genomic DNA preparation for Southern analysis. Statistical analysis was performed by use of an unpaired t test (unknown variance). Southern blotting was performed as described previously, using GFP cDNA as a probe (44, 51).
RESULTS
The XRCC2 Walker motif lysine residue is dispensable for XRCC2 HR function.
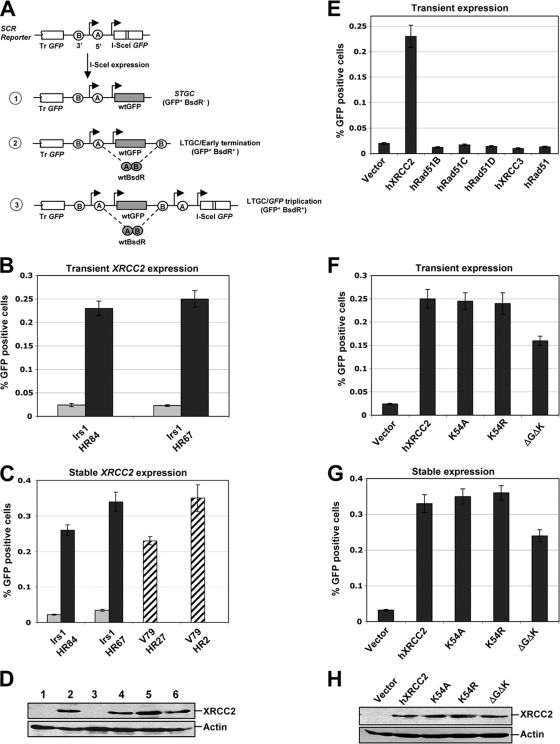
To study the role of XRCC2 in SCR regulation, we used XRCC2−/− irs1 cells containing randomly integrated single-copy SCR reporters to quantify HR/SCR in response to a site-specific chromosomal DSB induced by the I-SceI endonuclease (44, 51) (Fig. 1A). I-SceI-induced gene conversion generates wt GFP, with the GFP+ frequency being quantified by flow cytometry. HR-mediated STGC could arise by intrachromatid or interchromatid recombination, whereas LTGC, resulting in triplication of the GFP copies in the reporter, arises by SCR (Fig. 1A). Crossing-over between sister chromatids could also generate the “GFP triplication” outcome; however, this mechanism is suppressed in somatic cells (26, 44). We previously developed a method to quantify LTGC by means of a simple positive selection step (51). Two artificial exons of the BSD resistance gene bsdR were introduced in a “head-to-toe” orientation between the two GFP copies of an HR reporter. In the context of STGC (outcome 1) (Fig. 1A), corresponding to gene conversions of less than 1,031 bp, the SCR reporter remains unrearranged, and the cell is sensitive to BSD (BsdR−). When gene conversion extends beyond 1,031 bp, bsdR exon B is duplicated, allowing splicing between exon A of one cassette and exon B of the neighboring duplicated cassette to generate wt bsdR mRNA, and the cell becomes resistant to BSD (BsdR+) (Fig. 1A). Early termination of LTGC between 1.03 and 3.2 kb, i.e., prior to reaching the second GFP copy on the donor sister, is observed in a proportion of LTGC events (outcome 2) (Fig. 1A). The remaining LTGC products entail gene conversions of ≥3.2 kb, duplicating the entire BsdR cassette and triplicating the GFP copies (GFP triplication) (outcome 3) (Fig. 1A).
FIG. 1.
XRCC2 Walker motif lysine 54 residue is dispensable for its HR function. (A) HR/SCR reporter and I-SceI-induced repair products. I-SceI-induced gene conversion can resolve as STGC (1), LTGC/early termination (2), or LTGC/GFP triplication (3). Tr, truncated. (B) I-SceI-induced GFP+ frequencies in XRCC2−/− irs1 HR/SCR reporter cells. irs1 HR84 and HR67 cells were transiently cotransfected with I-SceI plasmid and either empty vector (gray bars) or hXRCC2 expression plasmid (black bars). Error bars, standard errors of the means. For the t test between wt XRCC2 and empty vector samples for each clone, P is <0.001. (C) I-SceI-induced GFP+ frequencies in irs1 HR clones stably expressing control vector (gray bars) or wt XRCC2 (black bars). For the t test between wt XRCC2-rescued and empty vector cells, P is <0.001. Striped bars, parental V79 HR27 and HR2 clones. (D) XRCC2 and actin protein levels in the cells shown in panel C. Lanes 1 and 3, empty vector in irs1 HR84 and HR67 cells; lanes 2 and 4, wt XRCC2 in the same two clones; lanes 5 and 6, XRCC2 levels in parental V79 HR27 and HR2 clones. (E) I-SceI-induced HR in irs1 HR67 following transient expression of hXRCC2 or indicated hRad51 paralog expression plasmids. For the t test between transfected hXRCC2 and all other samples, P is <0.003. (F) I-SceI-induced HR in irs1 HR67 cells transiently transfected with control vector, wt XRCC2, or XRCC2 K54A, K54R, or ΔG53ΔK54 mutant expression plasmids. For the t test between empty vector and wt XRCC2 or mutants, P is <0.002. (G) I-SceI-induced HR in irs1 HR67 cells stably expressing empty vector, wt XRCC2, or its mutants. For the t test between empty vector and wt XRCC2 or mutants, P is <0.001. (H) Abundance of XRCC2 and actin loading control for the experiment shown in panel G.
Multiple independent clones, each containing only one intact copy of the HR/SCR reporter, were established in irs1 XRCC2−/− and parental V79 hamster cells, as described in Materials and Methods. Induction of both GFP+ and BsdR+ cells was observed in these cells, following transient expression of plasmid-encoded I-SceI endonuclease. Note that, in the absence of I-SceI, the frequency of GFP+ events was usually zero and was always <0.001% (data not shown). To study the role of XRCC2 in HR, we transiently cotransfected individual reporter clones with the I-SceI expression vector and wt hXRCC2 expression vector or, in parallel, control empty vector. The frequency of I-SceI-induced GFP+ cells was low (∼ 0.025%) in two independent HR clones (irs1 HR84 and irs1 HR67) cotransfected with the control vector. However, coexpression of wt XRCC2 in the same clones displayed an increase of ∼10-fold (0.25%) in the efficiency of I-SceI-induced GFP+ cells, bringing HR to levels similar to those observed in parental V79 cells (Fig. 1B and C). To assess whether stable expression of XRCC2 rescues the HR deficiency in these cells, we generated stable pools of transfectants expressing either wt XRCC2 or the control empty vector (see Materials and Methods). Consistent with transient rescue experiments, stable expression of wt XRCC2 led to an increase of ∼10-fold in HR compared to the isogenic control vector cells (Fig. 1C).
To test whether expression of any of the other Rad51 paralogs, or Rad51 itself, could rescue the HR defect in irs1 cells, we transiently transfected irs1 HR67, in parallel, with a series of plasmids encoding human Rad51B, Rad51C, Rad51D, XRCC3, or Rad51 and compared I-SceI-induced HR with that for the same cell line transiently transfected with wt XRCC2. Interestingly, none of the other Rad51 paralogs, nor Rad51 itself, was able to complement the HR deficiency of irs1 cells (Fig. 1E). This indicates a specific, nonredundant role for XRCC2 in HR.
Rad51 paralogs contain a RecA/Rad51 family ATP-binding Walker motif. Previously, we noted that the Rad51C Walker A motif lysine residue is required for efficient HR (44). To define the role of the Walker A motif lysine residue in XRCC2 function, we analyzed Walker A motif mutants XRCC2 K54A, K54R, and ΔG53ΔK54 (i.e., lacking these two amino acids), comparing their function against that of wt XRCC2. Transient expression of either XRCC2 K54A or K54R in irs1 HR67 restored HR function as efficiently as wt XRCC2, while the XRCC2 ΔG53ΔK54 mutant showed a partial rescue of HR (Fig. 1F). Stable expression of these mutants in the same XRCC2−/− HR/SCR clone (irs1 HR67) gave similar results (Fig. 1G). The abundance of the stably expressed mutant XRCC2 proteins was equivalent to that of wt XRCC2 (Fig. 1H). We confirmed that the cDNAs used were correct by direct DNA sequencing of each strand of the relevant part of the XRCC2 plasmids (see Fig. S1 in the supplemental material). These surprising results demonstrate unequivocally that the ATP-binding Walker A motif lysine residue is dispensable for XRCC2 HR function.
XRCC2 regulates the balance between STGC and LTGC.
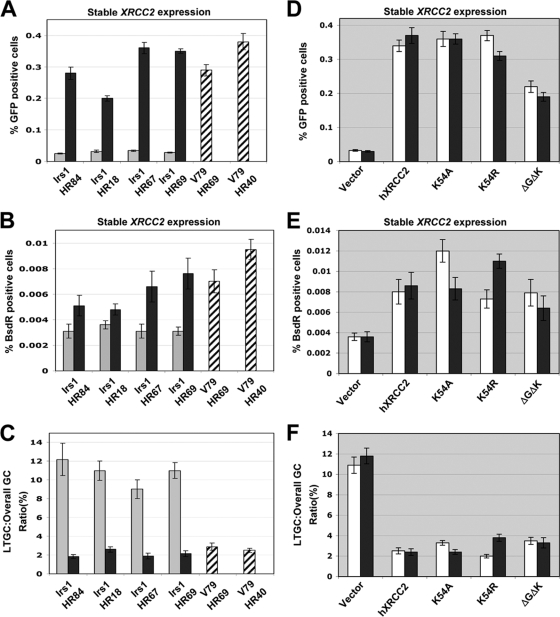
To test whether XRCC2 regulates long-tract SCR, we analyzed I-SceI-induced HR and SCR in irs1 (XRCC2−/−) HR/SCR clones 84, 18, 67, and 69, stably expressing either wt XRCC2 or control vectors, in parallel with parental V79 HR/SCR clones 69 and 40. I-SceI-induced overall gene conversion (overall GC) is measured by induction of GFP+ cells, whereas the SCR/LTGC outcome is measured by induction of BsdR+ GFP+ colonies. Since the I-SceI-induced BsdR+ GFP+ cells are a subset of all GFP+ events, the ratio of I-SceI-induced BsdR+ GFP+ frequency to overall I-SceI-induced GFP+ frequency (LTGC/overall GC) measures the probability that a given HR event will resolve as LTGC. Data were obtained and analyzed, as described in Materials and Methods, with correction for I-SceI transfection efficiency and plating efficiency. As noted previously, stable expression of wt XRCC2 in four independent clones (irs1 HR84, HR18, HR67, and HR69) increased the HR efficiency by ∼10-fold compared to that in the control vector cells, but expression of wt XRCC2 generated only a modest increase (∼2-fold) in the I-SceI-induced BsdR+ frequency (Fig. 2A and B). The ratio of I-SceI-induced BsdR+ GFP+ events to all I-SceI-induced GFP+ events (LTGC/overall GC) was 2 to 3% in cells expressing wt XRCC2, a value similar to the ratio observed in parental V79 clones (Fig. 2C). Strikingly, in four independent irs1 HR/SCR clones, this ratio was approximately fivefold higher (9 to 12%) in isogenic cultures expressing control empty vector (Fig. 2C). These results suggest that XRCC2 regulates the balance between STGC and LTGC during SCR.
FIG. 2.
XRCC2 regulates the balance between STGC and LTGC. (A) I-SceI-induced HR for indicated irs1 HR and parental V79 HR cells. Gray and black bars indicate stable expression of control vector or wt XRCC2, respectively. Striped bars indicate expression of V79 HR cells. For all irs1 HR clones, the t test between empty vector and hXRCC2-expressing samples shows a P value of <0.001. (B) Frequencies of I-SceI-induced BsdR+ colonies for the same experiment shown in panel A. (C) Ratio of I-SceI-induced BsdR+/GFP+ frequencies (LTGC/overall GC, expressed as a percentage) from the experiment shown in panels A and B. For all irs1 HR clones, the t test between stably expressing wt XRCC2 and empty vector samples shows a P value of <0.008. (D) I-SceI-induced GFP+ frequencies in irs1 HR67 (white bars) and irs1HR69 (black bars), stably expressing control vector, wt XRCC2 and K54 and ΔG53ΔK54 mutants. (E) I-SceI-induced BsdR+ frequencies for the experiment shown in panel D. (F) Ratio of I-SceI-induced BsdR+/GFP+ frequency from experiments shown in panels D and E. For the t test between empty vector and all other groups, P is <0.001.
The results obtained with XRCC2 are strongly reminiscent of our previous observations regarding Rad51C (44). The ability of wt Rad51C to reverse the LTGC bias in rad51C−/− cells is dependent on the conserved lysine residue in the Rad51C Walker A ATP-binding motif. To determine the role of XRCC2 ATP binding and hydrolysis in regulation of gene conversion tract length, we generated parallel pools of cells stably expressing Walker A motif lysine K54A, K54R, or ΔG53ΔK54 mutants in irs1 HR67 and HR69 clones. We compared I-SceI-induced GFP+ and BsdR+ GFP+ events in these parallel isogenic cultures, in comparison to the same cells expressing wt XRCC2 or control empty vector. Notably, each ATP mutant suppressed the ratio of LTGC/overall GC toward wt levels—almost as efficiently as wt XRCC2 (Fig. 2D to F). Thus, unlike Rad51C, the Walker A motif lysine residue of XRCC2 is dispensable for both its HR function and its LTGC suppression activity.
Qualitative alteration of LTGC products in XRCC2−/− irs1 cells.
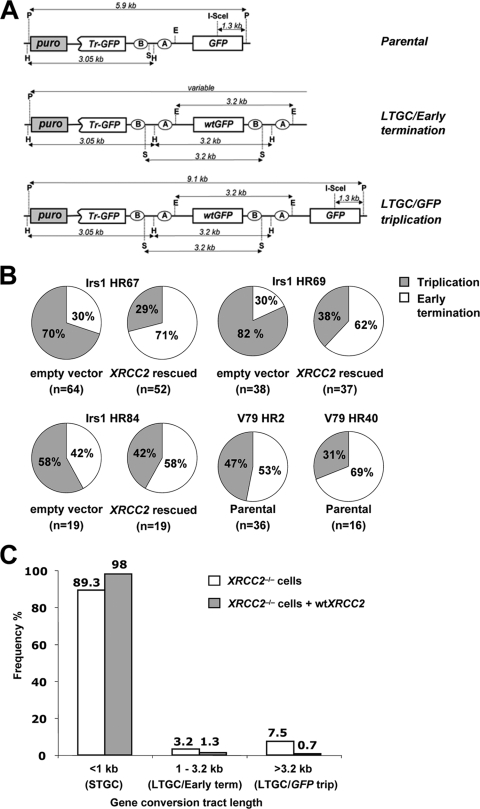
To test whether LTGC products are altered qualitatively in XRCC2−/− irs1 cells, we performed Southern analysis (with a GFP probe) to characterize the patterns of rearrangements within the SCR reporter from I-SceI-induced BsdR+ GFP+ colonies derived from irs1 cells expressing either wt XRCC2 or control vector. Figure 3A shows the expected restriction fragment sizes in genomic DNA Southern blots for the parental reporter, early terminating LTGC, and GFP triplication outcomes. GFP triplication implies a gene conversion tract length of ≥3.2 kb, whereas LTGC/early termination, which entails a duplication of the bsdR exon B but terminates prior to triplication of the GFP copies, corresponds to a tract length between 1.03 and 3.2 kb. To screen individual clones, we used parallel digests with either PstI alone or PstI plus I-SceI. This proved a reliable discriminator between the early termination and GFP triplication outcomes (see Fig. S2 in the supplemental material).
FIG. 3.
Southern analysis of LTGC events in irs1 cells. (A) Predicted rearrangements of SCR reporter in parental (top), LTGC/early termination (middle), and LTGC/GFP triplication (bottom) outcomes. Tr, truncated. (B) Relative frequencies of I-SceI-induced GFP triplication (gray) and early termination (white) LTGC events in irs1 HR clones expressing either empty vector or wt XRCC2 and in V79 HR2 and HR40 cells. The numbers of colonies analyzed are indicated in parentheses. (C) Frequency distribution of STGC, early termination, and GFP triplication events in irs1 HR67, HR69, and HR84 cells stably expressing control vector (white bars) or wt XRCC2 (gray bars). The data shown are calculated from those given in panel B and Fig. 2C. The numbers above each bar are the percentages of all gene conversions.
Southern analysis of I-SceI-induced BsdR+ GFP+ cells from XRCC2−/− irs1 cells expressing either wt XRCC2 or control empty vector displayed typical GFP triplication and early terminating LTGC events. In control vector-expressing XRCC2−/− irs1 HR67, HR69, and HR84 cells, 72% (87/121) of the I-SceI-induced LTGC events were GFP triplication outcomes, and 28% (34/121) entailed LTGC/early termination. Stable expression of wt XRCC2 in each of these clones altered the distribution in favor of early terminating LTGC events (Fig. 3B). In total, only 34% (37/108) of all the LTGC outcomes in the wt XRCC2-expressing cultures resolved as GFP triplication events, whereas 66% (71/108) resolved as early termination events (Fig. 3B). This difference was highly significant by χ2 analysis (P < 0.001). Analysis of LTGC events from two parental V79 HR2 and 40 clones revealed 60% (31/52) were early terminating events and 40% (21/52) were GFP triplication events. These results suggest that XRCC2 not only suppresses the ratio of LTGC/overall HR quantitatively (as shown above) but also skews LTGC qualitatively in favor of early termination (i.e., termination between 1.03 and 3.2 kb). This phenotype is strikingly similar to the observations we made previously regarding Rad51C (44). The frequency distribution of all gene conversions (i.e., STGC and LTGC), obtained by pooling data from three clones (irs1 HR67, -69, and -84) expressing either wt XRCC2 or control vector, is shown in Fig. 3C.
Differential regulation of STGC and LTGC by XRCC3.
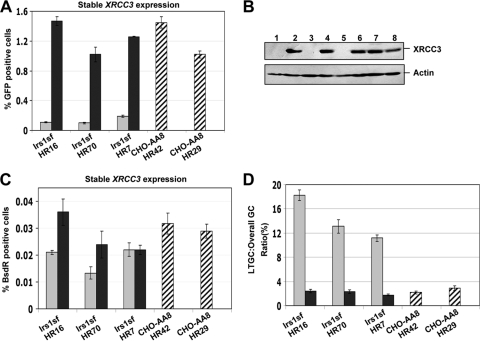
Brenneman et al. found that XRCC3−/− cells show a skewing in favor of longer gene conversion tracts compared to that of wt controls (6). By the nature of the assay system used (incorporation of mutationally “silent” restriction sites into the repaired chromosome during HR), this study was limited to analysis of gene conversions of up to ∼1,000 bp. To test whether XRCC3−/− cells also show a skewing in favor of gene conversions of >1 kb, we electroporated the HR/SCR reporter into irs1SF XRCC3−/− cells and identified clones carrying only one intact copy of the reporter, as described in Materials and Methods and previously (44, 51). Individual HR/SCR clones were tested for GFP+ and BsdR+ induction by I-SceI transfection. Cotransfection of I-SceI and wt hXRCC3 vectors in three independent HR/SCR XRCC3−/− clones (irs1SF HR16, HR70, and HR7) resulted in induction of I-SceI-induced GFP+ cells ∼10-fold greater than that seen in control I-SceI-transfected cells that received control empty vector in place of wt XRCC3. Indeed, expression of wt XRCC3 resulted in levels of I-SceI-induced HR comparable to those observed in parental CHO-AA8 HR42 and HR29 clones (see Fig. S3 in the supplemental material). Transient expression of Rad51B, Rad51C, Rad51D, XRCC2, or Rad51 in irs1SF HR16 failed to complement the HR deficiency in these cells, implying a specific and nonredundant role for XRCC3 in HR (see Fig. S4 in the supplemental material).
We studied HR and long-tract SCR in pools of irs1SF HR16, HR70, and HR7 stably transfected with either wt XRCC3 or control empty vector. Consistent with the transient rescue experiments described above, stable expression of wt XRCC3 in these clones caused an increase of 10-fold in I-SceI-induced HR compared to that of control isogenic populations lacking wt XRCC3 (Fig. 4A). Again, this rescue of function produced levels of HR similar to those observed in parental CHO-AA8 cells (Fig. 4A). In the same experiment, we compared the frequencies of I-SceI-induced BsdR+ cells for both control and wt XRCC3-rescued cells. Although there was an increase of ∼10-fold in the frequency of I-SceI-induced GFP+ events attributable to wt XRCC3 expression, wt XRCC3 produced only an increase of ∼2-fold in the frequency of I-SceI-induced BsdR+ events (Fig. 4C). Thus, the ratio of LTGC/overall GC, a measure of the probability of a gene conversion event resolving as LTGC, was about sixfold higher in control vector irs1SF HR16, HR70, or HR7 cells than in isogenic cultures expressing wt XRCC3 (Fig. 4D). This latter ratio of LTGC/overall GC (∼2%) for wt XRCC3-rescued cells was similar to that of parental CHO-AA8 cells (Fig. 4D). These results suggest that, like Rad51C (44) and XRCC2 (discussed above), XRCC3 is required for efficient gene conversion and also suppresses LTGC during SCR.
FIG. 4.
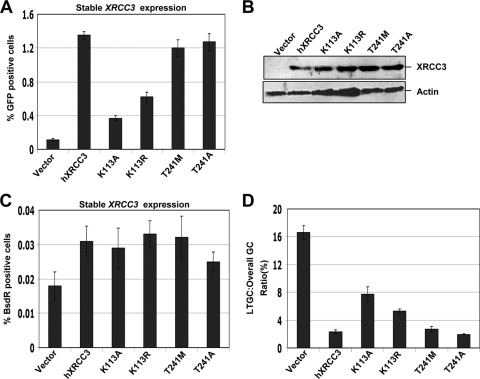
XRCC3 regulates the balance between STGC and LTGC. (A) I-SceI-induced GFP+ frequencies in irs1SF HR16, HR70, and HR7, stably expressing wt XRCC3 (black bars) or control vector (gray bars). Striped bars, parental CHO-AA8 HR42 and HR29 clones. For the t test among all the groups of HR clones expressing wt XRCC3 or empty vector, P is <0.01. (B) XRCC3 and actin protein levels for the experiment shown in panel A. Lanes 1, 3, and 5, empty vector-transfected cells in irs1SF HR16, HR70, and HR7 cells, respectively; lanes 2, 4, and 6, wt XRCC3 in the same three clones; lanes 7 and 8, XRCC3 levels in parental CHO-AA8 HR42 and HR29 clones. (C) Frequencies of I-SceI-induced BsdR+ colonies for the experiment shown in panel A. (D) Ratio of I-SceI-induced LTGC/overall GC, expressed as a percentage, from the experiment shown in panels A and B. For the t test between wt XRCC3-rescued and control HR cells in all three clones, P is <0.01.
Walker A motif lysine 113 but not cancer-associated threonine 241 is essential for XRCC3 HR/SCR function.
Given the differential requirements for ATP binding and hydrolysis by Rad51C and XRCC2, we asked whether the conserved Walker A motif lysine residue of XRCC3 is required for its HR and LTGC suppression activity. XRCC3 variant T241M has been shown to be positively associated with certain forms of bladder cancer (30, 34, 73, 81). However, this variant was found to be functional for overall GC (3). We asked whether this variant of XRCC3 affects long-tract SCR. We transiently transfected irs1SF HR16 cells with Walker A motif ATP-binding-defective (K113A), ATP hydrolysis-defective (K113R), or T241A mutant or T241M variant XRCC3, in parallel with I-SceI expression vector and the empty vector as controls and measured HR levels, as described in Materials and Methods. Consistent with previous results, XRCC3 T241M and T241A variants were proficient in I-SceI-induced HR (3). In contrast, transient expression of K113A or K113R mutants of XRCC3 only partially restored I-SceI-induced HR in irs1SF HR16 cells (see Fig. S5 in the supplemental material).
To study the impact of XRCC3 mutations on LTGC, we generated stable pools of transfectants expressing these mutants in irs1SF HR16 and compared both HR and LTGC in parallel, using stably expressed wt XRCC3 and the empty vector as positive and negative controls, respectively. Consistent with the above-noted transient rescue experiments, stable expression of XRCC3 T241M or T241A completely rescued HR, whereas XRCC3 K113A or K113R mutants only partially rescued HR (Fig. 5A). The corresponding BsdR+ frequency for the same experiment is shown in Fig. 5C. In irs1SF HR16 cells expressing the variant (T241M) or mutant (T241A) XRCC3 alleles, the ratio of I-SceI-induced LTGC/overall GC (BsdR+/GFP+) was ∼3%—a ratio similar to that observed in the same cells expressing wt XRCC3 (Fig. 5D). This indicates that these T241 variant XRCC3 alleles are not defective for either HR or LTGC suppression. In contrast, ATP-binding (K113A) and hydrolysis (K113R) mutants were partially defective for LTGC suppression (Fig. 5D).
FIG. 5.
Walker motif lysine residue but not cancer-associated threonine regulates XRCC3 HR/SCR function. (A) Frequency of I-SceI-induced HR in irs1SF HR16 stably expressing control vector, wt XRCC3, lysine 113, and threonine 241 mutants. For the t test between cultures expressing control vector and wt XRCC3 or threonine 241 mutants, P is <0.01; between vector and K113A, P is <0.0015; between vector and the K113R mutant, P is <0.01. (B) XRCC3 and actin protein levels for the experiment shown in panel A. (C) I-SceI-induced BsdR+ frequencies for the experiment shown in panel A. (D) Ratio of I-SceI-induced LTGC/overall GC, expressed as a percentage, from the experiment shown in panels A and B. For the t test between control vector and wt XRCC3 or threonine 241 mutants, P is <0.0025; between vector and K113A or K113R mutants, P is <0.0015.
Qualitative analysis of LTGC in XRCC3−/−cells.
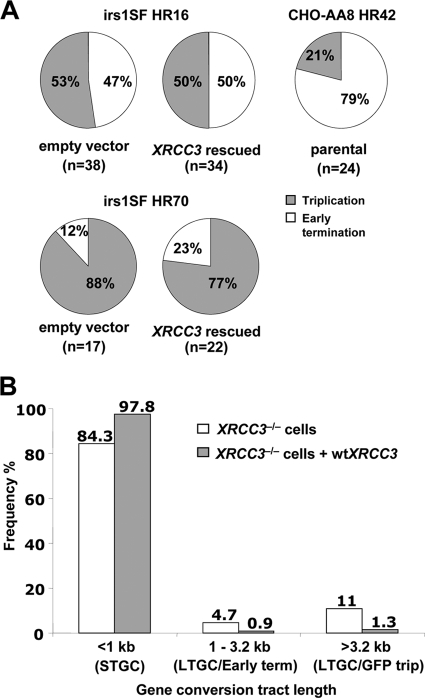
Inactivation of Rad51 paralog Rad51C or XRCC2 skews gene conversion in favor of LTGC both quantitatively and qualitatively (44; current study). To determine whether XRCC3−/− cells show a similar qualitative skewing of LTGC in favor of longer tracts, we used Southern blotting to quantify the relative abundance of early termination (gene conversion tract between 1.03 and 3.2 kb) and GFP triplication (gene conversion tract of ≥3.2 kb) outcomes of LTGC. We tested this by Southern blotting of individual I-SceI-induced BsdR+ GFP+ colonies derived from irs1SF HR16 and HR70 XRCC3−/− cells stably expressing either wt XRCC3 or the empty vector. Southern analysis revealed that 36% (20/55) were early termination and 64% were (35/55) GFP triplication events in control irs1SF HR16 and HR70 cells. In the isogenic clones expressing wt XRCC3, 39% (22/56) of the events were early terminating and 61% (34/56) entailed GFP triplication (Fig. 6A). Thus, in contrast to our studies of XRCC2 and Rad51C mutants, we did not observe a statistically significant shift in favor of early termination LTGC in XRCC3 null cells that were complemented with wt XRCC3. In parallel, we analyzed rearrangements within the SCR reporter in BsdR+ colonies from the parental CHO-AA8 HR42 clone. The frequency distribution of all gene conversions (STGC and LTGC) in the rescued and unrescued XRCC3−/− irs1SF HR16 and HR70 cells is shown in Fig. 6B. Parental cells revealed that 79% (19/24) were early terminating and 21% (5/24) were GFP triplication events (Fig. 6A). Notably, in both this study and our previous study of Rad51C, there is clone-to-clone variation in the balance between the early termination and GFP triplication outcomes (44). This suggests that the termination of LTGC is influenced by genetic locus and/or by clone-specific epigenetic factors.
FIG. 6.
Analysis of gene conversion tract lengths in irs1SF cells. (A) Relative frequencies of early termination (white) and GFP triplication (gray) events in irs1SF HR cells stably expressing wt XRCC3 or the control vector and in parental CHO-AA8 HR42 cells. (B) Frequency distribution of gene conversion tract lengths in irs1SF HR cells stably expressing the control vector (white bars) or wt XRCC3 (gray bars). The data shown are calculated from those given in panel A and Fig. 4C. The numbers above each bar are the percentages of all gene conversions.
DISCUSSION
The work described here reveals a novel function for the mammalian Rad51 paralog XRCC2 in limiting the extent of gene conversion between sister chromatids. One previous study concluded that XRCC2 has no impact on LTGC (27). However, this earlier study did not use a reporter capable of specifically selecting for LTGC events, and the number of LTGC events examined was therefore necessarily small. In this study, we used a reporter designed for the specific, positive selection of LTGC, allowing a level of quantitation not previously possible (51). Consistent with the original work of Brenneman et al., we also find that XRCC3 null cells show a bias in favor of longer gene conversions (6). This work implicates both the CX3 and BCDX2 complexes in facilitating the timely termination of gene conversion.
In agreement with previous reports, our studies indicate a shared role for Rad51 paralogs in facilitating the initial steps of HR. It is not known whether mammalian Rad51 paralogs show an epistatic relationship with respect to Rad51-catalyzed HR. However, chicken DT40 cells containing xrcc3 rad51d double mutants, in which both the BCDX2 and CX3 complexes are impaired, show a more severe sensitivity to camptothecin and cisplatin than rad51b rad51d double mutants, in which only the BCDX2 complex is dysfunctional (82). These results suggest that Rad51 paralogs, when present in a different complex, function at least in part independently in assisting Rad51-mediated HR. However, our attempts to deplete Rad51 paralogs in hamster cells and embryonic stem cells by using small interfering RNA were unsuccessful and resulted only in a mild HR defect (44; data not shown). This makes it impossible to use this system in its current form to study the potential synergy between CX3 and BCDX2 complexes in control of the balance between STGC and LTGC.
The molecular mechanisms by which Rad51 paralogs promote Rad51-mediated HR are not known. However, mammalian Rad51 paralogs have been proposed to play a role analogous to S. cerevisiae Rad55 and Rad57. Overexpression of S. cerevisiae RAD51 suppresses DSB-induced recombination defects in rad55 and rad57 mutants (22, 28). In contrast, the work described here and in our previous study shows that transient expression of other mammalian Rad51 paralogs or Rad51 itself is not sufficient to complement the gene conversion defect of XRCC2−/−, XRCC3−/−, or Rad51C−/− cells (44). These results suggest that each of these Rad51 paralogs has a nonredundant function in HR. Interestingly, although overexpression of S. cerevisiae RAD51 reverses a defect in DSB-induced HR in rad55 and rad57 mutants, RAD51 overexpression was not able to suppress the spontaneous SCR defect in rad57 mutants, suggesting a specialized role for Rad51 paralogs in the repair of daughter strand gaps that arise as a result of stalled replication forks (43). In this regard, mammalian XRCC3 has been shown to be required for replication fork progression in cells treated with DNA replication inhibitors (23). These results suggest that Rad51 paralogs have evolved with specialized roles to regulate both HR-mediated DSB repair and daughter strand gap repair.
The Rad51 paralogs possess a nucleotide binding Walker motif, which is highly conserved in the RecA/Rad51 family of proteins. Walker A motif lysine mutation in RecA or Rad51 results in a severe HR defect in E. coli and S. cerevisiae. Consistent with this, purified ATP-binding-competent but hydrolysis-defective Rad51 or RecA mutants, although able to support homologous pairing, are catalytically defective for strand exchange (53, 59, 65). Our analysis of Walker motif lysine mutants in XRCC2 and XRCC3 reveals an interesting divergence in the requirements for ATP binding and hydrolysis. Specifically, unlike Rad51C and XRCC3, each of which must both bind and hydrolyze ATP for optimal HR function, XRCC2-mediated HR and SCR functions are independent of its ATP-binding and ATP hydrolysis functions. This is consistent with a previous report demonstrating that XRCC2 Walker A motif lysine mutants produce a wt level of resistance to DNA-damaging agents when expressed in XRCC2−/− cells (46). Interestingly, Walker motif lysine mutation in S. cerevisiae Rad57 does not affect its function in HR. A similar mutation in S. cerevisiae Rad55, which forms a heterodimer with Rad57, shows an HR defect (28). In addition, the Rad51D Walker B but not A motif has been shown to be required for its HR function and interaction with other paralogs (80). Thus, across species, individual Rad51 paralogs show various degrees of dependence upon ATP binding and hydrolysis for their functions in HR. Vertebrate XRCC2 and ScRad57 appear to contribute to HR via an unconventional (ATP-independent) mechanism. Nonetheless, the ATP-binding and hydrolysis functions of these two Rad51 paralogs are evolutionarily conserved, the reasons for which are unclear at present.
A critical, as yet unanswered question is whether LTGC and STGC are products of a common SDSA-type mechanism or whether LTGC can arise by a distinct mechanism. In Drosophila melanogaster, SDSA can generate somatic gene conversions extending over several kilobases (1). Thus, a long gene conversion alone is not sufficient reason to invoke a distinct mechanism underlying LTGC. In the assay system we used, the distinction between STGC and LTGC is defined by the duplication of bsdR exon B, corresponding to a gene conversion of 1,031 bp. There is no a priori expectation that two hypothetically distinct mechanisms of gene conversion should, by chance, be divided perfectly into nonoverlapping sets by an arbitrary 1,031-bp cutoff point. In this regard, it is striking that the analyses of Rad51C, XRCC2, and XRCC3 point mutants, described here and in our previous work, reveal a strong quantitative impact on STGC but a modest effect (at most approximately twofold) on LTGC. Consequently, a Rad51 paralog mutant that is defective for overall HR reveals a proportionately increased probability of resolving as LTGC, as measured by the ratio between LTGC and overall HR. This could be interpreted as evidence that STGC and LTGC are mechanistically distinct and that Rad51 paralog dysfunction skews gene conversion in favor of LTGC by default. However, this interpretation is not supported by our detailed analysis of LTGC in Rad51C and XRCC2 mutants. In each of these cell types, expression of the relevant wt Rad51 paralog skews LTGC in favor of early termination (gene conversions between 1.03 and 3.2 kb), at the expense of the GFP triplication outcome (gene conversions of ≥3.2 kb). Thus, Rad51C and XRCC2 each exert a positive influence on the outcome of LTGC that is in addition to their roles in facilitating overall GC. These findings are reminiscent of the bias toward longer gene conversions in XRCC3−/− cells noted by Brenneman et al. (6) and suggest that at least a proportion of LTGCs arise by an SDSA-type mechanism similar to that governing STGC.
As was suggested previously in our work on Rad51C (44), XRCC2 and XRCC3 might promote displacement of the nascent strand during SDSA, thereby facilitating pairing with the second end of the DSB and ensuring timely termination of gene conversion. Defective Rad51 paralog function would impair nascent strand displacement, thereby favoring LTGC. In support of this model, it is notable that a Rad51C-containing protein complex has branch migration activity in vitro (35). Conceivably, such an activity might facilitate dissociation of the migrating D-loop and displacement of the nascent strand during SDSA. Given the involvement of XRCC2 in LTGC (this study) and its noninvolvement in resolution of Holliday junction structures (35, 77), it may be that CX3 acts independently of BCDX2 to resolve Holliday junctions but in concert with XRCC2 in branch migration. Indeed, Liu et al. reported a specific branch migration activity of BCDX2 (35). Presumably, D-loop migration is critical for the timely resolution of SDSA. Although D-loop migration may be related to Holliday junction branch migration, the precise relationship between these two activities remains to be determined (4). Interestingly, recent studies identified the Schizosaccharomyces pombe Rhp51 paralog Rhp57 as a regulator of both LTGC and crossing-over, suggesting that these specialized functions of Rad51 paralogs are evolutionarily conserved (2).
Despite the above-noted evidence linking mammalian LTGC to SDSA, some mammalian LTGCs may nonetheless arise by a distinct mechanism, such as gap repair with crossing-over (44) or BIR. Interestingly, recent work in yeast suggests that BIR can emerge as a by-product of a long-tract SDSA-type mechanism (63). Dissection of these hypotheses in mammalian cells will require the development of new tools to study LTGC.
Recent studies with S. cerevisiae and D. melanogaster indicate that aging is linked to DNA damage and repair (19, 50). DSBs are known to accumulate in aging and senescent cells, and homozygous mutations in DNA repair genes such as the Bloom's syndrome, Werner syndrome, or Fanconi anemia genes are associated with premature aging, age-related disease, and cancer predisposition (15). The rate of LOH increases with age in S. cerevisiae, and this is associated with increased BIR in aging cells (41). In aging flies, the balance between HR and nonhomologous end joining is skewed in favor of HR, with a bias toward LTGC in older males (50). These observations suggest that LTGC is a pathological outcome and that disequilibrium in the balance between short- and long-tract HR may contribute to aging and cancer. It will be interesting to determine whether the LTGC outcome is associated with aging in mammals and whether Rad51 paralogs regulate LOH between homologous chromosomes during aging and tumorigenesis.
Supplementary Material
Acknowledgments
We thank John Thacker and David Schild for generous gifts of cell lines and plasmids. We thank members of the Scully lab for critical comments and discussion. We thank K. Muniyappa for extending lab facilities and support.
This work was supported by NIH grants CA95175 and GM073894, an ACS Scholars award, a Leukemia and Lymphoma Society Scholar award, and an award from the Bloom's Syndrome Foundation.
Footnotes
Published ahead of print on 26 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, M. D., M. McVey, and J. J. Sekelsky. 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299265-267. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu, Y., Y. Tsutsui, T. Morishita, M. S. Siddique, Y. Kurokawa, M. Ikeguchi, F. Yamao, B. Arcangioli, and H. Iwasaki. 2007. Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J. 261352-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araujo, F. D., A. J. Pierce, J. M. Stark, and M. Jasin. 2002. Variant XRCC3 implicated in cancer is functional in homology-directed repair of double-strand breaks. Oncogene 214176-4180. [DOI] [PubMed] [Google Scholar]
- 4.Bachrati, C. Z., R. H. Borts, and I. D. Hickson. 2006. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 342269-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosco, G., and J. E. Haber. 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 1501037-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenneman, M. A., B. M. Wagener, C. A. Miller, C. Allen, and J. A. Nickoloff. 2002. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell 10387-395. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., and R. D. Kolodner. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 2381-85. [DOI] [PubMed] [Google Scholar]
- 8.Couedel, C., K. D. Mills, M. Barchi, L. Shen, A. Olshen, R. D. Johnson, A. Nussenzweig, J. Essers, R. Kanaar, G. C. Li, F. W. Alt, and M. Jasin. 2004. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 181293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 40437-41. [DOI] [PubMed] [Google Scholar]
- 10.D'Andrea, A. D., and M. Grompe. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 323-34. [DOI] [PubMed] [Google Scholar]
- 11.Davis, A. P., and L. S. Symington. 2004. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 242344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delacote, F., and B. S. Lopez. 2008. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: the trans-S double-strand break repair model. Cell Cycle 733-38. [DOI] [PubMed] [Google Scholar]
- 13.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26447-450. [DOI] [PubMed] [Google Scholar]
- 14.Elliott, B., C. Richardson, J. Winderbaum, J. A. Nickoloff, and M. Jasin. 1998. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 1893-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engels, W. R., D. Johnson-Schlitz, C. Flores, L. White, and C. R. Preston. 2007. A third link connecting aging with double strand break repair. Cell Cycle 6131-135. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson, D. O., and F. W. Alt. 2001. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene 205572-5579. [DOI] [PubMed] [Google Scholar]
- 17.French, C. A., C. E. Tambini, and J. Thacker. 2003. Identification of functional domains in the RAD51L2 (RAD51C) protein and its requirement for gene conversion. J. Biol. Chem. 27845445-45450. [DOI] [PubMed] [Google Scholar]
- 18.Godthelp, B. C., W. W. Wiegant, A. van Duijn-Goedhart, O. D. Scharer, P. P. van Buul, R. Kanaar, and M. Z. Zdzienicka. 2002. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 302172-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbunova, V., A. Seluanov, Z. Mao, and C. Hine. 2007. Changes in DNA repair during aging. Nucleic Acids Res. 357466-7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin, C. S., P. J. Simpson, C. R. Wilson, and J. Thacker. 2000. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Biol. 2757-761. [DOI] [PubMed] [Google Scholar]
- 21.Haber, J. E. 2006. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair (Amsterdam) 5998-1009. [DOI] [PubMed] [Google Scholar]
- 22.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 926925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry-Mowatt, J., D. Jackson, J. Y. Masson, P. A. Johnson, P. M. Clements, F. E. Benson, L. H. Thompson, S. Takeda, S. C. West, and K. W. Caldecott. 2003. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol. Cell 111109-1117. [DOI] [PubMed] [Google Scholar]
- 24.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3169-178. [DOI] [PubMed] [Google Scholar]
- 25.Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani, W. Carotenuto, G. Liberi, D. Bressan, L. Wan, N. M. Hollingsworth, J. E. Haber, and M. Foiani. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 4311011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 193398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, R. D., N. Liu, and M. Jasin. 1999. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401397-399. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, R. D., and L. S. Symington. 1995. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 154843-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadyk, L. C., and L. H. Hartwell. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132387-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuschel, B., A. Auranen, S. McBride, K. L. Novik, A. Antoniou, J. M. Lipscombe, N. E. Day, D. F. Easton, B. A. Ponder, P. D. Pharoah, and A. Dunning. 2002. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum. Mol. Genet. 111399-1407. [DOI] [PubMed] [Google Scholar]
- 31.Kuznetsov, S., M. Pellegrini, K. Shuda, O. Fernandez-Capetillo, Y. Liu, B. K. Martin, S. Burkett, E. Southon, D. Pati, L. Tessarollo, S. C. West, P. J. Donovan, A. Nussenzweig, and S. K. Sharan. 2007. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 176581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert, S., A. Watson, D. M. Sheedy, B. Martin, and A. M. Carr. 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121689-702. [DOI] [PubMed] [Google Scholar]
- 33.Lieber, M. R. 2008. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2831-5. [DOI] [PubMed] [Google Scholar]
- 34.Lindh, A. R., S. Rafii, N. Schultz, A. Cox, and T. Helleday. 2006. Mitotic defects in XRCC3 variants T241M and D213N and their relation to cancer susceptibility. Hum. Mol. Genet. 151217-1224. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Y., J. Y. Masson, R. Shah, P. O'Regan, and S. C. West. 2004. RAD51C is required for Holliday junction processing in mammalian cells. Science 303243-246. [DOI] [PubMed] [Google Scholar]
- 36.Llorente, B., C. E. Smith, and L. S. Symington. 2008. Break-induced replication: what is it and what is it for? Cell Cycle 7859-864. [DOI] [PubMed] [Google Scholar]
- 37.Malkova, A., M. L. Naylor, M. Yamaguchi, G. Ira, and J. E. Haber. 2005. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 25933-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masson, J. Y., M. C. Tarsounas, A. Z. Stasiak, A. Stasiak, R. Shah, M. J. McIlwraith, F. E. Benson, and S. C. West. 2001. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 153296-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McEachern, M. J., and J. E. Haber. 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 75111-135. [DOI] [PubMed] [Google Scholar]
- 40.McKinnon, P. J., and K. W. Caldecott. 2007. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 837-55. [DOI] [PubMed] [Google Scholar]
- 41.McMurray, M. A., and D. E. Gottschling. 2003. An age-induced switch to a hyper-recombinational state. Science 3011908-1911. [DOI] [PubMed] [Google Scholar]
- 42.McVey, M., M. Adams, E. Staeva-Vieira, and J. J. Sekelsky. 2004. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167699-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mozlin, A. M., C. W. Fung, and L. S. Symington. 2008. Role of the Saccharomyces cerevisiae Rad51 paralogs in sister chromatid recombination. Genetics 178113-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagaraju, G., S. Odate, A. Xie, and R. Scully. 2006. Differential regulation of short- and long-tract gene conversion between sister chromatids by Rad51C. Mol. Cell. Biol. 268075-8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Driscoll, M., and P. A. Jeggo. 2006. The role of double-strand break repair—insights from human genetics. Nat. Rev. Genet. 745-54. [DOI] [PubMed] [Google Scholar]
- 46.O'Regan, P., C. Wilson, S. Townsend, and J. Thacker. 2001. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem. 27622148-22153. [DOI] [PubMed] [Google Scholar]
- 47.Palmer, S., E. Schildkraut, R. Lazarin, J. Nguyen, and J. A. Nickoloff. 2003. Gene conversion tracts in Saccharomyces cerevisiae can be extremely short and highly directional. Nucleic Acids Res. 311164-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paques, F., W. Y. Leung, and J. E. Haber. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 182045-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Preston, C. R., C. Flores, and W. R. Engels. 2006. Age-dependent usage of double-strand-break repair pathways. Curr. Biol. 162009-2015. [DOI] [PubMed] [Google Scholar]
- 51.Puget, N., M. Knowlton, and R. Scully. 2005. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amsterdam) 4149-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rattray, A. J., B. K. Shafer, B. Neelam, and J. N. Strathern. 2005. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes Dev. 191390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rehrauer, W. M., and S. C. Kowalczykowski. 1993. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 2681292-1297. [PubMed] [Google Scholar]
- 54.Richardson, C., M. E. Moynahan, and M. Jasin. 1998. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 123831-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.San Filippo, J., P. Sung, and H. Klein. 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77229-257. [DOI] [PubMed] [Google Scholar]
- 56.Sartori, A. A., C. Lukas, J. Coates, M. Mistrik, S. Fu, J. Bartek, R. Baer, J. Lukas, and S. P. Jackson. 2007. Human CtIP promotes DNA end resection. Nature 450509-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scully, R., and D. M. Livingston. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharan, S. K., and S. G. Kuznetsov. 2007. Resolving RAD51C function in late stages of homologous recombination. Cell Div. 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shinohara, A., H. Ogawa, and T. Ogawa. 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69457-470. [DOI] [PubMed] [Google Scholar]
- 60.Shrivastav, M., L. P. De Haro, and J. A. Nickoloff. 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18134-147. [DOI] [PubMed] [Google Scholar]
- 61.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 212048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sigurdsson, S., S. Van Komen, W. Bussen, D. Schild, J. S. Albala, and P. Sung. 2001. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 153308-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, C. E., B. Llorente, and L. S. Symington. 2007. Template switching during break-induced replication. Nature 447102-105. [DOI] [PubMed] [Google Scholar]
- 64.Stark, J. M., and M. Jasin. 2003. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 23733-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sung, P. 1994. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 2651241-1243. [DOI] [PubMed] [Google Scholar]
- 66.Sung, P., and H. Klein. 2006. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 7739-750. [DOI] [PubMed] [Google Scholar]
- 67.Sweetser, D. B., H. Hough, J. F. Whelden, M. Arbuckle, and J. A. Nickoloff. 1994. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol. Cell. Biol. 143863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taghian, D. G., and J. A. Nickoloff. 1997. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol. 176386-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takata, M., M. S. Sasaki, E. Sonoda, T. Fukushima, C. Morrison, J. S. Albala, S. M. Swagemakers, R. Kanaar, L. H. Thompson, and S. Takeda. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 206476-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 175497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 212858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thacker, J. 2005. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 219125-135. [DOI] [PubMed] [Google Scholar]
- 74.Thacker, J., and M. Z. Zdzienicka. 2003. The mammalian XRCC genes: their roles in DNA repair and genetic stability. DNA Repair (Amsterdam) 2655-672. [DOI] [PubMed] [Google Scholar]
- 75.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108171-182. [DOI] [PubMed] [Google Scholar]
- 76.Vogelstein, B., and K. W. Kinzler. 2004. Cancer genes and the pathways they control. Nat. Med. 10789-799. [DOI] [PubMed] [Google Scholar]
- 77.Wang, R. C., A. Smogorzewska, and T. de Lange. 2004. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119355-368. [DOI] [PubMed] [Google Scholar]
- 78.Wang, X., G. Ira, J. A. Tercero, A. M. Holmes, J. F. Diffley, and J. E. Haber. 2004. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 246891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weinstock, D. M., C. A. Richardson, B. Elliott, and M. Jasin. 2006. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amsterdam) 51065-1074. [DOI] [PubMed] [Google Scholar]
- 80.Wiese, C., J. M. Hinz, R. S. Tebbs, P. B. Nham, S. S. Urbin, D. W. Collins, L. H. Thompson, and D. Schild. 2006. Disparate requirements for the Walker A and B ATPase motifs of human RAD51D in homologous recombination. Nucleic Acids Res. 342833-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winsey, S. L., N. A. Haldar, H. P. Marsh, M. Bunce, S. E. Marshall, A. L. Harris, F. Wojnarowska, and K. I. Welsh. 2000. A variant within the DNA repair gene XRCC3 is associated with the development of melanoma skin cancer. Cancer Res. 605612-5616. [PubMed] [Google Scholar]
- 82.Yonetani, Y., H. Hochegger, E. Sonoda, S. Shinya, H. Yoshikawa, S. Takeda, and M. Yamazoe. 2005. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res. 334544-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.