Abstract
Regulation of the Saccharomyces cerevisiae HO promoter has been shown to require the recruitment of chromatin-modifying and -remodeling enzymes. Despite this, relatively little is known about what changes to chromatin structure occur during the course of regulation at HO. Here, we used indirect end labeling in synchronized cultures to show that the chromatin structure is disrupted in a region that spans bp −600 to −1800 relative to the transcriptional start site. Across this region, there is a loss of canonical nucleosomes and a reduction in histone DNA cross-linking, as monitored by chromatin immunoprecipitation. The ATPase Snf2 is required for these alterations, but the histone acetyltransferase Gcn5 is not. This suggests that the SWI/SNF complex is directly involved in nucleosome removal at HO. We also present evidence indicating that the histone chaperone Asf1 assists in this. These observations suggest that SWI/SNF-related complexes in concert with histone chaperones act to remove histone octamers from DNA during the course of gene regulation.
The Saccharomyces cerevisiae HO gene encodes an endonuclease that generates a double-stranded break at the mating-type locus that allows the yeast to switch between a and α mating types (53). HO is transcribed transiently during the late G1 phase only in mother cells, but not in daughter cells (40). As such, it has been considered a paradigm for a developmentally and cell cycle-regulated gene in a relatively simple eukaryote.
Considerable progress has been made in defining the order of events that result in HO transcription. They are triggered by the dephosphorylation of the transcription factor Swi5 during late anaphase, which allows it to enter the nucleus (56). The HO promoter contains two binding sites for Swi5 1,300 and 1,800 bp upstream of the transcriptional start site within a region referred to as URS1. Binding of Swi5 and Pho2 to URS1 (8, 38) results in the recruitment of the SWI/SNF complex in mother cells (14). In daughter cells, the presence of the repressor Ash1 prevents SWI/SNF recruitment and subsequent stages in the activation of HO (14; reviewed in reference 12). The SAGA histone acetyltransferase complex is also recruited to URS1 and is required for HO transcription (14, 34). Chromatin immunoprecipitation (ChIP) studies suggest that both the SAGA complex itself and histone acetylation spread from URS1 to a second regulatory region,URS2, where the transcription factor SBF is recruited to a series of binding sites in the region from 100 to 700 bp upstream of the transcription start site (14, 34). SBF is in turn required for recruitment of the mediator complex and, subsequently, following reactivation of Cdk1, RNA polymerase II (13).
The involvement of chromatin-modifying and -remodeling enzymes in the activation of HO raises the possibility that the chromatin structure may be altered. In vitro, SWI/SNF-related complexes have been found to be capable of generating a range of different transitions in chromatin structure. These can involve nucleosome sliding, the unraveling of DNA from the surfaces of nucleosomes, destabilization of histone dimers, and transfer of entire octamers (17, 47). In vivo, there is evidence that SWI/SNF complexes can cause nucleosome sliding (36), increased accessibility to nucleases (28), and the depletion of some or all of the histone components of nucleosomes from regulatory and coding regions of genes (see Discussion).
HO transcription is strictly dependent on SWI/SNF; indeed, many of the genes encoding SWI/SNF components were identified in genetic screens dependent on this (43). This makes the HO promoter a useful system to study the changes to chromatin structure generated as a result of SWI/SNF action in vivo. Despite this, the nature of the change to chromatin structure that occurs during regulation of the HO promoter has not been described. Here, we use nuclease digestion and ChIP to monitor the structure of the HO promoter. We detect a transition in chromatin that involves five nucleosomes and is dependent on the catalytic action of the SWI/SNF complex and the histone chaperone Asf1.
MATERIALS AND METHODS
Yeast strains and media.
The strains K8124 (SWI2-myc18 Δash1 GAL-CDC20 Δcdc20), K8144 (Swi5-myc Δash1 GAL-CDC20 Δcdc20), K8145 (Swi5-myc Δash1 GAL-CDC20 Δcdc20 swi2-314), and K8134 (Swi2-myc Δash1 Δgcn5 GAL-CDC20 Δcdc20) have been described previously (14). H2B was myc tagged by a PCR-based strategy (30) inserting three mycs at the C terminus of Htb1 using the pYM4 template in strain W303 and was subsequently crossed with strain K8283 (Δash1 GAL-CDC20 Δcdc20), reported previously (11), resulting in strain TOH1012. The asf1Δ strain was initially obtained from Euroscarf (strain Y01310) but had the dominant resistance marker kanMX4 replaced with hygromycin by swapping the kanMX4 marker with hphMX4 by PCR-based one-step gene replacement, as previously described (20). The resulting strain was crossed with TOH1012, resulting in strain TOH1045. TOH1144 (nap1Δ) was generated by disruption of the NAP1 gene using the pAG32 template, which contains the hphMX4 dominant resistance marker (20) in K8283. All experiments were carried out at 30°C, and cell synchrony was achieved as previously described (14).
Chromatin analysis.
Spheroplast preparation and chromatin analysis using micrococcal nuclease, as well as Southern blotting and indirect end labeling, were performed as previously described (27), except that cells were treated with 1% formaldehyde at 25°C for 10 min to cross-link the chromatin prior to digestion. Cross-linking was quenched by the addition of 2.5 M glycine to a final concentration of 0.125 M, followed by incubation for a further 5 minutes. The cross-linked cells were subsequently washed three times with ice-cold Tris-buffered saline (20 mM Tris, pH 7.5, 120 mM NaCl) and then processed essentially as described previously (27). DNA was resuspended in 20 μl of Tris-EDTA buffer and digested with the restriction enzyme. DNA fragments were separated in a 20- by 25-cm gel electrophoresis system containing 1.5% agarose in 1× Tris-borate-EDTA buffer. The gels were normally run for 8 to 10 h at 80 V and were then stained with ethidium bromide to view total DNA. The gels were then washed with 500 ml of 1.5 M NaCl-0.5 M NaOH for 30 min and rinsed with distilled water. Then, the gel was treated with 500 ml of 1.5 M NaCl-0.5 M Tris-HCl, pH 7.0-1 mM EDTA for 45 min. The gel was blotted for 12 to 14 h against 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to an uncharged neutral nylon membrane, and the blot was rinsed with 2× SSC before being UV cross-linked at 1,200 J. The blot was then wetted with double-distilled water and kept airtight at −20°C until hybridization. Prehybridization with 20 ml 1.5× SSC-5× Denhardt's-0.1% sodium dodecyl sulfate (SDS) was carried out for an hour at 64°C. One hundred nanograms of DNA probe labeled with [α-32P]dCTP using the Amersham Rediprime II DNA-labeling system according to the manufacturer's instructions and 500 μl of 2.5-mg/ml sonicated salmon sperm were heated at 100°C for 5 min, cooled on ice, and then added to the prehybridized membrane and incubated overnight at 64°C. The probe DNA used for indirect end labeling adjacent to the NcoI site at the HO promoter was amplified using the primers CATGAAAGATTCATGAGATCTGACA and CGTGTATTTAGTTACATCACTTTTCG. The blot was washed once in 100 ml of prewarmed 2× SSC-0.1% SDS at 60°C for 30 min, followed by a wash in 100 ml of prewarmed 2× SSC-0.1% SDS at 64°C for 30 min, and rinsed in 100 ml of ambient-temperature 2× SSC-0.1% SDS and 100 ml of double-distilled water. It was then sealed in a thin plastic membrane and exposed to Amersham Biomax MS film at −80°C. The film was then developed and scanned/digitized at 100-μm resolution on a FLA-5100 imager, and image analysis was performed with AIDA software (Fujifilm).
ChIP.
Yeast cultures at a density of between 0.5 × 107 and 2 × 107 cells/ml were treated with 1% formaldehyde for 20 min at room temperature. Cross-linking was quenched by the addition of 2.5 M glycine to a final concentration of 0.125 M. The cross-linked cells were washed three times with ice-cold Tris-buffered saline and resuspended in 300 μl lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, and a mixture of protease inhibitors) and agitated with glass beads for 25 min at 4°C. Chromatin sonication was carried out using a Bioruptor ultrasonic cell disruptor (Diagenode) to produce sheared chromatin with an average fragment size of 200 bp. Immunoprecipitation was performed using magnetic beads (Dynal; 2 × 107 per sample). Dynabeads coated with rat monoclonal anti-mouse immunoglobulin G were used for immunoprecipitation of myc-tagged proteins using the 9E10 monoclonal antibody (Upstate). Dynabeads coated with protein A were used for immunoprecipitation using an anti-histone H3 antibody (ab1791; Abcam). The immunoprecipitation reactions, as well as the washing steps, were carried out as described previously (52a). PCRs were carried out in 50-μl reaction mixtures with 1/10 of the immunoprecipitated material and 1/100 of whole-cell extract under the following conditions: 94°C for 4 min and 20 cycles of 92°C for 30 s, 59°C for 30 s, and 68°C for 30 s. The PCR products were separated in 2% agarose gels, visualized with ethidium bromide, and digitized using a Fujifilm FLA-5100 phosphorimager. A set of primers (TCAAGAACACTCGCATTTACGGC and ACCTCACTTTCTGTCCCAGATACGA) that amplified an intergenic region at the HEM13 locus was used for normalization of the whole-cell extract and immunoprecipitated target values. The primers used for the HO open reading frame were GGTGAACCTGGTAGGTTAGATCCCA and CACAGACCAAGCATCCAAGCCA. The primers used for URS1 were CGTAAAAAGTTTGATTCGTGGCGG and CGATTTATCAAAGCACTCTGCGGTT. Similar observations were made using reverse transcription (RT)-PCR. For the Snf2p ChIPs, whole-cell extract and immunoprecipitated samples were produced as previously described, but data analysis was performed only by RT-PCR using the Applied Biosystems 7500 real-time PCR system. Signal normalization for H2B and H3 immunoprecipitations was also performed using oligonucleotide sets that amplified from the ACT1 promoter and open reading frame region, with the same results observed using the HEM13 primer set. The enrichment values shown in the figures are the ratios of enrichment with respect to HEM13 in the immunoprecipitation versus enrichment relative to HEM13 in genomic DNA.
Nucleosomal MNase protection.
Chromatin was not digested with a restriction enzyme following MNase digestion (see Fig. 4C), and the 757-bp probe used for Southern blotting was amplified using the primers GCACAAAAAAGGTACGTTAATTTCC and GACTAACTTAAAAACGAGGTCGTA.
FIG. 4.
Chromatin remodeling at HO involves the loss of canonical nucleosomes. (A) Schematic representation of the HO promoter. The solid black bars represent the probes used for Southern blot analysis. Nucleosome mapping involved preparing chromatin from the strain K8144 (ash1Δ SNF2) under conditions in which HO chromatin was in the repressed state (10 min following release) or the active state (25 min following release). The DNA fragment distribution of bulk chromatin could be seen by agarose gel electrophoresis followed by staining with ethidium bromide. (B) A ladder of nucleosomal repeats was detectable in chromatin from both the active and repressed conditions. To study the nucleosome contact at the HO promoter, fragments from agarose gels were transferred by Southern blotting and hybridized with a 722-bp probe spanning from bp −538 to −1260 within the HO URS region. (C) The loss of nucleosome length protection observed under activating conditions suggests that few nucleosomes remained capable of protecting 147 bp of DNA in this region. (D) Quantitative analysis of nucleosomal signal. Purified nucleosomal DNA was subjected to RT-PCR with primers specific for the nucleosome at bp −1000 to quantify loss of nucleosomal signal.
The quantitation of micrococcal-nuclease-digested chromatin (see Fig. 4D) was carried out in an Applied Biosystems 7500 real-time PCR system. PCRs were carried out in 25-μl reaction mixtures under the following conditions: 95°C for 10 min and 40 cycles of 92°C for 15 s, 59°C for 35 s, and 68°C for 25 s. Quantitation and normalization were carried out exactly as for the ChIPs but using the primers CGTAAAAAGTTTGATTCGTGGCGG and CGATTTATCAAAGCACTCTGCGGTT.
Measurement of HO mRNA.
For each strain to be analyzed, liquid cultures were grown to a cell density of 1 × 107 to 2 × 107 cells/ml. The cells (107) were resuspended in extraction buffer (0.5 M NaCl, 0.2 M Tris, pH 7.6, 0.01 M EDTA, 1% SDS) and snap-frozen by immersion in liquid nitrogen. An equal volume of acidic phenol chloroform prewarmed at 65°C was added, mixed vigorously using a vortex, and immediately snap-frozen in liquid nitrogen. The samples were then centrifuged in a benchtop centrifuge at 13,000 × g for 7 min and were subsequently reextracted with acidic phenol chloroform. The RNA was precipitated with ethanol, and the pellets were air dried and stored at −80°C. Quantitation was carried out using an Express quantitative RT-PCR kit (Invitrogen) with normalization against actin mRNA.
RESULTS
Characterization of chromatin structure at the HO locus.
In order to investigate the structure of chromatin at the HO locus, yeast nuclei were isolated and subjected to controlled digestion with micrococcal nuclease. The sites of increased cutting and protection were identified by indirect end labeling (26). We initially compared the chromatin structures in this region in wild-type and Snf2 mutant strains in asynchronous cultures. Although we observed nucleosomes positioned throughout the HO intergenic region, consistent with recent genome-wide studies (35) that included the same region, we did not observe Snf2-dependent changes in the chromatin structure (data not shown). One possible explanation for this is that, as the HO promoter is only active for a relatively short period of the cell cycle in mother cells, most of the cells in an asynchronous culture will not be expressing HO.
In order to investigate whether the chromatin structure is transiently rearranged during the course of activation at HO, we next studied the chromatin structure at HO in cultures that had been synchronized by the depletion of Cdc20. A system in which Cdc20 had been placed under the control of the Gal10 promoter was previously proven to be effective in studying the chromatin transition at HO (14). These cells arrest when grown in raffinose but can be released from this block when galactose is added as a carbon source. In order to test the synchronization procedure, the growth of cells following release from Cdc20 block was monitored by fluorescence-activated cell sorting, and the binding of Swi5 to URS1 was monitored by ChIP (see Fig. S1 in the supplemental material). Both cell division and the interaction of Swi5 occurred with the same timing that had been observed previously (13, 14), showing that the synchronization procedure was effective and occurred with the expected timing. In addition to synchronizing the culture, we also carried out analysis of the chromatin structure in a strain (K8144) in which the ASH1 gene had been deleted. This resulted in the HO gene being expressed in both mother and daughter cells and meant that we could study chromatin within a more homogeneous population.
The structure of chromatin at the HO regulatory region was monitored in cell cultures synchronized in this way by indirect end labeling. Figure 1A shows that at 0, 10, and 15 min following release from the Cdc20 block, a regular pattern of protection and cleavage was observed consistent with the presence of nucleosomes. Within this ladder, two more prominent hypersensitive sites within URS1 were observed in the regions that corresponded to previously characterized binding sites for the transcription factor Swi5 (52, 54). At 20, 25, and 30 min following release, a transition in chromatin structure involving five nucleosomes spanning URS1 and URS2 occurred. The nucleosomes that were altered are illustrated schematically in Fig. 1C. The timing of this transition in the chromatin structure is coincident with the recruitment of SWI/SNF (Fig. 1B) (14), suggesting that this complex might be involved in generating the alteration.
FIG. 1.
Detection of a chromatin transition during activation of HO. (A) Cultures of K8144 were synchronized in late anaphase by Cdc20 depletion, and samples taken at the indicated times following release from arrest were digested with MNase and subjected to indirect-end-labeling analysis. Ordered chromatin was detected up to 15 min from release. From 20 min on, a transition in chromatin structure was detected. The transition occurred in the region occupied by nucleosomes shaded light gray. Lane 1, Mw markers; lane 2, DNA that had been digested with NcoI and SphI; lane 3, DNA digested with NcoI and MNase; lanes 4 to 21, chromatin digested with NcoI and MNase (9, 18, and 37 U/ml) at the indicated times following release. (B) Snf2 is recruited to HO 20 min following release from Cdc20 arrest coincident with chromatin remodeling. Recruitment of Snf2 to URS1 was monitored by ChIP using the strain K8124, in which Snf2 is myc tagged. The error bars indicate standard deviations. (C) Schematic summary of the alterations to chromatin detected at HO. Nucleosomes within the region of disruption are shaded light gray.
Chromatin remodeling at HO is dependent on Snf2.
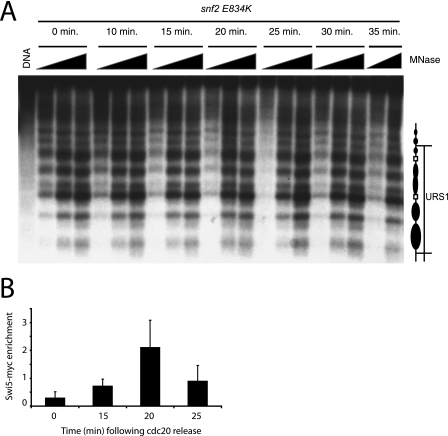
In order to test this further, the chromatin structure was characterized in a strain encoding a Snf2 protein with an E834K point mutation. Figure 2A shows that the swi2-314 allele resulted in the absence of the chromatin transition that was detected in the presence of wild-type Snf2 protein. This was not due to a delay in cell cycle progression in the swi2-314 strain, as the timing with which Swi5 was recruited was not greatly affected (Fig. 2B) (14) and no alteration to the chromatin structure could be detected in the mutant strain over an extended time course (see Fig. S2 in the supplemental material). This suggests that Snf2 or a protein acting subsequently to Snf2 is responsible for the transition in chromatin structure. Previously, it was shown that the SAGA histone acetyltransferase complex is recruited to HO subsequently to SWI/SNF. For this reason, we characterized the chromatin structure in strains in which the GCN5 gene had been deleted. The results, shown in Fig. 3, indicate that the chromatin transition still occurred despite the absence of Gcn5. As SAGA is required for subsequent steps in the activation of HO, we can conclude that neither the histone acetyltransferase activity of SAGA nor subsequent steps in the activation of this gene are required for the chromatin alteration detected.
FIG. 2.
The transition in chromatin structure is dependent on Snf2. (A) Cells from the strain K8145 encoding the snf2 E834K mutation were collected at the indicated times following release from anaphase arrest, digested with MNase, and subjected to indirect end labeling. No transition in the chromatin structure was observed in a strain with the snf2 E834K mutation. Chromatin structure was monitored using the NcoIF probe. (B) ChIP showing that Swi5 is recruited to HO in a K8145 strain bearing the snf2 E834K mutation with timing similar to that for SNF2 (see Fig. S1B in the supplemental material). The error bars indicate standard deviations.
FIG. 3.
Chromatin remodeling at HO occurs independently of Gcn5. (A and B) Cells from isogenic strains differing in the GCN5 locus (K8144 and K8134) were collected at the indicated times following release from anaphase arrest, digested with MNase, and subjected to indirect end labeling. The transition in the chromatin structure during activation of the promoter occurred in the absence of the Gcn5 acetyltransferase.
The chromatin transition at HO involves a loss of canonical nucleosomes.
Although the data presented in Fig. 1 indicate that a region between 500 and 1,500 bp upstream of the HO promoter is disrupted during the course of induction, the nature of the disruption is not clear. For example, the observed alterations to MNase digestion could result from nucleosomes being moved to different locations as a result of SWI/SNF action or a more dramatic alteration to nucleosome structure. In order to investigate this further, we next characterized the ability of nucleosomes at the HO promoter to protect 147-bp DNA fragments from digestion by micrococcal nuclease. Yeast nuclei from cultures before and after release from Cdc20 arrest were subjected to digestion with increasing amounts of micrococcal nuclease. Following the removal of proteins, this DNA could be separated on an agarose gel. Figure 4B shows that a significant proportion of the total yeast genomic DNA was digested into fragments of approximately 147 bp and that a ladder of multiples of this length could also be observed. This is consistent with the digestion of bulk yeast chromatin into DNA that is nucleosome length and multiples of nucleosome length. There was little change in this pattern when DNA was prepared from cells grown prior to and 30 min following release from Cdc20 arrest. In order to look at the nucleosome content of the HO regulatory region, a probe derived from the region in which we had detected an alteration by indirect end labeling (Fig. 3A) was used as a probe in a Southern blot of genomic micrococcal-nuclease-digested DNA. In contrast to the total DNA, the nucleosomal digestion pattern at the HO URS became far less distinct 30 min following release from Cdc20 arrest (Fig. 4C). This effect could be quantified by PCR using primers designed to amplify nucleosomes located at −1000 (Figure 4D). As the probe used in Fig. 4C spans 722 bp, repositioning of nucleosomes in this region would not be sufficient to result in a loss of nucleosomal signal. As a result, the reduction in the proportion of DNA giving rise to nucleosomal protection under activating conditions indicates a reduction in the content of intact nucleosomes over this region.
Chromatin remodeling at HO involves a loss of histone DNA contacts.
In order to monitor the extent to which histones retain in contact with the HO locus, ChIP assays were performed. Histone H2B contacts were monitored using a strain in which the gene encoding H2B had been tagged with a myc epitope. H2B occupancy at URS1 was found to drop approximately 20 min following release from Cdc20 arrest (Fig. 5B). Cross-linking of histone H3 was observed to be reduced with similar timing (Fig. 5B). This reduction in histone occupancy was specific to URS1, as it was not observed at other chromosomal regions, such as the HO coding region (Fig. 5C). As the reduction in histone cross-linking was coincident with the recruitment of Snf2 to HO (Fig. 1B), we next tested whether the alteration also occurred in a Snf2 mutant strain. Figure 5D shows that mutation of E834 to K within the Snf2 protein prevented histone loss.
FIG. 5.
The chromatin transition involves loss of DNA histone contacts. (A) Schematic representation of the HO promoter, including the positions of primers used for PCR. (B and C) DNA associated with myc-tagged H2B or the H3 C terminus (term) was enriched for by ChIP (TOH1012). The enrichment at either the URS (B) or coding region (C) was calculated relative to the HEM13 gene, which was used as a control in real-time PCR. (D) The reduction in histone contacts at the URS is dependent on Snf2, as it does not occur in a strain bearing the snf2 E834K mutation. The error bars indicate standard deviations.
Chromatin remodeling at HO also involves the histone chaperone Asf1.
In vitro, SWI/SNF has been observed to be more efficient at repositioning nucleosomes than at removing them from DNA in trans (57). This raises the possibility that an additional factor(s) assists SWI/SNF in the removal of nucleosomes at HO. In vitro, the histone chaperone Nap1 has been observed to accelerate the rate at which nucleosomes are disassembled (37). Our preliminary studies suggested that Nap1 does not contribute to remodeling at HO (data not shown). Another histone chaperone, Asf1, has been observed to be required for normal activation of the PHO5 and PHO8 genes (1). In concert, these observations made it of interest to investigate the role of histone chaperones in the remodeling of HO. Figure 6 shows that deletion of the histone chaperone Asf1 caused a reduction in remodeling at HO, as assessed by indirect end labeling (Fig. 6A). Furthermore, deletion of Asf1 also prevented the loss of histone contacts with URS1, as determined by ChIP (Fig. 6B). This important role for Asf1 in the chromatin transition at HO raises the issue of whether Asf1 is also required for HO transcription. Figure 6C shows that this is indeed the case and that the cell cycle-dependent transcription from HO is greatly reduced in an asf1Δ strain. The reduction in remodeling observed in the absence of Asf1 is unlikely to be due to a delay in cell cycle progression, as the timing of Swi5 recruitment, mitosis, and budding were not greatly delayed (see Fig. S3 in the supplemental material). Furthermore, no alteration to the chromatin structure at HO could be detected over an extended time course (see Fig. S3B in the supplemental material).
FIG. 6.
Nucleosome remodeling at HO requires Asf1. Indirect end labeling was used to monitor the chromatin structure at HO 0 min and 30 min following Cdc20 arrest using the NcoIF probe as described in the legend to Fig. 1. (A) Deletion of ASF1 (TOH1045) resulted in a reduction in the disruption of the chromatin structure at 30 min in comparison to the isogenic ASF1 plus TOH1064. (B) This was also evident as a failure to lose histone contacts, as assessed by H2B ChIP. (C) Real-time RT-PCR was used to measure the levels of the HO mRNA transcript. A reduction in cell cycle-regulated transcription was observed following deletion of ASF1. The error bars indicate standard deviations.
DISCUSSION
We have characterized a transition in chromatin structure occurring during the course of activation at the yeast HO promoter. It spans a region normally occupied by five nucleosomes. The remodeling event is likely to involve the loss of nucleosomes, as less DNA is cross-linked to histones and protected from nuclease digestion in the active conformation. However, the magnitude of the changes in cross-linking or nuclease sensitivity (two- to fourfold) raises the possibility that a proportion of nucleosomes remain intact within this region, as has been found to be the case during activation of the PHO5 promoter (7, 32). The chromatin transitions at PHO5 and HO also both involve the action of SWI/SNF and ASF1, and it is most likely that in both cases the outcome is the removal of a least a proportion of the nucleosomes from the remodeled region. However, our analysis of HO did not enable us to formally exclude the possibility that remodeling results in the generation of altered nucleosomes with reduced histone DNA contacts.
In order to study chromatin in a more homogeneous population of cells, we used a strain in which the Ash1 protein has been deleted. This causes the HO gene to be expressed in mother and daughter cells when otherwise it would be expressed only in mother cells. Use of this mutation has proven to be a useful means of dissecting the regulation of HO (14, 34); however, it remains open to the possibility that regulation may be different in the presence of Ash1. Indeed, there is evidence that while the SAGA complex is not required for SWI/SNF recruitment in the absence of Ash1, it does facilitate SWI/SNF recruitment when Ash1 is present (39). For this reason, we have attempted to characterize the chromatin transition in cell populations expressing Ash1. Unfortunately, we could not detect chromatin remodeling in such strains (data not shown). This could be because the nature of the remodeling event changes in the presence of Ash1 or because the proportion of cells with an active chromatin structure is reduced to a level at which we cannot detect changes.
It is notable that the changes we did detect were in the upstream regulatory region. This does not necessarily mean that additional changes could not occur in other regions of the gene. For example, if additional changes occurred but were very short lived, they might not be detected. Changes associated with transcription might be short lived, as active genes may be transcribed only in short bursts (11), an effect that may be especially significant at a gene such as HO that is transcribed at relatively low levels. It is notable that at highly transcribed genes, SWI/SNF has been observed to participate in the depletion of nucleosomes across the coding region (49, 51).
Genome-wide studies of nucleosome occupancy have revealed that many genes have a region of low nucleosome occupancy close to the transcriptional start site (5, 35, 44, 59). The mechanism by which this region is generated remains unclear. While there is some evidence that structural properties of the underlying DNA may not favor the accumulation of nucleosomes at these sites (2, 35, 48, 50), it also remains possible that other factors contribute to the removal of nucleosomes from these regions (3, 35, 55). The nucleosome depletion we detected here differs in that it occurs in an upstream regulatory region rather than at the promoter, involves several nucleosomes, and is not constitutive.
In this respect, the depletion we observed is more similar to that observed at the PHO5 promoter. Here, a compelling case has been made for the eviction of histones during the course of induction (6, 7, 32, 46). These observations are best interpreted in terms of a dynamic interplay between chromatin assembly and removal, resulting in net nucleosome depletion over a region spanning four nucleosomes. This results in an average of one nucleosome remaining present, but distributed unequally between four depleted locations (6, 25). In the case of PHO5, octamer removal is only partially dependent on SWI/SNF, as deletion of SNF2 causes a delay in the chromatin reconfiguration rather than completely preventing it (4). Partial dependence on SWI/SNF for nucleosome removal has also been observed in the GAL1,10 intragenic region (9) and for many genes induced upon heat shock (51).
At other genes, there is stronger dependence on SWI/SNF for chromatin remodeling, but at none of them has the transition in chromatin structure been characterized in so much detail as at PHO5. For example, at SUC2, there is a SWI/SNF-dependent alteration to the chromatin structure that has been shown to be independent of transcription (23, 49). At PHO8, the SAGA histone acetyltransferase complex is required for transient acetylation of histones prior to remodeling directed by SWI/SNF (45). Here, the remodeling event involves a loss of histone contacts and is facilitated by the action of Asf1 (1, 31). At FLO1, FLO11, and HIS3, SWI/SNF appears to move nucleosomes over a significant range (18, 19, 29), whereas at the human beta-interferon promoter, there is highly localized nucleosome repositioning (36).
Further studies have suggested that dynamic chromatin assembly and disassembly occur at many regulatory elements genome-wide (15, 24). Indeed, it has been observed that dependency on SWI/SNF is correlated with dynamic histone association (15). Although our study is restricted to the HO promoter, it is likely that SWI/SNF acts similarly at a subset of other yeast genes, where it may also act to destabilize nucleosomes (33, 49, 55).
The involvement of additional factors in facilitating the removal of histones by SWI/SNF also seems likely, given that SWI/SNF is more efficient at sliding nucleosomes than at removing them from DNA in vitro (57). In reactions catalyzed by the closely related enzyme RSC, the histone chaperone NAP-1 has been found to facilitate nucleosome transfer in vitro (37). However, here, we found that mutation of Nap1 does not affect chromatin remodeling at HO (data not shown). Instead, we found that the histone chaperone Asf1 plays a role in the removal of histones at HO (Fig. 6). This is reminiscent of the PHO5 and PHO8 promoters, where Asf1 contributes to octamer removal (1, 31). Although deletion of the ASF1 gene did reduce remodeling at HO, addition of affinity-purified native Asf1 to in vitro transfer reactions that included SWI/SNF had no significant effect on activity (data not shown). This suggests that additional factors may also be involved in remodeling at HO. One possibility is that the binding of transcription factors acts to promote the removal of nucleosomes at HO. In vitro studies indicate that histone eviction can be facilitated by transcription factor binding (22, 42), and there is good evidence that SWI/SNF interacts with a range of transcriptional activators (14, 41, 55, 58, 60). A second possibility is that modification of histones may potentate their removal. There is evidence that histone acetylation can facilitate the removal of nucleosomes in vitro (10, 16) and that in vivo, at least in some cases, histone acetylation precedes nucleosome removal by SWI/SNF (21, 51). Although at HO the removal of histone octamers is not dependent on SAGA, it could be that other modifying enzymes are capable of contributing to a signal for nucleosome removal in a way that is redundant with the action of SAGA. In this respect, it is noteworthy that H4 acetylation has been observed to occur early during the activation of HO (34).
Supplementary Material
Acknowledgments
We thank Kim Nasmyth and Tomo Tanaka for providing yeast strains and Nick Kent and Jane Mellor for advice on indirect end labeling. We also thank members of the Division of Gene Regulation for advice.
T.G. is a Wellcome Trust prize student. T.O.-H. is a Wellcome Trust Senior Research Fellow.
Footnotes
Published ahead of print on 26 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14657-666. [DOI] [PubMed] [Google Scholar]
- 2.Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton, S. C. Schuster, and B. F. Pugh. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446572-576. [DOI] [PubMed] [Google Scholar]
- 3.Badis, G., E. T. Chan, H. van Bakel, L. Pena-Castillo, D. Tillo, K. Tsui, C. D. Carlson, A. J. Gossett, M. J. Hasinoff, C. L. Warren, M. Gebbia, S. Talukder, A. Yang, S. Mnaimneh, D. Terterov, D. Coburn, A. L. Yeo, Z. X. Yeo, N. D. Clarke, J. D. Lieb, A. Z. Ansari, C. Nislow, and T. R. Hughes. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32878-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbaric, S., T. Luckenbach, A. Schmid, D. Blaschke, W. Horz, and P. Korber. 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 28227610-27621. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 111587-1598. [DOI] [PubMed] [Google Scholar]
- 7.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14667-673. [DOI] [PubMed] [Google Scholar]
- 8.Brazas, R. M., L. T. Bhoite, M. D. Murphy, Y. Yu, Y. Chen, D. W. Neklason, and D. J. Stillman. 1995. Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J. Biol. Chem. 27029151-29161. [DOI] [PubMed] [Google Scholar]
- 9.Bryant, G. O., V. Prabhu, M. Floer, X. Wang, D. Spagna, D. Schreiber, and M. Ptashne. 2008. Activator control of nucleosome occupancy in activation and repression of transcription. PloS Biol. 62928-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandy, M., J. L. Gutierrez, P. Prochasson, and J. L. Workman. 2006. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot. Cell 51738-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chubb, J. R., T. Trcek, S. M. Shenoy, and R. H. Singer. 2006. Transcriptional pulsing of a developmental gene. Curr. Biol. 161018-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosma, M. P. 2004. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep. 5953-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 71213-1220. [DOI] [PubMed] [Google Scholar]
- 14.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97299-311. [DOI] [PubMed] [Google Scholar]
- 15.Dion, M. F., T. Kaplan, M. Kim, S. Buratowski, N. Friedman, and O. J. Rando. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 3151405-1408. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira, H., J. Somers, R. Webster, A. Flaus, and T. Owen-Hughes. 2007. Histone tails and the H3 αN helix regulate nucleosome mobility and stability. Mol. Cell. Biol. 274037-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14165-173. [DOI] [PubMed] [Google Scholar]
- 18.Fleming, A. B., and S. Pennings. 2001. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 205219-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleming, A. B., and S. Pennings. 2007. Tup1-Ssn6 and Swi-Snf remodelling activities influence long-range chromatin organization upstream of the yeast SUC2 gene. Nucleic Acids Res. 355520-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 151541-1553. [DOI] [PubMed] [Google Scholar]
- 21.Govind, C. K., F. Zhang, H. Qiu, K. Hofmeyer, and A. G. Hinnebusch. 2007. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 2531-42. [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez, J. L., M. Chandy, M. J. Carrozza, and J. L. Workman. 2007. Activation domains drive nucleosome eviction by SWI/SNF. EMBO J. 26730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirschhorn, J. N., S. A. Brown, C. D. Clark, and F. Winston. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 62288-2298. [DOI] [PubMed] [Google Scholar]
- 24.Jamai, A., R. M. Imoberdorf, and M. Strubin. 2007. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25345-355. [DOI] [PubMed] [Google Scholar]
- 25.Jessen, W. J., S. A. Hoose, J. A. Kilgore, and M. P. Kladde. 2006. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat. Struct. Mol. Biol. 13256-263. [DOI] [PubMed] [Google Scholar]
- 26.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent, N. A., and J. Mellor. 1995. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 233786-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, Y., and D. J. Clark. 2002. SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc. Natl. Acad. Sci. USA 9915381-15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, Y., N. McLaughlin, K. Lindstrom, T. Tsukiyama, and D. J. Clark. 2006. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol. Cell. Biol. 268607-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15963-972. [DOI] [PubMed] [Google Scholar]
- 31.Korber, P., S. Barbaric, T. Luckenbach, A. Schmid, U. J. Schermer, D. Blaschke, and W. Horz. 2006. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 2815539-5545. [DOI] [PubMed] [Google Scholar]
- 32.Korber, P., T. Luckenbach, D. Blaschke, and W. Horz. 2004. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol. Cell. Biol. 2410965-10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102587-598. [DOI] [PubMed] [Google Scholar]
- 34.Krebs, J. E., M. H. Kuo, C. D. Allis, and C. L. Peterson. 1999. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 131412-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, W., D. Tillo, N. Bray, R. H. Morse, R. W. Davis, T. R. Hughes, and C. Nislow. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 391235-1244. [DOI] [PubMed] [Google Scholar]
- 36.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106685-696. [DOI] [PubMed] [Google Scholar]
- 37.Lorch, Y., B. Maier-Davis, and R. D. Kornberg. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 1033090-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBride, H. J., R. M. Brazas, Y. Yu, K. Nasmyth, and D. J. Stillman. 1997. Long-range interactions at the HO promoter. Mol. Cell. Biol. 172669-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitra, D., E. J. Parnell, J. W. Landon, Y. Yu, and D. J. Stillman. 2006. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol. Cell. Biol. 264095-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nasmyth, K. 1983. Molecular analysis of a cell lineage. Nature 302670-676. [DOI] [PubMed] [Google Scholar]
- 41.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 221615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owen-Hughes, T., R. T. Utley, J. Cote, C. L. Peterson, and J. L. Workman. 1996. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273513-516. [DOI] [PubMed] [Google Scholar]
- 43.Peterson, C. L., and I. Herskowitz. 1992. Characterization of the Yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68573-583. [DOI] [PubMed] [Google Scholar]
- 44.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122517-527. [DOI] [PubMed] [Google Scholar]
- 45.Reinke, H., P. D. Gregory, and W. Horz. 2001. A transient histone hyperacetylation signal marks nucleosomes for remodeling at the PHO8 promoter in vivo. Mol. Cell 7529-538. [DOI] [PubMed] [Google Scholar]
- 46.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 111599-1607. [DOI] [PubMed] [Google Scholar]
- 47.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7437-447. [DOI] [PubMed] [Google Scholar]
- 48.Satchwell, S. C., H. R. Drew, and A. A. Travers. 1986. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 191659-675. [DOI] [PubMed] [Google Scholar]
- 49.Schwabish, M. A., and K. Struhl. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 276987-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segal, E., Y. Fondufe-Mittendorf, L. Chen, A. Thastrom, Y. Field, I. K. Moore, J. P. Wang, and J. Widom. 2006. A genomic code for nucleosome positioning. Nature 442772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shivaswamy, S., and V. R. Iyer. 2008. Stress-dependent dynamics of global chromatin remodeling in yeast: dual role for SWI/SNF in the heat shock stress response. Mol. Cell. Biol. 282221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stillman, D. J., A. T. Bankier, A. Seddon, E. G. Groenhout, and K. A. Nasmyth. 1988. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 7485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Strahl-Bolshinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1183-93. [DOI] [PubMed] [Google Scholar]
- 53.Strathern, J. N., A. J. Klar, J. B. Hicks, J. A. Abraham, J. M. Ivy, K. A. Nasmyth, and C. McGill. 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31183-192. [DOI] [PubMed] [Google Scholar]
- 54.Tebb, G., T. Moll, C. Dowzer, and K. Nasmyth. 1993. SWI5 instability may be necessary but is not sufficient for asymmetric HO expression in yeast. Genes Dev. 7517-528. [DOI] [PubMed] [Google Scholar]
- 55.Venters, B. J., and B. F. Pugh. 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19360-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visintin, R., K. Craig, E. S. Hwang, S. Prinz, M. Tyers, and A. Amon. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2709-718. [DOI] [PubMed] [Google Scholar]
- 57.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400784-787. [DOI] [PubMed] [Google Scholar]
- 58.Yoon, S., H. Qiu, M. J. Swanson, and A. G. Hinnebusch. 2003. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol. Cell. Biol. 238829-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309626-630. [DOI] [PubMed] [Google Scholar]
- 60.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 132369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.