Abstract
Cleavage of histone pre-mRNAs at the 3′ end is guided by the U7 snRNP, which is a component of a larger 3′-end processing complex. To identify other components of this complex, we isolated proteins that stably associate with a fragment of histone pre-mRNA containing all necessary processing elements and a biotin affinity tag at the 5′ end. Among the isolated proteins, we identified three well-characterized processing factors: the stem-loop binding protein (SLBP), which interacts with the stem-loop structure upstream of the cleavage site, and both Lsm11 and SmB, which are components of the U7-specifc Sm ring. We also identified 3′hExo/Eri-1, a multifunctional 3′ exonuclease that is known to trim the 3′ end of 5.8S rRNA. 3′hExo primarily binds to the downstream portion of the stem-loop structure in mature histone mRNA, with the upstream portion being occupied by SLBP. The two proteins bind their respective RNA sites in a cooperative manner, and 3′hExo can recruit SLBP to a mutant stem-loop that itself does not interact with SLBP. UV-cross-linking studies used to characterize interactions within the processing complex demonstrated that 3′hExo also interacts in a U7-dependent manner with unprocessed histone pre-mRNA. However, this interaction is not required for the cleavage reaction. The region between the cleavage site and the U7-binding site interacts with three low-molecular-weight proteins, which were identified as components of the U7-specific Sm core: SmB, SmD3, and Lsm10. These proteins likely rigidify the substrate and function as the molecular ruler in determining the site of cleavage.
Replication-dependent histone pre-mRNAs are processed at the 3′ end by a single endonucleolytic cleavage that requires the U7 snRNP (12). Cleavage occurs between two sequence elements that are separated by approximately 15 nucleotides: a highly conserved stem-loop (SL) structure and a purine-rich histone downstream element (HDE). The resultant histone mRNA ends with the SL, followed by a 5-nucleotide single-stranded tail. The HDE interacts with the U7 snRNP, which is composed of an approximately 60-nucleotide U7 snRNA and an Sm ring. The Sm ring of the U7 snRNP differs from that found in spliceosomal snRNPs, as it contains Lsm10 and Lsm11 proteins in place of the SmD1 and SmD2 proteins. The remaining five components of the ring (SmB, SmD3, SmE, SmF, and SmG) are common to both types of snRNPs (31, 32). The U7 snRNP is recruited to histone pre-mRNA primarily through formation of a double-stranded RNA between the 5′ end of U7 snRNA and the HDE. The SL structure forms a tight complex with the SL binding protein (SLBP). SLBP also binds a 100-kDa zinc finger protein (ZFP100) that in turn interacts with Lsm11 and hence may additionally stabilize U7 snRNP on the HDE (12).
The cleavage reaction is catalyzed by CPSF-73 (14), a member of the metallo-β-lactamase family of mostly hydrolytic enzymes that utilize zinc ions during catalysis (2, 6). Following the initial endonucleolytic cleavage, CPSF-73 remains associated with the newly formed 5′ end and uses its processive 5′ exonuclease activity to degrade the downstream cleavage product (DCP) (38). This activity results in liberation of the U7 snRNP from the HDE and in vivo may additionally facilitate termination of transcription by RNA polymerase II. Cleavage of histone pre-mRNAs likely also requires CPSF-100, a close homologue of CPSF-73, and a protein of unknown function, symplekin (23). Interestingly, the same three proteins are also required for cleavage of pre-mRNAs resulting in formation of polyadenylated mRNAs (25). This surprising finding raises the question of how the two vastly different pre-mRNAs and their associated processing complexes recruit the same catalytic component to the cleavage site.
The major goal of this study was to identify new components and interactions within the processing machinery, including the previously proposed factor(s) that rigidifies the region of histone pre-mRNA spanning from the cleavage site to the HDE (33, 34). To achieve this goal, we used a 5′ biotin-tagged fragment of histone pre-mRNA to purify potential components of the 3′-end processing complex. We also carried out UV-cross-linking experiments and detected previously unreported interactions that occur between components of the processing complex and the RNA substrate.
MATERIALS AND METHODS
Preparation and labeling of RNA substrates.
The 86-nucleotide H2a and H1t pre-mRNA substrates were synthesized by T7 transcription, gel purified, treated with calf intestinal phosphatase, and labeled at the 5′ end with 32P by using T4 polynucleotide kinase (16). All other RNAs were synthesized by Dharmacon (Lafayette, CO).
In vitro processing and affinity purification of processing complexes.
Nuclear extracts were prepared from mouse myeloma cells, as described previously (16). A typical processing reaction mixture contained 0.05 pmol of a 5′-labeled RNA substrate and the following components in the final volume of 10 μl: 20 mM EDTA (pH 8), 15 mM Tris (pH 8), 15 mM glycerol, 75 mM KCl, and 0.375 mM dithiothreitol. The processing reaction was blocked by 100 ng of a 2′O-methyl oligonucleotide complementary to the first 15 nucleotides of the mammalian U7 snRNA (anti-mU7). A 2′O-methyl oligonucleotide directed to the first 17 nucleotides of the Drosophila U7 snRNA was used as a negative control (anti-dU7). Processing samples were incubated for 90 min at 32°C, treated for 60 min with proteinase K, and analyzed with 8%/7 M urea polyacrylamide gels. For isolation of processing complexes, the reaction mixture was scaled up 1,000-fold and contained 125 pmol of a 5′ biotinylated RNA substrate and 1.0 mg of Saccharomyces cerevisiae tRNA (Invitrogen). Unless otherwise indicated, the cleavage reaction was inhibited by adding NP-40 to give a final concentration of 0.2%. Each sample was preincubated on ice for 10 min, transferred to 32°C for an additional 10 min, and spun down to remove any precipitates. The supernatant was subsequently rotated for 2 h with 50 μl of streptavidin agarose beads (Sigma). The beads were collected by gentle spinning, rinsed twice with a processing buffer containing 0.2% NP-40, and rotated in the same buffer for 2 h. Proteins adsorbed on the beads were recovered by boiling in a sodium dodecyl sulfate (SDS)-containing dye, separated in an SDS-polyacrylamide gel, stained with Coomassie blue, and analyzed by mass spectrometry (MS). A small aliquot of the recovered proteins was additionally analyzed by Western blotting.
MS analysis.
In-gel digestion with porcine trypsin (Promega) was conducted as described previously (30). Extracted peptides were lyophilized and reconstituted in 5 μl of 50% methanol and 0.1% formic acid. Peptides were mixed 1:1 with a saturated solution of recrystallized α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 0.1% trifluoroacetic acid as the matrix and analyzed by matrix-assisted laser desorption ionization-time of flight MS using an ABI 4700 matrix-assisted laser desorption ionization-tandem time of flight mass spectrometer (Applied Biosystems Inc.). Mass spectra were collected automatically, and the 10 most intense peaks in the spectrum were analyzed by tandem MS (MS-MS). The combined MS and MS-MS spectra were searched against the mouse protein sequence database by using the MASCOT protein identification server (Matrix Science). Only those identifications with both a significant protein score (for MS) and a significant total ion score (for MS-MS) are reported.
Mobility shift assay.
The wild-type (WT) or mutant SL RNAs were labeled at the 5′ end and used as probes in the mobility shift assay, as described previously (13, 37). All proteins were expressed using the baculovirus expression system, premixed with the probe in the presence of 20 mM EDTA, and analyzed by electrophoresis with 6% native polyacrylamide gels.
UV-cross-linking and immunoprecipitation.
UV-cross-linking experiments were carried out as previously described (14). Processing reaction mixtures were prepared in a final volume of 20 μl and additionally contained 10 μg of yeast tRNA (Invitrogen). Following a 10-min incubation at 32°C, 15-μl aliquots were irradiated at room temperature with UV (1 J) while the remaining portion of each reaction mixture was incubated for an additional 80 min to control for processing efficiency. UV-irradiated samples were treated for 5 h with RNase T1, boiled in an SDS-containing dye, and directly analyzed by SDS-polyacrylamide gel electrophoresis. Immunoprecipitation experiments under native or denaturing conditions were carried out as described previously (14).
Antibodies.
Anti-mouse 3′hExo and anti-human Lsm11 antibodies were generated in rabbits by Pacific Immunology (Ramona, CA). The anti-mouse 3′hExo antibody was directed to the C-terminal peptide of mouse 3′hExo (PVEGAPAPQMPHSRK), whereas the anti-Lsm11 antibody was generated against bacterially expressed protein consisting of the N-terminal glutathione S-transferase followed by the first 170 amino acids of human Lsm11. The two antibodies were affinity purified using their respective antigens. The following primary antibodies were obtained from commercial sources: rabbit anti-human SNRPD3 (Atlas Antibodies), mouse monoclonal anti-human Lsm10 (Bio Matrix Research), and mouse monoclonal anti-human SmB (Sigma).
RESULTS
Affinity purification of stable processing complexes assembled on histone pre-mRNA.
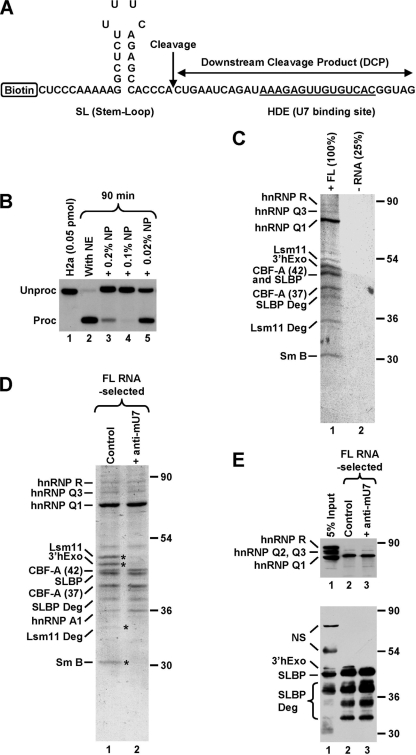
In vitro processing of histone pre-mRNA occurs without a detectable lag after the transfer of reaction mixtures from 0°C to 32°C (16, 20). This high rate makes isolating native processing complexes difficult since they are immediately disassembled following cleavage. To reduce the rate of the cleavage reaction, we used low concentrations of NP-40. This mild detergent at 0.2% reduces processing in vitro to less than 10% of the control efficiency (Fig. 1B, lane 3). Surprisingly, a concentration of 0.1% inhibits processing more strongly, while the lowest concentration of 0.02% shows only modest inhibition (Fig. 1B, lanes 4 and 5).
FIG. 1.
Affinity purification of a stable processing complex assembled on histone pre-mRNA. (A) Sequence of the 62-nucleotide FL RNA containing all elements required for 3′-end processing. The cleavage site is indicated with a vertical arrow, and the HDE that base pairs with the U7 snRNA is underlined. The biotin tag is located at the 5′ end of the RNA. (B) In vitro 3′-end processing of a 5′-labeled 86-nucleotide substrate RNA (0.05 pmol/reaction) that is nearly identical with the RNA shown in panel A but lacks the biotin tag and contains additional nucleotides at the 5′ and 3′ ends. Processing was carried out with a mouse nuclear extract (NE) under control conditions (lane 2) or in the presence of the indicated concentrations of NP-40. Lane 1 contains only the input RNA. (C) Coomassie-stained SDS-polyacrylamide gel containing proteins of a mouse nuclear extract adsorbed on streptavidin beads in the presence of 125 pmol of the biotinylated FL RNA (lane 1) or in its absence (lane 2). Processing complexes were formed in the presence of 0.2% NP-40 and subsequently purified on streptavidin beads. The sample lacking the RNA was four times smaller than the RNA-containing sample. Protein identities determined by MS are shown to the left, and size markers are to the right. In addition to FL proteins, MS identified a number of their degradation products. (D) Coomassie-stained SDS-polyacrylamide gel containing proteins of a mouse nuclear extract that associated with the biotinylated FL RNA (125 pmol) in the absence (lane 1) or presence (lane 2) of the anti-mU7 oligonucleotide. Protein bands present only in lane 1 are indicated with asterisks. (E) A fraction of material shown in D was analyzed by Western blotting with anti-hnRNP Q/R (top) or mixed antibodies to 3′hExo and SLBP (bottom). “NS” indicates cross-reacting proteins.
In order to affinity purify processing complexes, we scaled up the processing reaction mixture to 10 ml (an equivalent of 1,000 standard reaction mixtures carried out with a radioactive substrate). Of this volume, 75% was contributed by a nuclear extract prepared from ∼7.5 liters of mouse myeloma cell culture. The reaction mixture contained 2.5 μg (∼125 pmol) of a 62-nucleotide, 5′ biotinylated RNA that we refer to as the full-length (FL) RNA (Fig. 1A). This amount of RNA resulted in an approximately 2.5-fold-higher concentration of the substrate than in a standard processing reaction mixture (Fig. 1B). To measure the background of proteins nonspecifically associated with the beads, we prepared a smaller mock sample (25% volume) without the RNA substrate (Fig. 1C, lane 2). Both samples contained 0.2% NP-40, which effectively reduces the nonspecific background. Importantly, due to the presence of larger-than-normal amounts of the RNA substrate, this concentration of detergent resulted in virtually no processing after 60 min of incubation at 32°C (see Fig. S1, lane 9, in the supplemental material). To form and purify processing complexes, the two samples were incubated at 32°C for only 10 min, then placed on ice for an additional 10 min, and finally incubated with streptavidin beads. Proteins collected on the beads were resolved by SDS-polyacrylamide gel electrophoresis, detected by staining with Coomassie blue, and identified by MS.
The mock control without RNA produced almost no background (Fig. 1C, lane 2), whereas a number of distinct protein bands were detected in the sample containing the 5′ biotinylated RNA (Fig. 1C, lane 1). Remarkably, we observed a readily detectable amount of Lsm11, demonstrating that a significant fraction of the RNA substrate was associated with the U7 snRNP and hence not cleaved during the incubation time. In addition, we isolated stoichiometric amounts of the 30-kDa SmB protein that, together with Lsm11, is part of the U7-specific Sm ring. As expected, the FL RNA associated with SLBP, which binds to the SL sequence upstream of the cleavage site. In addition to proteins specific for histone pre-RNA, we isolated larger amounts of two general RNA binding proteins: hnRNP Q and CArG box-binding factor A (CBF-A). hnRNP Q, also known as Gry-rbp, is highly similar to hnRNP R and exists in three different isoforms that play a role in a number of RNA processing events, including splicing (28). CBF-A, also known as hnRNP A/B, exists in two alternatively spliced variants of 42 and 37 kDa and has been implicated in various cellular processes, including regulation of transcription on several genes (9). Finally, among the proteins associated with the FL RNA, we identified 3′hExo. We have previously shown that 3′hExo has a strong affinity to the SL sequence at the 3′ end of the mature histone mRNA but does not bind to the same sequence located internally, as it is in the FL RNA (13). Therefore, the affinity purification of 3′hExo was surprising.
To determine which of the identified proteins associate with the histone pre-mRNA in a U7-dependent manner, we repeated the pulldown experiment including a sample in which the nuclear extract was preincubated with 15 nmol of anti-mU7, a 2′O-methyl oligonucleotide complementary to the first 15 nucleotides of the mammalian U7 snRNA. The anti-mU7 oligonucleotide prevents U7 snRNP from binding to the HDE and completely blocks processing. As expected, both Lsm11 and SmB proteins were missing in the sample containing the anti-mU7 oligonucleotide (Fig. 1D, lane 2) and they were both readily detectable in the regular sample where the U7 snRNP was allowed to bind to the substrate RNA (Fig. 1D, lane 1). In addition to these two proteins, a third U7-dependent protein was detected after the gel was restained with silver. The identity of this protein has not been determined by MS, but its size of ∼15 kDa suggests that it was the SmD3 component of the of the U7-specific Sm ring (not shown). As expected, SLBP was found in both samples since binding of this protein to the SL is not dependent on the U7 snRNP. Binding of hnRNP Q and both variants of CBF-A to histone mRNA was also not affected by the presence of the anti-mU7 oligonucleotide. This was in contrast to what was found for 3′hExo, which was detected in stoichiometric amounts with Lsm11 and SLBP in the control sample but was missing in the sample containing the anti-mU7 oligonucleotide. This result suggested that 3′hExo, in addition to efficiently binding to the mature histone mRNA, can also be recruited to histone pre-mRNA in a U7-dependent manner.
The identities of SLBP, hnRNP Q, CBF-A, and 3′hExo as determined by MS were additionally confirmed by Western blotting (Fig. 1E). All proteins, with the exception of 3′hExo, were detected using antibodies readily recognizing both mouse and human proteins. The antibody against 3′hExo was originally generated against the C-terminal peptide of the human orthologue and only weakly cross-reacted with the mouse protein (Fig. 1E, bottom panel).
Mapping the interaction sites in histone pre-mRNA.
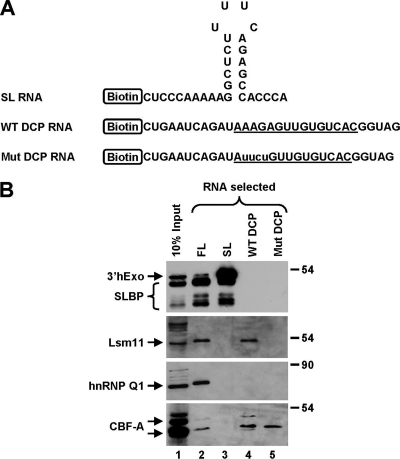
To determine what regions of the 62-nucleotide FL RNA interact with the isolated proteins and to analyze the specificity of each interaction, we used two shorter 5′ biotinylated RNAs that corresponded to the two halves generated by the cleavage reaction: the 31-nucleotide upstream product ending with the SL followed by the terminal ACCCA single-stranded tail and the 31-nucleotide DCP containing the HDE (Fig. 2A). In addition to the WT DCP RNA, we used a mutant DCP version in which the purine core of the HDE was changed from AAGA to UUCU. This mutation abolishes binding of the U7 snRNP to the DCP (14). Each substrate RNA (∼12.5 pmol) was incubated with 750 μl of a mouse nuclear extract in the presence of 0.2% NP-40 and in a total volume of 1.0 ml (the equivalent of 100 regular reaction mixtures), and bound proteins were analyzed by Western blotting. To increase the sensitivity of detection of 3′hExo, we generated an antibody directed to the last 15 amino acids of the mouse protein. The new antibody recognized a protein in a mouse nuclear extract of about 45 kDa, consistent with the molecular size of the mouse 3′hExo protein.
FIG. 2.
Mapping the interaction sites in histone pre-mRNA. (A) Sequences of biotinylated RNA halves. The U7-binding site is underlined, and nucleotide substitutions within the purine core of the mutant DCP RNA are indicated with lowercase letters. (B) Western blot analysis of affinity-purified complexes assembled on 5′ biotinylated RNAs (12.5 pmol), as indicated at the top of each lane. Processing complexes were formed in the presence of 0.2% NP-40 by using RNA substrates shown in panel A and subsequently purified on streptavidin beads. The presence of individual proteins bound to each RNA was detected by specific antibodies. In the top panel, anti-mouse 3′hExo and anti-SLBP antibodies were mixed. Lane 1 contains 10% of the extract used to assemble processing complexes on each RNA species.
Judging by Western analysis, a relatively small fraction of the input 3′hExo protein associated with the 62-nucleotide FL pre-mRNA (Fig. 2B, lane 2). In comparison, the SL RNA was bound by a much larger amount of 3′hExo, reflecting the high affinity of this protein to the mature 3′ end, particularly in the presence of SLBP (Fig. 2B, lane 3). Neither the WT nor the mutant versions of the DCP were associated with 3′hExo (Fig. 2B, lanes 4 and 5). SLBP was able to bind the FL RNA and the SL RNA (Fig. 2B, lanes 2 and 3). This is consistent with the fact that SLBP as a component of 3′-end processing machinery binds to the 5′ side of the SL structure in histone pre-mRNA and remains associated with the same site after cleavage. Like 3′hExo, SLBP did not bind to the DCP RNAs (Fig. 2B, lanes 4 and 5).
In contrast to 3′hExo and SLBP, Lsm11 as a component of U7 snRNP associated with the FL RNA and the WT DCP RNA (Fig. 2B, lanes 2 and 4) but not with the SL RNA (lane 3). Lsm11 was also not detected on the mutant DCP RNA, consistent with the inability of this mutant RNA to efficiently base pair with the U7 snRNA (Fig. 2B, lane 5). hnRNP Q associated only with the FL RNA (Fig. 2B, lane 2) and was not isolated with either the downstream (DCP) or the upstream (SL) cleavage products (Fig. 2B, lanes 3 and 4), suggesting that this protein requires the cleavage site and/or its flanking sequences for binding. On the other hand, the two variants of CBF-A associated only with the FL and the two DCP RNAs (Fig. 2B, lanes 2, 4, and 5), demonstrating that the binding was not affected by the UUCU mutation.
Cooperative binding of 3′hExo and SLBP to the SL RNA.
The above-mentioned experiment indicated that 3′hExo binds primarily to the mature 3′ end of histone mRNA, but a fraction of 3′hExo is also recruited in a U7-dependent manner to pre-mRNA prior to cleavage (see also below). Since interactions between proteins and RNA targets that are detected in vitro may not occur in vivo and simply result from redistribution of cell components following cell lysis (27), we looked for evidence that the binding of 3′hExo to the mature histone mRNA 3′ end is biologically relevant.
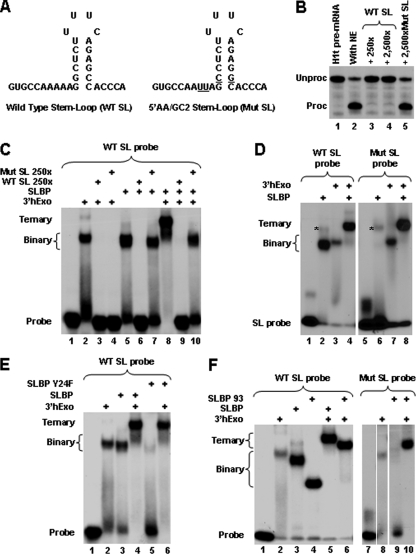
In vitro, 3′hExo and SLBP occupy two opposing sides of the SL structure at the 3′ end of the mature histone mRNA and each protein can bind to its target individually or simultaneously with the other (13, 37). One of the RNA binding determinants for 3′hExo is the single stranded ACCCA tail. SLBP binds to the 5′ side of the SL and requires two adenosines, located 2 and 3 nucleotides upstream of the stem (13). In addition, each protein requires specific nucleotides in the stem and in the loop, and mutating these nucleotides reduces or abolishes binding (3, 13). We mutated nucleotides critical for binding of SLBP to the SL RNA by inverting the second GC base pair in the stem and replacing the two essential adenosines located 2 and 3 nucleotides upstream of the stem with uridines (Fig. 3A). On the basis of our previous analysis, these mutations were expected to selectively abolish binding of SLBP to the SL without having a major effect on binding of 3′hExo (13). We first tested whether the resulting mutant SL RNA can bind SLBP and hence inhibit the in vitro processing reaction. Processing of the H1t pre-mRNA, which requires SLBP (16), was not affected by a 2,500 molar excess (relative to the level for the substrate RNA) of the mutant SL RNA (Fig. 3B, lane 5). At the same time, a 10-fold-lower concentration of the WT SL RNA was sufficient to fully inhibit processing of this substrate (Fig. 3B, lane 3). We conclude that the mutant SL RNA does not bind SLBP.
FIG. 3.
Cooperative binding of 3′hExo and SLBP to the SL RNA. (A) Sequences of the WT SL RNA and its mutant (Mut SL), containing nucleotide substitutions (underlined). (B) Ability of a molar excess of the WT and Mut SL RNA competitors to affect cleavage of the H1t histone pre-mRNA by sequestering SLBP present in the nuclear extract. Each competitor was mixed with the 5′-labeled H1t substrate and incubated with a mouse nuclear extract for 1 h. The input H1t pre-mRNA is shown in lane 1. (C) Ability of a molar excess of the WT and Mut SL RNA competitors to affect formation of binary and ternary complexes formed on the 5′-labeled WT SL probe by SLBP and/or 3′hExo. Each competitor was mixed with the 5′-labeled WT SL probe in the presence of 20 mM EDTA and incubated on ice for 30 min with baculovirus-expressed SLBP and/or 3′hExo. Complexes were analyzed with a 6% native polyacrylamide gel. (D) Ability of the 5′-end-labeled Mut SL RNA to form binary and ternary complexes with SLBP and/or 3′hExo. The Mut SL RNA probe was incubated on ice for 30 min with the baculovirus-expressed SLBP and/or 3′hExo in the presence of 20 mM EDTA, and complexes were analyzed with a 6% native polyacrylamide gel. A nonspecific complex formed by a contaminating protein present in the SLBP preparation (lanes 2 and 6) is indicated with an asterisk. (E) Ability of 3′hExo to recruit the Y24F SLBP mutant to the WT SL RNA, giving rise to a ternary complex containing all three components (lane 6). Formation of a binary and ternary complex containing the WT SLPB is shown for comparison in lanes 3 and 4. (F) Mapping of the region in SLBP required for the cooperative interaction with 3′hExo. The ability of the 93-amino-acid region of SLBP to form a ternary complex with 3′hExo and the Mut SL probe is shown (lane 10). Formation of a binary and ternary complex containing the WT SLPB and the 93-amino-acid SLBP mutant with the WT SL probe is shown in lanes 2 to 6.
We next analyzed the ability of the mutant SL RNA to compete with 5′-end-labeled WT SL RNA for binding to SLBP in the band shift assay. SLBP and 3′hExo each form stable complexes with the WT SL RNA probe that can be resolved with a native polyacrylamide gel (13, 37). 3′hExo has a larger mass than SLBP and consistently forms a complex with the WT SL RNA that migrates slightly more slowly than the SLBP-SL complex (Fig. 3C, compare lanes 2 and 5). Simultaneous binding of both proteins to their respective targets on the same molecule results in the formation of a ternary complex that migrates more slowly than either binary complex (Fig. 3C, lane 8). Formation of the binary complex between the 5′-labeled WT SL probe and 3′hExo was readily competed by a 250-fold molar excess of either the unlabeled WT or the mutant SL RNAs (Fig. 3C, lanes 3 and 4). Thus, the mutations in the mutant SL RNA do not significantly affect the interaction of the RNA with 3′hExo. In contrast, the formation of the binary complex between the 5′-labeled SL RNA and SLBP was unaffected by the mutant SL RNA (Fig. 3C, lane 7), further demonstrating that this mutant RNA does not interact with SLBP. The selective nature of the mutations introduced on binding of SLBP was additionally supported by the effect of the mutant SL competitor on the formation of the ternary complex. In the presence of a high molar excess of this competitor, the ternary complex was not formed, due to sequestration of 3′hExo. As a result, the WT SL probe formed a binary complex only with SLBP (Fig. 3C, lane 10).
To directly test the ability of the mutant SL RNA to bind 3′hExo and SLBP, we labeled this RNA at the 5′ end and used it as a probe in the band shift assay (Fig. 3D). As expected from the competition experiments, the mutant SL RNA did not form a stable binary complex with SLBP (Fig. 3D, compare lanes 2 and 6) while binding of 3′hExo to this RNA was unaffected (Fig. 3D, compare lanes 3 and 7). Remarkably, the mutant SL probe formed the ternary complex containing both SLBP and 3′hExo, and the efficiency of the assembly was the same as that for the WT SL probe (Fig. 3D, compare lanes 4 and 8). Thus, in the presence of 3′hExo, the mutant SL RNA becomes a suitable binding target for SLBP. This effect strongly suggests that the two proteins cooperate in recognizing their respective targets on the SL RNA.
We next tested whether 3′hExo can facilitate binding of a mutant version of SLBP to the WT SL RNA. A tyrosine at position 24 of the 73-amino-acid RNA binding domain (RBD) of SLBP plays an important role in the recognition of the SL RNA by SLBP (11), and replacement of this residue with an alanine reduces the binding efficiency to ∼15% relative to that of the WT protein (Fig. 3E, compare lanes 3 and 5). Importantly, the ternary complex containing 3′hExo and the Y24A SLBP mutant assembled on the WT SL probe with the same efficiency as did the ternary complex containing WT SLBP (Fig. 3E, compare lanes 4 and 6). Thus, 3′hExo alleviates the impaired interaction between SLBP and the SL RNA caused by mutations within either the target RNA or SLBP itself.
As determined by the directed yeast two-hybrid system, 3′hExo and SLBP do not interact unless the two proteins are coexpressed with the SL RNA (13). We reasoned that upon binding to the SL RNA, 3′hExo undergoes structural rearrangements and might actively recruit SLBP to the ternary complex by making direct protein-protein contacts that compensate for the impaired interactions of SLBP with the mutant SL RNA. To better understand the nature of this interaction and to map regions of SLBP necessary for the interaction, we tested various mutants of SLBP in the mobility shift assay. A 93-amino-acid portion of SLBP encompassing the 73-amino-acid RBD and the 20 amino acids following retains the full ability to bind WT SL RNA and to form the ternary complex with 3′hExo (Fig. 3F, lanes 4 and 6). This was expected since the RBD is the only region of SLBP that mediates binding to the RNA. We tested whether the 93-amino-acid region is sufficient for the cooperative interaction with 3′hExo by using the mutant SL RNA, which is not recognized by SLBP as a binding target. While the 93-amino-acid protein was unable to bind the mutant SL RNA (Fig. 3F, lane 9), the same deletion mutant of SLBP formed a stable ternary complex in the presence of 3′hExo (lane 10). Note that in contrast to SLBP and in agreement with our previous studies (37), 3′hExo binds to the SL RNA relatively weakly, as often indicated by the small amount of the fully formed binary complex and by the presence of a faster-migrating smear that results from the dissociation of the complex during electrophoresis (Fig. 3D and F, compare lanes 2 and 3). Yet, 3′hExo always forms a very tight and stable ternary complex in the presence of SLBP. Thus, the cooperativity in binding the SL RNA is mutual, with each protein stabilizing the binding of the other.
We additionally mutated either a 9-amino-acid region of the RBD that is not required for binding to the SL RNA (11) or the 20-amino-acid region that follows the RBD. Both SLBP mutants retained the ability to form a tight ternary complex with 3′hExo and the SL RNA (not shown). Thus, it is possible that the interaction between the two proteins is mediated by residues of SLBP that participate in RNA recognition.
U7-dependent interactions in the region between the cleavage site and the HDE.
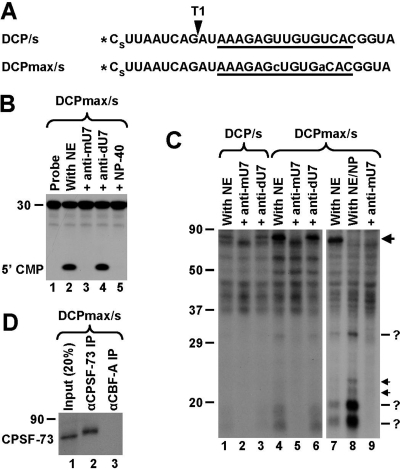
Studies with mutant histone pre-mRNAs containing various insertions between the cleavage site and the HDE suggested that this region becomes rigidified during processing by a component of the processing machinery and that binding of the U7 snRNP acts as a ruler to define the cleavage site (34). To identify proteins that interact with this region, we carried out UV-cross-linking studies using a 5′ labeled RNA that corresponds to the DCP. The 30-nucleotide RNA begins immediately 3′ of the cleavage site and, compared to the corresponding region in the natural H2a pre-mRNA, has the first guanosine replaced with uridine (Fig. 4A). As a result, digestion of the DCP RNA with RNase T1 generates a 9-nucleotide labeled fragment that spans almost the entire region between the 5′ end and the HDE. In addition, a phosphorothioate modification was placed within the first phosphodiester bond (Fig. 4A) to slow down catalysis and hence to allow UV cross-linking of transiently associated components of the processing complex (14). In a mouse nuclear extract, the DCP RNA is rapidly degraded to mononucleotides by the 5′-to-3′ exonuclease activity of CPSF-73, which depends on binding of the U7 snRNP to the HDE (38). In parallel to the DCP RNA, we used a related substrate containing a 2-nucleotide substitution within the HDE (Fig. 4A) that allowed for an uninterrupted 15-bp duplex with the U7 snRNA and thus more-efficient recruitment of the U7 snRNP. Both substrates, referred to as DCP/s and DCPmax/s, respectively, were degraded in a mouse nuclear extract with comparable efficiencies, and the degradation in each case was blocked by the presence of the anti-mU7 oligonucleotide or 0.1% NP-40 (Fig. 4B, lanes 3 and 5, and data not shown).
FIG. 4.
U7-dependent interactions in the region between the cleavage site and the HDE. (A) Sequence of the DCP/s and DCPmax/s substrates containing a single radioactive phosphate at the 5′ end (asterisk) and a phosphorothioate modification (s) between nucleotides 1 and 2. The HDE is underlined, and the site recognized by RNase T1 is indicated with an arrowhead. Nucleotide substitutions within the HDE of the DCPmax/s RNA that improve the base pair interaction with the U7 snRNA are indicated with lowercase letters. (B) In vitro degradation of the DCPmax/s RNA. Samples were incubated for 90 min at 32°C either under control conditions (lane 2) or in the presence of the indicated reagents (lanes 3 to 5). The length of the RNA substrate and the position of labeled mononucleotide are indicated to the left. (C) UV-cross-linking using the DCP/s (lanes 1 to 3) and DCPmax/s (lanes 4 to 9) RNAs. Samples were preincubated for 10 min at 32°C either under control conditions or in the presence of the indicated oligonucleotides. Samples in lanes 8 and 9 additionally contained 0.1% NP-40. Following incubation, samples were UV irradiated, treated with RNase T1, and separated in an SDS-polyacrylamide gel. The 80-kDa cross-link at the top of the gel is indicated with the large arrow. Low-molecular-weight U7-dependent cross-links are indicated with question marks (more-intense bands) and arrows (less-intense bands). (D) Immunoprecipitation (IP) of the 80-kDa cross-linked protein with anti-CPSF-73 antibody. A processing reaction mixture containing the 5′-labeled DCPmax/s substrate was UV-irradiated, treated with RNase T1, denatured by boiling in the presence of SDS, and incubated with the anti-CPSF-73 antibody (lane 2) or the anti-CBF-A antibody (lane 3). Precipitated proteins were resolved in an SDS-polyacrylamide gel and detected by autoradiography. The input (20%) is shown in lane 1.
The DCP/s and DCPmax/s RNAs were incubated with a mouse nuclear extract for 10 min at 32°C either under regular conditions or in the presence of the anti-mU7 oligonucleotide. The samples were subsequently UV irradiated, digested with an RNase T1, and analyzed with SDS gels for the presence of cross-linked, radioactively labeled proteins. A U7-dependent cross-link migrating at about 80 kDa was detected with both the DCP/s and the DCPmax/s RNAs (Fig. 4C, lanes 1 and 4), and its formation was sensitive to the presence of the anti-mU7 oligonucleotide (lanes 2 and 5) but unaffected by the presence of the control anti-dU7 oligonucleotide (lanes 3 and 6). The identity of this cross-linked protein was determined by immunoprecipitation to be CPSF-73 (Fig. 4D, lane 2). The cross-linking efficiency of CPSF-73 was much higher with the DCPmax/s RNA (Fig. 4C, compare lanes 1 and 4), likely reflecting the stronger binding of the U7 snRNP to the improved HDE and therefore more-efficient recruitment of CPSF-73 to the RNA substrate.
Interestingly, CPSF-73 was not the only protein cross-linked to the RNA in a U7-dependent manner. In the reaction mixture containing the DCPmax/s substrate, a group of three major U7-dependent cross-links and two minor cross-links were evident (Fig. 4C, lanes 4 and 6). The same major cross-links were also detected with the DCP/s substrate, although they were much less intense and visible only after longer exposure times (not shown). Since each radioactively labeled protein was covalently linked to a 9-nucleotide RNA fragment that corresponds to a mass of about 3 kDa, the approximate molecular masses of the major cross-linked proteins were estimated at 28 to 29 kDa, 16 to 17 kDa, and 14 to 15 kDa.
To determine whether cross-linking of the three major proteins depends on the ongoing cleavage reaction in the same fashion as does cross-linking of CPSF-73, we repeated the experiment in the presence of NP-40. Addition of this detergent to 0.1% almost completely blocks recruitment of CPSF-73 to the substrate (Fig. 4C, lane 8) and inhibits degradation of the DCP (Fig. 4B, lane 5), although it has no effect on binding of the U7 snRNP to the HDE (Fig. 2B, lane 4). Significantly, the presence of 0.1% NP-40, while inhibiting the degradation, greatly enhanced the intensity of all three major cross-links (Fig. 4C, lane 8), and their formation remained sensitive to the presence of the anti-mU7 oligonucleotide (lane 9).
Identification of U7-dependent cross-links.
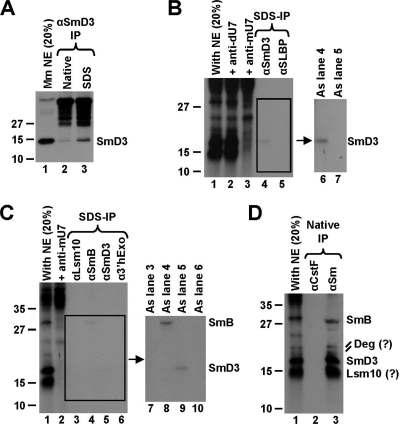
The sizes of the novel U7-dependent cross-linked proteins, ranging from approximately 15 to 30 kDa, and their interaction with the DCP in the presence of NP-40 suggested that these proteins may be components of the U7-specifc Sm ring. The 30 kDa cross-linked protein is likely SmB, which is the largest of the common Sm proteins associated with the U7 snRNA and was readily detected by MS in processing complexes assembled in the presence of NP-40 on the FL RNA (Fig. 1C and D). The fastest-migrating protein may in turn be Lsm10, while the protein of intermediate mobility is likely SmD3. The two minor cross-links migrating slightly over 20 kDa are likely degradation products of the SmB protein.
To determine the identities of the cross-linked proteins, we carried out immunoprecipitation experiments using antibodies specific to individual components of the U7-specific ring. A monoclonal antibody against SmD3 recognizes a major band at approximately 15 kDa by Western blot analysis, in agreement with the molecular mass of SmD3 (Fig. 5A, lane 1). As determined by the immunoprecipitation/Western assay, the antibody is not suitable for immunoprecipitation under native conditions (Fig. 5A, lane 2) but precipitates the SmD3 antigen with a low efficiency under denaturing conditions containing SDS (lane 3). Importantly, the presence of SDS has the advantage over the native conditions, as it completely disrupts the U7 Sm ring, thus allowing immunoprecipitation of the SmD3 protein alone rather than as a part of the entire complex. The antibody against SmD3 selectively precipitated the expected cross-linked protein (Fig. 5B, lanes 4 and 6, and C, lanes 5 and 9), and the amount of the precipitated material was consistent with the low efficiency of SmD3 precipitation (Fig. 5A, lane 3). An antibody directed against SLBP did not precipitate any radioactive protein (Fig. 5B, lanes 5 and 7), confirming the specificity of the anti-SmD3 precipitation. On the basis of these results, we conclude that the U7-dependent cross-linked protein migrating at 16 to 17 kDa is SmD3.
FIG. 5.
Identification of U7-dependent cross-links. (A) Immunoprecipitation (IP) of SmD3 from a mouse nuclear extract by a specific anti-SmD3 mouse monoclonal antibody under native (lane 2) or denaturing (lane 3) conditions. Lane 1 contains 20% of the extract used for each immunoprecipitation. (B) Immunoprecipitation of cross-linked proteins by the anti-SmD3 antibody. A processing reaction mixture containing the 5′-labeled DCPmax/s substrate was UV irradiated, treated with RNase T1, denatured by boiling in the presence of SDS, and incubated with the anti-SmD3 (lanes 4 and 6) or anti-SLBP (lanes 5 and 7) antibody. Precipitated proteins were resolved in an SDS-polyacrylamide gel and detected by autoradiography. The input (20%) is shown in lane 1. An image of the boxed gel fragment observed after a longer exposure time (lanes 4 and 5) is shown to the right. Cross-links formed in the presence of the anti-dU7 and the anti-mU7 oligonucleotides are shown in lanes 2 and 3, respectively. (C) Immunoprecipitation of cross-linked proteins by antibodies indicated at the top of each lane. The experiment was carried out as described for panel B. The input (20%) is shown in lane 1. An image of the boxed gel fragment observed after a longer exposure time (lanes 3 to 6) is shown to the right. Cross-links formed in the presence of the anti-mU7 oligonucleotide are shown in lane 2. (D) Immunoprecipitation of cross-linked proteins by the anti-Sm Y12 monoclonal antibody under native conditions. A monoclonal antibody against CstF-77 was used as a negative control (lane 2). The input (20%) is shown in lane 1.
By Western analysis, a monoclonal antibody against SmB recognizes a protein that migrates at about 28 kDa (not shown). This is consistent with the molecular mass of SmB and the position of this protein in Coomassie-stained gels containing components of the processing machinery (Fig. 1C and D). Under denaturing conditions, the same antibody selectively precipitated the largest cross-linked protein, thus confirming its identity as SmB (Fig. 5C, lanes 4 and 8). An antibody against 3′hExo used as a negative control did not precipitate any protein (Fig. 5C, lanes 6 and 10). To determine the identities of the smallest of the cross-linked proteins predicted to be Lsm10, we used an anti-Lsm10 monoclonal antibody. This antibody does not detect the endogenous protein on Western blots but was successfully used for detection of U7 snRNP and histone locus bodies in human cells by immunofluorescence (19). Under both native conditions (not shown) and denaturing conditions, the anti-Lsm10 antibody did not precipitate any radioactive protein (Fig. 5C, lanes 3 and 7).
We next investigated whether the smallest cross-linked protein is a component of the U7-snRNP by using the Y12 anti-Sm monoclonal antibody, which recognizes symmetrical dimethylarginines in Sm proteins (5) and under native conditions precipitates entire snRNPs. Under these conditions, the Y12 antibody precipitated all proteins cross-linked to the DCPmax/s substrate in a U7-dependent manner, including the minor putative SmB degradation products, although it failed to precipitate other proteins that associated with the same substrate independently of the U7 snRNP (Fig. 5D, lane 3). We conclude that the smallest cross-linked protein is a component of the U7 snRNP and, on the basis of its size, is likely Lsm10, which migrates immediately below SmD3 (32).
U7-dependent interaction in the SL region.
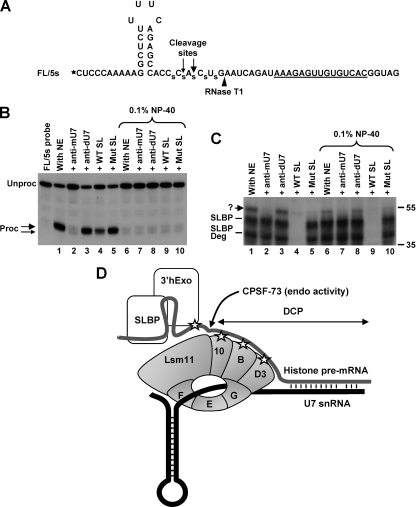
An intriguing observation from the above-mentioned cross-linking studies is that while the DCP RNA makes close contacts with SmD3, SmB, and Lsm10, we never detected a cross-link that would be consistent with the size of Lsm11. One possibility is that Lsm10 interacts with the region located immediately 3′ of the cleavage site that is present in the DCP RNA but that Lsm11 binds nucleotides 5′ of the cleavage site that are missing in the DCP RNA. We previously used a site-specifically labeled FL RNA substrate (FL/5s) that contained a single radioactive phosphate between the two last nucleotides of the stem and five phosphorothioate modifications in the region flanking the cleavage site (14). Interestingly, with this substrate, we observed the formation of a U7-dependent cross-link that migrated slightly more slowly than the cross-link formed with SLBP (14), a mobility consistent with the size of 3′hExo or Lsm11 (Fig. 1C and D).
To characterize this cross-link in more detail and to determine whether it was formed with 3′hExo or Lsm11, we used a similar substrate that lacked a guanosine upstream of the SL and contained a radioactive label at the 5′ end (Fig. 6A). After 90 min of incubation at 32°C, the labeled FL/5s RNA was processed with an efficiency of ∼60% (Fig. 6B, lane 1). This efficiency is significantly lower than the typical ∼95% observed with substrates lacking phosphorothioate modifications near the cleavage site (Fig. 1B, lane 2). Due to the presence of phosphorothioate modifications, the FL/5s pre-mRNA was additionally cleaved at a minor site 4 nucleotides past the stem. Processing of the FL/5s pre-mRNA responded to various RNA competitors in the same fashion as did the regular substrate, with the anti-mU7 preventing any cleavage (Fig. 6B, lane 2). The WT SL RNA competitor at a 250-fold molar excess reduced processing of the FL/5s pre-mRNA to 40% (Fig. 6B, lane 4). This is consistent with the fact that all derivatives of the H2a pre-mRNAs are only partially dependent on SLBP (16). The mutant SL RNA, which is unable to bind SLBP while retaining the ability to bind 3′hExo (Fig. 3), had no effect on the processing of this substrate (Fig. 6B, lane 5). This result further supports our initial observation (Fig. 3B) that 3′hExo is not required for 3′-end processing in vitro. We prepared the same set of reaction mixtures with 0.1% NP-40, which completely abolished processing of the FL/5s substrate (Fig. 6B, lanes 6 to 10).
FIG. 6.
U7-dependent interaction in the SL region. (A) Sequence of the FL/5s substrate. The 32P radioactive label at the 5′ end is represented by an asterisk, and the major and minor cleavage sites are indicated with large and small arrows, respectively. The site recognized by RNase T1 is indicated with an arrowhead, and phosphorothioate bonds are represented by “s.” (B) In vitro 3′-end processing of the FL/5s substrate labeled at the 5′ end. Samples were incubated for 90 min at 32°C under control conditions (lanes 1 and 6) or in the presence of a molar excess of the indicated competitors (lanes 2 to 5 and 7 to 10). Samples in lanes 6 to 10 additionally contained 0.1% NP-40. Note that the first lane containing the probe alone is not numbered, so the remaining lanes have the same numbers as the corresponding lanes in panel C. Products generated by cleavage at the major and minor cleavage sites are indicated with large and small arrows, respectively. (C) UV-cross-linking with the FL/5s substrate labeled at the 5′ end. A fraction of each sample shown in panel B, following 10 min of incubation at 32°C, was UV irradiated, treated with RNase T1, and separated in an SDS-polyacrylamide gel. The unknown cross-link is indicated with a question mark. (D) Hypothetical model depicting possible interactions in the stable processing complex assembled on histone pre-mRNA before cleavage. The top (gray) and bottom (black) lines represent histone pre-mRNA and U7 snRNA, respectively. Interactions detected by UV cross-linking are represented by asterisks. The site of cleavage by CPSF-73 is indicated. 3′hExo is actively recruited to the SL structure by the U7 snRNP and possibly by SLBP. After cleavage, 3′hExo tightly associates with the mature 3′ end bound by SLBP.
After a 10-min incubation at 32°C, an aliquot of each processing reaction mixture shown in Fig. 6B was UV irradiated, treated with RNase T1, and analyzed with an SDS-polyacrylamide gel for the presence of radioactively labeled cross-linked proteins. RNase T1 degraded the FL/5s RNA to a radioactive fragment containing the entire SL and extending 3 nucleotides past the cleavage site (Fig. 6A). Note that guanosines within the double-stranded stem are not recognized by the enzyme. Under regular reaction conditions, the 10-min incubation time was sufficient to process about 40% of the input RNA (see Fig. S2A in the supplemental material). After this time, strong cross-links were formed with SLBP and its slightly smaller degradation product (Fig. 6C, lane 1). The identity of these cross-links was confirmed by immunoprecipitation with an anti-SLBP antibody (not shown) (14). As expected, the two SLBP cross-links were competed off by a 250-fold molar excess of the WT SL RNA (Fig. 6C, lane 4) and remained unaffected by the presence of the anti-mU7 oligonucleotide (Fig. 6C, lane 2). In addition, we observed the formation of another cross-link, which migrated at ∼55 kDa, slightly above the SLBP cross-link, a mobility that was consistent with the size of either Lsm11 or 3′hExo. Formation of this previously unidentified cross-link was prevented by the anti-mU7 oligonucleotide and by a 250-fold molar excess of the WT SL RNA competitor (Fig. 6C, lanes 2 and 4, respectively). This behavior suggested that the 55-kDa cross-link is 3′hExo and results from the postcleavage binding of the protein to the upstream product terminating with the SL. Since the antibody against 3′hExo was not active in immunoprecipitations (not shown), we tested the identity of the 55 kDa cross-link by using the mutant SL RNA, which selectively binds 3′hExo. This RNA competitor had no effect on the processing reaction (Fig. 6B, lane 5) or the formation of the two SLBP cross-links but nearly completely eliminated the 55-kDa cross-link (Fig. 6C, lane 5), strongly suggesting that it indeed is 3′hExo.
Interestingly, in the presence of 0.1% of NP-40, we still detected the 55 kDa cross-link, and its formation was prevented by the anti-mU7 oligonucleotide (Fig. 6C, lanes 6 and 7). Note that there is no detectable processing of the FL/5s substrate after 90 min of incubation in the presence of 0.1% NP-40 (Fig. 6B, compare lanes 6 and 7; see also Fig. S2B in the supplemental material) and that samples were UV irradiated after 10 min of incubation. This result indicates that 3′hExo is actively recruited to histone pre-mRNA prior to cleavage in a manner dependent on the U7 snRNP, thus confirming our initial observations from the affinity purification experiments. In addition, the lack of any cross-link consistent with the size of Lsm11 suggests that this component of the U7-specific Sm ring, in contrast to Lsm10, SmB, and SmD3, does not contact the RNA substrate during 3′-end processing.
DISCUSSION
As a result of studies of both yeast and mammalian cells, the mechanism of 3′-end processing by cleavage/polyadenylation is relatively well understood (8, 40). The macromolecular assembly catalyzing this process is conserved throughout eukaryotes and consists of at least 12 protein components that form a complex network of interactions with each other and sequence elements in the pre-mRNA (25). Metazoan histone pre-mRNAs are cleaved at the 3′ end by a distinct process for which many details remain unknown. So far, only five factors have been identified through both biochemical and genetic studies (12), and two of them are common with cleavage/polyadenylation: CPSF-73, which functions as the 3′ endonuclease (14, 26), and symplekin, which plays a yet uncharacterized role in both processing reactions (23). The three remaining factors include SLBP, which binds the SL structure upstream of the cleavage site; the U7 snRNP, which interacts with the HDE downstream of the cleavage site; and ZFP100, which is believed to bridge the two flanking factors. The two processing reactions likely also share CPSF-100, which physically interacts with CPSF-73 (15) and may function as a regulatory subunit of this endonuclease (24). This list is almost certainly incomplete, and further studies are required both for identification of new processing factors and for determination of how they interact with each other within the processing complex to carry out the endonucleolytic cleavage. In this study, we analyzed the composition of a stable processing complex assembled on histone pre-mRNA in the presence of low concentrations of NP-40 that inhibit the cleavage reaction. We also used UV-cross-linking to detect and characterize previously undescribed interactions within the processing complex.
Histone pre-mRNA stably associates with known processing factors.
Among the proteins identified by MS as components of a stable, catalytically inactive complex were known processing factors (SLBP and two proteins of the U7 snRNP, Lsm11 and SmB). In addition, silver staining detected a protein with a molecular size consistent with the third protein of the U7 snRNP (SmD3), although its identity has not been confirmed by MS. Most notable was the absence of other factors that are known or believed to function in 3′-end processing of histone pre-mRNAs (CPSF-73, CPSF-100, symplekin, and ZFP100). Association of these proteins with the histone pre-mRNA may be sensitive to the presence of NP-40, transient in nature, and/or substoichiometric in comparison to what was found for the most abundant and stable components (SLBP and the U7 snRNP).
In addition to processing-specific proteins, we isolated larger amounts of hnRNP Q (28) and CBF-A (9), two multifunctional and abundant proteins involved in various aspects of RNA and DNA metabolism. hnRNP Q seems to interact near the cleavage site, whereas CBF-A binds to the DCP, and this interaction is not abolished by mutating the AAGA purine core. We also isolated smaller amounts of a third general RNA binding protein, hnRNP A1 (Fig. 1C). Our attempts to sufficiently deplete these proteins from mouse nuclear extracts by specific antibodies were unsuccessful, so the importance of these proteins for 3′-end processing of histone pre-mRNAs remains to be determined. However, we favor the possibility that binding of all three hnRNPs to histone pre-mRNAs is not biologically relevant (or at most plays only an auxiliary role) and simply results from using an excess of the substrate in the pulldown experiments.
Role for 3′hExo in histone mRNA metabolism?
The most intriguing result of the affinity purification experiments was the isolation of 3′hExo. The association of 3′hExo is primarily a postprocessing event resulting from a very strong affinity of this protein to the mature 3′ end of histone mRNA. An important role in binding 3′hExo to the cleaved histone mRNA is played by the cooperation in binding between SLBP and 3′hExo. The cooperation is mutual, with 3′hExo allowing SLBP to bind a mutant version of the SL RNA and vice versa, with SLBP stabilizing the relatively weak interaction of 3′hExo with the SL RNA. Since 3′hExo and SLBP do not interact in the absence of the SL RNA (13), the most likely mechanism of the cooperative binding involves a direct protein-protein interaction triggered by the association of each protein with its respective RNA site. By using various mutants of SLBP, we narrowed down the region of SLBP involved in this interaction to the RBD.
Interestingly, both affinity purification and cross-linking experiments suggest that 3′hExo is recruited to the processing complex in a U7-dependent manner prior to cleavage of histone pre-mRNA. The observation that 3′hExo can be sequestered from nuclear extracts by the mutant SL RNA without causing any reduction in the efficiency of cleavage indicates, however, that 3′hExo is not an essential processing factor. An attractive possibility is that 3′hExo is initially actively recruited to the histone pre-mRNA and following cleavage is rapidly transferred to the mature 3′ end, to which it has a strong affinity. The mechanism by which 3′hExo is recruited to histone pre-mRNA is unknown. One possibility is that the base pairing between the U7 snRNA and the HDE promotes structural changes in histone pre-mRNA (21) that stimulate an interaction of 3′hExo with the internally located SL. Alternatively, 3′hExo may be stabilized on the SL in histone pre-mRNA by interactions with a component of the U7 snRNP and SLBP.
In mammalian cells, 3′hExo has been detected in the cytoplasm, the nucleoplasm, and the nucleolus (37), likely performing a plethora of tasks in these compartments, including 3′-end trimming of the 5.8S rRNA in the nucleolus (1, 17). More recently, 3′hExo has been also shown to localize to promyelocytic leukemia (PML) bodies (19). The function of PML bodies is unclear, although they have been implicated in many distinct processes, including cell stress response, cycle regulation, DNA repair, and transcription (7). Interestingly, in S phase, PML bodies double and become more dynamic, suggesting a role in monitoring chromatin integrity during DNA replication (10), a function closely related to histone mRNA metabolism. PML bodies often communicate with Cajal bodies (35). A subset of Cajal bodies, which harbor factors involved in processing of histone pre-mRNAs, are found close to histone loci (18, 19). We believe that the localization data, together with the active recruitment of 3′hExo to the processing complex and the cooperation with SLBP in binding to the mature histone mRNA 3′ end, support the notion that 3′hExo has a biological relevance to in vivo histone mRNA biogenesis and/or metabolism. Interestingly, the cytoplasmic histone mRNAs typically contain three nucleotides after the SL (29), while cleavage occurs five nucleotides past the SL, indicating that the last two nucleotides are removed during the transition from the nucleus to the cytoplasm. This trimming resembles a similar reaction in vitro, with recombinant SLBP and 3′hExo bound to the SL (13), and further suggests that these two proteins also bind the mature 3′ end of histone mRNA in vivo.
One obvious potential role for 3′hExo, an active 3′ exonuclease, was in the rapid degradation of histone mRNAs that occurs at the end of S phase or after arrest of DNA replication. However, recent data indicate that the degradation of histone mRNAs under each circumstance is mediated by other proteins (29). Thus, how 3′hExo functions in histone mRNA metabolism remains to be elucidated.
The region upstream of the HDE interacts with proteins of the U7-specific Sm ring.
Cross-linking experiments revealed that the region located between the cleavage site and the HDE interacts in a U7-dependent manner with a set of three proteins, ranging in size from approximately 15 to 30 kDa. The identities of two cross-linked proteins were determined by immunoprecipitation as SmB and SmD3. On the basis of the electrophoretic mobility, the smallest of the three proteins is likely Lsm10. This interpretation is further supported by the predicted position of Lsm10 in the U7-specific ring. As demonstrated by structural studies of the conventional Sm ring, the SmD1 protein occupies the position adjacent to SmB (22). Since, in the U7-specific ring, Lsm10 substitutes for SmD1, it can be assumed that Lsm10, together with SmD3 and SmB, forms a continuous platform that tightly associates with the pre-mRNA region spanning from the cleavage site to the HDE. Importantly, this configuration would place Lsm10 close to the cleavage site, potentially explaining the mechanism of the U7-dependent molecular ruler that functions during 3′-end processing to determine the cleavage site in histone pre-mRNA (33, 34). Histone pre-mRNA is likely to bind to the top surface of the U7-specific Sm ring. The existence of a secondary RNA binding site in this part of the ring, in addition to the known binding site in the internal cavity, was shown for a hexamer of the Pyrococcus abyssi Sm1 protein in a complex with RNA (36).
Mechanistically, binding of the Sm ring to the region upstream of the HDE may help in stabilizing the U7 snRNP on the pre-mRNA substrate. A similar role is played by the SmD3, SmB, and SmD1 proteins of the U1 snRNP during pre-mRNA splicing (39). These proteins, through their positively charged C-terminal regions, interact with nucleotides in the vicinity of the 5′ splice, thus allowing more-stable association of the U1 snRNP with intron-containing pre-mRNAs. In addition to stabilizing the U7 snRNP, the three proteins may also rigidify the region upstream of the HDE or appropriately expose the cleavage site. The former function was previously postulated for an unknown component of the processing machinery, perhaps a protein of the U7 snRNP (33) or symplekin (23).
We recently showed that truncated DCP substrates containing 7 or fewer nucleotides upstream of the HDE instead of the typical number of 9 to 11 nucleotides (4) are not degraded by the 5′ exonuclease activity of CPSF-73, although they bind the U7 snRNP with normal efficiency (38). Presumably, the truncated DCP substrates contain insufficient space 5′ of the HDE to bind the Sm ring of the U7 snRNP and therefore to recruit (or activate) CPSF-73.
Surprisingly, in spite of testing a variety of specifically labeled histone pre-mRNAs and conditions, we were unable to detect a cross-link that might correspond to Lsm11. We conclude that this component of the Sm ring, in contrast to the three other Sm proteins, is unlikely to tightly contact the substrate and probably plays a more specific role in processing, possibly directly or indirectly recruiting CPSF-73 to histone pre-mRNA.
A model depicting the predicted interactions between Sm proteins and histone pre-mRNA during 3′-end processing and the proposed configuration of the Sm ring in relation to the substrate and the cleavage site is shown in Fig. 6D. In this model, the cleavage site is flanked by Lsm10 and Lsm11, the two unique components of the U7-specifc Sm core that are essential for processing (31). The interactions with the three components of the Sm complex that occur downstream of the cleavage site (Lsm10, SmD3, and SmB) are perfectly suited to serve as a molecular ruler to determine the site of the endonucleolytic attack by CPSF-73, which is located at a fixed distance from the HDE (33, 34).
Supplementary Material
Acknowledgments
We thank the following researchers for providing antibodies: G. Dreyfuss (University of Pennsylvania) for anti-hnRNP Q antibody, J. Dean (Imperial College of Science, Technology and Medicine, London, United Kingdom) for anti-CBF-A, and C. MacDonald (Texas Tech University Health Sciences Center) for anti-CstF-77 antibody.
This work was supported by NIH grant GM29832.
Footnotes
Published ahead of print on 26 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ansel, K. M., W. A. Pastor, N. Rath, A. D. Lapan, E. Glasmacher, C. Wolf, L. C. Smith, N. Papadopoulou, E. D. Lamperti, M. Tahiliani, J. W. Ellwart, Y. Shi, E. Kremmer, A. Rao, and V. Heissmeyer. 2008. Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat. Struct. Mol. Biol. 15523-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L. 1999. An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol. 169-91. [PubMed] [Google Scholar]
- 3.Battle, D. J., and J. A. Doudna. 2001. The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA 7123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond, U. M., T. A. Yario, and J. A. Steitz. 1991. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 51709-1722. [DOI] [PubMed] [Google Scholar]
- 5.Brahms, H., J. Raymackers, A. Union, F. de Keyser, L. Meheus, and R. Luhrmann. 2000. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 27517122-17129. [DOI] [PubMed] [Google Scholar]
- 6.Callebaut, I., D. Moshous, J. P. Mornon, and J. P. De Villartay. 2002. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 303592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ching, R. W., G. Dellaire, C. H. Eskiw, and D. P. Bazett-Jones. 2005. PML bodies: a meeting place for genomic loci? J. Cell Sci. 118847-854. [DOI] [PubMed] [Google Scholar]
- 8.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 112755-2766. [DOI] [PubMed] [Google Scholar]
- 9.Dean, J. L., G. Sully, R. Wait, L. Rawlinson, A. R. Clark, and J. Saklatvala. 2002. Identification of a novel AU-rich-element-binding protein which is related to AUF1. Biochem. J. 366709-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dellaire, G., R. W. Ching, H. Dehghani, Y. Ren, and D. P. Bazett-Jones. 2006. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J. Cell Sci. 1191026-1033. [DOI] [PubMed] [Google Scholar]
- 11.Dominski, Z., J. A. Erkmann, J. A. Greenland, and W. F. Marzluff. 2001. Mutations in the RNA binding domain of stem-loop binding protein define separable requirements for RNA binding and histone pre-mRNA processing. Mol. Cell. Biol. 212008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominski, Z., and W. F. Marzluff. 2007. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene 396373-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominski, Z., X. Yang, H. Kaygun, and W. F. Marzluff. 2003. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol. Cell 12295-305. [DOI] [PubMed] [Google Scholar]
- 14.Dominski, Z., X. C. Yang, and W. F. Marzluff. 2005. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 12337-48. [DOI] [PubMed] [Google Scholar]
- 15.Dominski, Z., X. C. Yang, M. Purdy, E. J. Wagner, and W. F. Marzluff. 2005. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 251489-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominski, Z., L. X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 193561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabel, H. W., and G. Ruvkun. 2008. The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat. Struct. Mol. Biol. 15531-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gall, J. G. 2003. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 4975-980. [DOI] [PubMed] [Google Scholar]
- 19.Ghule, P. N., Z. Dominski, X. C. Yang, W. F. Marzluff, K. A. Becker, J. W. Harper, J. B. Lian, J. L. Stein, A. J. Van Wijnen, and G. S. Stein. 2008. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc. Natl. Acad. Sci. USA 10516964-16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gick, O., A. Kramer, W. Keller, and M. L. Birnstiel. 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 51319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaeger, S., F. Martin, J. Rudinger-Thirion, R. Giege, and G. Eriani. 2006. Binding of human SLBP on the 3′-UTR of histone precursor H4-12 mRNA induces structural rearrangements that enable U7 snRNA anchoring. Nucleic Acids Res. 344987-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khusial, P., R. Plaag, and G. W. Zieve. 2005. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci. 30522-528. [DOI] [PubMed] [Google Scholar]
- 23.Kolev, N. G., and J. A. Steitz. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 192583-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolev, N. G., T. A. Yario, E. Benson, and J. A. Steitz. 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 91013-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel, C. R., Y. Bai, and L. Tong. 2008. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 651099-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandel, C. R., S. Kaneko, H. Zhang, D. Gebauer, V. Vethantham, J. L. Manley, and L. Tong. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mili, S., and J. A. Steitz. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 101692-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mourelatos, Z., L. Abel, J. S. Yong, N. Kataoka, and G. Dreyfuss. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 205443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullen, T. E., and W. F. Marzluff. 2008. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2250-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker, C. E., M. R. Warren, D. R. Loiselle, N. N. Dicheva, C. O. Scarlett, and C. H. Borchers. 2005. Identification of components of protein complexes. Methods Mol. Biol. 301117-151. [DOI] [PubMed] [Google Scholar]
- 31.Pillai, R. S., M. Grimmler, G. Meister, C. L. Will, R. Luhrmann, U. Fischer, and D. Schumperli. 2003. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 172321-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillai, R. S., C. L. Will, R. Lührmann, D. Schümperli, and B. Müller. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 205470-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scharl, E. C., and J. A. Steitz. 1996. Length suppression in histone messenger RNA 3′-end maturation: processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc. Natl. Acad. Sci. USA 9314659-14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scharl, E. C., and J. A. Steitz. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 132432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun, J., H. Xu, S. H. Subramony, and M. D. Hebert. 2005. Interactions between coilin and PIASy partially link Cajal bodies to PML bodies. J. Cell Sci. 1184995-5003. [DOI] [PubMed] [Google Scholar]
- 36.Thore, S., C. Mayer, C. Sauter, S. Weeks, and D. Suck. 2003. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J. Biol. Chem. 2781239-1247. [DOI] [PubMed] [Google Scholar]
- 37.Yang, X. C., M. Purdy, W. F. Marzluff, and Z. Dominski. 2006. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J. Biol. Chem. 28130447-30454. [DOI] [PubMed] [Google Scholar]
- 38.Yang, X. C., K. D. Sullivan, W. F. Marzluff, and Z. Dominski. 2009. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol. Cell. Biol. 2931-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, D., N. Abovich, and M. Rosbash. 2001. A biochemical function for the Sm complex. Mol. Cell 7319-329. [DOI] [PubMed] [Google Scholar]
- 40.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.