Abstract
Diminished cyclooxygenase 2 (COX-2) expression in fibroblasts, with a resultant defect in the production of the antifibrotic mediator prostaglandin E2, plays a key role in the pathogenesis of idiopathic pulmonary fibrosis (IPF). Here, we have characterized the molecular mechanism. We found that COX-2 mRNA levels in fibroblasts from patients with IPF (F-IPF) were significantly lower than those in fibroblasts from nonfibrotic lungs (F-NL) after transforming growth factor β1 and interleukin-1β treatment but that COX-2 mRNA degradation rates were similar, suggesting defective transcription. A reporter gene assay showed that there were no clear differences between F-IPF and F-NL in transcription factor involvement and activation in COX-2 gene transcription. However, a chromatin immunoprecipitation assay revealed that transcription factor binding to the COX-2 promoter in F-IPF was reduced compared to that in F-NL, an effect that was dynamically linked to reduced histone H3 and H4 acetylation due to decreased recruitment of histone acetyltransferases (HATs) and increased recruitment of transcriptional corepressor complexes to the COX-2 promoter. The treatment of F-IPF with histone deacetylase (HDAC) inhibitors together with cytokines increased histone H3 and H4 acetylation. Both HDAC inhibitors and the overexpression of HATs restored cytokine-induced COX-2 mRNA and protein expression in F-IPF. The results demonstrate that epigenetic abnormality in the form of histone hypoacetylation is responsible for diminished COX-2 expression in IPF.
Chromatin structural changes, including alterations in the histone acetylation/deacetylation balance, have been reported to occur in cancer cells, where they may contribute to carcinogenesis (33). Here, we describe for the first time a defect in the epigenetic control of an antifibrotic gene in a fibrotic lung disorder. Idiopathic pulmonary fibrosis (IPF) is a progressive and lethal fibrotic lung disorder with a 5-year survival rate of less than 50% (22). IPF is characterized by inflammatory injury and irreversible fibrosis of the lung parenchyma; however, its pathogenesis is poorly understood. While steroids and other immunosuppressive agents serve as the standard treatment for IPF, they have proved to be inadequate (35). Thus, no effective therapy is currently available, and novel therapeutic strategies based on a more complete understanding of the pathogenesis of IPF are clearly needed (35). Fibroblast proliferation and excessive collagen production are the most important pathological hallmarks of IPF, which leads to dramatic changes in the lung architecture and progressive respiratory insufficiency. Fibroblast proliferation and collagen production are regulated by a complex interaction between profibrotic and antifibrotic mediators. Among the identified mediators, the cytokine transforming growth factor β1 (TGF-β1) and the lipid mediator prostaglandin E2 (PGE2) have been recognized as potent profibrotic and antifibrotic mediators, respectively, and are therefore critical in IPF pathogenesis (4, 12).
PGE2, a major eicosanoid product of lung fibroblasts (19), has been shown to inhibit lung fibroblast proliferation, reduce collagen levels by inhibiting the synthesis of collagen mRNA, and decrease fibroblast chemotaxis (30, 31, 34) and is thus an autocrine mediator that controls fibroblast cellular overactivation. PGE2 is produced from endogenous arachidonic acid via the cyclooxygenase (COX) pathway. COX exists in two isoforms: COX-1, the constitutive housekeeping isoform, and COX-2, inducible by inflammatory stimuli (12, 14, 15). These stimuli include TGF-β1 (28), tumor necrosis factor alpha (54), interleukin-1β (IL-1β), lipopolysaccharide, and phorbol myristate acetate (58), and thrombin (48). COX-2 induction by mediators and cytokines present in the inflammatory milieu of the lung may therefore represent an important mechanism by which fibroblasts can increase their capacity for PGE2 synthesis and thereby limit cellular proliferation and collagen synthesis. A defect in this homeostatic process may promote or sustain fibrosis in the lung. Indeed, studies have shown that although fibroblasts from IPF patients (F-IPF) and fibroblasts from nonfibrotic lungs (F-NL) have identical eicosanoid profiles and COX-1 protein expression levels, F-IPF synthesize significantly less PGE2 at the baseline than F-NL (28, 58). Moreover, the ability of F-IPF to release PGE2 in response to a variety of inducers is significantly impaired compared with that of F-NL due to the diminished abilities of these cells to upregulate COX-2 mRNA and protein expression (58). There is also a significant inverse correlation between the PGE2-synthetic capacity of F-IPF and the degree of fibrosis of the lung tissue from which the F-IPF were obtained (58). Consistent with results from studies of humans, COX-2-deficient mice are more susceptible to pulmonary fibrogenesis than COX-1-deficient and wild-type mice (6, 28) and the overexpression of COX-2 in the lungs of mice leads to an increase in PGE2 production by fibroblasts, accompanied by a decrease in fibroblast proliferation (27). Levels of PGE2 in the lavage fluids and the amounts produced by lavage fluid macrophages obtained from patients with IPF are reduced compared with those from control subjects (7). Furthermore, no COX-1 and COX-2 immunoreactivity in the fibroblastic foci and reduced COX-1 and COX-2 immunoreactivity in bronchiolar epithelial cells of IPF lungs have also been observed previously (42), and IL-1β significantly increases COX-2 expression in lung tissues from control subjects but not in those from IPF patients (59), further suggesting that diminished COX-2 expression may be a generalized abnormality in pulmonary cells of IPF patients. Thus, the decreased capacities of F-IPF to upregulate COX-2 expression and COX-2-derived PGE2 synthesis in the presence of increasing levels of profibrotic mediators such as TFG-β1 (9, 32) may result in a loss of the normal feedback inhibitory mechanisms, lead to unopposed fibroblast proliferation and collagen synthesis, and contribute to the pathogenesis of IPF.
The COX-2 gene is an immediate-early gene, and its expression is subject to multilevel regulation through both transcriptional and posttranscriptional mechanisms. Like the transcription of other inducible genes, that of the COX-2 gene is controlled by transcription factor activation and binding to recognition sequences on the gene promoter, as well as by changes in chromatin structure. Human COX-2 is encoded by a 7.5-kb genomic DNA segment with 10 exons (62). The 5′-end-flanking promoter region of human COX-2 contains a canonical TATA box and multiple regulatory elements, including two putative NF-κB binding sites, one CCAAT/enhancer binding protein (C/EBP) binding site, and one cyclic AMP response element (CRE) (52). Previous studies of the human COX-2 promoter by us and others have demonstrated that COX-2 expression is critically governed by different transcription factors, including CRE binding protein (CREB) (39, 60), C/EBP (21), activator protein 1 (AP-1) (50), and NF-κB (25), in a highly cell type-specific and stimulus-specific manner. In this study, we aimed to compare F-IPF with F-NL to explore the molecular mechanisms underlying the diminished COX-2 expression in F-IPF in response to TGF-β1 and IL-1β.
In quiescent cells, genomic DNA is wrapped around histones to form nucleosomes, restricting transcriptional access to the DNA. When cells are stimulated with extracellular mediators, histones in the chromatin undergo an array of posttranslational modifications to regulate gene transcription (23, 26). Histone acetylation on lysine residues by histone acetyltransferases (HATs) and histone deacetylation by histone deacetylases (HDACs) are associated with transcriptional activation and repression, respectively (23, 26). Several transcription coactivators, including p300, CREB binding protein (CBP), p300/CBP-associated factor (PCAF), and general control nonderepressible 5 (GCN5), have been found to possess intrinsic HAT activity (reviewed in reference 13), and at least 13 HDACs have been identified. HDAC complexes are generally recruited to transcription factors by “bridging” factors, such as nuclear receptor corepressor (NCoR), SMRT, co-RE1-silencing transcription factor (CoREST), and mammalian SIN3 homolog A (mSin3a). NCoR exists in core repression complexes with HDAC3. CoREST was identified initially as a corepressor for the RE1-silencing transcription factor (REST) (3, 10, 46). The CoREST complex and the mSin3a complex contain, in addition to CoREST and mSin3a, respectively, HDAC1 and HDAC2 (5, 24, 57, 64), providing a mechanism to mediate the repression of transcription. Evidence that these different HDAC complexes display different specificities for histone tails, targeting them to hypoacetylated chromatin, is emerging (56, 63).
Chromatin structural changes, including alterations in the histone acetylation/deacetylation balance, may contribute to carcinogenesis (33). We have shown previously that induced COX-2 gene transcription in human airway smooth muscle cells is closely associated with increased histone H4 acetylation (39). However, whether alterations in the histone acetylation/deacetylation balance result in the repression of COX-2 transcription in IPF is unknown. Here, we found that COX-2 gene transcription in F-IPF was defective compared to that in F-NL, due to deficient histone H3 and H4 acetylation as a result of decreased recruitment of HATs and increased recruitment of the NCoR, CoREST, and mSin3a transcriptional corepressor complexes to the COX-2 promoter. The results demonstrate that defective histone acetylation is responsible for diminished COX-2 gene transcription in IPF.
MATERIALS AND METHODS
Fibroblast cell culture.
F-IPF and F-NL from the explanted lungs of patients with IPF who underwent lung transplantation at the University of Pittsburgh Medical Center and from normal lung tissues obtained from organ donors under a protocol approved by the University of Pittsburgh Institutional Review Board (43) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin, streptomycin, and an antimycotic agent as described previously (43). Because alterations in eicosanoid profiles have been reported to accompany the serial passage of fibroblasts (44), F-IPF and F-NL cells (six cell lines each) were used at passages 5 and 6, respectively, to ensure purity and maintain the differences present in vivo, although there is evidence that the differences in maximal COX activity between F-NL and F-IPF can persist through the 12th passage (58). Comparisons of the responses of F-IPF and F-NL to TGF-β1 and IL-1β were used throughout the study.
COX-2 protein/mRNA expression and PGE2 production.
COX-2 protein expression in resting cells and cytokine-stimulated cells was examined by Western blotting as reported previously (39-41). mRNA expression was analyzed by real-time quantitative PCR (qPCR) with an MX3000P system (Stratagene) using Excite mastermix and Sybr green (Biogene, Cambridge, United Kingdom) and the following primers for the COX-2 gene: sense, 5′-GGAACACAACAGAGTATGCG-3′, and antisense, 5′-AAGGGGATGCCAGTGATAGA-3′ (Eurofins MWG Operon, Ebersberg, Germany). The housekeeping gene for β2-microglobulin (β-2M) was used to confirm the loading of equal amounts of RNA and to calculate the relative expression of specific gene transcripts by normalization as we described previously (38). The PGE2 concentration in the culture medium was measured by a commercially available enzyme-linked immunosorbent assay kit (Cayman Chemicals, MI).
COX-2 mRNA stability.
To assess whether COX-2 gene expression is posttranscriptionally regulated and whether this process is defective in F-IPF, we performed an actinomycin D (Act D) chase experiment as described previously (37) to analyze COX-2 mRNA stability. COX-2 mRNA levels in both F-IPF and F-NL at different time points after the addition of Act D (5 μg/ml) were measured by qPCR.
Transient transfection, DNA constructs, and reporter gene assays.
The COX-2-firefly luciferase reporter constructs (generated from pGL3 basic) containing the following different human COX-2 promoter fragments have been described in detail previously (8): C2.1 (positions −917 to +49 relative to the transcription start site; 966 bp), Dra (−625 to +49; 674 bp), Sty (−358 to +49; 407 bp), Alu (−190 to +49; 239 bp), Rsa (−86 to +49; 135 bp), and the Sty fragment with mutations in the NF-κB (−224 to −214), C/EBP (−132 to −124), or CRE (−59 to −53) site or both the NF-κB and CRE sites. The 5× CRE-luciferase construct (5× CRE Luc) was kindly donated by Steve Rees (GlaxoSmithKline); the 5× NF-κB construct (pGL3.6 kappaB.BG.Luc) was kindly donated by Robert Newton (University of Calgary, Canada) and was described previously (2); the C/EBP cis reporting system (pC/EBP-Luc) was obtained from Stratagene (La Jolla, CA). The internal control Renilla luciferase reporter construct pRL-SV40 was obtained from Promega (Southampton, United Kingdom). All transient transfections were conducted by using FuGene HD according to the recommended protocol of the manufacturer (Roche Molecular Biochemicals, East Sussex, United Kingdom). A total of 3.5 × 104 human F-NL or F-IPF cells were seeded into each well of 24-well plates. When the cells were 90% confluent after 72 h of growth, the cells were serum starved for 18 h prior to incubation with DMEM containing complexes of firefly luciferase reporter plasmids (0.4 μg/ml) and FuGene HD (2 μl), together with Renilla luciferase reporter plasmids at 4 ng/well as an internal control. After 18 h of incubation, the transfected cells were incubated with either IL-1β (1 ng/ml) or TGF-β1 (10 ng/ml) (both from R&D) for 4 h. The cells were then washed with phosphate-buffered saline and lysed in 100 μl of reporter lysis buffer (Promega). Firefly luciferase activity from the testing reporters and Renilla luciferase activity from the internal control reporter were measured by using the dual-luciferase reporter assay system (Promega) with a MicroLumatPlus LB96V automatic microplate luminometer (Berthold Technologies, Herts, United Kingdom). Relative luciferase activity values were obtained by normalizing the firefly luciferase activity against the internal control Renilla luciferase activity. The amounts of change (n-fold) were obtained by comparing relative luciferase activities from experimental groups against those from the respective controls. Data from Renilla luciferase activity showed that there was no difference in transfection efficiency between F-NL and F-IPF. The PCAF expression vector pCXFLAG-PCAF and the empty vector pCX were obtained from T. Kouzarides (Cambridge University, Cambridge, United Kingdom); the CBP and p300 expression vectors pCMVb-CBP and pCMVb-p300 and the empty vector pCMVb were obtained from D. Chakravarti (University of Pennsylvania, Philadelphia). Transfection with expression vectors was performed using TransFast transfection reagent according to the procedures recommended by the manufacturer (Promega Corporation, Madison, WI). Briefly, F-IPF cells were grown to 80% confluence in a six-well plate. Up to 4 μg of expression vectors or empty vectors was added to TransFast reagent in a ratio of 2:1, and the preparation was mixed and incubated at room temperature for 0.25 h. The transfection mixture was added to the six-well plates in a total volume of 1 ml of culture medium, and the plates were incubated for 1 h, after which the culture medium was made up to 3 ml and cells were incubated for 24 h. Cells were then serum starved for 24 h before being treated with IL-1β (1 ng/ml) for 4 and 24 h for analyses of COX-2 mRNA and protein expression, respectively.
ChIP assay.
To detect the in vivo binding of transcription factors to the human COX-2 promoter and other chromatin events in the native chromatin environment, a chromatin immunoprecipitation (ChIP) assay was conducted by using a ChIP-IT express kit as described by the manufacturer (Active Motif, Rixensart, Belgium), with some modifications. Briefly, human F-IPF and F-NL cells were seeded into 150-cm2 flasks, grown to confluence, and serum starved for 18 h. After treatment with either IL-1β (1 ng/ml) or TGF-β1 (2 ng/ml), protein-DNA complexes were fixed with 1% formaldehyde in DMEM. The fixed cells were washed and lysed in complete protease inhibitor cocktail and sonicated on ice. After centrifugation at 500 × g for 12 min, one portion of the chromatin supernatant was used as a chromatin input control, and the remains were subdivided into aliquots and then incubated with no antibody or a nonimmune rabbit immunoglobulin G (IgG) antibody (Santa Cruz, CA) as negative controls or with specific IgG antibodies against NF-κB p65 (New England Biolabs, Herts, United Kingdom); C/EBPβ, CREB-1, p300, CBP, PCAF, GCN5, CoREST, NCoR, and mSin3a (Santa Cruz); and acetylated and nonacetylated histones H3 and H4 (Millipore Corporation, CA) overnight at 4°C. The immunoprecipitated antibody-protein-chromatin complexes were collected by using magnetic particles, which were washed successively with low-salt and high-salt buffers and then eluted with elution buffer. DNA was digested with 10 mg/ml of proteinase K (Sigma) for 1 h at 45°C. The DNA was then extracted with phenol-chloroform, and the purified DNA pellet was resuspended in H2O and subjected to qPCR amplification with the following primers designed specifically for the COX-2 promoter region (positions −299 to +6): forward, 5′-AAGACATCTGGCGGAAACC-3′, and reverse, 5′-ACAATTGGTCGCTAACCGAG-3′ (39). To determine the specificity of the ChIP assay, distal controls upstream and downstream of the COX-2 promoter were applied to analyze changes in histone acetylation with primers designed specifically for different regions of the COX-2 gene: forward, 5′-TCAGCCCAACTGCTTATGTG-3′, and reverse, 5′-GGGAGTCATCTCGGTGTGAT-3′, for the region from −12360 to −12142; forward, 5′-CCCAACAAATTTCAGACGCT-3′, and reverse, 5′-TACATTTGGGATGCTGGTCA-3′, for the region from −2440 to 2206; forward, 5′-AAGTGGGTGCCATACTCAGC-3′, and reverse, 5′-GAGAAGGCTTCCCAGCTTTT-3′, for the region from +1727 to +2093; and forward, 5′-CTTCCATCTCCAAGACCCAA-3′, and reverse, 5′-TCTTCCTGCTAGGCTACCCA-3′, for the region from +21087 to +21307. The amounts of COX-2 promoter DNA that were present in the bound (immunoprecipitated) fractions were calculated relative to the input control by using the 2−ΔΔCT method, where ΔΔCT is the difference between the threshold cycle (CT) for the bound fraction and the CT for the input fraction. The amounts of COX-2 promoter DNA present in both nonantibody and nonimmune rabbit IgG negative control immunoprecipitates (IPs) were minimal and markedly smaller than those in the specific-antibody IPs. The associations of acetylated histones H3 and H4 with the COX-2 DNA were further normalized relative to the association of total histones H3 and H4 with the COX-2 DNA.
HDAC activity assay.
To detect global HDAC activity, nuclear extraction from F-IPF and F-NL cells was performed by using the CelLytic NuCLEAR extraction kit according to the instructions of the manufacturer (Sigma). An HDAC colorimetric detection assay was performed with 30 μg of the nuclear extract in a two-step procedure carried out in a microtiter plate according to instructions of the manufacturer (Millipore Corporation, CA). In the first step, samples were incubated with the HDAC assay substrate, allowing the deacetylation of the substrate. Next, the activator solution released p-nitroanilide from the deacetylated substrate or standard. HDAC activity was expressed as the absorbance at 405 nm.
HDAC inhibitor study.
To assess the role of HDACs in COX-2 repression in F-IPF, two general HDAC inhibitors were used. Suberoylanilide hydroxamic acid (SAHA) was purchased from Axxora (Nottingham, United Kingdom), and LBH589 (panobinostat) was generously provided by Peter Atadja (Novartis Pharmaceuticals). Confluent and serum-starved F-IPF cells were pretreated with or without SAHA (10 μM) or LBH589 (10 nM) for 1 h and then incubated with or without IL-1β (1 ng/ml) or TGF-β1 (2 ng/ml) for 4 and 24 h. Samples were then collected for analyses of histone H3 and H4 acetylation at the COX-2 promoter by a ChIP assay, COX-2 mRNA expression by qPCR, and COX-2 protein expression by Western blotting.
Statistical analyses.
Data were expressed as means ± standard errors of the means (SEM) of n determinations. Statistical analyses were performed by using GraphPad Prism software (version 4). An unpaired two-tailed Student t test was used to determine the significant differences between the means; P values of <0.05 were accepted as statistically significant.
RESULTS
COX-2 protein expression and PGE2 production.
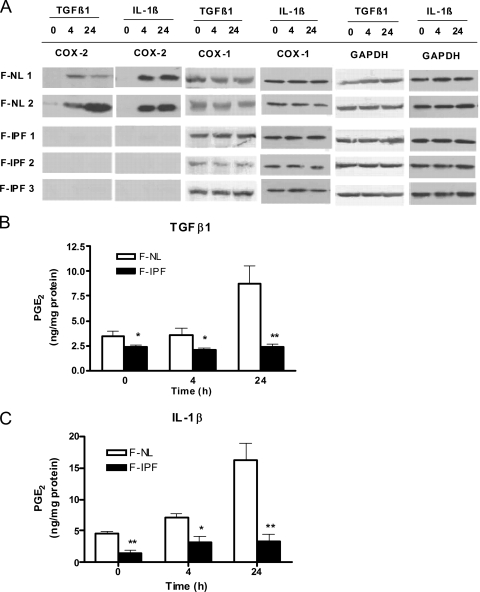
To confirm that COX-2 protein expression in F-IPF was reduced, we first treated both F-IPF and F-NL with TGF-β1 (2 ng/ml) and IL-1β (1 ng/ml) for 4 and 24 h and measured COX-2 protein by Western blotting. We found that both TGF-β1 and IL-1β failed to induce COX-2 protein expression in F-IPF at both 4 and 24 h; in contrast, both cytokines induced COX-2 protein expression in F-NL at 4 h, and the effect remained at 24 h (Fig. 1A). The expression of the housekeeping isoform COX-1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) in both F-IPF and F-NL was unchanged after cytokine treatment, and no difference between the cell groups was observed (Fig. 1A). All six F-IPF cell lines showed similarly diminished COX-2 expression levels compared to those in all six F-NL cell lines. The results shown in Fig. 1A are from a representative experiment. As PGE2 is a major product of COX-2 function, we also measured PGE2 production after cytokine treatment. The levels of PGE2 production in F-IPF in response to both TGF-β1 and IL-1β were significantly lower than those in F-NL, particularly after 24 h of treatment (Fig. 1B and C), correlating with the corresponding COX-2 protein levels in F-IPF and F-NL. The results thus confirmed that these F-IPF cell lines exhibited significantly diminished COX-2 expression and PGE2 production compared to those in F-NL in response to both TGF-β1 and IL-1β and were suitable for the proposed study to explore the underlying molecular mechanisms.
FIG. 1.
COX-2 protein expression and PGE2 production in F-NL and F-IPF in response to TGF-β1 and IL-1β. (A) Confluent and serum-starved cells were incubated with TGF-β1 (2 ng/ml) or IL-1β (1 ng/ml) for 4 and 24 h prior to the collection of total cell lysates for Western blot analyses of COX-2, COX-1, and the loading control GAPDH. The results shown are representative of data from three experiments. (B and C) Cell culture media from the above-mentioned experiments were also collected for the determination of PGE2 concentrations by an enzyme-linked immunosorbent assay. The results are expressed as means ± SEM of data from three separate experiments performed in duplicate or triplicate. *, P < 0.05, and **, P < 0.01 compared to results for corresponding F-NL.
COX-2 mRNA expression and stability.
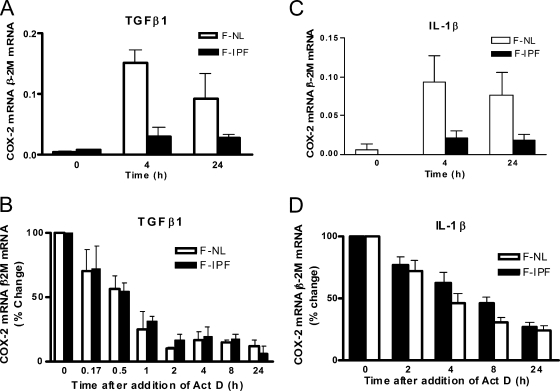
To assess whether COX-2 mRNA expression was impaired in F-IPF, we treated both F-IPF and F-NL with TGF-β1 (2 ng/ml) for 4 and 24 h and measured COX-2 mRNA by real-time reverse transcriptase PCR (RT-PCR). As shown in Fig. 2A and C, both TGF-β1 and IL-1β induced a small increase in COX-2 mRNA in F-IPF treated for 4 and 24 h compared with that in untreated cells; however, the levels were significantly lower than those in F-NL. To explore whether the reduced level of COX-2 mRNA in F-IPF was due to decreased mRNA stability, we performed the Act D chase experiment. We found that TGF-β1-induced COX-2 mRNA degraded faster than IL-1β-induced COX-2 mRNA; however, there was no difference in COX-2 mRNA degradation between F-IPF and F-NL (Fig. 2B and D). The results strongly suggest that defective transcriptional mechanisms are responsible for the diminished COX-2 expression in F-IPF.
FIG. 2.
TGF-β1- and IL-1β-induced COX-2 mRNA expression and stability in F-NL and F-IPF. (A and C) Confluent and serum-starved cells were incubated with either TGF-β1 (2 ng/ml) or IL-1β (1 ng/ml) for 4 and 24 h. (B and D) Confluent and serum-starved cells were incubated with either TGF-β1 (2 ng/ml) or IL-1β (1 ng/ml) for 4 h prior to incubation with the general transcription inhibitor Act D (5 μg/ml) for the indicated times. Total RNA was then isolated, and the levels of COX-2 and internal control β-2M mRNAs were determined by quantitative RT-PCR. The results are calculated as the ratio of COX-2 mRNA to β-2M mRNA (A and C) and the percent change of the ratio relative to the control value (that for 0 h) (B and D) and are expressed as means ± SEM of results from three separate experiments performed in duplicate or triplicate.
Transcription factor involvement.
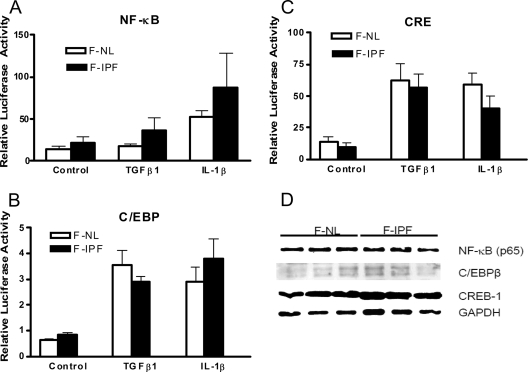
It has been demonstrated previously that COX-2 gene transcription is critically governed by different transcription factors in a highly cell type- and stimulus-specific manner. To determine the promoter region essentially required for TGF-β1- and IL-1β-induced COX-2 transcription, we transfected F-IPF and F-NL cells with reporter constructs containing various lengths of the human COX-2 promoter as described previously (8) and depicted in Fig. S1A in the supplemental material. We found that the promoter region downstream of bp −358 (in the Sty construct), containing binding sites for NF-κB, C/EBP, and CRE, was essential in COX-2 gene transcription in response to TGF-β1 and IL-1β and that there was no significant difference in COX-2 promoter activity between F-IPF and F-NL (see Fig. S1B and C in the supplemental material). To identify the transcription factors responsible for TGF-β1- and IL-1β-induced COX-2 transcription, the promoter activity of the Sty fragment (bp −358 to +49) was compared to those of constructs with site-directed mutations. The results showed that similar regulatory elements were required for human COX-2 transcription in human F-IPF and F-NL cells in response to TGF-β1 and IL-1β and that the CRE, C/EBP, and NF-κB sites acted cooperatively to achieve optimal COX-2 transcription in lung fibroblasts (see Fig. S2 in the supplemental material). To confirm that TGF-β1 and IL-1β could activate transcription factors that bind to NF-κB, C/EBP, and CRE sites in F-NL and F-IPF, a reporter gene assay using constructs each carrying five copies of the consensus sequence of one of the three elements was conducted. Following incubation with TGF-β1 and IL-1β, increases in the activities of all three reporter constructs over the control level in both F-IPF and F-NL were observed, with no significant difference between the cell groups (Fig. 3A to C). The levels of expression of the transcription factors (NF-κB p65, C/EBPβ, and CREB-1) that bind to the three elements was also analyzed by Western blotting, and no clear difference between F-NL and F-IPF was observed (Fig. 3D). These results strongly suggest that the activation and expression of the transcription factors required for COX-2 gene transcription in F-IPF are unaltered compared to those in F-NL.
FIG. 3.
Transcription factor expression and cytokine-induced activation in F-NL and F-IPF. (A to C) Serum-starved F-NL and F-IPF at 90% confluence in 24-well plates were cotransfected with a Renilla luciferase internal control reporter construct at 4 ng/well and pGL3.6 kappaB.BG. Luc (A), 5× CRE Luc (B), or pC/EBP-Luc (C) at 0.4 μg/well by using FuGene HD transfection reagent as described in Materials and Methods. Cells were then stimulated without or with TGF-β1 (10 ng/ml) or IL-1β (1 ng/ml) for 4 h. The luciferase activities from firefly and Renilla reporters were assayed by using the dual-luciferase reporter assay system, and the relative luciferase activity was obtained by normalizing the firefly luciferase activity against the internal control Renilla luciferase activity. The results are expressed as mean ± SEM of data from three separate experiments performed in triplicate. (D) Total cell lysates from confluent and serum-starved F-NL and F-IPF were also collected for Western blot analyses of CREB-1, C/EBPβ, NF-κB p65, and the loading control GAPDH as described in Materials and Methods.
Native binding of transcription factors to the COX-2 promoter.
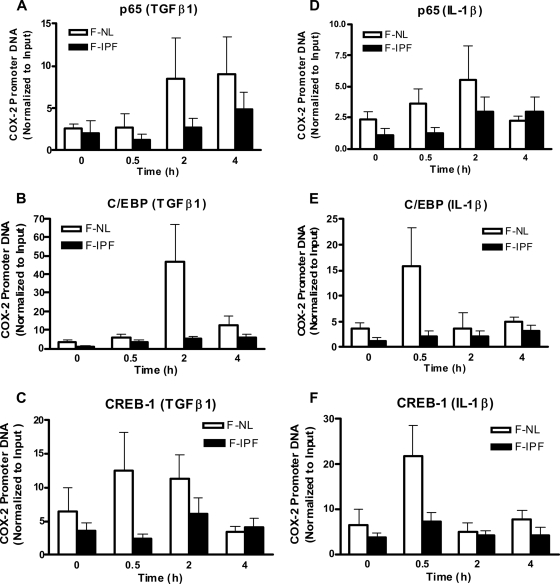
Since the activation and expression of transcription factors were not altered in F-IPF and therefore could not contribute to the diminished COX-2 transcription in IPF, we then went on to investigate whether the binding of transcription factors to the COX-2 promoter in the native chromatin environment was reduced in IPF by performing a ChIP assay using antibodies against the transcription factors NF-κB (p65), C/EBPβ, and CREB-1, which bind to the NF-κB, C/EBP, and CRE sites of the COX-2 promoter, respectively. After treatment of the cells with TGF-β1 (2 ng/ml) and IL-1β (1 ng/ml) for up to 4 h, PCR amplifications were conducted with a fixed amount of antibody-immunoprecipitated DNA by using the specific primer pairs encompassing the region of the human COX-2 promoter from position −299 to +6. IPs derived from F-NL cells with an antibody to p65, CREB-1, or C/EBPβ resulted in different patterns of enrichment of the COX-2 promoter DNA in a time- and stimulus-specific manner after TGF-β1 and IL-1β treatment (Fig. 4). TGF-β1 induced marked increases in p65, C/EBPβ, and CREB-1 binding to the COX-2 promoter in F-NL, with maximum increases observed at 4, 2, and 0.5 h after stimulation, respectively, and the effect decreased thereafter (Fig. 4A to C). IL-1β treatment for 0.5 h induced marked increases in p65, C/EBPβ, and CREB-1 binding to the COX-2 promoter in F-NL, and the effect decreased for C/EBPβ and CREB-1 thereafter, but a further increase in p65 binding was observed at 2 h after treatment (Fig. 4D to F). In contrast, TGF-β1- and IL-1β-induced p65 (Fig. 4A and D), C/EBPβ (Fig. 4B and E), and CREB-1 (Fig. 4C and F) binding to the COX-2 promoter was markedly lower in F-IPF than in F-NL. The results demonstrate that the binding of these transcription factors to the COX-2 promoter in the native chromatin environment was significantly impaired in F-IPF, thus providing an explanation for the diminished COX-2 transcription in IPF.
FIG. 4.
TGFβ1- and IL-1β-induced native transcription factor binding to the human COX-2 promoter in F-NL and F-IPF. Confluent and serum-starved F-NL and F-IPF cells in 150-cm2 flasks were incubated with TGF-β1 (2 ng/ml) (A to C) or IL-1β (1 ng/ml) (D to F) for the times indicated. The protein-DNA complexes were cross-linked by formaldehyde treatment, and chromatin pellets were extracted and sonicated. The transcription factors NF-κB (A and D), C/EBPβ (B and E), and CREB-1 (C and F) were immunoprecipitated with specific antibodies, and the associated COX-2 promoter DNA was amplified by real-time PCR as described in Materials and Methods. The results are normalized relative to the input control and are the means ± SEM of data for three separate F-NL and F-IPF cell lines analyzed in duplicate.
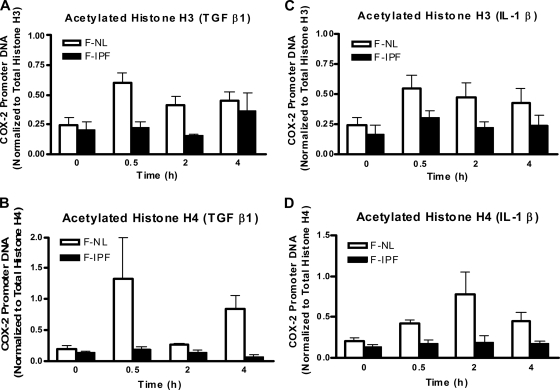
Histone H3 and H4 acetylation at the COX-2 promoter.
As histone acetylation and deacetylation are closely associated with active and repressive chromatin states and increased and decreased transcription factor binding to specific gene promoters, respectively, we anticipated that the impaired transcription factor binding to the COX-2 promoter in F-IPF might be due to restricted access to the promoter as a result of reduced histone acetylation at the COX-2 promoter. We therefore analyzed histone H3 and H4 acetylation at the COX-2 promoter site by a ChIP assay. Both TGF-β1 and IL-1β cause marked increases of histone H3 and H4 acetylation at the COX-2 promoter in F-NL, with different time patterns, after 0.5 to 4 h of treatment (Fig. 5). Histone H3 and H4 acetylation at the COX-2 promoter in F-IPF was markedly reduced or absent compared to that in F-NL, although modest increases at some time points after cytokine treatment were observed (Fig. 5). The results suggest that histone acetylation at the COX-2 promoter in F-IPF is insufficient compared to that in F-NL. To determine the specificity of the ChIP assay, distal controls upstream and downstream of the COX-2 promoter were applied to analyze changes in histone acetylation in F-NL with primers designed specifically for different regions of the COX-2 gene. Two regions upstream (−2440 to −2206 and −12360 to −12142) and two regions downstream (+1727 to +2206 and +21087 to +21307) of the minimum COX-2 promoter sequence were selected. Basal histone H3 and H4 acetylation was observed at almost all regions in control cells; however, after cytokine treatment, marked increases of histone H3 and H4 acetylation were observed only in the minimum COX-2 promoter region (see Fig. S3 in the supplemental material), indicating that cytokine-induced histone acetylation occurs specifically at the COX-2 promoter region.
FIG. 5.
TGFβ1- and IL-1β-induced histone H3 and H4 acetylation at the human COX-2 promoter site in F-NL and F-IPF. Confluent and serum-starved F-NL and F-IPF cells in 150-cm2 flasks were incubated with TGF-β1 (2 ng/ml) (A and B) or IL-1β (1 ng/ml) (C and D) for the times indicated. The protein-DNA complexes were cross-linked by formaldehyde treatment, and chromatin pellets were extracted and sonicated. Acetylated histones H3 (A and C) and H4 (B and D) and total histones H3 and H4 were immunoprecipitated with specific antibodies. The associated COX-2 promoter DNA was amplified by real-time PCR, and the amounts of COX-2 promoter DNA in acetylated-histone IPs were calculated and further normalized relative to the amount in total-histone IPs as described in Materials and Methods. The results are means ± SEM of data for three separate F-NL and F-IPF cell lines tested in duplicate.
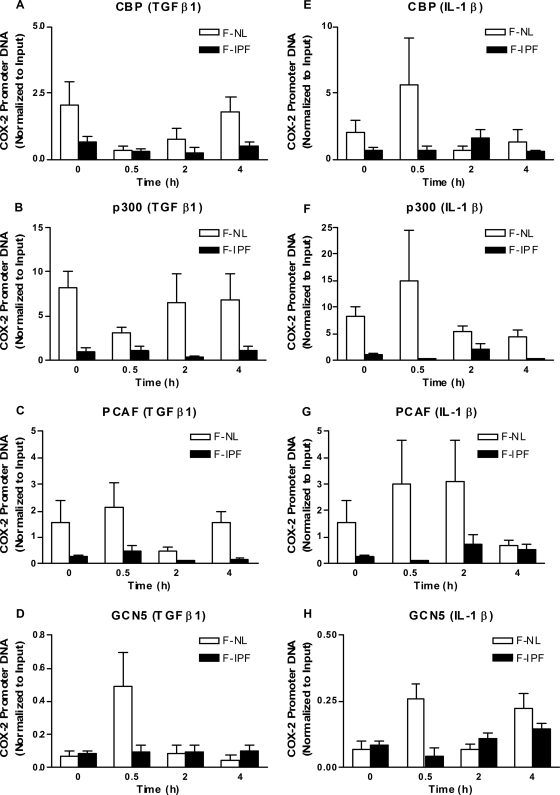
HAT/HDAC recruitment to the COX-2 promoter.
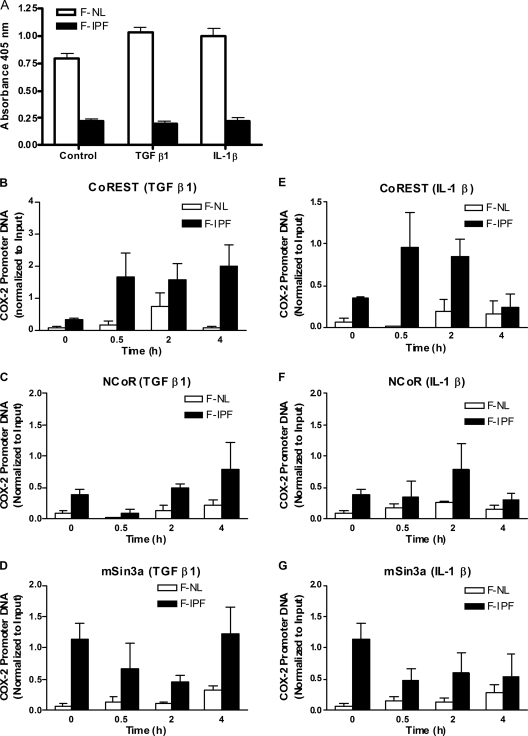
Since histone acetylation and deacetylation are regulated by HATs and HDACs, respectively, we anticipated that the insufficient histone acetylation in F-IPF was due to reduced recruitment of HATs and/or increased recruitment of HDACs. We went on to analyze this possibility by using a ChIP assay, focusing on HATs that are known to be expressed in lung fibroblasts and are thought to be important, such as CBP, p300, PCAF, and GCN5. TGF-β1 treatment for 0.5 h caused a marked increase of GCN5 (and, to a lesser extent, PCAF) association with the COX-2 promoter in F-NL compared to the control level (at 0 h) but had no effect on CBP and p300 association with the promoter (Fig. 6A to D), whereas IL-1β treatment for 0.5 h caused a marked increase of CBP, p300, PCAF, and GCN5 association with the COX-2 promoter (Fig. 6E to H). In contrast, the levels of association of all four HATs with the COX-2 promoter after TGF-β1 and IL-1β treatment were consistently lower in F-IPF than in F-NL, although modest increases were observed at some time points (Fig. 6). We then examined whether global HDAC activity in F-IPF was different from that in F-NL by using a colorimetric detection assay. As shown in Fig. 7A, global HDAC activity in the nuclear extracts from F-NL was markedly higher than that in the extracts from F-IPF without stimulation, and treatment with TGF-β1 and IL-1β for 4 h slightly increased HDAC activity in F-NL but not in F-IPF, suggesting that repressed COX-2 transcription in F-IPF is not caused by increased HDAC activity. Since HDACs are usually associated with transcription-inhibitory complexes such as the CoREST, NCoR, and mSin3a complexes, which makes it difficult to detect the direct association of HDACs with specific gene promoters (5, 24, 64), we then analyzed the association of the major proteins of the three complexes, CoREST, NCoR, and mSin3a, with the COX-2 promoter. As shown in Fig. 7B to G, without stimulation, the degrees of association of CoREST, NCoR, and mSin3a with the COX-2 promoter in F-IPF were markedly higher than those in F-NL. Treatment with TGF-β1 and IL-1β increased the association of CoREST and NCoR with the COX-2 promoter in both F-IPF and F-NL and of mSin3a in F-NL with different time patterns; however, the levels of association of CoREST, NCoR, and mSin3a in F-IPF were consistently higher than those in F-NL (Fig. 7B to G). The results strongly suggest indirectly that despite the lower global HDAC activity in F-IPF than in F-NL, there is a correlation between increased HDAC recruitment to the COX-2 promoter and COX-2 repression in F-IPF.
FIG. 6.
TGF-β1- and IL-1β-induced HAT association with the human COX-2 promoter site in F-NL and F-IPF. Confluent and serum-starved F-NL and F-IPF cells in 150-cm2 flasks were incubated with TGF-β1 (2 ng/ml) (A to D) or IL-1β (1 ng/ml) (E to H) for the times indicated. The protein-DNA complexes were cross-linked by formaldehyde treatment, and chromatin pellets were extracted and sonicated. The HATs CBP (A and E), p300 (B and F), PCAF (C and G), and GCN5 (D and H) were immunoprecipitated with specific antibodies, and the associated COX-2 promoter DNA was amplified by real-time PCR as described in Materials and Methods. The results are normalized relative to the input control and are means ± SEM of data for three separate F-NL and F-IPF cell lines tested in duplicate.
FIG. 7.
Global HDAC activity and transcriptional corepressor complex association with the human COX-2 promoter in F-NL and F-IPF. (A) Confluent and serum-starved F-NL and F-IPF cells in 150-cm2 flasks were incubated without or with TGF-β1 (2 ng/ml) or IL-1β (1 ng/ml) for 4 h. Nuclear extraction was performed and HDAC activity was measured with 30 μg of nuclear extract as described in Materials and Methods. The results are expressed as means ± SEM of data for three separate F-NL and F-IPF cell lines analyzed in duplicate. (B to G) Confluent and serum-starved F-NL and F-IPF cells in 150-cm2 flasks were incubated with TGF-β1 (2 ng/ml) (B to D) or IL-1β (1 ng/ml) (E to G) for the times indicated. The protein-DNA complexes were cross-linked by formaldehyde treatment, and chromatin pellets were extracted and sonicated. CoREST (B and E), NCoR (C and F), and mSin3a (D and G) were immunoprecipitated with specific antibodies, and the associated COX-2 promoter DNA was amplified by real-time PCR as described in Materials and Methods. The results are normalized relative to the input control and are means ± SEM of results for three separate F-NL and F-IPF cell lines analyzed in duplicate.
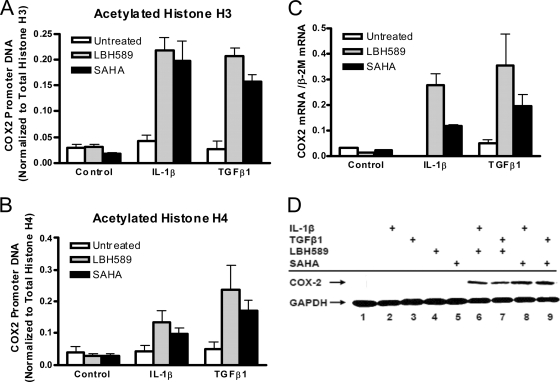
Effect of HDAC inhibitors and HAT overexpression on COX-2 transcription.
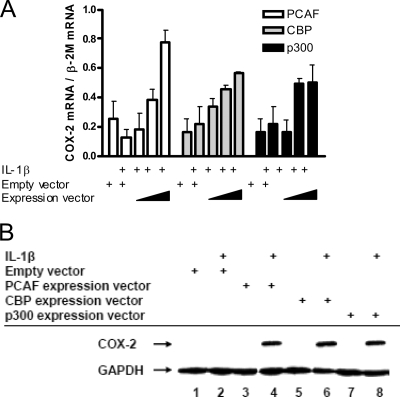
To determine whether there was a direct link between increased HDAC recruitment to the COX-2 promoter and COX-2 repression in F-IPF, we first examined the effects of two HDAC inhibitors, SAHA and LBH589, on histone acetylation at the COX-2 promoter and COX-2 mRNA and protein expression in F-IPF. A preliminary study of COX-2 mRNA expression showed concentration-dependent effects within the range of 0.1 to 100 μM for SAHA and 0.1 to 10 nM for LBH589 (data not shown), and based on this finding, 10 μM SAHA and 10 nM LBH589 were chosen for further studies. When F-IPF cells were treated with the inhibitors alone, no effect on histone H3 and H4 acetylation compared to that in untreated cells was observed; however, when the cells were treated with the inhibitors and IL-1β or TGF-β1, a marked increase in histone H3 and H4 acetylation at the COX-2 promoter compared to that in cells treated with cytokines alone was observed (Fig. 8A and B). Treatment of the cells with the inhibitors and IL-1β or TGF-β1 also induced marked inductions of COX-2 mRNA expression (at 4 h) (Fig. 8C) and protein expression (at 24 h) (Fig. 8D) compared to those in cells treated with cytokines alone. We then examined whether the overexpression of HATs in F-IPF cells could have effects similar to those of HDAC inhibitors. As shown in Fig. 9A, the transfection of the cells with empty vectors (2 μg) had no effect on IL-1β-treated cells (4 h) relative to untreated cells; however, transfection with vectors expressing PCAF, CBP, and p300 (1, 1.5, and 2 μg for all) markedly increased IL-1β-induced COX-2 mRNA expression in a concentration-dependent manner compared to that in cells treated with IL-1β alone, which had no effect. Expression vectors on their own also had no effect (data not shown). The transfection of the cells with vectors expressing PCAF, CBP, and p300 (4 μg for all) also markedly increased IL-1β-induced COX-2 protein expression (at 24 h), whereas IL-1β and expression vectors alone had no effect (Fig. 9B). The results show that COX-2 transcription in response to cytokine stimulation in F-IPF can be restored by either the inhibition of HDAC activity or the overexpression of HATs and therefore strongly suggest that histone hypoacetylation at the COX-2 promoter is directly linked to diminished COX-2 expression in IPF.
FIG. 8.
Effect of HDAC inhibitors on histone H3 and H4 acetylation at the human COX-2 promoter and COX-2 expression in F-IPF. Confluent and serum-starved F-IPF cells in 150-cm2 flasks were incubated without or with (+) SAHA (10 μM) or LBH589 (10 nM) for 1 h before being treated without or with (+) TGF-β1 (2 ng/ml) or IL-1β (1 ng/ml) for a further 4 h (A to C) or 24 h (D) as indicated. The protein-DNA complexes were then cross-linked by formaldehyde treatment, and chromatin pellets were extracted and sonicated. Acetylated histones H3 (A) and H4 (B) and total histones H3 and H4 were immunoprecipitated with specific antibodies. (A and B) The associated COX-2 promoter DNA was amplified by real-time PCR, and the amounts of COX-2 promoter DNA in acetylated-histone IPs were calculated and further normalized relative to the amount in total-histone H3 and H4 IPs, respectively, as described in Materials and Methods. The results are means ± SEM of data for three separate F-IPF cell lines analyzed in duplicate. (C) Total RNA was then isolated, and mRNA levels for COX-2 and the internal control β-2M were determined by quantitative RT-PCR. The results were calculated as the ratio of COX-2 mRNA and β-2M mRNA and are expressed as means ± SEM of data from three separate experiments performed in duplicate. (D) Total cell lysates were then collected for Western blot analyses of COX-2 and the loading control GAPDH. The results shown are representative of those from two experiments.
FIG. 9.
Effect of HDAC inhibitors on histone H3 and H4 acetylation at the human COX-2 promoter and COX-2 expression in F-IPF. F-IPF at 80% confluence in six-well plates were transfected with either empty vectors or vectors expressing PCAF, CBP, or p300 (4 μg/well) by using TransFast transfection reagent as described in Materials and Methods. Cells were then serum starved for 24 h before being treated without or with (+) IL-1β (1 ng/ml) for 4 h (A) and 24 h (B). (A) Total RNA was then isolated, and mRNA levels for COX-2 and the internal control β-2M were determined by quantitative RT-PCR. The results were calculated as the ratio of COX-2 mRNA and β-2M mRNA and are expressed as means ± SEM of data from three separate experiments performed in duplicate. (B) Total cell lysates were then collected for Western blot analyses of COX-2 and the loading control GAPDH. The results shown are representative of those from two experiments.
Collectively, data from the present study demonstrate that COX-2 gene transcription by TGF-β1 and IL-1β in F-NL requires chromatin remodeling via histone H3 and H4 acetylation by HATs (CBP, p300, GCN5, and PCAF). However, HAT recruitment to the COX-2 promoter in F-IPF is significantly reduced compared to that in F-NL, whereas the association of the CoREST and mSin3a transcriptional corepressor complexes, which consist of HDAC1 and HDAC2, and the NCoR complex, which consists of HDAC3, with the COX-2 promoter in F-IPF is markedly increased compared to that in F-NL, resulting in insufficient acetylation of histone H3 and H4 at the COX-2 promoter and decreased transcription factor binding to the COX-2 promoter, eventually leading to diminished COX-2 transcription in F-IPF.
DISCUSSION
The major findings of our present study are that defective histone acetylation due to reduced recruitment of HATs and increased recruitment of the HDAC-containing corepressor complexes to the COX-2 promoter prevents activated transcription factors from binding to the COX-2 promoter, resulting in diminished COX-2 gene transcription in IPF. This is the first description of an epigenetic abnormality causing dysregulated gene expression in pulmonary fibrosis.
The expression of COX-2 is a key element in various pathophysiological processes, including inflammation (55), cardiovascular disease (18), tissue remodeling (11), and cancer (20). Although transcriptional regulation of human COX-2 has been studied in other cell systems, it is not clear how the COX-2 gene is regulated transcriptionally by external stimuli such as TGF-β1 and IL-1β in human lung fibroblasts and, in particular, why COX-2 expression is repressed in F-IPF, leading to reduced PGE2 production. Evidence that different mechanisms are involved in altered COX-2 expression in different diseases is accumulating. For instance, aberrant methylation and histone deacetylation at the COX-2 promoter are closely associated with absent COX-2 expression in a subset of gastric cancers (29), whereas human antigen R, which promotes mRNA stabilization (47), is associated with increased COX-2 expression in ovarian carcinoma (14). The 3′ untranslated region of COX-2 mRNA contains multiple copies of an adenylate- and uridylate-rich (AU-rich) element (16). This element, which is present within the 3′ untranslated regions of many proto-oncogene and cytokine mRNAs, confers posttranscriptional control of expression by acting as an mRNA instability determinant (61) or as a translation-inhibitory element (45). Since cytokines upregulate COX-2 expression in lung fibroblasts both transcriptionally and posttranscriptionally (15) and previous studies have demonstrated a failure to increase steady-state mRNA levels in F-IPF upon cytokine stimulation (28, 58), indicating reduced transcription rates and/or enhanced turnover rates for COX-2 transcripts, it is likely that key processes in COX-2 transcriptional and/or posttranscriptional regulation are defective in F-IPF. We therefore looked at COX-2 mRNA expression and stability in F-IPF compared to those in F-NL and found that the COX-2 mRNA levels in F-IPF were markedly lower than those in F-NL but that the degrees of mRNA stability in F-IPF and F-NL were comparable regardless of the cytokines used. The results thus suggest that defective transcriptional regulation may be mainly responsible for the diminished COX-2 expression in IPF. The kinetic difference in COX-2 mRNA degradation between TGF-β1 and IL-1β shows the involvement of different signaling pathways, resulting in IL-1β-induced COX-2 mRNA's being more stable than TGF-β1-induced COX-2 mRNA.
The diminished COX-2 expression in F-IPF in response to unrelated inducers such as cytokines, phorbol myristate acetate, and lipopolysaccharide strongly suggests that the defect resides not at the level of receptors or the immediate postreceptor signaling mechanisms, but at more distal, efferent steps in enzyme synthesis. We then investigated transcription factor involvement and activation in both F-IPF and F-NL. By applying reporter gene assays, we demonstrated that F-IPF and F-NL utilized the same set of regulatory elements and transcription factors (NF-κB, C/EBP, and CREB) for cytokine-induced COX-2 promoter activity and that there were no differences between F-IPF and F-NL in transcription factor protein expression and cytokine-induced COX-2 promoter activity. The results indicate that postreceptor signaling up to the stage of transcription factor activation in COX-2 transcription is not impaired in F-IPF and therefore could not contribute to the diminished COX-2 expression in IPF. The results are also consistent with the finding that not all inducible genes are repressed in F-IPF.
Since the binding of transcription factors to the COX-2 promoter in the reporter gene assay, unlike that in the native chromatin environment, is not regulated by histone modifications and chromatin states, we anticipated that the binding of transcription factors to the COX-2 promoter in the native chromatin environment in F-IPF was impaired and went on to investigate this possibility by using a ChIP assay. We found that the binding of transcription factors NF-κB, C/EBPβ, and CREB-1 to the COX-2 promoter in the native chromatin environment in F-IPF was significantly impaired, thus resulting in reduced COX-2 gene transcription and providing an explanation for the diminished COX-2 expression in IPF. It is well established that the binding of activated transcription factors to specific inducible gene promoter sites is tightly controlled by the chromatin state as a result of histone modifications, particularly the balance between histone acetylation and deacetylation (1, 26). We have reported previously that induced COX-2 gene transcription in human airway smooth muscle cells is closely associated with histone H4 acetylation at the COX-2 promoter site (39). The acetylation of histones at the COX-2 promoter site may be attributed to the recruitment of coactivators with intrinsic HAT activity, such as CBP, p300, GCN5, and PCAF, all of which have been demonstrated previously to be able to acetylate both histones H3 and H4 (13). We therefore anticipated that the impaired transcription factor binding to the COX-2 promoter in F-IPF might be due to restricted access to the promoter as a result of decreased HAT recruitment and/or increased HDAC recruitment and consequently reduced histone acetylation at the COX-2 promoter site. We indeed demonstrated in this study that even though similar regulatory elements and transcription factors were involved in cytokine-induced COX-2 transcription in F-NL and F-IPF, COX-2 transcription in F-IPF was reduced due to decreased histone acetylation as a result of the reduced recruitment of HATs to the COX-2 promoter. There was a clear difference between TGF-β1 and IL-1β in specific HAT recruitment to the COX-2 promoter in F-NL, i.e., TGF-β1 recruited PCAF and GCN5, whereas IL-1β recruited all four HATs tested. This difference, like the difference in COX-2 mRNA degradation, may reflect different signaling pathways utilized by the two different cytokines. Interestingly, we revealed that global HDAC activity in F-IPF was markedly lower than that in F-NL under both unstimulated and stimulated conditions, suggesting that reduced COX-2 transcription in F-IPF is not due to increased global HDAC activity and that the transcription of some other genes in F-IPF may be increased as a result of decreased HDAC activity. In addition, we also showed that the association of the CoREST and mSin3a transcriptional repressor complexes (containing HDAC1 and HDAC2) and the NCoR complex (containing HDAC3) with the COX-2 promoter in F-IPF was markedly greater than that in F-NL both with and without stimulation, suggesting that increased recruitment of HDACs to the COX-2 promoter, rather than increased global HDAC activity, contributes to histone hypoacetylation and the repression of COX-2 transcription in F-IPF. This result is consistent with the recent finding that NCoR functions as a repressor for COX-2 gene transcription (49). We also used specific antibodies to look at the recruitment of individual HDACs to the COX-2 promoter. However, the data (not shown) did not reveal a clear and distinctive pattern of association in both F-NL and F-IPF. This may be because HDACs are usually associated with transcriptional corepressor complexes such as the CoREST and NCoR complexes, which makes it difficult to detect the direct association of HDACs with specific gene promoters (5, 24, 64). To further demonstrate the direct link between histone hypoacetylation and COX-2 repression, we applied two well-characterized HDAC inhibitors, SAHA and LBH589 (36). We found that both restored histone acetylation at the COX-2 promoter and COX-2 mRNA and protein expression in F-IPF in response to cytokine stimulation, confirming that histone hypoacetylation as a result of locally increased HDAC activity at the COX-2 promoter is responsible for COX-2 repression in IPF. As reduced HAT recruitment may also be responsible for histone hypoacetylation at the COX-2 promoter in IPF, we also tried the overexpression of HATs to enhance HAT recruitment to the COX-2 promoter to counter increased local HDAC activity. We found that HAT overexpression restored the expression of COX-2 mRNA and protein in F-IPF cells in response to IL-1β, confirming that locally reduced HAT recruitment to the COX-2 promoter is also responsible for histone hypoacetylation and, consequently, COX-2 repression in IPF. The finding that both HDAC inhibitors and HAT overexpression had no effect on their own suggests that the restoration of COX-2 expression in F-IPF requires not only HDAC inhibition and HAT recruitment to induce histone acetylation at the promoter but also cytokine stimulation to induce transcription factor activation and promoter binding to initiate gene transcription.
Evidence of links between DNA methylation and histone hypoacetylation is accumulating, with DNA methylation and the hypoacetylation of histones H3 and H4 being frequently associated with silent genes. In addition, several proteins that bind specifically to methylated DNA are associated with complexes that include HDACs (17, 29). Specific to COX-2, it has been reported previously that a subset of colorectal and gastric cancers exhibit the methylation of multiple CpG islands, which mediate transcriptional repression by recruiting HDACs (29, 53). Therefore, the involvement of DNA methylation and other histone modifications, methylation in particular, in diminished COX-2 expression in IPF cannot be excluded. In fact, histone methylation may be necessary for DNA methylation to occur, which leads ultimately to transcriptional silencing (51). Further studies will be needed to establish the link between histone methylation, DNA methylation, and histone hypoacetylation in diminished COX-2 transcription in IPF.
In summary, the findings of our present study demonstrated that epigenetic abnormality in the form of histone hypoacetylation as a result of decreased recruitment of the transcription coactivators with intrinsic HAT activity and increased recruitment of the transcription corepressor complexes containing HDACs to the COX-2 promoter is responsible for the diminished COX-2 gene transcription observed in IPF. These findings demonstrate the involvement of defective histone acetylation in dysregulated gene expression in fibrotic lung diseases and may help our understanding of the pathogenesis of fibrotic diseases and the identification of novel molecular targets for therapeutic purposes.
Supplementary Material
Acknowledgments
This work was supported by the British Lung Foundation (grant number P06-4) and the Medical Research Council (grant number G0600890).
Footnotes
Published ahead of print on 1 June 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Adcock, I. M., L. Tsaprouni, P. Bhavsar, and K. Ito. 2007. Epigenetic regulation of airway inflammation. Curr. Opin. Immunol. 19694-700. [DOI] [PubMed] [Google Scholar]
- 2.Allport, V. C., D. M. Slater, R. Newton, and P. R. Bennett. 2000. NF-kappaB and AP-1 are required for cyclo-oxygenase 2 gene expression in amnion epithelial cell line (WISH). Mol. Hum. Reprod. 6561-565. [DOI] [PubMed] [Google Scholar]
- 3.Andres, M. E., C. Burger, M. J. Peral-Rubio, E. Battaglioli, M. E. Anderson, J. Grimes, J. Dallman, N. Ballas, and G. Mandel. 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 969873-9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atamas, S. P., and B. White. 2003. Cytokine regulation of pulmonary fibrosis in scleroderma. Cytokine Growth Factor Rev. 14537-550. [DOI] [PubMed] [Google Scholar]
- 5.Ballas, N., E. Battaglioli, F. Atouf, M. E. Andres, J. Chenoweth, M. E. Anderson, C. Burger, M. Moniwa, J. R. Davie, W. J. Bowers, H. J. Federoff, D. W. Rose, M. G. Rosenfeld, P. Brehm, and G. Mandel. 2001. Regulation of neuronal traits by a novel transcriptional complex. Neuron 31353-365. [DOI] [PubMed] [Google Scholar]
- 6.Bonner, J. C., A. B. Rice, J. L. Ingram, C. R. Moomaw, A. Nyska, A. Bradbury, A. R. Sessoms, P. C. Chulada, D. L. Morgan, D. C. Zeldin, and R. Langenbach. 2002. Susceptibility of cyclooxygenase-2-deficient mice to pulmonary fibrogenesis. Am. J. Pathol. 161459-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borok, Z., A. Gillissen, R. Buhl, R. F. Hoyt, R. C. Hubbard, T. Ozaki, S. I. Rennard, and R. G. Crystal. 1991. Augmentation of functional prostaglandin E levels on the respiratory epithelial surface by aerosol administration of prostaglandin E. Am. Rev. Respir. Dis. 1441080-1084. [DOI] [PubMed] [Google Scholar]
- 8.Bradbury, D. A., R. Newton, Y. M. Zhu, H. El-Haroun, L. Corbett, and A. J. Knox. 2003. Cyclooxygenase-2 induction by bradykinin in human pulmonary artery smooth muscle cells is mediated by the cyclic AMP response element through a novel autocrine loop involving endogenous prostaglandin E2, E-prostanoid 2 (EP2), and EP4 receptors. J. Biol. Chem. 27849954-49964. [DOI] [PubMed] [Google Scholar]
- 9.Broekelmann, T. J., A. H. Limper, T. V. Colby, and J. A. McDonald. 1991. Transforming growth factor beta 1 is present at sites of extracellular matrix gene expression in human pulmonary fibrosis. Proc. Natl. Acad. Sci. USA 886642-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong, J. A., J. Tapia-Ramirez, S. Kim, J. J. Toledo-Aral, Y. Zheng, M. C. Boutros, Y. M. Altshuller, M. A. Frohman, S. D. Kraner, and G. Mandel. 1995. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80949-957. [DOI] [PubMed] [Google Scholar]
- 11.Cipollone, F., C. Prontera, B. Pini, M. Marini, M. Fazia, D. De Cesare, A. Iezzi, S. Ucchino, G. Boccoli, V. Saba, F. Chiarelli, F. Cuccurullo, and A. Mezzetti. 2001. Overexpression of functionally coupled cyclooxygenase-2 and prostaglandin E synthase in symptomatic atherosclerotic plaques as a basis of prostaglandin E(2)-dependent plaque instability. Circulation 104921-927. [DOI] [PubMed] [Google Scholar]
- 12.Coker, R. K., and G. J. Laurent. 1998. Pulmonary fibrosis: cytokines in the balance. Eur. Respir. J. 111218-1221. [DOI] [PubMed] [Google Scholar]
- 13.de la Cruz, X., S. Lois, S. Sanchez-Molina, and M. A. Martinez-Balbas. 2005. Do protein motifs read the histone code? Bioessays 27164-175. [DOI] [PubMed] [Google Scholar]
- 14.Denkert, C., W. Weichert, S. Pest, I. Koch, D. Licht, M. Kobel, A. Reles, J. Sehouli, M. Dietel, and S. Hauptmann. 2004. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res. 64189-195. [DOI] [PubMed] [Google Scholar]
- 15.Diaz, A., K. P. Chepenik, J. H. Korn, A. M. Reginato, and S. A. Jimenez. 1998. Differential regulation of cyclooxygenases 1 and 2 by interleukin-1 beta, tumor necrosis factor-alpha, and transforming growth factor-beta 1 in human lung fibroblasts. Exp. Cell Res. 241222-229. [DOI] [PubMed] [Google Scholar]
- 16.Dixon, D. A., C. D. Kaplan, T. M. McIntyre, G. A. Zimmerman, and S. M. Prescott. 2000. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3′-untranslated region. J. Biol. Chem. 27511750-11757. [DOI] [PubMed] [Google Scholar]
- 17.Dobosy, J. R., and E. U. Selker. 2001. Emerging connections between DNA methylation and histone acetylation. Cell. Mol. Life Sci. 58721-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubois, R. N., S. B. Abramson, L. Crofford, R. A. Gupta, L. S. Simon, L. B. Van De Putte, and P. E. Lipsky. 1998. Cyclooxygenase in biology and disease. FASEB J. 121063-1073. [PubMed] [Google Scholar]
- 19.Elias, J. A. 1988. Tumor necrosis factor interacts with interleukin-1 and interferons to inhibit fibroblast proliferation via fibroblast prostaglandin-dependent and -independent mechanisms. Am. Rev. Respir. Dis. 138652-658. [DOI] [PubMed] [Google Scholar]
- 20.Fang, H. Y., T. S. Lin, J. P. Lin, Y. C. Wu, K. C. Chow, and L. S. Wang. 2003. Cyclooxygenase-2 in human non-small cell lung cancer. Eur. J. Surg. Oncol. 29171-177. [DOI] [PubMed] [Google Scholar]
- 21.Gorgoni, B., M. Caivano, C. Arizmendi, and V. Poli. 2001. The transcription factor C/EBPbeta is essential for inducible expression of the cox-2 gene in macrophages but not in fibroblasts. J. Biol. Chem. 27640769-40777. [DOI] [PubMed] [Google Scholar]
- 22.Gross, T. J., and G. W. Hunninghake. 2001. Idiopathic pulmonary fibrosis. N. Engl. J. Med. 345517-525. [DOI] [PubMed] [Google Scholar]
- 23.Hebbes, T. R., A. L. Clayton, A. W. Thorne, and C. Crane-Robinson. 1994. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 131823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Humphrey, G. W., Y. Wang, V. R. Russanova, T. Hirai, J. Qin, Y. Nakatani, and B. H. Howard. 2001. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 2766817-6824. [DOI] [PubMed] [Google Scholar]
- 25.Inoue, H., and T. Tanabe. 1998. Transcriptional role of the nuclear factor kappa B site in the induction by lipopolysaccharide and suppression by dexamethasone of cyclooxygenase-2 in U937 cells. Biochem. Biophys. Res. Commun. 244143-148. [DOI] [PubMed] [Google Scholar]
- 26.Ito, K., P. J. Barnes, and I. M. Adcock. 2000. Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol. Cell. Biol. 206891-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins, G., S. L. Hart, R. J. Hodges, Q. H. Meng, C. Kinnon, G. J. Laurent, and R. J. McAnulty. 2002. Cyclooxygenase-2 overexpression, using an integrin-targeted gene delivery system (the LID vector), inhibits fibroblast proliferation in vitro and leads to increased prostaglandin E2 in the lung. Chest 121102S-104S. [DOI] [PubMed] [Google Scholar]
- 28.Keerthisingam, C. B., R. G. Jenkins, N. K. Harrison, N. A. Hernandez-Rodriguez, H. Booth, G. J. Laurent, S. L. Hart, M. L. Foster, and R. J. McAnulty. 2001. Cyclooxygenase-2 deficiency results in a loss of the anti-proliferative response to transforming growth factor-beta in human fibrotic lung fibroblasts and promotes bleomycin-induced pulmonary fibrosis in mice. Am. J. Pathol. 1581411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikuchi, T., F. Itoh, M. Toyota, H. Suzuki, H. Yamamoto, M. Fujita, M. Hosokawa, and K. Imai. 2002. Aberrant methylation and histone deacetylation of cyclooxygenase 2 in gastric cancer. Int. J. Cancer 97272-277. [DOI] [PubMed] [Google Scholar]
- 30.Kohyama, T., R. F. Ertl, V. Valenti, J. Spurzem, M. Kawamoto, Y. Nakamura, T. Veys, L. Allegra, D. Romberger, and S. I. Rennard. 2001. Prostaglandin E2 inhibits fibroblast chemotaxis. Am. J. Physiol. Lung Cell. Mol. Physiol. 281L1257-L1263. [DOI] [PubMed] [Google Scholar]
- 31.Korn, J. H., P. V. Halushka, and E. C. LeRoy. 1980. Mononuclear cell modulation of connective tissue function: suppression of fibroblast growth by stimulation of endogenous prostaglandin production. J. Clin. Investig. 65543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroki, S., A. Ohta, N. Sueoka, O. Katoh, H. Yamada, and M. Yamaguchi. 1995. Determination of various cytokines and type III procollagen aminopeptide levels in bronchoalveolar lavage fluid of the patients with pulmonary fibrosis: inverse correlation between type III procollagen aminopeptide and interferon-gamma in progressive patients. Br. J. Rheumatol. 3431-36. [DOI] [PubMed] [Google Scholar]
- 33.Lafon-Hughes, L., M. V. Di Tomaso, L. Mendez-Acuna, and W. Martinez-Lopez. 2008. Chromatin-remodelling mechanisms in cancer. Mutat. Res. 658191-214. [DOI] [PubMed] [Google Scholar]
- 34.Liu, X., R. S. Ostrom, and P. A. Insel. 2004. cAMP-elevating agents and adenylyl cyclase overexpression promote an antifibrotic phenotype in pulmonary fibroblasts. Am. J. Physiol. Cell Physiol. 286C1089-C1099. [DOI] [PubMed] [Google Scholar]
- 35.Lynch, J. P., III, and W. J. McCune. 1997. Immunosuppressive and cytotoxic pharmacotherapy for pulmonary disorders. Am. J. Respir. Crit. Care Med. 155395-420. [DOI] [PubMed] [Google Scholar]
- 36.Marks, P. A., and R. Breslow. 2007. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2584-90. [DOI] [PubMed] [Google Scholar]
- 37.Nie, M., L. Corbett, A. J. Knox, and L. Pang. 2005. Differential regulation of chemokine expression by peroxisome proliferator-activated receptor gamma agonists: interactions with glucocorticoids and β2-agonists. J. Biol. Chem. 2802550-2561. [DOI] [PubMed] [Google Scholar]
- 38.Nie, M., A. J. Knox, and L. Pang. 2005. β2-Adrenoceptor agonists, like glucocorticoids, repress eotaxin gene transcription by selective inhibition of histone H4 acetylation. J. Immunol. 175478-486. [DOI] [PubMed] [Google Scholar]
- 39.Nie, M., L. Pang, H. Inoue, and A. J. Knox. 2003. Transcriptional regulation of cyclooxygenase 2 by bradykinin and interleukin-1β in human airway smooth muscle cells: involvement of different promoter elements, transcription factors, and histone H4 acetylation. Mol. Cell. Biol. 239233-9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang, L., and A. J. Knox. 1997. Effect of interleukin-1 beta, tumour necrosis factor-alpha and interferon-gamma on the induction of cyclo-oxygenase-2 in cultured human airway smooth muscle cells. Br. J. Pharmacol. 121579-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang, L., M. Nie, L. Corbett, R. Donnelly, S. Gray, and A. J. Knox. 2002. Protein kinase C-epsilon mediates bradykinin-induced cyclooxygenase-2 expression in human airway smooth muscle cells. FASEB J. 161435-1437. [DOI] [PubMed] [Google Scholar]
- 42.Petkova, D. K., C. A. Clelland, J. E. Ronan, S. Lewis, and A. J. Knox. 2003. Reduced expression of cyclooxygenase (COX) in idiopathic pulmonary fibrosis and sarcoidosis. Histopathology 43381-386. [DOI] [PubMed] [Google Scholar]
- 43.Pilewski, J. M., L. Liu, A. C. Henry, A. V. Knauer, and C. A. Feghali-Bostwick. 2005. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am. J. Pathol. 166399-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polgar, P., and L. Taylor. 1980. Alterations in prostaglandin synthesis during senescence of human lung fibroblasts. Mech. Ageing Dev. 12305-310. [DOI] [PubMed] [Google Scholar]
- 45.Rajagopalan, L. E., and J. S. Malter. 1996. Turnover and translation of in vitro synthesized messenger RNAs in transfected, normal cells. J. Biol. Chem. 27119871-19876. [DOI] [PubMed] [Google Scholar]
- 46.Schoenherr, C. J., and D. J. Anderson. 1995. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 2671360-1363. [DOI] [PubMed] [Google Scholar]
- 47.Sengupta, S., B. C. Jang, M. T. Wu, J. H. Paik, H. Furneaux, and T. Hla. 2003. The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J. Biol. Chem. 27825227-25233. [DOI] [PubMed] [Google Scholar]
- 48.Sokolova, E., Z. Grishina, F. Buhling, T. Welte, and G. Reiser. 2005. Protease-activated receptor-1 in human lung fibroblasts mediates a negative feedback downregulation via prostaglandin E2. Am. J. Physiol. Lung Cell. Mol. Physiol. 288L793-L802. [DOI] [PubMed] [Google Scholar]
- 49.Subbaramaiah, K., and A. J. Dannenberg. 2007. Cyclooxygenase-2 transcription is regulated by human papillomavirus 16 E6 and E7 oncoproteins: evidence of a corepressor/coactivator exchange. Cancer Res. 673976-3985. [DOI] [PubMed] [Google Scholar]
- 50.Subbaramaiah, K., D. T. Lin, J. C. Hart, and A. J. Dannenberg. 2001. Peroxisome proliferator-activated receptor gamma ligands suppress the transcriptional activation of cyclooxygenase-2. Evidence for involvement of activator protein-1 and CREB-binding protein/p300. J. Biol. Chem. 27612440-12448. [DOI] [PubMed] [Google Scholar]
- 51.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414277-283. [DOI] [PubMed] [Google Scholar]
- 52.Tazawa, R., X. M. Xu, K. K. Wu, and L. H. Wang. 1994. Characterization of the genomic structure, chromosomal location and promoter of human prostaglandin H synthase-2 gene. Biochem. Biophys. Res. Commun. 203190-199. [DOI] [PubMed] [Google Scholar]
- 53.Toyota, M., L. Shen, M. Ohe-Toyota, S. R. Hamilton, F. A. Sinicrope, and J. P. Issa. 2000. Aberrant methylation of the cyclooxygenase 2 CpG island in colorectal tumors. Cancer Res. 604044-4048. [PubMed] [Google Scholar]
- 54.Vancheri, C., M. A. Sortino, V. Tomaselli, C. Mastruzzo, F. Condorelli, G. Bellistri, M. P. Pistorio, P. L. Canonico, and N. Crimi. 2000. Different expression of TNF-alpha receptors and prostaglandin E2 production in normal and fibrotic lung fibroblasts: potential implications for the evolution of the inflammatory process. Am. J. Respir. Cell Mol. Biol. 22628-634. [DOI] [PubMed] [Google Scholar]
- 55.Vane, J. R., J. A. Mitchell, I. Appleton, A. Tomlinson, D. Bishop-Bailey, J. Croxtall, and D. A. Willoughby. 1994. Inducible isoforms of cyclooxygenase and nitric-oxide synthase in inflammation. Proc. Natl. Acad. Sci. USA 912046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vermeulen, M., M. J. Carrozza, E. Lasonder, J. L. Workman, C. Logie, and H. G. Stunnenberg. 2004. In vitro targeting reveals intrinsic histone tail specificity of the Sin3/histone deacetylase and N-CoR/SMRT corepressor complexes. Mol. Cell. Biol. 242364-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Westendorf, J. J. 2006. Transcriptional co-repressors of Runx2. J. Cell. Biochem. 9854-64. [DOI] [PubMed] [Google Scholar]
- 58.Wilborn, J., L. J. Crofford, M. D. Burdick, S. L. Kunkel, R. M. Strieter, and M. Peters-Golden. 1995. Cultured lung fibroblasts isolated from patients with idiopathic pulmonary fibrosis have a diminished capacity to synthesize prostaglandin E2 and to express cyclooxygenase-2. J. Clin. Investig. 951861-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xaubet, A., J. Roca-Ferrer, L. Pujols, J. Ramirez, J. Mullol, A. Marin-Arguedas, A. Torrego, J. M. Gimferrer, and C. Picado. 2004. Cyclooxygenase-2 is up-regulated in lung parenchyma of chronic obstructive pulmonary disease and down-regulated in idiopathic pulmonary fibrosis. Sarcoidosis Vasc. Diffuse Lung Dis. 2135-42. [PubMed] [Google Scholar]
- 60.Xie, W., B. S. Fletcher, R. D. Andersen, and H. R. Herschman. 1994. v-src induction of the TIS10/PGS2 prostaglandin synthase gene is mediated by an ATF/CRE transcription response element. Mol. Cell. Biol. 146531-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu, N., C. Y. Chen, and A. B. Shyu. 1997. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol. 174611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yokoyama, C., and T. Tanabe. 1989. Cloning of human gene encoding prostaglandin endoperoxide synthase and primary structure of the enzyme. Biochem. Biophys. Res. Commun. 165888-894. [DOI] [PubMed] [Google Scholar]
- 63.Yoon, H. G., Y. Choi, P. A. Cole, and J. Wong. 2005. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 981454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.