Abstract
The transcription factor RUNX-1 plays a key role in megakaryocyte differentiation and is mutated in cases of myelodysplastic syndrome and leukemia. In this study, we purified RUNX-1-containing multiprotein complexes from phorbol ester-induced L8057 murine megakaryoblastic cells and identified the ets transcription factor FLI-1 as a novel in vivo-associated factor. The interaction occurs via direct protein-protein interactions and results in synergistic transcriptional activation of the c-mpl promoter. Interestingly, the interaction fails to occur in uninduced cells. Gel filtration chromatography confirms the differentiation-dependent binding and shows that it correlates with the assembly of a complex also containing the key megakaryocyte transcription factors GATA-1 and Friend of GATA-1 (FOG-1). Phosphorylation analysis of FLI-1 with uninduced versus induced L8057 cells suggests the loss of phosphorylation at serine 10 in the induced state. Substitution of Ser10 with the phosphorylation mimic aspartic acid selectively impairs RUNX-1 binding, abrogates transcriptional synergy with RUNX-1, and dominantly inhibits primary fetal liver megakaryocyte differentiation in vitro. Conversely, substitution with alanine, which blocks phosphorylation, augments differentiation of primary megakaryocytes. We propose that dephosphorylation of FLI-1 is a key event in the transcriptional regulation of megakaryocyte maturation. These findings have implications for other cell types where interactions between runx and ets family proteins occur.
Over the past 2 decades, a number of transcription factors/cofactors have been identified that play essential roles in megakaryocytic differentiation. These include GATA-1 (46, 57), GATA-2 (4), Friend of GATA-1 (FOG-1) (55), NF-E2 p45 (47), mafG and mafK (39), SCL/Tal1 (30), GABPα (41), FLI-1 (17, 49), ZBP-89 (62), and RUNX-1 (14, 18). Yet, how these transcription factors act together to coordinate terminal megakaryocytic maturation remains incompletely understood. Moreover, there is increasing evidence that terminal megakaryocyte maturation is coordinated with localization at vascular sinusoidal niches within the bone marrow (1, 21, 26). How signaling events related to these spatial cues, as well as more-traditional cytokine-mediated transduction pathways, intersect with these key megakaryocyte transcriptional regulators also remains unclear.
The transcription factor RUNX-1 belongs to a family of proteins that share a conserved 128-amino-acid runt homology domain, which mediates DNA binding and interaction with the cofactor CBF-β (for a review, see reference 20). RUNX-1−/− mice die between embryonic day 12.5 (E12.5) and E13.5 due to central nervous system hemorrhage and failure of all definitive hematopoiesis (38, 59). The latter cause of death is due to a defect in the emergence of hematopoietic stem cells from the aorta-gonadal-mesonephros region during embryogenesis (31, 34, 64). Conditional knockout studies of mice demonstrate a specific role for RUNX-1 in megakaryocyte differentiation during adult stages of hematopoiesis (14, 18). RUNX-1-deficient megakaryocytes have hypolobulated nuclei, underdeveloped cytoplasm, low DNA ploidy, and enhanced replating activity in semisolid medium culture assays. Haploinsufficiency of CBF-β also perturbs megakaryopoiesis in mice (54). These findings indicate that RUNX-1/CBF-β is required for terminal megakaryocyte maturation.
Germ line mutations in RUNX-1 cause familial platelet disorder with the propensity to develop acute myelogenous leukemia (FPD/AML), a rare autosomal dominant disorder characterized by quantitative and qualitative platelet defects and a high incidence of developing myelodysplastic syndrome (MDS) and leukemia (40, 48). Acquired monoallelic RUNX-1 mutations occur in about 15% of cases of de novo MDS, particularly those that progress to AML (5, 16, 32). Biallelic mutations have been identified in a subset of FAB M0 AMLs (44). Although many of the mutations in these disorders occur within the runt domain and affect DNA and/or CBF-β binding, other mutations occur outside of these regions and have incompletely understood mechanistic effects.
In this study, we purified RUNX-1-containing multiprotein complexes from 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced L8057 murine megakaryoblastic cells in order to further understand RUNX-1 activity during megakaryopoiesis and to potentially gain insight into how some RUNX-1 mutations may contribute to MDS and AML. We identify the ets transcription factor FLI-1 as a novel direct RUNX-1 binding partner and show that it synergizes with RUNX-1 in megakaryocyte gene transcriptional reporter assays. We show that the interaction depends in part on the dephosphorylation of serine 10 in FLI-1, which occurs during megakaryocyte terminal maturation. Perturbation of RUNX-1-FLI-1 interactions by mutation of serine 10 to the phosphomimetic residue aspartic acid impairs differentiation of primary murine megakaryocytes in a dominant-negative fashion. We also show that two MDS-associated RUNX-1 mutations result in a loss of FLI-1 interaction. Collectively, these findings indicate that physical interaction between RUNX-1 and FLI-1, regulated in part by phosphorylation of FLI-1 Ser10, plays an important role in normal megakaryocyte terminal maturation. Dysregulation of this interaction may contribute to impaired hematopoiesis in some cases of myelodysplastic syndrome.
MATERIALS AND METHODS
Plasmid construction.
The generation of the pEFbirA-V5-His and pEF-FLAG-Biotag vectors has previously been described (62). The cDNA encoding murine RUNX-1 (isoform 3; NM_001111023.1), excluding the start ATG, was cloned in-frame into the pEF-FLAG-Biotag vector to generate an amino-terminal FLAG-birA recognition motif fusion molecule. For generation of the glutathione S-transferase (GST) fusion proteins, full-length cDNAs encoding murine FLI-1 and RUNX-1, or sequences encoding the runt domain of Runx-1 alone, were cloned into the pGEX-5x1 vector (GE Healthcare). cDNAs encoding murine full-length RUNX-1, the runt domain of RUNX-1, FLI-1, and CBF-β were cloned into pGADT7 vector (Clontech) for in vitro transcription/translation assays. For coimmunoprecipitation (co-IP) experiments, the cDNA for murine GATA-1 was cloned into the pEF-FLAGpgkpuropA vector, and the cDNA for RUNX-1 was cloned into pEF-V5-His vector (Invitrogen). The cDNA for murine FLI-1 was cloned into both expression vectors. For the murine c-mpl promoter reporter construct, −1,992 bp upstream of the transcriptional start site of murine c-mpl gene was cloned into the firefly vector pGL4.10 (Promega). For retroviral expression, the cDNA encoding murine FLI-1 was cloned into MSCV-IRES-hCD4 vector (63).
Cell culture and transfection.
Cells were cultured in 5% CO2 at 37°C in a humidified tissue culture incubator. All tissue culture medium was supplemented with 100 U/ml of penicillin-streptomycin and 2 mM l-glutamine. L8057 cells were cultured as previously described (19) and induced to differentiate with 50 nM tetradecanoyl phorbol acetate (TPA) (Sigma) for 3 days. The generation of L8057 cells stably expressing birA and FLAG-biotin-tagged RUNX-1 (FLAG-BioRUNX-1) followed the procedures described previously (62). 293T cells, PLAT-E cells, and primary fetal liver cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (heat inactivated). COS-7 cells were cultured in low-glucose DMEM supplemented with 10% fetal calf serum (heat inactivated). 293T cells, COS-7 cells, and PLAT-E cells were transfected using FuGene 6 reagent (Roche), according to the manufacturer's instructions.
RUNX-1 multiprotein complex purification and proteomic analysis.
The methods for purification of biotinylated transcriptional factor complexes and mass spectrometry (MS) of associated proteins were performed as described previously (62). Briefly, L8057 clones stably expressing birA and FLAG-biotin-tagged RUNX-1, or birA alone, were induced with TPA (50 nM) for 3 days. Crude nuclear extracts were prepared and dialyzed into BC139K buffer, centrifuged, and precleared with agarose beads. The FLAG-biotin-tagged RUNX-1 complexes were immunoprecipitated first using anti-FLAG M2 agarose beads (Sigma). The beads and bound material were washed with BC139K buffer and eluted by four successive incubations with excess FLAG peptide (0.1 mg/ml) (Sigma). The eluted material was then incubated with streptavidin (SA) agarose beads (Invitrogen), washed, and centrifuged. The pellet was boiled with sodium dodecyl sulfate (SDS) sample buffer and loaded onto a 10% SDS-polyacrylamide gel electrophoresis (PAGE) gel. After being stained with colloidal Coomassie blue (Invitrogen), the lanes were divided into three sections and cut into ∼1-mm cubes. The samples were washed, dehydrated with acetonitrile, dried, rehydrated, and digested with sequencing-grade trypsin (Promega). Peptides were extracted and separated on a nanoscale reverse-phase high-performance liquid chromatography capillary column. As each peptide was eluted, they were subjected to electrospray ionization and fed into an LTQ linear ion trap MS (Thermo Scientific). Eluting peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein or translated nucleotide databases with the acquired fragmentation pattern by using the software program SEQUEST (Thermo Scientific) (11).
Co-IP assays.
Several different co-IP assays were performed in this study, as follows. (i) Endogenous RUNX-1 or FLI-1 pulldown assays were performed with anti-RUNX-1 antibody (H-65; Santa Cruz) or anti-FLI-1 antibody (C-19; Santa Cruz) using the Pierce ProFound co-IP kit by following the manual instructions, except that IP buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.5], 0.1% NP-40, 1 mM EDTA, 10% [vol/vol] glycerol, 1:1,000 dilution of protease inhibitor cocktail [P8340; Sigma], and 1 mM dithiothreitol) was used instead of coupling buffer for the co-IP. (ii) Small-scale tandem FLAG-SA purification of RUNX-1 was performed as described above, except DNase I (1 μg/ml), RNase A (1 μg/ml), and ethidium bromide (50 μg/ml) were added to the nuclear extract before and during the pulldown experiment. (iii) COS-7 or 293T cells were transiently cotransfected with plasmids expressing either FLAG- or V5-tagged protein constructs using FuGene6 (Roche) transfection reagents.
GST pulldown experiments.
GST, GST-FLI-1, GST-RUNX-1, or GST-runt constructs were transformed into BL21 competent cells. Positive colonies were grown in LB medium until reaching an optical density at 600 nm reading of up to 0.6 and then induced with 500 μM IPTG (isopropyl-β-d-thiogalactopyranoside) at 20°C for 2 h. Cell pellets were snap-frozen in liquid nitrogen and stored overnight at −80°C. Cells pellets were then resuspended in HEMGT-150 buffer (150 mM KCl, 25 mM HEPES [pH 7.9], 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 0.1% Tween 20) with a 1:100 dilution of protease inhibitor cocktail (Sigma) and 100 μg/ml lysozyme (Amersham Biosciences), incubated on ice for 30 min, and sonicated three times for 30 s each using 60% duty at power lever 4 with an ultrasonic processor (W-380; Heat Systems Ultrasonics). A sample of the supernatant was examined by SDS-PAGE and stained with colloidal Coomassie blue along with serial dilutions of bovine serum albumin. In vitro translation of FLI-1, RUNX-1, runt domain of RUNX-1, or CBF-β with [35S]methionine incorporation was performed by following the manufacturer's instructions (TNT coupled reticulocyte lysate systems; Promega). One microgram of GST or GST fusion protein and 15 μl of 35S-labeled FLI-1, RUNX-1, runt, or CBF-β were incubated in HEMGT-150 buffer on a rotating wheel overnight at 4°C. Glutathione Sepharose 4B beads (GE Healthcare) were added and incubated for an additional 3 h at 4°C on a rotating wheel. The beads were washed four times with HEMGT-150 buffer for 15 min each, boiled in SDS sample buffer, and loaded onto an SDS-PAGE gel. The gels were then stained with colloidal Coomassie blue, dried in a gel drier (Bio-Rad), and exposed with Kodak BioMax MS film.
Sephacryl S-400 gel filtration chromatography and 2-D gel electrophoresis.
Crude nuclear extracts were prepared from uninduced or induced (with 50 nM TPA for 3 days) L8057 cells. Sephacryl S-400 gel filtration chromatography assays were performed as previously described (62). Fractions containing the large or small FLI-1 complexes were pooled separately. The proteins were concentrated and prepared for two-dimensional (2-D) gel electrophoresis using the ReadyPrep cleanup kit (Bio-Rad). Samples were incubated with 11-cm IPG strips (pH 3 to 10) overnight at room temperature. Isoelectric focusing was performed according to the manufacturer's instructions. Focused samples were separated in a second dimension by SDS-PAGE on a 4 to 20% gradient acrylamide gel. Western blot analysis of the 2-D gel was then performed using anti-FLI-1 antibodies (C-19; Santa Cruz).
Posttranslational modification analysis.
For posttranslational modification analysis, endogenous native FLI-1 protein from both induced and uninduced L8057 cells was immunopurified using anti-FLI-1 antibody (C-19; Santa Cruz) coupled to CNBr-activated Sepharose beads (Amersham Biosciences). Alternatively, SA affinity purification of biotinylated RUNX-1 from induced BBR cells was performed to enrich RUNX-1-associated FLI-1. Eluted material was separated by SDS-PAGE (Criterion XT precast gel; Bio-Rad) and stained with colloidal Coomassie blue. Selected bands around 50 kDa were excised, in-gel digested with trypsin, and analyzed for posttranslational modification by MS. In brief, the digestions were analyzed by on-line microscale capillary reversed-phase high-performance liquid chromatography hyphenated to a linear ion trap MS (LTQ; Thermo Scientific). Samples were loaded onto an in-house-packed 100-μm (inside diameter) by 15-cm C18 column (5-μm and 100-Å Magic C18; Michrom Bioresources) and separated at approximately 500 nl/min, with 30-min linear gradients from 5 to 40% acetonitrile in 0.4% formic acid. After each survey spectrum, the six most intense ions per cycle were selected for fragmentation/sequencing. All MS datasets were searched against a combined International Protein Index mouse protein sequence database using the Mascot search engine (version 2.1.04; Matrix Science).
Luciferase reporter assays.
The luciferase reporter assay system (Promega) was used in this study. In brief, luc2-c-mpl was cotransfected with RUNX-1/CBF-β, FLI-1, and/or GATA-1/FOG-1 expression constructs into COS-7 cells using FuGene 6 reagent by following the manufacturer's instructions. After 48 h, cell lysates were prepared, and firefly luciferase activity was measured with a luminometer (Thermo LabSystems). A total of 20 μl of pooled cell lysates from the same transfection group were boiled with SDS sample buffer and separated by SDS-PAGE. After electrophoresis and membrane transfer, Western blotting was performed with anti-FLAG-horseradish peroxidase (HRP) antibody (Sigma) (for FLAG-tagged CBF-β, FLI-1, GATA-1, and FOG-1) or anti-V5-HRP (for V5-tagged RUNX-1) antibody (Invitrogen).
ChIP assays.
Primary murine fetal liver cells were harvested from E13.5 C57BL/6 mouse embryos and cultured in the presence of 1% thrombopoietin (TPO) conditional medium (56) for 4 days, and mature megakaryocytes were enriched by the discontinuous bovine serum albumin density gradient, as previously described (7). Murine L8057 megakaryoblastic cells were induced with 50 nM TPA for 3 days. GATA-1 antibody (N-6), Fli-1 antibody (c-19), normal rabbit serum, and normal rat serum were purchased from Santa Cruz, and Runx-1 antibody (catalog no. 39000) was purchased from Active Motif. Chromatin immunoprecipitation (ChIP) was performed as previously described, with modifications (65). Dynabeads protein A (100.02D; Invitrogen) was used instead of Sepharose protein A beads. For GATA-1 ChIP, beads were incubated with rabbit anti-rat immunoglobulin G(H+L) [IgG(H+L)] (Jackson Research Laboratories, Inc.) before incubation with the antibody. Antibodies were incubated with beads for 3 h before being incubated with sonicated chromatin overnight, and two washes with 0.25 M LiCl were added before the washes with Tris-EDTA. Precipitated DNA was quantified using real-time PCR and a QuantiTect SYBR green PCR kit (Qiagen) on an iCycler system (Bio-Rad). The level of enrichment relative to a negative control region located 7.5 kb upstream of the c-mpl promoter transcriptional start site was calculated using the threshold cycle method (61). The PCR primer sequences are as follows: c-mpl, 5′-CTG CCA ACA GAA GGC TCA TG-3′ (forward) and 5′-CTG TCA GAT ACA GCC CCA CGT-3′ (reverse); GPIIb, 5′-ACC CCT CTG CCC AGT ATT GA-3′ (forward) and 5′-AAT GCT GTG ATG TGG CCA AG-3′ (reverse); and Upstream (−7.5 kb), 5′-GAACTCCAGTTCCGGAGGAT-3′ (forward) and 5′-GTGTGCCCTTGAGTCCTTGT-3′ (reverse).
Retroviral infection of primary megakaryocytes.
E13.5 fetal livers from C57BL/6 mice were harvested. Single-cell suspensions were cultured in DMEM containing 10% fetal calf serum (heat inactivated) and 1% TPO- and 1% Kit ligand-conditioned medium for 24 h. The medium containing fresh retrovirus particles with MIhCD4, MIhCD4-FLI-1, MIhCD4-FLI-1 S10A, or MIhCD4-FLI-1 S10D packaged in PLAT-E cells was placed into RetroNectin (Takara, Japan)-precoated plates, according to the manufacturer's instructions. Primary fetal liver cells were transferred to the plate and centrifuged at 600 × g for 90 min at room temperature in the presence of 6 mg/ml Polybrene (Sigma). A second infection was performed 24 h later. After 2 days, primary fetal liver cells were placed back in culture with DMEM containing 10% fetal calf serum and 1% TPO-conditioned medium for an additional 12 days. Medium was changed every 3 days. Cells were then stained with CD41-fluorescein isothiocyanate (BD Pharmingen), CD42b-PE (Emfret), human CD4 (hCD4)-biotin, and SA-allophycocyanin (BD Pharmingen) antibody. CD41+/CD42b+ cells from the hCD4+ cell populations were evaluated by a FACSCalibur flow cytometer (BD Biosciences). All experiments involving mice were approved by the Animal Care and Use Committees at Children's Hospital Boston.
RESULTS
RUNX-1 multiprotein complex purification from megakaryocytic cells.
The murine megakaryoblastic cell line L8057 was chosen as a source of material because it can be expanded to a large cell number and can be chemically induced to differentiate using TPA (19). Metabolic biotin tagging of RUNX-1 was utilized to facilitate multiprotein complex purification via SA affinity chromatography (6, 62). Clonal L8057 cell lines were generated that stably express the bacterial biotin ligase birA alone or birA and a recombinant form of RUNX-1 (murine isoform 3) containing a 23-amino-acid birA recognition motif fused to its amino terminus. A FLAG epitope tag was placed amino terminally to the birA recognition motif to allow for tandem affinity purification (Fig. 1A). A clone was chosen that expresses the recombinant RUNX-1 at levels similar to that of endogenous RUNX-1 isoform 3, as shown by Western blot analysis using an anti-RUNX-1 antibody (Fig. 1A, left). (Note that the recombinant protein migrates just slightly slower than the endogenous protein [upper endogenous band] in this Western blot.) Stripping this blot and reprobing it with SA-conjugated HRP reveals in vivo biotinylation of the recombinant RUNX-1 in the experimental, but not control (birA alone), cells (Fig. 1A, right).
FIG. 1.
RUNX-1 multiprotein complex purification from induced murine megakaryoblastic L8057 cells. (A) Metabolic biotin tagging of RUNX-1 in L8057 cells. Schematic diagram (top) shows coexpression of birA and biotin-tagged RUNX-1 in L8057 cells. The biotin acceptor lysine is indicated by an asterisk. A FLAG epitope tag is included amino terminally to the birA recognition motif. Western blot (WB) analysis of nuclear extracts from stable L8057 clonal lines expressing birA alone or birA and recombinant-tagged RUNX-1 using anti-RUNX-1 antibody (bottom left) and SA-conjugated HRP (SA-HRP) (bottom right). Two endogenous RUNX-1 isoforms are indicated, with the larger corresponding to murine isoform 3. Recombinant RUNX-1 is indicated running slightly slower than endogenous isoform 3. (B) Schematic diagram showing anti-FLAG-SA tandem affinity purification of FLAG-BioRUNX-1-containing complexes. a-FLAG-agarose, anti-FLAG-agarose. (C) Colloidal Coomassie blue-stained SDS-PAGE gel of purified material from representative tandem affinity purification.
Tandem anti-FLAG immunoaffinity and SA affinity purification of crude nuclear extracts from TPA-induced cells was performed, and the copurified proteins were separated by SDS-PAGE (Fig. 1B and C). Proteins from the entire lane were digested in situ with trypsin and identified by microcapillary liquid chromatography-tandem mass spectrometry (LC− MS-MS) using the SEQUEST database search algorithm. Proteins also identified in samples from control cells (expressing birA alone) were considered nonspecific interactions. Results from five independent experiments are shown in Table 1. In addition to RUNX-1 and its previously reported interacting partners, CBF-β (37, 60), GATA-1 (10, 58), Sin3A (28), PRMT1 (66), and PML (33), we reproducibly and specifically isolated multiple peptides corresponding to the ets family transcription factor FLI-1 (see Fig. S1 in the supplemental material for identified peptides). Experimental variation in the number of peptides obtained for each of these proteins likely reflects differences in the amount of input material used, purification strategy, and sampling issues related to the LC−MS-MS analysis.
TABLE 1.
Partial list of RUNX-1-associated proteins identified by whole-lane MS
| Protein | No. of proteins identified in the indicated exptsf
|
Total no. of peptidesc | Total no. of peptides in controld | ||||
|---|---|---|---|---|---|---|---|
| Expt 1a | Expt 2a | Expt 3b | Expt 4b | Expt 5b | |||
| RUNX-1 | 67 | 25 | 9 | 3 | 19 | 123 | 0 |
| CBF-β | 32 | 2 | 2 | 3 | 3 | 42 | 1 |
| GATA-1 | 1 | 0 | 2 | 0 | 0 | 3 | 1 |
| PML | 20 | 6 | 0 | 0 | 0 | 26 | 0 |
| PRMT-1 | 0 | 0 | 4 | 3 | 0 | 7 | 0 |
| Sin3A | 0 | 0 | 3 | 0 | 0 | 3 | 0 |
| FLI-1 | 22 | 0 | 5 | 3 | 1 | 31 | 0 |
| Total no. of proteins | 183 | 46 | 358 | 243 | 73 | ||
| Total no. of specific proteinse | 152 | 14 | 219 | 143 | 22 | ||
Tandem anti-FLAG-SA affinity purification.
Single SA affinity purification.
Experiment-to-experiment variation in the number of peptides obtained for each of the proteins likely reflects differences in the amount of input material used, purification strategy, and/or sampling and detection issues related to the LC−MS-MS analysis. The total number of peptides corresponding to each protein from five independent purifications is shown.
From cells expressing birA alone.
Proteins not identified in cells expressing birA alone.
For each experiment, the total number of milligrams of input protein used was as follows: experiment 1, 288 mg; experiment 2, 120 mg; experiment 3, 170 mg; experiment 4, 55 mg; experiment 5, 100 mg.
Validation of RUNX-1-FLI-1 interactions and mapping of binding domains.
FLI-1 was first identified as the product of a gene activated by Friend viral complex insertion in murine erythroleukemia cells (2). It plays essential roles in megakaryopoiesis and vasculogenesis during normal development in mice (17, 49). Haploinsufficiency of FLI-1 causes macrothrombocytopenia in patients with Paris-Trousseau or Jacobsen syndrome, a constitutional 11q23 deletion disorder (17, 45). FLI-1 physically interacts with GATA-1 and acts synergistically with GATA-1 and FOG-1 to activate megakaryocyte-specific genes, such as αIIb (9, 61).
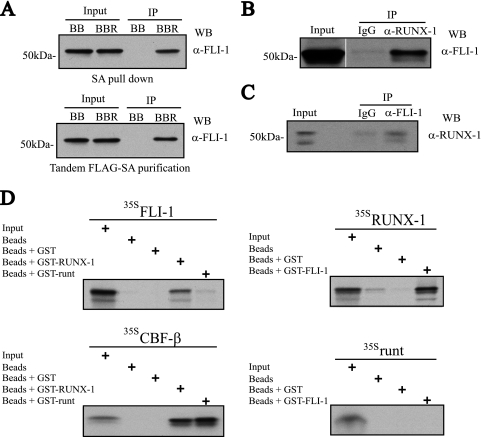
Western blot analysis of the FLAG-BioRUNX-1 single SA or tandem FLAG-SA affinity copurified proteins was first performed to validate the MS findings. As shown in Fig. 2A, a band at the expected molecular mass for FLI-1 (∼51 kDa) was observed from the experimental cells but not from control cells expressing the empty FLAG-biotinylation vector, confirming the MS results. Co-IP of endogenous proteins in induced L8057 cells using anti-RUNX-1 (Fig. 2B) or anti-FLI-1 (Fig. 2C), but not control IgG, as the immunoprecipitating antibody confirmed physical association of the native proteins. Treatment of the extracts with DNase, RNase, and ethidium bromide (50 μg/ml) (25) or the use of a non-DNA binding RUNX-1 mutant protein (RUNX-1R174A) (53) failed to disrupt binding (see Fig. 5A), suggesting that the interaction occurs via protein-protein contacts rather than through possible DNA or RNA-linked intermediates. In order to test for direct protein-protein interactions, GST pulldown experiments were performed on recombinant proteins produced in bacterial cells or by in vitro transcription and translation in the presence of [35S]methionine. As shown in Fig. 2D, 35S-labeled FLI-1 binds to GST-RUNX-1, and 35S-labeled RUNX-1 binds to GST-FLI-1, but neither binds to GST beads alone. The RUNX-1 runt domain by itself fails to appreciably interact with FLI-1 in these assays, although it binds to the control CBF-β, demonstrating specificity of the experimental system. It also suggests that the FLI-1-interacting region of RUNX-1 lies outside of the runt domain.
FIG. 2.
Physical interaction between RUNX-1 and FLI-1. (A) FLI-1 Western blot (WB) analysis of single SA (top)- or tandem anti-FLAG-SA (bottom)-purified FLAG-BioRUNX-1 complexes from TPA-induced L8057 cells. BB, birA plus empty FLAG-biotinylation vector; BBR, birA plus FLAG-BioRUNX-1 expression vector; α-, anti-; IP, immunoprecipitation. (B) Co-IP of endogenous RUNX-1 and FLI-1 from TPA-induced L8057 cells. Immunoprecipitation with normal rabbit IgG or anti-RUNX-1 antibody, and Western blot with anti-FLI-1 antibody. A 2% input is shown. (C) Immunoprecipitation with normal rabbit IgG or anti-FLI-1 antibody, and Western blot with anti-RUNX-1 antibody. A 2% input is shown. (D) Direct RUNX-1-FLI-1 protein-protein interactions. In vitro transcribed and translated [35S]methionine-labeled (35S) FLI-1, RUNX-1, runt domain, or CBF-β was incubated with uncoupled Sepharose beads or beads coupled with bacterially produced GST, GST-RUNX-1, GST-FLI-1, or GST-runt domain, as indicated. The beads were washed, and eluted material was separated by SDS-PAGE. An autoradiogram of the gel is shown. Ten percent of the 35S-labeled input protein is included.
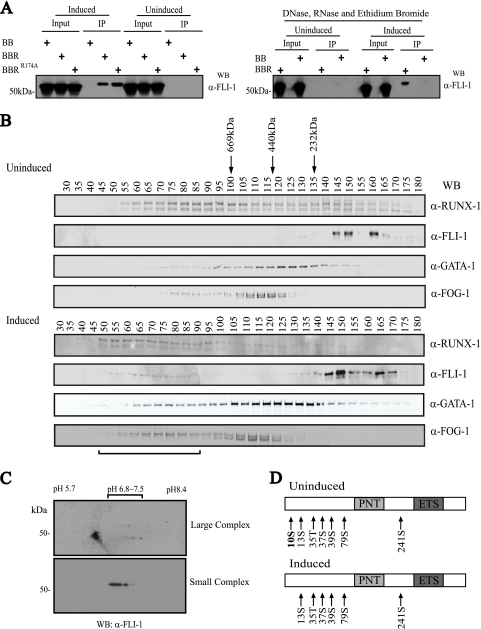
FIG. 5.
Posttranslational modification of FLI-1 correlates with assembly of a large multiprotein complex. (A) Differentiation-dependent and DNA-/RNA-independent interaction between RUNX-1 and FLI-1. (Left) Western blot (WB) for FLI-1 of eluted material, following tandem anti-FLAG-SA affinity purification of FLAG-BioRUNX-1 multiprotein complexes from TPA-induced (3 days) or uninduced L8057 cells. BB, birA and empty FLAG-biotinylation vector; BBR, birA and FLAG-BioRUNX-1; BBRR174A, birA and non-DNA binding mutant FLAG-BioRUNX-1R174A. (Right) The same tandem affinity purification as that for the left panel, but nuclear extracts were treated with DNase, RNase, and ethidium bromide (50 μg/ml) prior to and during co-IP. α-, anti-. (B) Sephacryl 400-gel filtration chromatography of crude nuclear extracts from uninduced (top) and 3-day TPA-induced (bottom) L8057 cells. Western blot analysis of every fifth fraction for RUNX-1, FLI-1, GATA-1, and FOG-1 is shown. Migration positions of globular molecular weight standards are indicated. The void volume corresponds to fractions 19 to 20. Horizontal bar at bottom indicates fractions containing the “large” FLI-1 complex. (C) 2-D gel electrophoresis and anti-FLI-1 Western blot of pooled fractions 66 and 71 (large complex) (top) and pooled fractions 156 and 161 (small complex) (bottom) from the gel filtration chromatography shown in panel B, bottom. (D) Summary of MS phosphorylation analysis of immunopurified FLI-1 from uninduced and induced L8057 cells. Detected phosphorylated residues are indicated. PNT, pointed domain.
More-detailed interaction domain mapping was performed by co-IP experiments using epitope-tagged proteins expressed in 293T cells (Fig. 3). For FLI-1, a region encompassing the ETS DNA binding domain region (amino acids 281 to 361) is sufficient and necessary for its interaction with RUNX-1, although some residues in the carboxy-terminal region (amino acids 361 to 452) contribute to binding. For RUNX-1, regions carboxy terminal to the runt domain that include the negative regulatory DNA binding domain (amino acids 179 to 291) or activation domain (amino acids 292 to 370) are sufficient for interaction with FLI-1. The inputs for some of the smaller constructs in both cases were not visualized by Western blotting. This may reflect loss of the protein from the bottom of the gel or poor retention in the nitrocellose due to their small size. However, this does not alter the overall conclusions of the mapping study.
FIG. 3.
Mapping of RUNX-1 and FLI-1 interaction domains. (A) Mapping of FLI-1 domains involved in RUNX-1 interaction. (Left) Schematic diagram of constructs used in the right panel. (Right) 293T cells were cotransfected with the FLAG-tagged FLI-1 constructs and V5-tagged RUNX-1. Nuclear extracts were immunoprecipitated (IP) with anti-FLAG antibody, and the copurified material was analyzed by Western blotting (WB) using the indicated antibodies. Input represents 2.5% of the starting material. α-, anti-; Vec, vector; NRBD, negative regulatory DNA binding domain; AD, activation domain; PNT, pointed domain. (B) Mapping of RUNX-1 domains involved in FLI-1 interaction. (Left) Schematic diagram of constructs used in the right panel. (Right) 293T cells were cotransfected with V5-tagged RUNX-1 constructs and FLAG-tagged FLI-1. Nuclear extracts were immunoprecipitated with anti-V5 antibody, and the copurified material was analyzed by Western blot analysis using the indicated antibodies. Input represents 2.5% of the starting material.
Functional synergism between RUNX-1 and FLI-1 in megakaryocyte gene transcription.
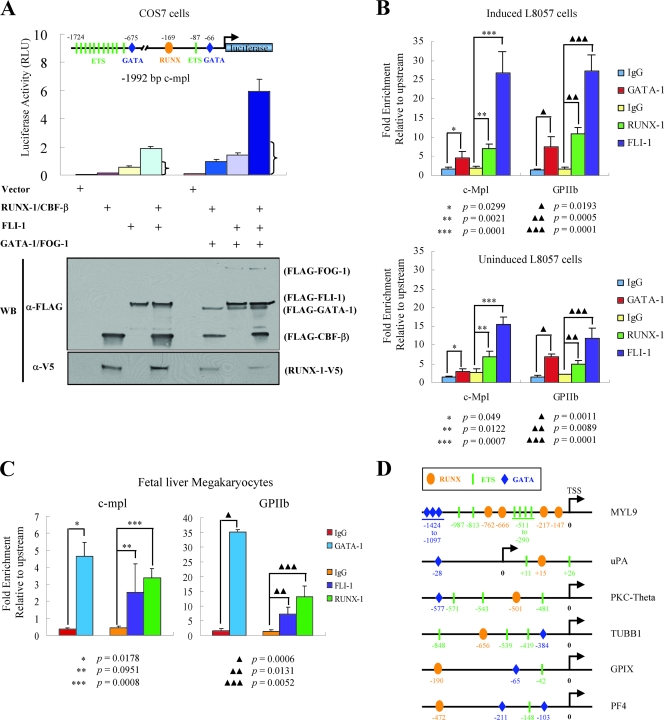
We next examined whether RUNX-1 and FLI-1 function synergistically in megakaryocyte-specific gene regulation. Transcriptional reporter assays were performed, using the c-mpl promoter as a model (61). A construct was generated consisting of firefly luciferase cDNA under the control of 1,992 bp of DNA sequence located 5′ to the murine c-mpl transcriptional start site. This region contains a RUNX binding motif as well as several ETS and GATA consensus sites (Fig. 4A). The reporter construct and expression plasmids for RUNX-1/CBF-β and FLI-1 were transiently transfected into COS-7 cells, and the level of firefly luciferase activity was measured. Expression levels of the constructs were equivalent among the different samples, indicating similar transfection efficiencies (Fig. 4A). The combination of RUNX-1/CBF-β and FLI-1 produced significantly greater activity than either RUNX-1/CBF-β or FLI-1 alone, indicating a synergistic effect (31.2-fold-greater activity than that of the vector alone for the combination of RUNX-1/CBF-β/FLI-1 versus 2.2-fold greater for RUNX-1/CBF-β or 9.6-fold greater for FLI-1 by themselves). The same pattern was evident when GATA-1 and FOG-1 were included, but the overall transcriptional activity levels were even greater (75-fold-greater activity for the combination versus 11.9-fold greater for RUNX-1/CBF-β or 17.7-fold greater for FLI-1 by themselves) (Fig. 4A).
FIG. 4.
Functional synergy between FLI-1 and RUNX-1. (A) Transcriptional reporter assays of the c-mpl promoter. A reporter construct consisting of 1,992 bp of the murine c-mpl promoter linked to the firefly luciferase cDNA was cotransfected with an empty expression plasmid or plasmids expressing RUNX-1-V5 and/or FLAG-CBF-β, FLAG-FLI-1, FLAG-GATA-1, and FLAG-FOG-1, as indicated. The positions of the RUNX, ETS, and GATA binding motifs relative to the natural transcriptional start site are indicated in the diagram of the reporter construct. RUNX, oval; ETS, rectangle; GATA, diamond. Luciferase activity was measured after 48 h and is indicated on the y axis. The mean ± standard error of the mean from three independent transfections is indicated. The bracket indicates the expected level if RUNX-1/CBF-β and FLI-1 acted additively (sum of the activation of RUNX-1/CBF-β and FLI-1 alone). Western blot (WB) analysis showing expression levels of the various proteins (10% of whole-cell lysate) from one representative experiment is displayed at the bottom. RLU, relative light units; α-, anti-. (B) Quantitative ChIP assays showing occupancy of the c-mpl and GPIIb promoters by GATA-1, RUNX-1, and FLI-1 in induced (TPA, 50 nM, for 72 h) (top) and uninduced (bottom) L8057 cells. The levels of enrichment at the c-mpl and GPIIb promoters using anti-GATA-1, anti-RUNX-1, or anti-FLI-1 antibodies or control rat (for GATA-1) or rabbit (for RUNX-1 and FLI-1) IgG are shown relative to a negative control region located −7.5 kb upstream of the c-mpl promoter transcriptional start site. Results represent the mean ± standard deviation from three independent experiments. (C) Quantitative ChIP assay for GATA-1, RUNX-1, and FLI-1 occupancy of the c-mpl and GPIIb promoters in mature primary fetal liver-derived megakaryocytes and as described in panel B. The results show the mean from three independent experiments for RUNX-1 and FLI-1 and two independent experiments for GATA-1, ± standard deviation. (D) Schematic diagram showing the presence of putative RUNX, ETS, and GATA factor binding motifs in the promoters of the genes for human MYL9, uPA, PKC-theta, β1-tubulin (TUBB1), GPIX, and rat PF4.
ChIP assays in induced L8057 cells (Fig. 4B, top) and primary murine fetal liver megakaryocytes (Fig. 4C) show occupancy of c-mpl as well as the GPIIb promoters by both RUNX-1 and FLI-1 (and GATA-1) in vivo. The occurrence of nearby RUNX and ETS factor binding motifs is also found in the promoters of other megakaryocyte-specific genes, such as MYL9, uPA, PKC-theta, β1-tubulin, GPIX, and PF4 (Fig. 4D). Several of these genes have been found to be reduced in expression in a patient with FPD/AML (51, 52) or have been previously implicated as a direct RUNX-1 target gene (23). Collectively, these data suggest that RUNX-1 and FLI-1 act together to directly regulate megakaryocyte-specific genes.
Differentiation-dependent interactions between RUNX-1 and FLI-1.
The interaction experiments described so far were performed with L8057 cells that were treated with TPA for 72 h. Surprisingly, when we examined uninduced L8057 cells, little if any binding between RUNX-1 and FLI-1 was observed, even though both proteins are abundantly expressed (Fig. 5A). The differentiation-dependent interaction between RUNX-1 and FLI-1 is also evident by gel filtration chromatography assays (Fig. 5B). In uninduced cells, FLI-1 migrates as relatively small complexes of <232 kDa, and RUNX-1 migrates as broad peaks, with regions of enrichment at >669 kDa and <232 kDa. However, after induction with TPA, a small proportion of the FLI-1 population elutes as a much larger complex (>669 kDa) and comigrates with a portion of RUNX-1. A subpopulation of GATA-1 and FOG-1 molecules also shifts to the larger peak fractions containing both RUNX-1 and FLI-1 upon TPA stimulation. These findings are consistent with the assembly of a large multiprotein complex involving RUNX-1, FLI-1, GATA-1, and FOG-1 upon TPA-induced differentiation of L8057 cells. Consistent with this, ChIP assays show enhanced enrichment, particularly for FLI-1, at the c-mpl and GPIIb promoters after TPA induction of L8057 cells (Fig. 4B, compare top and bottom panels).
TPA-induced dephosphorylation of FLI-1.
The reason for the enhanced FLI-1 and RUNX-1 interaction in induced L8057 cells, relative to uninduced cells, could be due to a TPA-dependent posttranslational modification of one or more of the complex components. Since the interaction is easily detectable in non-TPA-treated heterologous cells (Fig. 3), this suggests that RUNX-1-FLI-1 interactions are actively inhibited in uninduced L8057 cells. In order to examine potential changes in FLI-1 modifications that correlate with its participation in the large multiprotein complex, gel filtration fractions containing the large or small FLI-1 complexes were pooled separately, concentrated, and examined by 2-D gel electrophoresis and Western blot analysis. As shown in Fig. 5C, the majority of FLI-1 from the large complex migrates as a smaller and more-basic species compared to FLI-1 from the small complex.
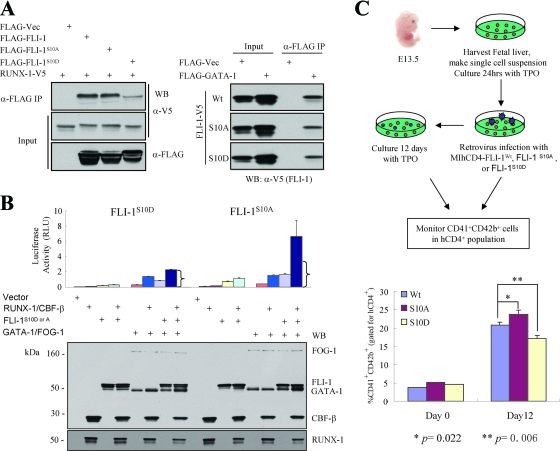
Since dephosphorylation could explain part of these migration differences, phosphorylation analysis of endogenous FLI-1 from uninduced rather than induced L8057 cells was performed. Phosphorylation of seven sites (Ser10, Ser13, Thr35, Ser37, Ser39, Ser79, and Ser241) was detected by MS on FLI-1 from uninduced cells (Fig. 5D; see also Fig. S2 in the supplemental material). However, only six of these sites (Ser13, Thr35, Ser37, Ser39, Ser79, and Ser241) were detected from induced cells. This raised the possibility that dephosphorylation of Ser10 may facilitate interaction between FLI-1 and RUNX-1. In order to test this, mutant FLI-1 molecules were generated that replace serine at codon 10 with either alanine, to block phosphorylation, or aspartic acid, to mimic constitutive phosphorylation. As shown in Fig. 6A and Fig. S3 in the supplemental material, FLI-1S10A interacts with RUNX-1 about as well as the wild-type molecule in COS-7 cells. In contrast, FLI-1S10D has markedly reduced binding. The effects are specific to RUNX-1, since FLI-1S10D binds GATA-1 equivalent to wild-type FLI-1 or FLI-1S10A (Fig. 6A).
FIG. 6.
Requirement of FLI-1 Ser10 dephosphorylation for efficient RUNX-1-FLI-1 interaction and megakaryocytic differentiation. (A) FLI-1 S10D mutation disrupts RUNX-1-FLI-1 interaction (left) but does not interfere with FLI-1-GATA-1 interaction (right). (Left) The empty FLAG vector or FLAG-tagged wild-type FLI-1, FLI-1S10A, or FLI-1S10D mutants were coexpressed with RUNX-1-V5 in COS-7 cells, as indicated. (Right) The empty FLAG vector or FLAG-tagged GATA-1 was cotransfected with V5-tagged wild-type FLI-1, FLI-1S10A, or FLI-1S10D mutants, as indicated. In both experiments, immunoprecipitation (IP) was performed using an anti-FLAG antibody from crude nuclear extracts, and copurified material was analyzed by Western blotting, using anti-V5-HRP or anti-FLAG-HRP. A 2.5% input is shown. Representative Western blots are shown. See Fig. S3 in the supplemental material for repeat co-IP experiments and co-IP in the reverse direction. Vec, vector; α-, anti-. (B) Loss of FLI-1/RUNX-1 synergistic c-mpl activation by FLI-1S10D substitution. Transcriptional reporter assays were performed as described in the legend to Fig. 4, except FLAG-FLI-1S10A and FLAG-FLI-1S10D were substituted for FLAG-tagged wild-type FLI-1. Brackets indicate the expected activity level if RUNX-1/CBF-β and the FLI-1 mutants were acting additively only. The bottom panel shows Western blot analysis of all the expressed proteins; 10% input of whole-cell lysis is shown. RLU, relative light units. (C) Dominant-negative effect of FLI-1S10D on differentiation of primary megakaryocytes. (Top) Schematic diagram showing experimental design. (Bottom) Percentage of CD41+/CD42b+ double-positive cells (after gating for human CD4+ population) immediately after the 2-day retroviral infection (day 0) or 12 days after retroviral infection. Results represent the mean ± standard error of the mean from three independent experiments. P values (Student's t test) were calculated relative to the wild-type overexpression sample. Wt, wild type.
We next used these mutant FLI-1 molecules in transcriptional synergy assays of FLI-1 and RUNX-1. As shown in Fig. 6B, FLI-1S10A synergistically activates the c-mpl promoter with RUNX-1 in COS-7 cells, similar to that observed for wild-type FLI-1 (Fig. 4A). In contrast, FLI-1S10D fails to synergize. Western blot analysis of cell lysates shows equivalent expression levels of the transfected constructs in the different samples. In addition, proper nuclear localization of FLI-1S10D was confirmed by indirect immunofluorescence microscopy (data not shown). Collectively, these findings suggest that dephosphorylation of FLI-1 at Ser10 facilitates the assembly of a complex with RUNX-1 (and likely with GATA-1/FOG-1) and activation of genes involved in megakaryocyte maturation. High evolutionary conservation of serine 10 of FLI-1 and surrounding amino acid sequences is consistent with a critical regulatory role for this portion of the molecule (see Fig. S4 in the supplemental material).
Effect of FLI-1 Ser10 phosphorylation on primary megakaryocyte differentiation.
In order to examine the functional significance of FLI-1 Ser10 phosphorylation in a more physiologic setting, wild-type FLI-1 or the mutants FLI-1S10A and FLI-1S10D were retrovirally overexpressed in primary fetal liver megakaryocyte precursors, and terminal maturation was assessed by flow cytometric analysis for cell surface coexpression of CD41 and CD42b after culturing in the presence of TPO (Fig. 6C). The retroviral vectors coexpress human CD4 on the cell surface to track transduced cells. Immediately after the 2-day retroviral transduction, levels of double-positive CD41+/CD42b+ cells were relatively low and not significantly different between the different samples (after gating for hCD4+ cells). After 12 days of additional culture, the percentage of CD41+/CD42b+ cells increased in all samples, but the cells overexpressing FLI-1S10A had a greater number (P of <0.02; Student's t test) and cells overexpressing Fli-1S10D had a lower number (P of <0.01; Student's t test) of double-positive cells compared to that of the wild-type molecule. There were an insufficient number of cells to confirm equivalent expression of the different constructs at the protein level. However, similar levels of coexpressed hCD4 were found in each of the transduced samples (see Fig. S5 in the supplemental material). Examination of total FLI-1 mRNA transcripts in the total hCD4+ cell population after 3 days of culture indicate an increase of ∼7- to 15-fold in Fli-1 transcripts over endogenous levels (after normalization using β-actin mRNA levels), but no significant difference in expression among the different constructs (see Fig. S5 in the supplemental material). No significant differences in overall protein stability of the different mutants were apparent when expressed in COS-7 cells (Fig. 6A). We conclude that FLI-1S10A and FLI-1S10D act in a dominant fashion to enhance and inhibit megakaryocyte maturation, respectively, when overexpressed in primary murine fetal liver megakaryocytes.
DISCUSSION
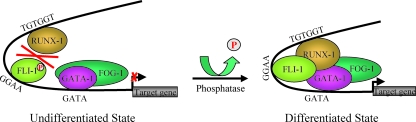
In this study, we demonstrate differentiation-dependent direct physical interactions between RUNX-1 and the ets transcription factor FLI-1 in megakaryocytic cells and show that they result in synergistic transcriptional activation of a megakaryocyte-specific gene. Moreover, we identify a key dephosphorylation step for FLI-1 that facilitates RUNX-1-FLI-1 interactions and leads to enhanced megakaryocyte differentiation in primary fetal liver megakaryocytes. Collectively, our findings suggest that a dephosphorylation-dependent interaction between RUNX-1 and FLI-1 may be a key event in activation of transcriptional programs of terminal megakaryocyte maturation (Fig. 7).
FIG. 7.
Model of differentiation-dependent dephosphorylation of FLI-1 and subsequent FLI-1/RUNX-1/GATA-1/FOG-1 multiprotein complex formation during megakaryocytic differentiation. P, phosphate group at serine 10.
Although we cannot exclude the possibility that the serine 10 mutations we utilized have effects on megakaryopoiesis unrelated to alterations in phosphorylation and/or RUNX-1 binding, this seems like a less likely possibility. First, the mutant proteins have stability, nuclear localization, and GATA-1 binding activity similar to those of wild-type FLI-1. Second, replacement of serine 10 by alanine and aspartic acid produces opposite functional consequences, which is difficult to explain if they were nonspecifically affecting protein structure.
FLI-1 was previously shown to physically interact with GATA-1 and synergistically transactivate megakaryocyte-specific genes such as αIIb with GATA-1 and FOG-1 (9, 61). Several groups have also reported direct interactions between GATA-1 and RUNX-1 (10, 58, 62). Our data using bacterially produced and in vitro transcribed/translated protein products argue that direct physical interactions also occur independently between FLI-1 and RUNX-1. This suggests that a large macromolecular enhancesome complex, involving RUNX-1, CBF-β, GATA-1, FOG-1, and FLI-1, assembles during megakaryocyte development and is stabilized by multiple protein-protein contacts. This model is supported by our findings of comigration of these components on gel filtration chromatography from induced L8057 cells (Fig. 5B), cooccupancy of the c-mpl and GPIIb promoters in primary megakaryocytes (Fig. 4C), and maximal transcriptional activation of the c-mpl promoter only when all five components are present (Fig. 4A).
Direct interactions between RUNX-1 and other ets family transcription factors, including MEF, ELF-1, NERF-2, ETS-1, and PU.1, have been previously described (13, 15, 22, 29, 42). Using a candidate gene approach, Mao et al. reported the interaction between RUNX-1 and FLI-1 in vitro (29). Our findings document this interaction in the context of native multiprotein complexes from megakaryocytic cells and demonstrate their functional importance in megakaryocyte development. Moreover, they uncover the differentiation-dependent nature of these interactions in megakaryopoiesis.
Functional activation of RUNX-1 by interaction with ETS-1 has been previously examined in the context of lymphocyte development (13, 15, 22). Our data provide additional support for the general principle of combinatorial control mechanisms between runx and ets family transcription factors. In the earlier studies, interaction between RUNX-1 and ETS-1 was shown to induce cooperative activity by simultaneously relieving DNA binding autoinhibition of both molecules. These interactions involve the ETS DNA binding domain and adjacent region of ETS-1 and the negative regulatory DNA binding region of RUNX-1. Our mapping studies show involvement of overlapping comparable regions in FLI-1 and RUNX-1 (Fig. 3). It is therefore likely that similar conformational changes and enhanced DNA binding occur upon FLI-1-RUNX-1 interactions during megakaryocyte development.
Pang et al. showed that FLI-1 and the related ets factor GABPα have overlapping but stage-specific effects on megakaryopoiesis, with GABPα involved in gene regulation during early megakaryocyte development and FLI-1 predominantly affecting late stages (41). The poor interaction between FLI-1 and RUNX-1 in uninduced cells could therefore be due to competition by GABPα. However, we have not been able to demonstrate significant interaction between GABPα and RUNX-1 in uninduced L8057 cells (H. Huang and A. Cantor, unpublished observation), suggesting that other mechanisms are likely involved.
How phosphorylation of serine 10 of FLI-1 inhibits interactions with RUNX-1 is unclear, given that we mapped the interacting domain of FLI-1 to reside in the carboxy-terminal region of the molecule (amino acids 281 to 452). It is possible that serine 10 is in close proximity to the binding region in 3-D space and/or that phosphorylation of serine has more-indirect effects on protein folding. Structural studies involving the entire FLI-1 molecule should provide important insights into this mechanism.
The importance of FLI-1 Ser10 dephosphorylation on RUNX-1-FLI-1 interactions begs the following questions: what kinase(s) is responsible for the phosphorylation in immature cells, and what phosphatase(s) is responsible for removing it during differentiation? Interestingly, Liu et al. showed that RUNX-1 recruits the Ca2+-/calmodulin-dependent protein phosphatase calcineurin to activate ETS-1 during T-cell receptor stimulation (27). This involves the removal of a phosphate group(s) located within the amino-terminal 250-amino-acid residues of ETS-1 that can be phosphorylated by glycogen synthase kinase-3β (GSK-3β) in vitro. Whether similar enzymes are involved in RUNX-1-FLI-1 synergy during megakaryocyte development remains to be determined. Binding of SDF-1 to its receptor CXCR4, an important event in the chemoattraction of megakaryocytes to the bone marrow sinusoidal vessel, causes a dose-dependent release of calcium from intracellular stores (1, 8). Whether these two processes are linked to the regulation of RUNX-1-FLI-1 interactions in megakaryocyte development will require further studies. However, such a scenario could help coordinate megakaryocyte maturation, with spatial cues within the bone marrow.
Germ line mutations in RUNX-1 cause FPD/AML, and acquired mutations are found in de novo MDS and FAB M0 AML. We tested several reported mutations that reside within the FLI-1 interacting domain for their effect on FLI-1 binding. None of the missense mutations we have tested to date disrupt RUNX-1-FLI-1 interactions, based on co-IP assays of overexpressed proteins in COS-7 cells. However, the loss of FLI-1 interaction was observed in two mutations that generate C-terminal truncated molecules, RUNX-1Y254X and RUNX-1R293X (see Fig. S6 in the supplemental material). Although we cannot conclude that a loss of RUNX-1-FLI-1 interactions is the cause of the abnormal hematopoiesis in these patients, it may be a potential contributing factor.
Megakaryocytic and erythroid cells are derived from a common bipotential progenitor cell (MEPs). FLI-1 promotes megakaryocytic differentiation and simultaneously represses erythroid differentiation (12). This is thought to be due to antagonism of the erythroid-specific transcription factor EKLF (3, 12, 50). Like FLI-1, RUNX-1 and CBF-β expression decreases as multipotent erythroid-megakaryocyte progenitor cells differentiate along the erythroid pathway (24, 35). Our findings of physical and functional complexes between FLI-1 and RUNX-1/CBF-β suggest that they may act cooperatively in determining cell fate choice of MEPs for the megakaryocyte lineage.
Transcriptional networks involving RUNX-1, FLI-1, ETS-1, and GATA-2 have recently been described in the control of hematopoietic stem cell emergence during embryogenesis (36, 43). In addition to binding GATA-1, RUNX-1 also efficiently interacts with GATA-2 (H. Huang and A. Cantor, unpublished observation). Our findings in this report may therefore have parallels in hematopoietic stem cell development, including phosphorylation-/dephosphorylation-dependent protein-protein interactions among these key transcription factors.
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIH (R01HL082952) and a Junior Faculty Scholar Award from the American Society of Hematology (A.B.C.).
We thank Nancy Speck for the murine RUNX-1 cDNA, Yaacov Ben-David for the murine FLI-1 cDNA, Ross Tomaino and Steven Gygi at the Taplin Mass Spectrometry Facility at Harvard Medical School for assistance with proteomics, and Zhe Li for critical review of the manuscript.
Footnotes
Published ahead of print on 26 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Avecilla, S. T., K. Hattori, B. Heissig, R. Tejada, F. Liao, K. Shido, D. K. Jin, S. Dias, F. Zhang, T. E. Hartman, N. R. Hackett, R. G. Crystal, L. Witte, D. J. Hicklin, P. Bohlen, D. Eaton, D. Lyden, F. de Sauvage, and S. Rafii. 2004. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nat. Med. 1064-71. [DOI] [PubMed] [Google Scholar]
- 2.Ben-David, Y., E. B. Giddens, K. Letwin, and A. Bernstein. 1991. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 5908-918. [DOI] [PubMed] [Google Scholar]
- 3.Bouilloux, F., G. Juban, N. Cohet, D. Buet, B. Guyot, W. Vainchenker, F. Louache, and F. Morle. 2008. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood 112576-584. [DOI] [PubMed] [Google Scholar]
- 4.Chang, A. N., A. B. Cantor, Y. Fujiwara, M. B. Lodish, S. Droho, J. D. Crispino, and S. H. Orkin. 2002. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl. Acad. Sci. USA 999237-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christiansen, D. H., M. K. Andersen, and J. Pedersen-Bjergaard. 2004. Mutations of AML1 are common in therapy-related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood 1041474-1481. [DOI] [PubMed] [Google Scholar]
- 6.de Boer, E., P. Rodriguez, E. Bonte, J. Krijgsveld, E. Katsantoni, A. Heck, F. Grosveld, and J. Strouboulis. 2003. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 1007480-7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drachman, J. G., D. F. Sabath, N. E. Fox, and K. Kaushansky. 1997. Thrombopoietin signal transduction in purified murine megakaryocytes. Blood 89483-492. [PubMed] [Google Scholar]
- 8.Durig, J., N. G. Testa, and C. M. Heyworth. 1999. Distinct biological effects of macrophage inflammatory protein-1alpha and stroma-derived factor-1alpha on CD34+ hemopoietic cells. Stem Cells 1762-71. [DOI] [PubMed] [Google Scholar]
- 9.Eisbacher, M., M. L. Holmes, A. Newton, P. J. Hogg, L. M. Khachigian, M. Crossley, and B. H. Chong. 2003. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol. Cell. Biol. 233427-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elagib, K. E., F. K. Racke, M. Mogass, R. Khetawat, L. L. Delehanty, and A. N. Goldfarb. 2003. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 1014333-4341. [DOI] [PubMed] [Google Scholar]
- 11.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5976-989. [DOI] [PubMed] [Google Scholar]
- 12.Frontelo, P., D. Manwani, M. Galdass, H. Karsunky, F. Lohmann, P. G. Gallagher, and J. J. Bieker. 2007. Novel role for EKLF in megakaryocyte lineage commitment. Blood 1103871-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goetz, T. L., T. L. Gu, N. A. Speck, and B. J. Graves. 2000. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol. Cell. Biol. 2081-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Growney, J. D., H. Shigematsu, Z. Li, B. H. Lee, J. Adelsperger, R. Rowan, D. P. Curley, J. L. Kutok, K. Akashi, I. R. Williams, N. A. Speck, and D. G. Gilliland. 2005. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu, T. L., T. L. Goetz, B. J. Graves, and N. A. Speck. 2000. Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1). Mol. Cell. Biol. 2091-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada, H., Y. Harada, H. Niimi, T. Kyo, A. Kimura, and T. Inaba. 2004. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood 1032316-2324. [DOI] [PubMed] [Google Scholar]
- 17.Hart, A., F. Melet, P. Grossfeld, K. Chien, C. Jones, A. Tunnacliffe, R. Favier, and A. Bernstein. 2000. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity 13167-177. [DOI] [PubMed] [Google Scholar]
- 18.Ichikawa, M., T. Asai, T. Saito, G. Yamamoto, S. Seo, I. Yamazaki, T. Yamagata, K. Mitani, S. Chiba, H. Hirai, S. Ogawa, and M. Kurokawa. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10299-304. [DOI] [PubMed] [Google Scholar]
- 19.Ishida, Y., J. Levin, G. Baker, P. E. Stenberg, Y. Yamada, H. Sasaki, and T. Inoue. 1993. Biological and biochemical characteristics of murine megakaryoblastic cell line L8057. Exp. Hematol. 21289-298. [PubMed] [Google Scholar]
- 20.Ito, Y. 2008. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 9933-76. [DOI] [PubMed] [Google Scholar]
- 21.Junt, T., H. Schulze, Z. Chen, S. Massberg, T. Goerge, A. Krueger, D. D. Wagner, T. Graf, J. E. Italiano, Jr., R. A. Shivdasani, and U. H. von Andrian. 2007. Dynamic visualization of thrombopoiesis within bone marrow. Science 3171767-1770. [DOI] [PubMed] [Google Scholar]
- 22.Kim, W. Y., M. Sieweke, E. Ogawa, H. J. Wee, U. Englmeier, T. Graf, and Y. Ito. 1999. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 181609-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopf, E., and R. Miskin. 2005. A RUNX/AML-binding motif residing in a novel 13-bp DNA palindrome may determine the expression of the proximal promoter of the human uPA gene. J. Thromb. Haemost. 32057-2064. [DOI] [PubMed] [Google Scholar]
- 24.Kundu, M., and P. P. Liu. 2003. Cbf beta is involved in maturation of all lineages of hematopoietic cells during embryogenesis except erythroid. Blood Cells Mol. Dis. 30164-169. [DOI] [PubMed] [Google Scholar]
- 25.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 896958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larson, M. K., and S. P. Watson. 2006. A product of their environment: do megakaryocytes rely on extracellular cues for proplatelet formation? Platelets 17435-440. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H., M. Holm, X. Q. Xie, M. Wolf-Watz, and T. Grundstrom. 2004. AML1/Runx1 recruits calcineurin to regulate granulocyte macrophage colony-stimulating factor by Ets1 activation. J. Biol. Chem. 27929398-29408. [DOI] [PubMed] [Google Scholar]
- 28.Lutterbach, B., J. J. Westendorf, B. Linggi, S. Isaac, E. Seto, and S. W. Hiebert. 2000. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275651-656. [DOI] [PubMed] [Google Scholar]
- 29.Mao, S., R. C. Frank, J. Zhang, Y. Miyazaki, and S. D. Nimer. 1999. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias. Mol. Cell. Biol. 193635-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikkola, H. K., J. Klintman, H. Yang, H. Hock, T. M. Schlaeger, Y. Fujiwara, and S. H. Orkin. 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421547-551. [DOI] [PubMed] [Google Scholar]
- 31.Mukouyama, Y., N. Chiba, T. Hara, H. Okada, Y. Ito, R. Kanamaru, A. Miyajima, M. Satake, and T. Watanabe. 2000. The AML1 transcription factor functions to develop and maintain hematogenic precursor cells in the embryonic aorta-gonad-mesonephros region. Dev. Biol. 22027-36. [DOI] [PubMed] [Google Scholar]
- 32.Nakao, M., S. Horiike, Y. Fukushima-Nakase, M. Nishimura, Y. Fujita, M. Taniwaki, and T. Okuda. 2004. Novel loss-of-function mutations of the haematopoiesis-related transcription factor, acute myeloid leukaemia 1/runt-related transcription factor 1, detected in acute myeloblastic leukaemia and myelodysplastic syndrome. Br. J. Haematol. 125709-719. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, L. A., P. P. Pandolfi, Y. Aikawa, Y. Tagata, M. Ohki, and I. Kitabayashi. 2005. Physical and functional link of the leukemia-associated factors AML1 and PML. Blood 105292-300. [DOI] [PubMed] [Google Scholar]
- 34.North, T., T. L. Gu, T. Stacy, Q. Wang, L. Howard, M. Binder, M. Marin-Padilla, and N. A. Speck. 1999. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 1262563-2575. [DOI] [PubMed] [Google Scholar]
- 35.North, T. E., T. Stacy, C. J. Matheny, N. A. Speck, and M. F. de Bruijn. 2004. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells 22158-168. [DOI] [PubMed] [Google Scholar]
- 36.Nottingham, W. T., A. Jarratt, M. Burgess, C. L. Speck, J. F. Cheng, S. Prabhakar, E. M. Rubin, P. S. Li, J. Sloane-Stanley, A. S. J. Kong, and M. F. de Bruijn. 2007. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 1104188-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa, E., M. Inuzuka, M. Maruyama, M. Satake, M. Naito-Fujimoto, Y. Ito, and K. Shigesada. 1993. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology 194314-331. [DOI] [PubMed] [Google Scholar]
- 38.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84321-330. [DOI] [PubMed] [Google Scholar]
- 39.Onodera, K., J. A. Shavit, H. Motohashi, M. Yamamoto, and J. D. Engel. 2000. Perinatal synthetic lethality and hematopoietic defects in compound mafG::mafK mutant mice. EMBO J. 191335-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Owen, C. J., C. L. Toze, A. Koochin, D. L. Forrest, C. A. Smith, J. M. Stevens, S. C. Jackson, M. C. Poon, G. D. Sinclair, B. Leber, P. R. Johnson, A. Macheta, J. A. Yin, M. J. Barnett, T. A. Lister, and J. Fitzgibbon. 2008. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood 1124639-4645. [DOI] [PubMed] [Google Scholar]
- 41.Pang, L., H. H. Xue, G. Szalai, X. Wang, Y. Wang, D. K. Watson, W. J. Leonard, G. A. Blobel, and M. Poncz. 2006. Maturation stage-specific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood 1082198-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrovick, M. S., S. W. Hiebert, A. D. Friedman, C. J. Hetherington, D. G. Tenen, and D. E. Zhang. 1998. Multiple functional domains of AML1: PU.1 and C/EBPalpha synergize with different regions of AML1. Mol. Cell. Biol. 183915-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pimanda, J. E., K. Ottersbach, K. Knezevic, S. Kinston, W. Y. Chan, N. K. Wilson, J. R. Landry, A. D. Wood, A. Kolb-Kokocinski, A. R. Green, D. Tannahill, G. Lacaud, V. Kouskoff, and B. Gottgens. 2007. Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl. Acad. Sci. USA 10417692-17697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preudhomme, C., D. Warot-Loze, C. Roumier, N. Grardel-Duflos, R. Garand, J. L. Lai, N. Dastugue, E. Macintyre, C. Denis, F. Bauters, J. P. Kerckaert, A. Cosson, and P. Fenaux. 2000. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2 alpha B gene in Mo acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood 962862-2869. [PubMed] [Google Scholar]
- 45.Raslova, H., E. Komura, J. P. Le Couedic, F. Larbret, N. Debili, J. Feunteun, O. Danos, O. Albagli, W. Vainchenker, and R. Favier. 2004. FLI1 monoallelic expression combined with its hemizygous loss underlies Paris-Trousseau/Jacobsen thrombopenia. J. Clin. Investig. 11477-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shivdasani, R. A., Y. Fujiwara, M. A. McDevitt, and S. H. Orkin. 1997. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 163965-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shivdasani, R. A., M. F. Rosenblatt, D. Zucker-Franklin, C. W. Jackson, P. Hunt, C. J. Saris, and S. H. Orkin. 1995. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell 81695-704. [DOI] [PubMed] [Google Scholar]
- 48.Song, W. J., M. G. Sullivan, R. D. Legare, S. Hutchings, X. Tan, D. Kufrin, J. Ratajczak, I. C. Resende, C. Haworth, R. Hock, M. Loh, C. Felix, D. C. Roy, L. Busque, D. Kurnit, C. Willman, A. M. Gewirtz, N. A. Speck, J. H. Bushweller, F. P. Li, K. Gardiner, M. Poncz, J. M. Maris, and D. G. Gilliland. 1999. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23166-175. [DOI] [PubMed] [Google Scholar]
- 49.Spyropoulos, D. D., P. N. Pharr, K. R. Lavenburg, P. Jackers, T. S. Papas, M. Ogawa, and D. K. Watson. 2000. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 205643-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Starck, J., N. Cohet, C. Gonnet, S. Sarrazin, Z. Doubeikovskaia, A. Doubeikovski, A. Verger, M. Duterque-Coquillaud, and F. Morle. 2003. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol. Cell. Biol. 231390-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, L., J. R. Gorospe, E. P. Hoffman, and A. K. Rao. 2007. Decreased platelet expression of myosin regulatory light chain polypeptide (MYL9) and other genes with platelet dysfunction and CBFA2/RUNX1 mutation: insights from platelet expression profiling. J. Thromb. Haemost. 5146-154. [DOI] [PubMed] [Google Scholar]
- 52.Sun, L., G. Mao, and A. K. Rao. 2004. Association of CBFA2 mutation with decreased platelet PKC-theta and impaired receptor-mediated activation of GPIIb-IIIa and pleckstrin phosphorylation: proteins regulated by CBFA2 play a role in GPIIb-IIIa activation. Blood 103948-954. [DOI] [PubMed] [Google Scholar]
- 53.Tahirov, T. H., T. Inoue-Bungo, H. Morii, A. Fujikawa, M. Sasaki, K. Kimura, M. Shiina, K. Sato, T. Kumasaka, M. Yamamoto, S. Ishii, and K. Ogata. 2001. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell 104755-767. [DOI] [PubMed] [Google Scholar]
- 54.Talebian, L., Z. Li, Y. Guo, J. Gaudet, M. E. Speck, D. Sugiyama, P. Kaur, W. S. Pear, I. Maillard, and N. A. Speck. 2007. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFbeta dosage. Blood 10911-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsang, A. P., Y. Fujiwara, D. B. Hom, and S. H. Orkin. 1998. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 121176-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villeval, J. L., K. Cohen-Solal, M. Tulliez, S. Giraudier, J. Guichard, S. A. Burstein, E. M. Cramer, W. Vainchenker, and F. Wendling. 1997. High thrombopoietin production by hematopoietic cells induces a fatal myeloproliferative syndrome in mice. Blood 904369-4383. [PubMed] [Google Scholar]
- 57.Vyas, P., K. Ault, C. W. Jackson, S. H. Orkin, and R. A. Shivdasani. 1999. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood 932867-2875.10216081 [Google Scholar]
- 58.Waltzer, L., G. Ferjoux, L. Bataille, and M. Haenlin. 2003. Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J. 226516-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 933444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, S., Q. Wang, B. E. Crute, I. N. Melnikova, S. R. Keller, and N. A. Speck. 1993. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 133324-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, X., J. D. Crispino, D. L. Letting, M. Nakazawa, M. Poncz, and G. A. Blobel. 2002. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 215225-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woo, A. J., T. B. Moran, Y. Schindler, S. K. Choe, N. B. Langer, M. R. Sullivan, Y. Fujiwara, B. H. Paw, and A. B. Cantor. 2008. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Mol. Cell. Biol. 282675-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie, H., M. Ye, R. Feng, and T. Graf. 2004. Stepwise reprogramming of B cells into macrophages. Cell 117663-676. [DOI] [PubMed] [Google Scholar]
- 64.Yokomizo, T., M. Ogawa, M. Osato, T. Kanno, H. Yoshida, T. Fujimoto, S. Fraser, S. Nishikawa, H. Okada, M. Satake, T. Noda, and Y. Ito. 2001. Requirement of Runx1/AML1/PEBP2alphaB for the generation of haematopoietic cells from endothelial cells. Genes Cells 613-23. [DOI] [PubMed] [Google Scholar]
- 65.Yu, M., M. Wan, J. Zhang, J. Wu, R. Khatri, and T. Chi. 2008. Nucleoprotein structure of the CD4 locus: implications for the mechanisms underlying CD4 regulation during T cell development. Proc. Natl. Acad. Sci. USA 1053873-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, X., V. Jankovic, A. Gural, G. Huang, A. Pardanani, S. Menendez, J. Zhang, R. Dunne, A. Xiao, H. Erdjument-Bromage, C. D. Allis, P. Tempst, and S. D. Nimer. 2008. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 22640-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.