Abstract
Mass spectrometry with or without pre-analysis peptide fractionation can be used to decipher the residues on proteins where oxidative modifications caused by peroxynitrite, singlet oxygen and electrophilic lipids have occurred. Peroxynitrite nitrates tyrosine and tryptophan residues on the surface of actin. Singlet oxygen, formed by the interaction of UVA light with tryptophan, can oxidize neighboring cysteine, histidine, methionine, tyrosine and tryptophan residues. Dose-response inactivation by 4-hydroxynonenal (4HNE) of human bile acid CoA: amino acid N-acyltransferase (hBAT) and the cytosolic brain isoform of creatine kinase (CKBB) is associated with site-specific modifications. Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR MS) using nanoLC-electrospray ionization-mass spectrometry (ESI-MS) or direct infusion-ESI-MS with gas phase fractionation identified 14 4HNE adducts on hBAT and 17 on CKBB, respectively. At 4HNE concentrations in the physiological range, one member of the catalytic triad of hBAT (His362) was modified; for CKBB, although all four residues in the active site that were modifiable by 4HNE were ultimately modified, only one, Cys283, occurred at physiological concentrations of 4HNE. These results suggest that future in vivo studies should carefully assess the critical sites that are modified rather than using antibodies that do not distinguish between different modified sites.
Introduction
Post-translational modifications (PTM) of proteins occur extensively throughout their lifetime in a cell. For certain proteins, rather than the addition of a modifying group such as a phosphate, the PTM may be the removal of an N-terminal amino acid (e.g., methionine), a longer N-terminal peptide, or the conversion of a C-terminal glycine to a C-terminal amidate. Similarly, some proteins are translated as a single polypeptide but are then digested by specific proteases to release individual and very bioactive proteins. Examples include chromogranin A in the brain (1) and the polypeptide containing the capsid protein in the HIV virus (2). Another large group of PTMs are those formed enzymatically, such as phosphorylation (3, 4), N- (5) and O-glycosylation (6), lysine N-methylation (7), and N-acetylation (8). These proteins are also then subject to modification by enzymes that remove the PTMs, i.e., phosphatases, glycosidases (9), lysine N-demethylases (10) and acyl hydrolases (11).
The remaining group of protein PTMs result from chemically reactive species generated during different levels and types of oxidative stress which may be exacerbated by inflammatory conditions in infectious and chronic diseases (Table 1). Activation of neutrophils and other monocytes leads to the generation of a respiratory burst and the formation of superoxide anion radical (O2−·) (12). This oxidant species can undergo two main reactions: the first is a chemical reaction with another radical, nitric oxide (NO·), to form peroxynitrite (ONO2−) and the second, a catalytic one, with superoxide dismutase to form oxygen and hydrogen peroxide (H2O2) (13, 14). Peroxynitrite reacts with tyrosine and tryptophan residues to form nitrotyrosine (15) and nitrotryptophan (16–19). It may also act as an oxidizing agent, and modify cysteine residues. Hydrogen peroxide is also an oxidizing agent and under certain circumstances converts protein cysteine sulfhydryl groups to sulfenic (SOH), sulfinic (SO2H) and sulfonic (SO3H) acids (20–22). It may also be converted by neutrophil myeloperoxidase to hypochlorous acid (HOCl) that in turn reacts with nitrite to form nitryl chloride; we have previously shown that this increases the chlorination of a tyrosine-like residue in polyphenols (23, 24). Interestingly, conversion of peroxynitrite to nitryl chloride blocks nitration of tyrosine groups (25). Another oxidizing species is singlet oxygen (1O2) which is generated following the impact of ultraviolet light with tryptophan residues (26) and during the respiratory burst in neutrophils (27) – the former occurs in the lens of the eye and may be the basis of cataract formation. Singlet oxygen has a very short lifetime in a solution containing a protein and reacts with cysteine, histidine, methionine, tyrosine and tryptophan residues (28, 29). Singlet oxygen also reacts with polyunsaturated lipids to generate electrophilic lipid products such as malondialdehyde and 4-hydroxynonenal (4HNE) (30). These electrophilic lipids react with lysine, arginine and N-terminal amino acids to form Schiff bases, as well as with cysteine, lysine and histidine groups to form Michael adducts (31). Since Michael adducts are formed by the reaction of the unsaturated bond with the amino group, the aldehyde group remains unreacted. The resulting PTM is an example of a protein-associated carbonyl.
Table 1.
Oxidative post-translational modifications of amino acids
| Amino acid | Modification | Increase in molecular weight (Da) |
|---|---|---|
| Arginine | 4HNE 2-Pentylpyrrole | 120.0939 |
| Cysteine | Oxidation (SOH) | 15.9949 |
| Oxidation (SO2H) | 31.9898 | |
| Oxidation (SO3H) | 47.9847 | |
| S-nitrosylation | 28.9902 | |
| 4HNE Michael adduct | 156.1150 | |
| Dehydrated Michael adduct | 138.1045 | |
| Histidine | Oxidation | 15.9949 |
| 4HNE Schiff’s base | 138.1045 | |
| 4HNE Michael adduct | 156.1150 | |
| Lysine | 4HNE Schiff’s base | 138.1045 |
| 4HNE Michael adduct | 156.1150 | |
| Methionine | Oxidation | 15.9949 |
| Oxidation | 31.9898 | |
| Tryptophan | Oxidation | 15.9949 |
| Oxidation | 31.9898 | |
| Nitration | 44.9851 | |
| Tyrosine | Oxidation | 15.9949 |
| Oxidation | 31.9898 | |
| Nitration | 44.9851 |
Biology of oxidative stress
It has long been appreciated that oxidative stress is part of the etiology of many chronic diseases, including cardiovascular disease, diabetes, arthritis, autoimmune disease and many neurodegenerative diseases. Oxidation of low-density lipoprotein (LDL) is well recognized as a biomarker of cardiovascular disease. Its failure to be metabolized leads to its accumulation in foam cells. However, there are many other proteins that are also undergoing oxidative post-translational modifications. Proteins containing carbonyl groups can be reacted with 2,4-dinitrophenylhydrazine to form 2,4-dinitrophenylhydrazones (DNP) (32). These proteins can be separated by 2-dimensional gel electrophoresis and detected by Western blotting with an anti-DNP antibody and visualized by a secondary antibody coupled to a fluorescent probe (32). We have used a similar reagent (biotin hydrazide) to visualize protein carbonyls in livers of normal and ApoE−/− mice (33). Nitration can be monitored as well by Western blotting with anti-nitrotyrosine antibodies (34); similarly, there are anti-4HNE antibodies to detect proteins where 4HNE adducts have formed (35).
The antibody-based, Western blot procedures mentioned above contain two implicit assumptions – the first is that the antibody detects all modified groups of a given type, for instance all the nitrotyrosine residues or all the 4HNE-amino acid adducts. Since the antibody is raised against specific nitrotyrosine-containing peptides, it cannot be guaranteed that it reacts equally with each one, given that the neighboring amino acid residues are so different. Similarly, one has to ask the question, do anti-4HNE antibodies distinguish between Schiff bases and Michael adducts?
The effect of oxidative post-translational modifications
Aside from the structural issues, a much deeper, second assumption awaits us – that an oxidative modification is deleterious to the function of the protein. This seems at first thought a reasonable assumption; however, since oxidative stress is an unavoidable consequence of living in an oxygen-rich atmosphere, intuitively it seems that low levels of oxidation could have no effect or could even have benefit, subtly altering a protein’s properties. Consistent with this latter thought process, in vitro data have shown that mild oxidation lowers kinetic barriers for HDL remodeling, thereby improving its ability to take up cholesterol (36).
Rationale for the use of mass spectrometry in studying oxidative stress
Mass spectrometry approaches to the study of proteins have developed at an amazing speed over the past 20 years. Electrospray ionization (ESI) (37) and matrix-assisted laser desorption ionization (MALDI) (38) are soft ionization procedures that enable intact peptides and proteins to go into the gas phase. Any oxidant-induced changes in the chemistry of a protein (or a peptide derived from it) will be accompanied by changes in its mass. By using proteases to break the protein into smaller peptide pieces, the amino acid residue(s) where the oxidative modification has occurred can be determined. The changes in mass for peptides containing specific modifications are shown in Table 1.
Models of oxidative stress
Two models of oxidative stress for PTM analysis by mass spectrometry are described here: the first is the reaction of the electrophilic lipid 4HNE with human bile acid CoA:amino acid N-acyltransferase (hBAT) and with human brain isoform of creatine kinase (CK-BB). The second is the reaction of singlet oxygen with αB-crystallin as a product of ultraviolet light exposure. Site-specific PTM analysis was performed on a linear quadrupole ion trap (LTQ) Fourier transform ion cyclotron resonance hybrid mass spectrometer (LTQ FT; Thermo Fisher Scientific, San Jose, CA) with chip-based direct infusion-electrospray ionization (ESI) and/or nanoLC-ESI.
Modification experiments
Oxidative modifications of recombinant human CK-BB and αB-crystallin were analyzed by direct infusion with a fully automated monolithic silicon microchip-based electrospray interface, the TriVersa NanoMate (Advion, Ithaca, NY) with gas phase mass selection (39). Prior to MS analysis, recombinant CK-BB (10 µM, Sigma-Aldrich, St. Louis, MO) was incubated with increasing amounts of 4HNE (10, 30, 100 and 300 µM) for 2 h at 37°C, as described previously (39). Excess 4HNE was removed by treatment with 1 mM histidine. Recombinant human αB-crystallin, expressed in E. coli. and purified using ion exchange and gel filtration as described elsewhere (40), was exposed to 50 mJ/cm2 of UVA light (320–400 nm) over a period of 2 h. The samples were immersed in an ice-water bath to maintain the temperature at approximately 5°C. The modified proteins were digested overnight with trypsin and chymotrypsin as described above. Aliquots (10 µL) of digested CK-BB and αB-crystallin were loaded onto C18 ZipTip columns (Millipore, Billerica, MA). Peptides were eluted with 15 µL of a 4:1 acetonitrile/water solution in 0.1% formic acid and then diluted 1:1 with 0.1% formic acid. The NanoMate was set to load 5 µL of sample (8 pmol) which was electrosprayed by applying a 1.9 kV spray voltage and a 0.3 psi nitrogen head pressure to the sample tip to obtain a constant spray for 20–30 min. The capillary temperature, capillary voltage, and tube lens voltage were set to 150°C, 20 V, and 100 V, respectively. The LTQ FT mass spectrometer was operated in a “top three” data-dependent acquisition mode with gas phase mass selection. The mass spectrometer was set to perform FT-ICR MS full scans for the determination of the three most abundant precursor ions with the use of Xcalibur. For each of the three most abundant ions, a FT-ICR MS single-ion monitoring (SIM) scan was performed followed by tandem mass spectrometry in the LTQ ion trap. The cycle time for the full scan followed by successive FT-ICR SIM and LTQ MS/MS scans for the three most abundant ions was approximately 2.1 s. Gas phase mass selection, also termed gas phase fractionation (41), was performed in the ion trap with four mass window selections: a 3 min FT-ICR MS scan (m/z 225–500), a 7 min FT-ICR MS scan (m/z 450–800), a 3 min FT-ICR MS scan (m/z 750–1200), and a 1 min FT-ICR MS scan (m/z 1150–2000) for a total method acquisition time of 14 min. The resolution was set to 100,000 for FT-ICR MS full scans and to 50,000 for FT-ICR MS SIM scans. Dynamic exclusion was enabled after a repeat count of three for the duration of the method. This method was validated against the LC-ESI method described above as well as by LC-ESI QTOF MS/MS as described for 4HNE and cytochrome c in Isom et al. (31).
Recombinant human bile acid CoA:amino acid N-acyltransferase (hBAT) was purified as described previously (33). A 1.6 µM solution was reacted with 8, 16, 32, 64 and 128 µM 4HNE at 4°C for 1 h and excess 4HNE removed by adjusting the reaction mixture to 1 mM histidine (33). The modified protein was digested overnight with sequencing grade trypsin or chymotrypsin in 25 mM NH4HCO3 buffer (pH 8.0) (at 37°C for trypsin and at room temperature for chymotrypsin). The samples were analyzed by nanoLC (Eksigent; Dublin, CA) on a 15 cm × 75 µm i.d. reverse-phase C18 column with a linear gradient of 5–95% acetonitrile in 0.1% formic acid at a flow rate of 200 nl.min−1. Eluted tryptic and chymotryptic peptides were electrosprayed at 2 kV. Peptide fragmentation was induced by collision-induced dissociation (CID) in the ion trap, and fragment ions were also analyzed in the ion trap. The LTQ FT mass spectrometer was operated in a “top three” data-dependent acquisition mode. The mass spectrometer was set to switch between an FT-ICR MS full scan (m/z 200 – 2,000) followed by successive FT-ICR MS single-ion monitoring scans and LTQ MS/MS scans of the three most abundant precursor ions in the FT-ICR MS full scan as determined by the Xcalibur software (Thermo Fisher Scientific). Dynamic exclusion was enabled after a repeat count of three for a period of 90 s.
LTQ FT MS/MS data, from both LC-ESI and direct infusion-ESI runs, were searched against a custom FASTA sequence databases containing the protein of interest as well as nine unrelated human proteins, as a negative control, with the TurboSEQUEST algorithm within Bioworks 3.2 (Thermo Fisher Scientific). Monoisotopic precursor and fragment ion masses were searched with a mass tolerance of 2 ppm and 1 Da, respectively. For identification of 4HNE-modified peptides, the TurboSEQUEST searches were amended to search for the mass additions of 156.1150, 138.1045, and 120.0939 for Michael, Schiff base, and 2-pentylpyrrole adducts, respectively. For identification of oxidized αB-crystallin peptides, the TurboSEQUEST searches were amended to search for the mass additions of 15.9949, 31.9898, and 47.9844. Additionally, FT-ICR MS spectra were extracted from each sample data set for manual identification of modifications based on high mass accuracy. The modified peptides were manually validated by their absence in the unmodified FT-ICR MS spectra; a mass accuracy cutoff of 2 ppm was used.
Detecting peptide modifications
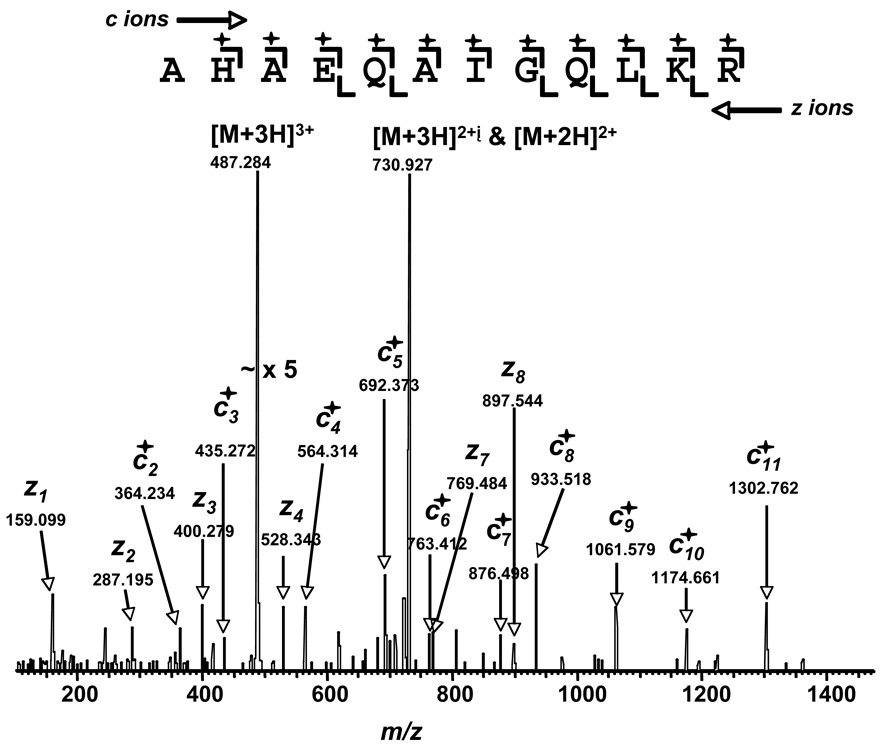
4HNE forms multiple adducts on both CK-BB (Table 2) and hBAT (Table 3) (33, 39). In both cases, the principal sites of adduct formation are histidine residues, as Michael adducts. Even the lysine adducts on hBAT are mostly Michael adducts, as are the cysteine groups on CK-BB. It appears that 4HNE forms more Michael adducts (+m/z 156.1150) than Schiff bases (+m/z 138.1045), although this may be a reflection of the instability of the latter. In one case, an apparent Schiff base was formed on the histidine residue in the hBAT tryptic peptide 335AH*AEQAIGQLKR346 as shown in the ECD mass spectrum (Fig. 1). However, this 138.1045 change in mass of the peptide may be a result of in-source dehydration as noted previously (42).
Table 2.
Summary of 4HNE modifications of CK-BB identified at varying 4HNE concentrations
| Modified | Concentration (µM) | |||||
|---|---|---|---|---|---|---|
| amino acid | 5 | 10 | 30 | 100 | 500 | 3000 |
| H7 | SB | MAB/SAB | MA/SA | MA/SA | ||
| H26 | M | MB | M | MA | ||
| H29 | MA | MA | ||||
| K45 | M | |||||
| H66 | M | MA | ||||
| K86 | M | M | ||||
| H97 | MB | MA | MA | |||
| K101 | M | MA | ||||
| C141 | MB | MAB | M | MA | ||
| C145 | MB | MB | M | MA | ||
| K177 | MA | |||||
| H191 | MB | M | MA | |||
| H219 | MB | MA/S | MA/S | |||
| H234 | S | MA/SA | MA/SA | |||
| K247 | M/S | |||||
| C254 | MC | MC | MB/SB | MB/SB | M/S | M/SA |
| H276 | MA | MA | ||||
| C283 | MB | MB | MAB | M | MA | |
| H296 | SB | M/S | M/S | |||
| H305 | M | M | ||||
| K313 | M | |||||
| K358 | M | |||||
| K381 | M | |||||
M (+156.1150) or S (+138.0145) were peptide modifications detected by direct infusion ESI-FT-ICR-MS; MA or SA were detected using nanoLC-ESI-Qtof-MS at 100–3000 µM 4HNE and direct infusion ESI-FT-ICR-MS; MB or SB were detected only using nanoLC-ESI-FT-ICR-MS and direct infusion ESI-FT-ICR-MS at 5–100 µM 4HNE; MC was detected using nanoLC-ESI-FT-ICR-MS.
Table 3.
Summary of 4HNE modifications* of hBAT identified at varying 4HNE concentrations
| 4HNE concentration (µM) | |||||
|---|---|---|---|---|---|
| Modified peptide sequence | 8 | 16 | 32 | 64 | 128 |
| 1MIQLTATPVSALVDEPVHIR20 | H18 | H18 | |||
| 51RANEFGEVDLNHASSLGGDMYMGVHPMGLFWSLKPEK86 | H62 | H62, H74 | H62, H74 | H62, H74 | H62, H74 |
| 271HGQIHQPLPHSAQL284 | H271, H280 | H271, H280 | H271, H275 | H271, H275 | H271, H275 |
| H280 | H280 | H280 | |||
| 316AQGQFLFIVGEGDKTINSK334 | K329, K334 | K329, K334 | K329, K334 | K329, K334 | K329, K334 |
| 335AHAEQAIGQLKR346 | H336 | H336 | H336 | H336 | H336 |
| 350NNWTLLSYPGAGHLIEPPYSPLCCASTTHDLR381 | H362 | H362 | H362 | H362 | H362, C372 |
| C373, H378 | |||||
| 381RLHWGGEVIPHAAAQEHAWK400 | H397 | H397 | H397 | H397 | |
all these modifications were Michael adducts
Figure 1.
ECD LTQ FT MS/MS spectrum of hBAT peptide 335AH*AEQAIGQLKR346. The spectrum contains a rich series of N-terminal c ions and C-terminal z ions. The c2 ion has a m/z 364.234. The expected unmodified value for this ion is m/z 226.130. The difference is 138.104, the expected value for a Schiff base adduct, or a dehydrated Michael adduct. All the other cn ions (n>2) ( ) were increased by the same amount.
) were increased by the same amount.
In the case of CK-BB, modifications of all four 4HNE-modifiable residues in the active site were detected. Modification of the active site Cys283 was identified at all concentrations of 4HNE where inactivation of enzymatic activity was observed (Table 2 and Figure 2). At 5 µM 4HNE there was no significant inhibition of CK-BB activity; nonetheless, there was modification of a non-active site cysteine (Cys254). This demonstrates the importance of mass spectrometry in PTM analysis as some modifications are not deleterious in effect. In addition, in order to detect the Cys254 4HNE modification it was necessary to use nanoLC-ESI-FT-ICR-MS to obtain sufficient sensitivity. This modification was not detected using direct infusion-ESI and gas phase fractionation. As observed for His336 on hBAT (Fig. 1), several of the histidine residues (His7, His 219, H234 and His296) on CK-BB formed 4HNE Michael adducts (+156.1150) as well as potential dehydrated Michael adducts (+138.1045). A similar pair of 4HNE adducts were observed for Cys254 (Table 2).
Figure 2.
4HNE-induced inactivation of hBAT (□) and CK-BB (♦). Residual hBAT and CK-BB activities following incubation with 4HNE at varying concentrations are dose-dependent. Data are mean values of three independent experiments.
Besides oxidation by electrophilic lipids such as 4HNE, proteins are also prone to oxidation by UV light induced oxidants such as 1O2 and H2O2. The crystallin proteins in the lens of the eye do not undergo any turnover from birth to death (43). Since they are exposed to UVA light (320–400 nm) throughout life and to increasing amounts of UVB light (280–320 nm) with age, the oxidative changes can be significant and lead to a loss of function of the chaperone activities of these proteins (40). Analysis of tryptic peptides revealed that recombinant human αB-crystallin exposed to 50 mJ/cm2 for 2 h is oxidized on methionine and tryptophan residues in the N-terminal region (Met1 and Trp9) and the region responsible for chaperone function (Met68 and Trp60) (44). Interestingly, several isobaric forms of the tryptic peptides were observed. For 1MDIAIHHPWIR11 and 57APSWFDTGLSEMR69, there were two mono-oxygenated isomers, three di-oxygenated isomers and two tri-oxygenated isomers. The tandem mass spectrum of the triply oxygenated 57APSWFDTGLSEMR69 peptide is shown in Fig. 3. The y2-y7 fragment ions all indicate that Met68 is modified by a single oxygen atom, whereas the jump in mass between b3 and b4 (218 Da) for Trp60 is due to two oxygen atoms (186 + 2 × 16).
Figure 3.
LTQ tandem mass spectrum of the triply oxidized αB-crystallin peptide 57APSWFDTGLSEMR69. The b4 ion (m/z 474.2) is 218.1 bigger than the b3 ion (m/z 256.1), showing that the Trp residue contains two oxygen atoms (186.1 + 2 × 16). Similarly, the y2 ion (m/z 322.1) indicates that one oxygen atom has been added to the expected ion (m/z 306.1). Fragment ions that had undergone addition of oxygen to the Met residue (I) and/or the Trp residue (:) are marked.
4HNE modifications of hBAT were analyzed by nanoLC-ESI-FT-ICR due to sample limitations. Functionally, its enzyme activity was completely inhibited at 32 µM (Fig. 2). A total of fourteen modifications were observed at the different concentrations of 4HNE (Table 2). Seven of these modifications (His62, His271, His280, Lys329, Lys334, His336 and His362) were detected at the lowest concentration tested (8 µM; 1:5 molar ratio of hBAT:4HNE). Only one (His362) was a member of the catalytic triad (Cys235:Asp328:His362) (45). Because of their proximity to Asp328, the modifications at Lys329, Lys334 and His336 may have also contributed to changes in hBAT activity.
Summary
Oxidative posttranslational modification of proteins is widespread. In normal tissues proteins often exist in multiple, electrophoretically resolvable forms, many of which are oxidized states. It remains to be determined whether all oxidative modifications are necessarily deleterious to the function of the protein. In a recent study, mild oxidation of HDL with H2O2 increased its ability to abstract cholesterol from macrophages (36). In contrast, oxidation with HOCl resulted in a greater oxidant stress and loss of function (36). Thus whether a particular oxidative modification is deleterious may be dependent on the specific protein, and the dose, as well as the exact chemical nature of the modification.
The versatility, high mass accuracy and high mass resolving capabilities of modern FT-ICR MS instruments are well suited to the identification and characterization of oxidative post-translational modifications.
If an investigator has access to purified, recombinant protein, direct infusion automated nanoelectrospray is a very convenient way to examine the effects of a wide range of concentrations of the oxidative agent. In the present study with human CKBB, 8 pmol of protein was consumed in analysis performed at 6 different concentrations of 4HNE. In the direct infusion method, the mass range is divided in four parts – the very high mass resolution means that even peptides that have the same nominal mass can be resolved. Our direct infusion method made use of gas-phase rather than chromatographic fractionation and is accordingly much faster. Furthermore, since each nozzle of the electrospray chip is only used once, there is no possibility of carryover that can be a difficulty in LC analytical approaches. It is also possible to do real time dynamic exclusion of previously observed peptides in order to examine low intensity ones. However, when the amount of protein is restricted, or to detect modifications occurring at a very low level, nanoLC-ESI-FT-ICR is recommended (Table 3). In the nanoLC approach, the sample is concentrated by the LC, elutes in a smaller volume and hence has better signal-to-noise than direct infusion. The limitations of nanoLC are the longer time that it takes for each analysis (90 min per LC run versus 15 min per infusion for each concentration). NanoLC-ESI-Qtof was less sensitive than either FT-ICR technique (Table 3); however, newer instruments in this category may not be as limited.
Finally, although mass spectrometry can be very effective in studying oxidative posttranslational modifications, it should be borne in mind that it cannot in a MSMS experiment establish with certainty which isomers of tryptophan residues are formed; however, an ion trap method based on MSn may be suitable since it would enable further fragmentation of isobaric MS2 daughter ions.
The structural analysis of protein oxidative modifications will continue to be enhanced by future refinement of MS instrumentation. The most meaningful studies will result from a combination of high resolution MS and functional studies, where the oxidative modifications are correlated with functional effects. Besides LDL (36), there may be other proteins whose functions are enhanced by oxidative modifications. It will be important to identify others in this category to balance the presumption that all modifications are deleterious.
Acknowledgments
Support for the UAB Center for Nutrient-Gene Interaction in Cancer Prevention was provided by a grant-in-aid (U54 CA100949, S. Barnes, PI) from the National Cancer Institute. Support for research on botanicals and dietary supplements and their effects on protein oxidation at the Purdue University-University of Alabama at Birmingham Botanical Center for Age-related Disease was provided by a grant (P50 AT00477, Connie M. Weaver, PI) from the National Center for Complementary and Alternative Medicine and the NIH Office of Dietary Supplements. S. Eliuk and H. Kim were supported by funds from the US Agency for International Development, Kikkoman Corporation, and the Cranberry Institute to H. Kim. Support for the research on bile acid amino acid amidation was provided by a grant (R01 DK46390, S. Barnes PI) from the National Institute for Diabetes, Digestive and Kidney Diseases. Support for research on UV light-induced damage to lens crystallins was provided by a grant from the Alabama EyeSight Foundation. Funds for the purchase of the mass spectrometers were provided by the National Center for Research Resources (S10 RR17261 S. Barnes, PI). Additional support for the operation of the UAB Biomedical FT-ICR laboratory was provided by the Supporters of the UAB Comprehensive Cancer Center.
Literature
- 1.Helle KB, Corti A, Metz-Boutigue MH, Tota B. The endocrine role for chromogranin A: a prohormone for peptides with regulatory properties. Cell. Mol. Life Sci. 2007;64:2863–2886. doi: 10.1007/s00018-007-7254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich H, Josephs SF, Duran ER, Rafalski JA, Whitehorn EA, Baumeister K, Ivanoff L, Petteway SR, Pearson MI, Lautenberger JA, Papas TS, Ghrayeb J, Chang NT, Gallo RC, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 3.Johnson SA, Hunter T. Kinomics: methods for deciphering the kinome. Nat. Methods. 2005;2:17–25. doi: 10.1038/nmeth731. [DOI] [PubMed] [Google Scholar]
- 4.Ptacek J, Snyder M. Charging it up: global analysis of protein phosphorylation. Trends Genet. 2006;22:545–554. doi: 10.1016/j.tig.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Medzihradszky KF. Characterization of protein N-glycosylation. Methods Enzymol. 2005;405:116–138. doi: 10.1016/S0076-6879(05)05006-8. [DOI] [PubMed] [Google Scholar]
- 6.Peter-Katalinić J. Methods in Enzymology: O-glycosylation of proteins. Methods Enzymol. 2005;405:139–171. doi: 10.1016/S0076-6879(05)05007-X. [DOI] [PubMed] [Google Scholar]
- 7.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu. Rev. Biophys. Biomol. Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasan S, Hottiger MO. Histone acetyl transferases: a role in DNA repair and DNA replication. J Mol Med. 2002;80:463–474. doi: 10.1007/s00109-002-0341-7. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki T, Funakoshi Y. Free N-linked oligosaccharide chains: formation and degradation. Glycoconj. J. 2006;23:291–302. doi: 10.1007/s10719-006-6975-x. [DOI] [PubMed] [Google Scholar]
- 10.Cheng X, Zhang X. Structural dynamics of protein lysine methylation and demethylation. Mutat. Res. 2007;618:102–115. doi: 10.1016/j.mrfmmm.2006.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrier J, Durand A, Giardina T, Puigserver A. Catabolism of intracellular N-terminal acetylated proteins: involvement of acylpeptide hydrolase and acylase. Biochimie. 2005;87:673–685. doi: 10.1016/j.biochi.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Imlay JA. Cellular Defenses Against Superoxide and Hydrogen Peroxide. Anu Rev Biochem. 2008 Jan 2; doi: 10.1146/annurev.biochem.77.061606.161055. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 14.Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 15.Beckman JS, Barnes S, Kirk MC, Freeman B, Coward L. Nitrated Proteins and Peptides. In: Caprioli R, editor. Encyclopedia of Mass Spectrometry. Volume 2. NY: Elsevier; 2005. pp. 232–242. [Google Scholar]
- 16.Alvarez B, Rubbo H, Kirk M, Barnes S, Freeman BA, Radi R. Peroxynitrite-dependent tryptophan nitration. Chem. Res. Toxicol. 1996;9:390–396. doi: 10.1021/tx950133b. [DOI] [PubMed] [Google Scholar]
- 17.Taylor SW, Fahy E, Murray J, Capaldi RA, Ghosh SS. Oxidative post-translational modification of tryptophan residues in cardiac mitochondrial proteins. J. Biol. Chem. 2003;278:19587–19590. doi: 10.1074/jbc.C300135200. [DOI] [PubMed] [Google Scholar]
- 18.Sala A, Nicolis S, Roncone R, Casella L, Monzani E. Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical nitrating agents. Eur. J. Biochem. 2004;271:2841–2852. doi: 10.1111/j.1432-1033.2004.04219.x. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki T, Mower HF, Friesen MD, Gilibert I, Sawa T, Ohshima H. Nitration and nitrosation of N-acetyl-L-tryptophan and tryptophan residues in proteins by various reactive nitrogen species. Free Radic. Biol. Med. 2004;37:671–681. doi: 10.1016/j.freeradbiomed.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 20.Skorey K, Ly HD, Kelly J, Hammond M, Ramachandran C, Huang Z, Gresser MJ, Wang Q. How does alendronate inhibit protein-tyrosine phosphatases? J. Biol. Chem. 1997;272:22472–22480. doi: 10.1074/jbc.272.36.22472. [DOI] [PubMed] [Google Scholar]
- 21.Carballal S, Radi R, Kirk MC, Barnes S, Freeman BA, Alvarez B. Sulfenic acid formation in human serum albumin by hydrogen peroxide and peroxynitrite. Biochemistry. 2003;42:9906–9914. doi: 10.1021/bi027434m. [DOI] [PubMed] [Google Scholar]
- 22.Kishikawa M, Nakanishi T, Shimizu A, Yoshino M. Detection by mass spectrometry of highly increased amount of S-sulfonated transthyretin in serum from a patient with molybdenum cofactor deficiency. Pediatr. Res. 2000;47:492–494. doi: 10.1203/00006450-200004000-00013. [DOI] [PubMed] [Google Scholar]
- 23.D’Alessandro T, Prasain J, Botting NP, Moore R, Darley-Usmar VM, Patel RP, Barnes S. Polyphenols, inflammatory response, and cancer prevention: chlorination of isoflavones by human neutrophils. J Nutr. 2003;133:3773S, 3777S. doi: 10.1093/jn/133.11.3773S. [DOI] [PubMed] [Google Scholar]
- 24.Boersma BJ, D’Alessandro T, Benton MR, Kirk M, Wilson LS, Prasain J, Botting NP, Barnes S, Darley-Usmar VM, Patel RP. Neutrophil myeloperoxidase chlorinates soy isoflavones and enhances their antioxidant properties. Free Radic Biol Med. 2003;35:1417–1430. doi: 10.1016/j.freeradbiomed.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Whiteman M, Siau JL, Halliwell B. Lack of tyrosine nitration by hypochlorous acid in the presence of physiological concentrations of nitrite. Implications for the role of nitryl chloride in tyrosine nitration in vivo. J Biol Chem. 2003;278:8380–8384. doi: 10.1074/jbc.M211086200. [DOI] [PubMed] [Google Scholar]
- 26.Steinbeck MJ, Khan AU, Karnovsky MJ. Intracellular singlet oxygen generation by phagocytosing neutrophils in response to particles coated with a chemical trap. J. Biol. Chem. 1992;267:13425–13433. [PubMed] [Google Scholar]
- 27.Davies MJ, Truscott RJ. Photo-oxidation of proteins and its role in cataractogenesis. J. Photochem. Photobiol. B. 2001;63:114–125. doi: 10.1016/s1011-1344(01)00208-1. [DOI] [PubMed] [Google Scholar]
- 28.Davies MJ. The oxidative environment and protein damage. Biochim. Biophys. Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Rodriguez ME, Guo M, Kenney ME, Oleinick NL, Anderson VE. Oxidative modification of cytochrome c by singlet oxygen. Free Radic. Biol. Med. 2008;44:1700–1711. doi: 10.1016/j.freeradbiomed.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonarduzzi G, Arkan MC, Başağa H, Chiarpotto E, Sevanian A, Poli G. Lipid oxidation products in cell signaling. Free Radic Biol Med. 2000;28:1370–1378. doi: 10.1016/s0891-5849(00)00216-1. [DOI] [PubMed] [Google Scholar]
- 31.Isom AL, Barnes S, Wilson L, Kirk M, Coward L, Darley-Usmar V. Modification of cytochrome c by 4-hydroxy-2-nonenal: evidence for histidine, lysine, and arginine-aldehyde adducts. J. Am. Soc. Mass Spectrom. 2004;15:1136–1147. doi: 10.1016/j.jasms.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 32.Kim H, Eliuk S, Deshane J, Meleth S, Sanderson T, Pinner A, Robinson G, Wilson L, Kirk M, Barnes S. 2D gel proteomics: an approach to study age-related differences in protein abundance or isoform complexity in biological samples. Methods Mol Biol. 2007;371:349–391. doi: 10.1007/978-1-59745-361-5_24. [DOI] [PubMed] [Google Scholar]
- 33.Shonsey EM, Eliuk SM, Johnson MS, Barnes S, Falany CN, Darley-Usmar VM, Renfrow MB. Inactivation of human liver bile acid CoA:amino acid N-acyltransferase by the electrophilic lipid, 4-hydroxynonenal. J. Lip Res. 2008;49:282–294. doi: 10.1194/jlr.M700208-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Beckman JS, Chen J, Ischiropoulos H, Crow JP. Oxidative chemistry of peroxynitrite. Methods Enzymol. 1994;233:229–240. doi: 10.1016/s0076-6879(94)33026-3. [DOI] [PubMed] [Google Scholar]
- 35.Waeg G, Dimsity G, Esterbauer H. Monoclonal antibodies for detection of 4-hydroxynonenal modified proteins. Free Radic. Res. 1996;25:149–159. doi: 10.3109/10715769609149920. [DOI] [PubMed] [Google Scholar]
- 36.Gao X, Jayaraman S, Gursky O. Mild oxidation promotes and advanced oxidation impairs remodeling of human high-density lipoprotein in vitro. J Mol Biol. 2008;376:997–1007. doi: 10.1016/j.jmb.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 38.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 39.Eliuk SM, Renfrow MB, Shonsey E, Kirk MC, Barnes S, Kim H. Active site modification of cytosolic creatine kinase by 4-hydroxy-2-nonenal correlates with reduction in activity. Chem Res. Toxicol. 2007;20:1260–1268. doi: 10.1021/tx7000948. [DOI] [PubMed] [Google Scholar]
- 40.Mafia K, Gupta R, Kirk MC, Wilson L, Srivastava OP, Barnes S. UV-A-induced structural and functional changes in human lens deamidated αB-crystallin. Mol. Vision. 2008;14:234–248. [PMC free article] [PubMed] [Google Scholar]
- 41.Davis MT, Spahr CS, McGinley MD, Robinson JH, Bures EJ, Beierle J, Mort J, Yu W, Luethy R, Patterson SD. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. II. Limitations of complex mixture analyses. Proteomics. 2001;1:108–117. doi: 10.1002/1615-9861(200101)1:1<108::AID-PROT108>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, Minkler PE, Sayre LM. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2—(E)-nonenal. Chem Res. Toxicol. 2003;16:901–911. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- 43.Lynnerup N, Kjeldsen H, Heegaard S, Jacobsen C, Heinemeier J. Radiocarbon dating of the human eye lens crystallins reveal proteins without carbon turnover throughout life. PLoS ONE. 2008;3:e1529. doi: 10.1371/journal.pone.0001529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sreelakshmi Y, Sharma KK. Recognition sequence 2 (residues 60–71) plays a role in oligomerization and exchange dynamics of alphaB-crystallin. Biochemistry. 2005;44:12245–12252. doi: 10.1021/bi051005h. [DOI] [PubMed] [Google Scholar]
- 45.Sfakianos M, Wilson L, Sakalian M, Falany CN, Barnes S. Conserved residues in the putative catalytic triad of human bile acid Coenzyme A: amino acid N-acyltransferase. J. Biol. Chem. 2002;277:47270–47275. doi: 10.1074/jbc.M207463200. [DOI] [PubMed] [Google Scholar]