Abstract
Horizontal gene transfer has been occasionally mentioned in eukaryotic genomes, but such events appear much less numerous than in prokaryotes, where they play important functional and evolutionary roles. In yeasts, few independent cases have been described, some of which corresponding to major metabolic functions, but no systematic screening of horizontally transferred genes has been attempted so far. Taking advantage of the synteny conservation among five newly sequenced and annotated genomes of Saccharomycetaceae, we carried out a systematic search for HGT candidates amidst genes present in only one species within conserved synteny blocks. Out of 255 species-specific genes, we discovered 11 candidates for HGT, based on their similarity with bacterial proteins and on reconstructed phylogenies. This corresponds to a minimum of six transfer events because some horizontally acquired genes appear to rapidly duplicate in yeast genomes (e.g. YwqG genes in Kluyveromyces thermotolerans and serine recombinase genes of the IS607 family in Saccharomyces kluyveri). We show that the resulting copies are submitted to a strong functional selective pressure. The mechanisms of DNA transfer and integration are discussed, in relation with the generally small size of HGT candidates. Our results on a limited set of species expand by 50% the number of previously published HGT cases in hemiascomycetous yeasts, suggesting that this type of event is more frequent than usually thought. Our restrictive method does not exclude the possibility that additional HGT events exist. Actually, ancestral events common to several yeast species must have been overlooked, and the absence of homologs in present databases leaves open the question of the origin of the 244 remaining species-specific genes inserted within conserved synteny blocks.
Introduction
The transfer of genetic information between organisms normally separated by reproductive barriers, a process now known as horizontal (or lateral) gene transfer (HGT or LGT), was for long time considered limited to specific systems such as, for example, transducing viruses or bacteriophages (reviewed in [1]). With the rapidly increasing number of genome sequences, examples of horizontally transferred genes accumulated, especially for bacterial genomes where they play important functional and evolutionary roles [2], [3]. The role of HGT in eukaryotic evolution was generally regarded as more limited if one excludes their ancestral organelle endosymbioses, but is now gaining greater attention with the increasing number of well supported cases, many of which with significant functional implications [4]. Although the majority of such cases concerns protists with phagotrophic life style [5], significant examples have recently been reported for fungal genomes, especially plant pathogens [6] or species living in complex microbial populations such as rumen [7]. Horizontal transfer has also been proposed for a variety of non-infective selfish genetic elements irregularly found in fungal species, such as plasmids, mycoviruses, mobile group I introns and their encoded homing endonucleases, and even transposons [8]. It is also debated as the possible origin of clusters of genes encoding secondary metabolite enzymes [8]–[10].
Among fungi, hemiascomycetous yeasts represent a homogeneous, monophyletic subdivision in which numerous genomes have been sequenced [11]–[13], including several isolates of Saccharomyces cerevisiae [14]–[18 and www.broad.mit.edu], one of the most extensively studied eukaryotic genome. Beside the selfish genetic elements mentioned above, few genes of putative bacterial origin were recognized in the genomes of several hemiascomycetous yeasts such as Eremothecium (Ashbya) gossypii [19], Kluyveromyces lactis, Debaryomyces hansenii, Yarrowia lipolytica [20], S. cerevisiae [15], [19], [21]–[23], Dekkera bruxellensis [24] or Candida parapsilosis [25]. Most of them encode metabolic enzymes that may play important physiological roles in the adaptation of the host species. Perhaps the most spectacular case so far is the acquisition of a bacterial gene encoding di-hydroorotate dehydrogenase (possibly from a Lactococcus) by an ancestor of all Saccharomycetaceae, forming the URA1 gene encoding the cytoplasmic enzyme active even in anaerobic condition, while the ancestral URA9 gene encoding the strictly aerobic mitochondrial enzyme was secondarily lost in the Saccharomyces sensu stricto and a few other species [22]. Independent transfers of the same bacterial function to distinct eukaryotic clades seem to have occurred repeatedly in yeasts and other organisms, often in replacement of ancestral eukaryotic genes lost during evolution [21], [23], [25].
Despite the well-documented above examples, cases of horizontal gene transfer in yeasts, and in fungi in general, remain anecdotal. A reason for this may be that genes of foreign origin were not systematically sought for in available genome sequences. Another reason is that, despite suggestive signatures such as distinctive nucleotide composition or biased codon usage, the gold standard for identifying HGT remains phylogenetic incongruence of the suspected gene(s) with respect to the accepted species phylogeny. This discriminative criterion transfers the burden of proof to the proper taxonomic sampling of the sequenced species across phylogenies, a problem rarely solved at present. In yeasts for example, extensive genomic studies have focused on S. cerevisiae and the human pathogen C. albicans, and their close relatives, leaving the broad evolutionary range of other hemiascomycetes relatively unexplored [11], [12]. In a recent work, the genomes of five protoploid species of Saccharomycetaceae, belonging to four distinct clades that separated from S. cerevisiae before its genome duplication, have been analyzed and compared [13]. This set consists of three newly sequenced genomes, Zygosaccharomyces rouxii, Kluyveromyces (Lachancea) thermotolerans and Saccharomyces (Lachancea) kluyveri, and two previously published ones, K. lactis [20] and E. gossypii [26]. It is thought to reflect the ancestral genome of the Saccharomycetaceae family. Despite their broad evolutionary distances, distinct metabolic properties and habitat, the genomes of these species have numerous conserved blocks of synteny within which individual, lineage-specific gene insertion or loss can be examined. We used this criterion to identify inserted genes among which, after analysis, some proved to represent novel cases of HGT from bacterial origins. The presence of these genes suggests that HGT may be more frequent and functionally important than usually suspected and, consequently, may play a significant role in genome evolution.
Results
Conservation of synteny and identification of putative gene transfers
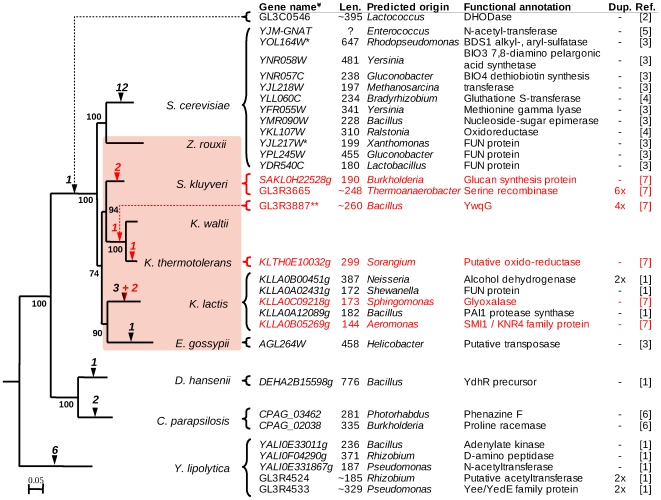
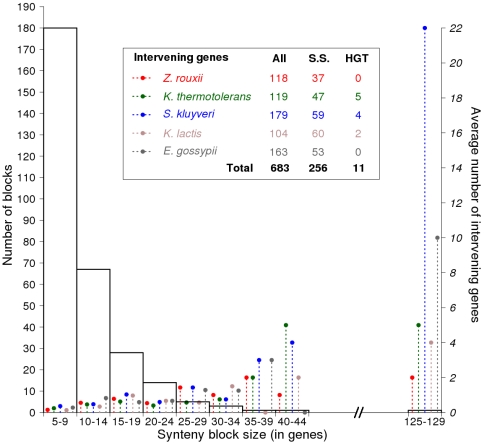
Analysis of synteny among five protoploid genomes of the Saccharomycetaceae family (a phylogeny of these species among other sequenced hemiascomycetes is illustrated by Figure 1) has revealed a striking conservation of gene order, and most often orientation in the different pairwise comparisons [13]. Synteny blocks of 14 to 26 genes on average, cover over 80% of all protein-coding genes. We extended here the analysis of synteny block conservation to all five genomes simultaneously (Materials and Methods). A total of 300 synteny blocks, ranging in size from 5 to 42 anchor points was found (Figure 2), with the notable exception of a block made of 127 anchor points (This block is discussed in [27]). The number of blocks decreases as their size increase, and only a few blocks with more than 30 anchor points are found. In total, conserved synteny blocks common to all five species cover ca. 65% of each genome among which we could examine the insertion of novel genes in one species compared to the others. To do so, we established a list of “intervening genes”, inserted within a block in one of the five species without altering its overall conservation (i.e. flanking genes remain unchanged in order and almost always in orientation). They are more numerous in S. kluyveri and E. gossypii. The number of intervening genes is found roughly proportional to the block size (Figure 2). Out of the 300 synteny blocks examined, we initially found 682 intervening genes. Their translation products were individually compared to all known hemiascomycetous genomes in order to test their uniqueness or find similarities (Materials and Methods). A total of 427 genes have ectopic homologs in the other yeast species and were, therefore, considered as resulting from internal chromosomal rearrangements. Among the remaining 255 species-specific genes, 244 have no significant hit (according to parameters, Materials and Methods), and thus could not be further studied (See Discussion and Supplementary Table S1). The remaining 11 gene products show significant sequence similarity with bacterial proteins and are, therefore, candidates for HGT. The candidates include four single intervening genes (one in K. thermotolerans, two in K. lactis and one in S. kluyveri), four members of a multi-gene family in K. thermotolerans, and three members of a multi-gene family in S. kluyveri. These HGT candidates, together with other previously published cases in hemiascomycetous yeasts, are summarized in Figure 1.
Figure 1. Phylogenetic tree of the Saccharomycetaceae, with published HGT cases.
The tree was constructed by maximum likelihood using PHYML, from alignments of conserved protein families with only one member per species and corresponding to true orthologs as defined by SONS [13]. Alignments were performed using the MAFFT algorithm and further cleaned with Gblocks before concatenation (53 families, 19144 residues). Bootstrap values are indicated next to the nodes. Triangles represent cases of HGT (black are published cases and red are cases discussed in this paper). References: (1) Dujon et al. 2004; (2) Gojkovic et al. 2004; (3) Hall et al. 2005; (4) Hall and Dietrich, 2007; (5) Wei et al. 2007; (6) Fitzpatrick et al. 2008; (7) This work. Len.: length in amino-acids (for multigene families, the average is shown). Dup.: Number of copies of transferred gene. FUN: unknown function. ¥ In cases of duplication and/or ancestral HGT, the Génolevures families replace individual gene names. * Highly similar orthologs of S. cerevisiae genes YOL164W and YJL217W have been identified in K. thermotolerans, suggesting more ancestral HGT events. ** The gene is present in four copies in K. thermotolerans, but only once in K. waltii.
Figure 2. Size distribution of conserved synteny blocks among the five protoploid Saccharomycetaceae, and intervening gene numbers.
Synteny block common to all 5 protoploid Saccharomycetaceae species (inset) were classified by their number of anchoring points (abscissa, Materials and Methods). The number of blocks in each size category is referred to the scale on the left axis. The mean number of intervening genes per species and per block category is represented by dotted lines (scale on the right axis), and total figure for each species are in inset. All: total number of intervening genes. S-S: total number of species-specific intervening genes. HGT: total number of horizontally transferred genes.
Single horizontally acquired genes
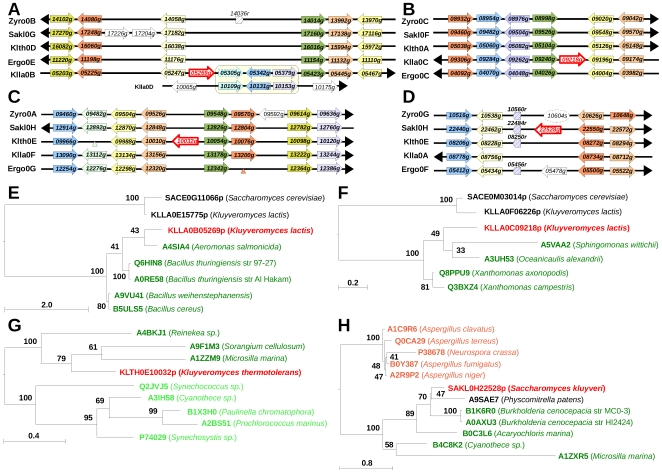
The K. lactis KLLA0B05269g gene, coding a 144 amino-acid long protein, is contained in a conserved synteny block made of 42 anchor points (one of the largest conserved synteny block among our five protoploid Saccharomycetaceae, Figure 3A) and shares no similarity with any yeast, fungal or eukaryotic protein presently known. Instead, it shares high sequence similarity with proteins of Aeromonas salmonicida (53% amino-acid identity, Table 1) and various Bacillus species (including B. thuringiensis, B. cereus, and B. weihenstephanensis, 42–49% identity, see alignment in Supplementary Figure S1A). None of these proteins is annotated, except the B. cereus protein annotated as a member of the SMI1/KNR4 family, involved in the regulation of the cell wall synthesis in yeast [28]. The SMI1 protein of S. cerevisiae (YGR229C) has homologs in all hemiascomycetous yeasts, including K. lactis (KLLA0E15775p protein) but those are 3- to 4-fold longer in sequences than KLLA0B05269g and share only limited sequence identity with it (average 22%, Table 1). As expected, the tree reconstructed with S. cerevisiae and K. lactis SMI1 proteins shows that the KLLA0B05269g gene product is closer to bacterial proteins than to the yeast SMI1 homologs (Figure 3E), suggesting a recent acquisition of this gene in the K. lactis lineage from a bacterium. Its actual function remains to be established. Interestingly, the KLLA0B05269g gene is flanked in its distal side by a segmental duplication of 5.5 kb with chromosome D (Figure 3A). This segment involves three genes, two of which being annotated as pseudogenes on chromosome B. The contiguity of the HGT candidate with the segmental duplication (only ∼2 bp) raises the question of a possible link between the two events (Discussion).
Figure 3. Single HGT candidates in K. lactis, K. thermotolerans and S. kluyveri.
(A) Part of the conserved synteny block surrounding the KLLA0B05269g gene (previously reported in [20]). Only protein coding and tRNA genes are shown. Orthologous genes are colored, intervening genes are white, tRNA genes are indicated by short hatched arrows, pseudogenes are identified by dotted lines. Arrows represent gene orientation. The segmental duplication between chromosomes B and D of K. lactis is shown at the bottom of the figure. KLLA0D10109g is similar to S. cerevisiae YOR120W GCY1, KLLA0D10131g similar to S. cerevisiae YKL221W MCH2, and KLLA0D10153g and KLLA0B05379g are slightly similar to S. cerevisiae YKL222C protein of unknown function. (B) Part of the conserved synteny block surrounding the KLLA0C09218g gene. Same legend as (A). (C) Part of the conserved synteny block surrounding KLTH0E10032g gene. Same legend as (A). Triangles represent single gene species-specific deletions. (D) Part of the conserved synteny block surrounding SAKL0E22528g. Same legend as (A). ZYRO0G10604s is the centromere of chromosome G in Z. rouxii (dotted oval). Note the local inversion of two genes in Z. rouxii. (E) Phylogenetic tree reconstructed from sequence alignment of 81 sites of the KLLA0B05269g product, bacterial proteins (green) and products of S. cerevisiae SMI1 gene and its K. lactis homolog (YGR229C/SACE0G11066p and KLLA0E15775p, in black). Bootstrap values are indicated next to the nodes and branch length scale is shown at bottom left. (F) Phylogenetic tree reconstructed from sequence alignment of 65 sites of the KLLA0C09218g product, bacterial proteins (green) and the S. cerevisiae glyoxalase I gene (YML004C/SACE0M03014p) and its K. lactis homolog (KLLA0F06226p). Same legend as (B). (G) Phylogenetic tree reconstructed from sequence alignment of 85 sites of the KLTH0E10032g product and bacterial proteins (dark green, cyanobacteria in light green). Same legend as (B). (H) Phylogenetic tree reconstructed from sequence alignment of 72 sites of the SAKL0H22528g product, bacterial and moss proteins (green) and Pezizomycotina proteins (light red). Same legend as (B).
Table 1. Pairwise sequence identity between single HGT candidate products, best bacterial hits and yeast/fungal similarly annotated proteins.
| Query gene | Génolevures/Uniprot annotation | Species | Reference* | Length (aa) | Identity (%)** |
| KLLA0B05269g (144 amino-acids) | hypothetical protein ASA_0453 | Aeromonas salmonicida | A4SIA4 | 143 | 53 |
| hypothetical protein BT9727_2262 | Bacillus thuringiensis str. 97–27 | Q6HIN8 | 142 | 49 | |
| hypothetical protein BALH_2200 | Bacillus thuringiensis str. Al Hakam | A0RE58 | 142 | 48 | |
| hypothetical protein BcerKBAB4_2135 | Bacillus weihenstephanensis | A9VU41 | 143 | 44 | |
| SMI1/KNR4 family protein BCAH1134_3336 | Bacillus cereus | B5ULS5 | 134 | 42 | |
| SMI1 homolog | Kluyveromyces thermotolerans | KLTH0C01760p | 516 | 28 | |
| SMI1 homolog | Saccharomyces kluyveri | SAKL0G15466p | 515 | 26 | |
| ADL139W – SMI1 protein | Eremothecium gossypii | ERGO0D06072p | 657 | 26 | |
| SMI1 homolog | Zygosaccharomyces rouxii | ZYRO0B15180p | 619 | 24 | |
| YGR229C – SMI1 protein | Saccharomyces cerevisiae | SACE0G11066p | 505 | 24 | |
| KLLA0E15862g a – KNR4/SMI1 family protein | Kluyveromyces lactis | KLLA0E15775p | 535 | 23 | |
| KLLA0C09218g (173 amino-acids) | hypothetical protein XAC0586 | Xanthomonas axonopodis pv. Citri str. 306 | Q8PPU9 | 164 | 44 |
| putative glyoxalase/bleomycin resistance protein/dioxygenase XCV0638 | Xanthomonas campestris pv. Vesicatoria str. 85–10 | Q3BXZ4 | 131 | 41 | |
| hypothetical protein OA2633_07919 | Oceanicaulis alexandrii HTCC2633 | A3UH53 | 131 | 39 | |
| glyoxalase/bleomycin resistance protein/dioxygenase Swit_2866 | Sphingomonas wittichii RW1 | A5VAA2 | 127 | 39 | |
| Glyoxalase I homolog | Saccharomyces kluyveri | SAKL0A02244p | 341 | 25 | |
| KLLA0F06226p | Kluyveromyces lactis | KLLA0F06226p | 338 | 24 | |
| YML004C - Glyoxalase I | Saccharomyces cerevisiae | SACE0M03014p | 326 | 25 | |
| Glyoxalase I homolog | Zygosaccharomyces rouxii | ZYRO0D09064p | 347 | 28 | |
| Glyoxalase I homolog | Kluyveromyces thermotolerans | KLTH0A05896p | 346 | 26 | |
| KLTH0E10032g (299 amino-acids) | Hypothetical protein Sce1221 | Sorangium cellulosum So ce56 | A9F1M3 | 292 | 37 |
| Putative uncharacterized protein M23134_00952 | Microscilla marina ATCC 23134 | A1ZZM9 | 277 | 27 | |
| Putative NADH-ubiquinone oxido-reductase MED297_07641 | Reinekea sp. MED297 | A4BKJ1 | 284 | 29 | |
| NAD-dependent epimerase/dehydratase family protein CYA_1049 | Synechococcus sp. JA-3-3Ab | Q2JVJ5 | 318 | 27 | |
| Putative uncharacterized protein CY0110_10507 | Cyanothece sp. CCY 0110 | A3IH58 | 325 | 27 | |
| Putative chaperon-like protein for quinone binding in Photosystem II ycf39 | Paulinella chromatophora | B1X3H0 | 320 | 24 | |
| Putative chaperon-like protein for quinone binding in Photosystem II A9601_13281 | Prochlorococcus marinus AS9601 | A2BS51 | 320 | 26 | |
| Ycf39 protein | Synechocystis sp. PCC 6803 | P74029 | 219 | 22 | |
| SAKL0H22528g (190 amino-acids) | Predicted protein PHYPADRAFT_163804 | Physcomitrella patens | A9SAE7 | 248 | 38 |
| MoeA domain protein Bcenmc03_4042 | Burkholderia cenocepacia str MC0-3 | B1K6R0 | 674 | 38 | |
| MoeA domain protein Bcen2424_3480 | Burkholderia cenocepacia str HI2424 | A0AXU3 | 693 | 38 | |
| Putative uncharacterized protein M23134_06735 | Microscilla marina ATCC 23134 | A1ZXR5 | 194 | 32 | |
| Protein involved in beta-1-3-glucan synthesis Cyan7425DRAFT_0799 | Cyanothece sp. | B4C8K2 | 180 | 31 | |
| Putative uncharacterized protein AM1_4879 | Acaryochloris marina str MBIC 11017 | B0C3L6 | 165 | 31 | |
| 1,3-beta-glucan biosynthesis protein, putative ACLA_009040 | Aspergillus clavatus | A1C9R6 | 517 | 34 | |
| Catalytic activity: UDP-glucose+((1)) An17g02120 | Aspergillus niger str CBS 513.88/FGSC A1513 | A2R9P2 | 525 | 33 | |
| Putative uncharacterized protein ATEG_09455 | Aspergillus terreus str NIH 2624 | Q0CA29 | 531 | 32 | |
| 1,3-beta-glucan biosynthesis protein, putative AFUB_053320 | Aspergillus fumigatus A1163 | B0Y387 | 515 | 32 | |
| Glucan synthesis regulatory protein gs-1 | Neurospora crassa | P38678 | 532 | 30 |
References are taken from Uniprot database (http://www.uniprot.org/) for bacterial proteins and from Génolevures (http://www.genolevures.org/) for yeast proteins
Sequence identity is measured on total query sequence length.
KLLA0E15862g corresponds to the new Génolevures annotation KLLA0E15775g.
A second HGT candidate also found in K. lactis, KLLA0C09218g, encodes a 173 amino-acid long protein and is located in a 6 anchor point synteny block (Figure 3B). Applying the same methodology, we found sequence similarity of its product with proteins from diverse bacterial genomes, including Xanthomonas axonopodis, X. campestris, Oceanicaulis alexandrii and Sphingomonas wittichii (Table 1 and alignment in Supplementary Figure S1B). Two of these proteins are annotated as putative glyoxalases/dioxygenases. Again, a glyoxalase I gene exists in S. cerevisiae (YML004C) and has homologs in other hemiascomycetous yeasts (including K. lactis KLLA0F06226p protein), but such genes are 2-fold longer in sequences, and show limited similarity with our HGT candidate (average 19%, Table 1). The phylogenetic tree reconstructed shows that the KLLA0C09218g gene product is closer to bacterial proteins than to the yeast glyoxalase I proteins (Figure 3F), suggesting another acquisition by HGT in the K. lactis lineage. The function of this gene remains to be determined.
In K. thermotolerans, one single HGT candidate, KLTH0E10032g, encoding a 299 amino-acid long protein, was found in a conserved synteny block made of 12 anchor points (Figure 3C). Its product shares sequence similarity with proteins from the myxobacterium Sorangium cellulosum (36% identity, 51% similarity, Table 1 and alignment in Supplementary Figure S1C), the cyanobacterium Microscilla marina and the aquatic gamma-proteobacterium Reinekea sp., the latter being annotated as a putative NADH-ubiquinone oxido-reductase. Lower similarity levels are also found with other cyanobacterial proteins (with an average of 22% pairwise identity), and with flavin reductases from mammals (from M. musculus, R. norvegicus, E. caballus, B. taurus, C. familiaris and H. sapiens, with an average of 14% identity, not shown). The limited conservation of KLTH0E10032g gene product with bacterial proteins (except Sorangium cellulosum) raises the question of its origin. If it actually originates from bacteria through HGT, it is possible that we are missing the actual donor group (it might not be represented in Uniprot database) or that its sequence has rapidly diverged after transfer. The low bootstrap values associated with the tree are consistent with the second hypothesis (Figure 3G).
Our last single HGT candidate was found in S. kluyveri, SAKL0H22528g. It encodes a 190 amino-acid long protein, located in a conserved synteny block made of 11 adjacent anchor points (Figure 3D). Its translation product shares similarity with bacterial proteins from Burkholderia cenocepacia (38% identity, Table 1 and alignment in Supplementary Figure S1D), Acaryochloris marina, Cyanothece sp. and Microscilla marina (average of 31% identity). The B. cenocepacia protein, even if 3-fold longer, is particularly well aligned in its C-terminal part with our HGT candidate sequence. Interestingly, the SAKL0H22528g product also shares similarity with a predicted protein of the Funariaceae moss Physcomitrella patens (38% pairwise identity, Table 1 and alignment in Supplementary Figure S1D), not explicitly annotated. Our HGT candidate, as the latter protein, aligns only with the C-terminal part of MoeA domain protein from B. cenocepacia (Supplementary Figure S1D), overlapping the SMI1 domain involved in cell wall synthesis. This could be explained by an independent HGT from the pathogenic B. cenocepacia to the moss [29]. No significant similarity was found with any yeast protein, but weakly similar proteins involved in glucan synthesis exist in Neurospora crassa and Aspergillus species. They show however poorly aligned sequences with our HGT candidate. All hemiascomycetous yeast proteins involved in glucan synthesis or its regulation are 3- to 10-fold longer in sequences, and show very poor alignments with SAKL0H22528p (not shown). The reconstructed phylogenetic tree shows that SAKL0H22528p is more closely related to bacterial proteins than to proteins known to act in glucan synthesis in the Pezizomycotina (Figure 3H), and suggests that it is a good candidate for horizontal transfer, although its original function still has to be refined.
Duplicated horizontally acquired genes
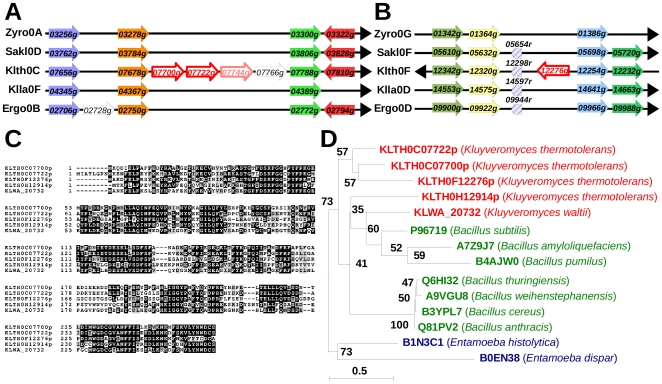
Interestingly, we also found horizontally acquired genes present in several copies in yeast genomes, forming families of paralogs. One such case is observed in K. thermotolerans, where KLTH0C07700g and KLTH0C07722g genes form a pair of tandem paralogs, encoding proteins of ca. 260 amino-acids, inserted in a conserved synteny block made of 12 anchor points (Figure 4A). Note that four genes are inserted in this interval, and that the product of KLTH0C07744g gene shows weak similarity to the products of the tandem pair (not shown), and may represent a diverged repeat of the same tandem array. Two other paralogs to these genes exist on other chromosomes of K. thermotolerans. KLTH0F012276g is contained in another conserved synteny block made of 7 anchor points (Figure 4B). KLTH0H12914g is not in a conserved synteny block. All four genes form a species-specific protein family among our yeasts (Génolevures family GL3R3887) that shares 38 to 48% identity with proteins from various Bacillus species, annotated as YwqG (Table 2 and Figure 4C). These proteins also share similarity with proteins of two Entamoeba species, a parasitic genus in which diverse prokaryotic gene transfers have already been described [30]. The dN/dS ratio calculated on aligned sequences of this family (Materials and Methods) shows strong functional pressure on these genes (with average dN/dS values of 0.23, Supplementary Table S2), indicating that they are expressed in yeast and submitted to selection. Interestingly, a 259 amino-acid long protein predicted from the genome of K. waltii (KLWA_20732), a close relative to K. thermotolerans, belongs to this family. This suggests that the transfer from bacteria occurred in the ancestor of these two yeast species. The K. waltii gene is not syntenic to any of the four genes of K. thermotolerans, suggesting transfer to ectopic location or duplications followed by gene loss. The presence of large intergenes in both species, opposite to the HGT genes where corresponding regions are aligned (Supplementary Figure S2) suggest that the duplications arose before the speciation of the two species, and were subsequently lost during evolution. However, the possibility of independent transfer of the same bacterial gene to K. thermotolerans and K. waltii cannot be formally excluded.
Figure 4. Duplicated HGT candidates in K. thermotolerans.
(A) Part of the conserved synteny block surrounding KLTH0C07700g and KLTH0C07722g tandem genes. Same legend as Figure 3A. (B) Part of the conserved synteny block surrounding KLTH0F12276g gene. Same legend as Figure 3A. (C) Sequence alignment of proteins from K. thermotolerans and K. waltii. (D) Phylogenetic tree reconstructed from sequence alignment of 100 sites of K. thermotolerans protein family members, K. waltii gene, bacterial (green) and amoebal (blue) proteins. Bootstrap values are indicated next to the nodes and branch length scale is shown at bottom left.
Table 2. Pairwise sequence identity between K. thermotolerans protein family, K. waltii protein and best bacterial and amoebal hits.
| Identity (%)** | ||||||||
| Génolevures/Uniprot annotation | Species | Reference* | Length (aa) | KLTH0C07700g | KLTH0C07722g | KLTH0F12276g | KLTH0H12914g | KLWA_20732 |
| KLTH0C07700p | K. thermotolerans | - | 259 | - | - | - | - | - |
| KLTH0C07722p | K. thermotolerans | - | 267 | 64 | - | - | - | - |
| KLTH0F12276p | K. thermotolerans | - | 259 | 52 | 53 | - | - | - |
| KLTH0H12914p | K. thermotolerans | - | 264 | 48 | 50 | 44 | - | - |
| KLWA_20732a | K. waltii | - | 259 | 52 | 51 | 43 | 48 | - |
| YwqG | B. subtilis | P96719 | 261 | 49 | 48 | 41 | 47 | 52 |
| YwqG | B. amyloliquefaciens str FZB42 | A7Z9J7 | 262 | 44 | 46 | 43 | 47 | 48 |
| YwqG | B. pumilus ATCC 7061 | B4AJW0 | 271 | 46 | 46 | 41 | 44 | 45 |
| Putative uncharacterized protein BT9727_2468 | B. thuringiensis subsp konkukian | Q6HI32 | 271 | 47 | 46 | 37 | 41 | 45 |
| Putative uncharacterized protein BAS2507 | B. anthracis | Q81PV2 | 271 | 48 | 45 | 37 | 41 | 44 |
| Putative uncharacterized protein BcerKBAB4_2451 | B. weihenstephanensis str KBAB4 | A9VGU8 | 271 | 44 | 45 | 36 | 40 | 42 |
| YwqG | B. cereus AH1134 | B5UWG9 | 271 | 44 | 42 | 35 | 38 | 43 |
| Putative uncharacterized protein EDI_035210 | E. dispar SAW760 | B0EN38 | 284 | 35 | 35 | 36 | 34 | 32 |
| Putative uncharacterized protein EHI_001040 | E. histolytica HM-1:IMSS | B1N3C1 | 267 | 30 | 31 | 30 | 31 | 29 |
References are taken from Uniprot database (URL: http://www.uniprot.org/) for bacterial and Entamoeba proteins and from Génolevures (URL: http://www.genolevures.org/) for yeast proteins.
Sequence identity is measured on total query sequence length.
K. waltii gene KLWA_20732 sequence was taken from annotation published in Kellis et al. (2004).
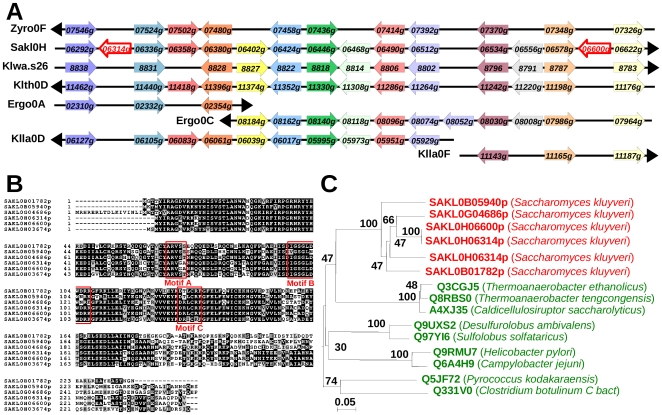
Another case of amplification of horizontally acquired genes was found in S. kluyveri. Three genes, SAKL0B01782g, SAKL0G04686g and SAKL0H06600g, detected according to our method (Figure 5A) are members of a family also including SAKL0B05940g, SAKL0H03674g and SAKL0H06314g. The latter gene falls in the same synteny block as SAKL0H06600g (Figure 5A), but was not originally detected in our method because of a synteny breakpoint in the K. lactis genome (not shown in Figure 5A). The predicted gene products of this family share 56 to 100% amino-acid identity between themselves (Table 3 and Figure 5B). Interestingly, the 100% identity between SAKL0H06314g and SAKL0H06600g, separated by 12 other genes along the same chromosome, extends to 113 and 138 nt in promoter and terminator regions, respectively, indicating a recent duplication event. The three genes not identified from our method are located in conserved synteny blocks restricted to fewer species including those of the Lachancea clade (Figure 5A and Supplementary Figure 3). The products of these six genes have no significant similarity in eukaryotes so far, thus they form a species-specific protein family in S. kluyveri (Génolevures family GL3R3665). Instead, they share similarity with putative serine recombinases of the IS607 family found in bacteria and archaea (Table 3). The most similar ones (27–31% identity) are in Thermoanaerobacter ethanolicus, T. tengcongensis and Caldicellulosiruptor saccharolyticus, three species of Firmicutes (Table 3 and Figure 5C). These proteins are composed of a DNA binding domain in their N-terminal part, that is highly conserved in all six S. kluyveri proteins (first 50 amino-acids, Figure 5B) and a catalytic domain covers almost the rest of their sequence [31], [32]. Motifs A, B and C of the catalytic domain of the serine recombinases [33] are conserved in S. kluyveri proteins with, however, two exceptions: the catalytic serine, at position 71 in motif A (Figure 5B), is replaced by a glycine in SAKL0H06314p and SAKL0H06600p, which should abolish the activity of these proteins, and the Arg-132 in motif C is replaced by a threonine residue in SAKL0B01782p and SAKL0B05940p. The dN/dS ratio between members of the six gene family shows a bias towards synonymous against non-synonymous mutations (dN/dS values of 0.19 on average, Supplementary Table S2), suggesting a functional selective pressure on these genes. Because the serine recombinases are part of mobile elements in prokaryotes, we looked for the presence/absence of the 6 HGT genes in different strains of S. kluyveri isolated from various parts of the world (Supplementary Table S3) using PCR primers in the two flanking genes. Thus, we did not observe any polymorphism for five out of the six HGT genes in all studied strains. Indeed, PCR results for the SAKL0H03674g gene suggests that this locus is highly polymorphic, as revealed by the absence of amplicon in five strains, the presence of two amplicons in two other strains (CBS6626 and CBS10368), and of an insertion in another strain (CBS10369, Supplementary Table S4). Note that this chromosomal region is particularly rearranged (Supplementary Figure S3C). Nevertheless, the transposase activity of this protein family in yeast remains to be demonstrated.
Figure 5. Duplicated HGT candidates in S. kluyveri.
(A) Chromosomal region surrounding SAKL0H06314g and SAKL0H06600g and conserved syntenic regions of the other protoploid genomes including K. waltii. Same legend as Figure 3A. (B) Sequence alignment of the six putative horizontally transferred serine recombinases of S. kluyveri. Red rectangles indicate the conserved motifs (A, B and C) within the catalytic domain according to Grindley et al. [33]. (C) Phylogenetic tree built from protein alignment of the six S. kluyveri proteins and of the closest serine recombinase from archaea (Q9UXS2, Q97YI6 and Q5JF72) and bacteria especially Firmicutes (Q3CGJ5, Q8RBS0, A4XJ35 and Q331V0) and proteobacteria (Q9RMU7 and Q6A4H9). After horizontal gene transfer, the first putative serine recombinase in S. kluyveri has been successively duplicated in the genome. The presence of identical genes (SAKL0H06314g and SAKL0H06600g) suggests a recent duplication event or the result of a conversion event. Bootstrap values are indicated next to the nodes.
Table 3. Pairwise sequence identity between K. kluyveri protein family and best bacterial and archaeal hits.
| Identity (%)** | |||||||||
| Génolevures/Uniprot annotation | Species | Reference* | Length (aa) | SAKL0B01782p | SAKL0B05940p | SAKL0H03674p | SAKL0H06314p | SAKL0H06600p | SAKL0G04686p |
| SAKL0B01782p | S. kluyveri | - | 237 | - | - | - | - | - | - |
| SAKL0B05940p | S. kluyveri | - | 253 | 59 | - | - | - | - | - |
| SAKL0H03674p | S. kluyveri | - | 249 | 57 | 56 | - | - | - | - |
| SAKL0H06314p | S. kluyveri | - | 248 | 56 | 57 | 61 | - | - | - |
| SAKL0H06600p | S. kluyveri | - | 248 | 56 | 57 | 61 | 100 | - | - |
| SAKL0G04686p | S. kluyveri | - | 254 | 56 | 62 | 57 | 62 | 62 | - |
| Regulatory protein, MerR:Resolvase | T. ethanolicus | Q3CGJ5 | 197 | 29 | 28 | 27 | 27 | 27 | 29 |
| Predicted site-specific integrase-resolvase | T. tengcongensis | Q8RBS0 | 196 | 29 | 28 | 27 | 27 | 27 | 28 |
| Resolvase, N-terminal domain | C. saccharolyticus | A4XJ35 | 201 | 29 | 28 | 28 | 28 | 28 | 29 |
| Putative resolvase | D. ambivalens | Q9UXS2 | 194 | 25 | 24 | 24 | 24 | 24 | 24 |
| First ORF in transposon ISC1904 | S. solfataricus | Q97YI6 | 192 | 25 | 23 | 23 | 24 | 24 | 24 |
| Putative transposase OrfA | H. pylori | Q9RMU7 | 217 | 26 | 24 | 24 | 25 | 25 | 24 |
| Putative uncharacterized protein OrfA | C. jejuni | Q6A4H9 | 212 | 26 | 23 | 22 | 23 | 23 | 23 |
| Predicted site-specific integrase/resolvase | P. kodakaraensis | Q5JF72 | 209 | 27 | 23 | 24 | 22 | 22 | 25 |
| Putative IS transposase (OrfA) | C. botulinum C bacteriophage | Q331V0 | 219 | 24 | 25 | 25 | 23 | 23 | 26 |
References are taken from Uniprot database (http://www.uniprot.org/) for bacteria and archae and from Génolevures (http://www.genolevures.org/) for yeast proteins.
Sequence identity is measured on total query sequence length.
Discussion
Cases of horizontal gene transfers have been previously reported in hemiascomycetous yeast genomes. Here, we have exploited the remarkable synteny conservation among five distantly related yeast species to systematically screen for the presence of species-specific insertion of genes. Using this strategy, we identified 15 novel genes of HGT origin (11 intervening genes and four additional family members), representing a minimum of 6 independent transfer events that occurred in 3 distinct species. This increases the number of previously published cases of HGT in yeasts by 50%. As of today, HGT genes have been found in almost all yeast species (except Z. rouxii) in which they were sought for, suggesting that this mechanism is more frequent than usually imagined. Given the restrictive method used in this work, it is likely that other cases of HGT were missed either because they did not fall into conserved synteny blocks or because they were ancestral to several lineages and, therefore, not retained as “species-specific” genes. As an illustration of this, the URA1 gene, previously shown to have been acquired by an ancestor of S. cerevisiae and S. kluyveri [22], has syntenic orthologs in K. thermotolerans and K. lactis, that were, therefore, considered as anchor points in our synteny blocks and not as intervening genes. We also identified highly similar orthologs of S. cerevisiae HGT genes YOL164W and YJL217W in K. thermotolerans, suggesting more ancestral HGT events. Another reason limiting the discovery of HGT genes is the present content of databases. The collection of 244 species-specific intervening genes remaining without homology in databases is puzzling (see Supplementary Table S1). It is possible that some of them correspond to HGT from non-sequenced group of organisms. With the development of high-throughput sequencing technologies providing new sequences of environmental or non-cultivated species, one can hope that the number of trans-kingdom homologs will increase.
From the total number of HGT identified today among Hemiascomycetes (Figure 1), yeasts follow the amoeba where up to 152 HGT candidates were found in Trichomonas vaginalis [4], but are far ahead of Metazoa where only a few HGT events were described so far (e.g. in the Nematode, ICL and MS genes subsequently fused [34], and in Ciona intestinalis, cellulose synthase presumed to have been transferred in the early Urochordates ancestor [35]).
Our results extend the idea that HGT genes can rapidly duplicate in their novel host. One case of duplicated HGT gene was previously reported in K. lactis, and two cases in Y. lipolytica, but were not analyzed further [20]. We show here that duplications occurred in K. thermotolerans and S. kluyveri, forming families of up to six genes. Although the latter case concerns a bacterial transposase, it is unlikely that its duplication in yeast results from its activity because no polymorphism is observed among the tested strains, and two copies have a mutation in the catalytic domain. Outside yeasts, other cases of duplicated horizontally acquired genes were described in amoeba [36], [37]. Such duplications suggest that HGT genes are functional in their host and submitted to selective pressures as judged from low dN/dS ratios. Similar figures are also found for the previously published cases of duplicated HGT in hemiascomycetous yeasts (respectively dN/dS values of 0.03 and 0.13 on average for K. lactis and Y. lipolytica, Supplementary Table S2).
As judged from database annotations, horizontally acquired genes of yeasts correspond to a large variety of functions, primarily concerning cellular metabolism (Figure 1). In one previously reported example, the successive integrations of HGT genes of the biotin biosynthesis pathway (BIO3 and BIO4) from diverse bacterial origins into S. cerevisiae genome argue for the reconstruction of a previously lost function [23].
The transfer of genes from bacteria to yeasts raises questions about the mechanism involved in foreign DNA uptake and integration into chromosomes. Trans-kingdom conjugation has been observed between E. coli and S. cerevisiae cells [38]. Bacterial conjugation, however, involves long DNA segments, while we always observe single-gene insertions. Transformation of yeast cells by exogenous DNA is an other possibility. In the laboratory, specific treatments are needed to increase the frequency of transformation to a measurable level. But very rare events can play important role within large populations and long evolutionary time scales. Fragments of mitochondrial DNA can integrate chromosomes at double-strand breaks (DSB) [39] and several such fragments (NUMTs) are present in yeast genomes [40]. It is possible that HGT would be similarly facilitated by chromosomal DSBs. Remarkably, however, NUMTs are essentially found outside or at the border of conserved synteny blocks. The fact that horizontally acquired genes tend to be smaller than the average yeast genes (median of 248±96 codons compared to 410±11, Figure 1) is consistent with transformation by short DNA fragments. Two exceptions nevertheless exist, BDS1 in S. cerevisiae (647 amino-acids) and YdhR in D. hansenii (776 amino-acids). Cases of introgression of large DNA fragments have been mentioned among the Saccharomyces sensu stricto complex [41], [42], and even from more distantly related yeasts, e.g. Zygosaccharomyces bailii into S. cerevisiae [43]. In addition, some beer strains are hybrids between S. cerevisiae and S. kudriavzevii [44], [45], in which introgressed chromosomal fragments can be exchanged. Cases of horizontal gene transfers between yeasts have also been identified through phylogenetic incongruence of the gene tree (e.g. the DAL5 transporter family in [46]).
Finally, as our screening method tolerated small local rearrangements within conserved synteny blocks (Materials and Methods), we are able to examine whether insertion of HGT genes is accompanied or not by other local rearrangements. An intriguing contiguity between the HGT gene KLLA0B05269g and a segmental duplication with chromosome D is observed in K. lactis, but we cannot decide whether the two events are concomitant or not. We also observe the presence of tRNA genes next to HGT genes (Figures 3A,D, 4B, S2A,C,D and S3B,C), sometimes associated with relics of transposable elements (Long-Terminal Repeats, LTRs, Supplementary Figures S2C,D and Figures S3 B,C). Other cases of intervening genes are also observed in the vicinity (Figures 3D, 4A, S2A,B and S3C,D), suggesting that these regions may be more susceptible to rearrangements, possibly because they are more susceptible to meiotic double-strand breaks than others, as previously reported in S. cerevisiae chromosome III [47], [48].
Materials and Methods
Protoploid yeast genomes
Sequences and annotations used in this work were taken from the Génolevures website (http://www.genolevures.org/), and published by Dietrich et al. (Eremothecium (Ashbya) gossypii genome) [26], Dujon et al. (Kluyveromyces lactis genome) [20] and the Génolevures Consortium (Zygosaccharomyces rouxii, Saccharomyces (Lachancea) kluyveri and Kluyveromyces (Lachancea) thermotolerans genomes) [13]. The new genera Lachancea has been introduced by Kurtzmann (2003) [49]. The Kluyveromyces waltii genome used for some comparisons has been annotated by Kellis et al. [50]. Protein families taken from Génolevures website were previously defined from systematic comparisons of complete predicted proteomes from nine hemiascomycetous species [51]. Orthologs for the five protoploid Saccharomycetaceae were identified from protein families using gene neighborhood conservation [52].
Synteny block construction
Construction of synteny blocks conserved within the genomes of Z. rouxii, S. kluyveri, K. thermotolerans, K. lactis and E. gossypii is based on the physical adjacency of sets of orthologous genes along chromosomes, controlled by two parameters: the minimum number of orthologous genes common to all 5 species, used as anchor points, and the maximum number of tolerated non-orthologous genes between two adjacent anchor points. Adjacency was deduced from sequence-derived chromosome maps as annotated by the Génolevures Consortium (http://www.genolevures.org/). Note that tandem gene repeats are considered as equivalent to a single gene by our method. We set the two parameters to 5 minimum anchor points and 25 maximum intervening genes, by extension of a previous work [13]. Note that only 8 cases of more than 10 consecutive intervening genes were actually found within synteny blocks, with a maximum of 18 intervening genes in a raw. Finally, we used annotated tRNA genes to consolidate existing synteny blocks (annotated using tRNAscan [53] for K. waltii draft genome).
Identification of putative HGT
Within synteny blocks, we extracted as “intervening genes” for further analysis only those that are present in one species and absent in all four others. For all intervening genes, we checked for the possible presence of homologs at ectopic location in other yeast species. Remaining species-specific intervening genes were finally compared to the NR nucleotide database of NCBI (release 10.5, ftp://ftp.ncbi.nih.gov) using Blastx tool [54], without filter of low complexity sequence, and with the default threshold of e-value of 10. Best hits were extracted for each gene, eliminating the gene itself. This list was finally filtered manually to find significant hits with proteins belonging to any other species but yeasts, by applying a threshold of 1.0E−6 to the e-value and a minimum of 15% identity with compared sequence.
HGT characterization
Sequences were aligned using MAFFT program [55], alignments were curated using Gblocks tool (version 0.91b) [56], removing gaps and saturated positions and thus keeping only informative sites. Identity percentages were calculated over the total query length. Phylogenetic trees were inferred from sequence divergence, using PHYML tool with a JTT substitution model corrected for heterogeneity among sites by a gamma-law distribution using 4 categories of substitution rates, proportion of invariable sites and the alpha parameter of the gamma-law distribution optimized according to the data (version 3.0) [57], validated by 100 aLRT replicates [58]. The resulting trees were drawn using Treedyn [59] or NJplot [60] programs. ClustalW [61] tool was used for global alignment of single HGT candidates with their best bacterial hits. Gblocks, PHYML and Treedyn programs are those of Phylogeny.fr web server [62].
Calculation of dN/dS ratio
We used the number of non-synonymous over synonymous mutations as a measure of sequence divergence of paralogous copies of HGT family members. The program yn00 of the PAML package [63] has been used with default parameters. This program detects non-synonymous and synonymous sites (respectively N and S) within a protein family, and then counts for each protein pair the number of non-synonymous mutations by non-synonymous sites and synonymous mutations by synonymous site (respectively dN and dS).
Experimental data
PCR amplifications were performed to detect the presence/absence of the six putative serine recombinase genes in the genomes of the different S. kluyveri strains (Supplementary Table S3). Ploidy of the strains is taken from [27]. Primers were designed in the two flanking genes and synthesized by Eurogentec (Seraing, Belgium). Their characteristics are listed in Supplementary Table S5. Reactions were performed in a final volume of 25 µl in an Applied Biosystems thermocycler (Courtaboeuf, France) using ex-Taq DNA polymerase from Takara (France) in the recommended buffer and about 50 ng of genomic DNA as a template. The following conditions were used: an initial denaturation of 2 min at 94°C followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and elongation at 72°C for 2 min, and a final elongation at 72°C for 2 min. PCR products were electrophoresed on 1% agarose gel, migrated in TAE 1X (ca. 100V), colored with BET and visualized through UV.
Supporting Information
Alignments of single HGT candidates in K. lactis, K. thermotolerans and S. kluyveri. These alignments have been produced using ClustalW tool (Materials and Methods). (A) Sequence alignment of KLLA0B05269p with bacterial proteins. Aeromonas salmonicida protein is annotated as hypothetical protein, and Bacillus cereus protein as member of the SMI1/KNR4 family. (B) Sequence alignment of KLLA0C09218p with bacterial proteins. Xanthomonas axonopodis and Oceanicaulis alexandrii proteins are annotated as hypothetical, Xanthomonas campestris protein as putative glyoxalase/bleomycin resistance protein/dioxygenase, and Sphingomonas wittichii protein as glyoxalase/bleomycin resistance protein/dioxygenase. The N-terminal extension in our candidate sequence with respect to bacterial sequences goes beyond a conserved in-frame methionine, and could be explained by an alternative upstream start codon or an incorrect annotation. (C) Sequence alignment of KLTH0E10032p with the Sorangium cellulosum protein, annotated as hypothetical. The sequences share 36% identity, as measured on total length of query protein. (D) Sequence alignment of SAKL0H22528p with bacterial and moss proteins. Physcomitrella patens protein is predicted, and Burkholderia cenocepacia strain MC0-3 protein is annotated as MoeA domain protein.
(0.54 MB PPT)
Part of conserved syntenic regions surrounding HGT candidates in K. thermotolerans and K. waltii. Gaps in syntenic chromosomal regions supports a possible loss of genes. (A) Syntenic region of K. waltii HGT candidate (KLWA_20732) in K. thermotolerans and S. kluyveri. Orthologous genes, inferred from sequence similarity for K. waltii, are colored, intervening genes are white, and tRNA genes are indicated by short hatched arrows. Arrows represent gene orientation. Note that the scale is not respected. We observe a well conserved synteny between all three species of the Lachancea clade, with a large intergenic region at the location corresponding to KLWA_20732 in K. thermotolerans, but not in S. kluyveri. (B) Syntenic region of K. thermotolerans tandem pair HGT candidates in K. waltii and S. kluyveri. Same legend as (A). Grey gene KLWA_23011 has an ectopic homolog. Note that KLTH0C07744g shows poor similarity with the genes of the tandem pair, and thus may also represent a diverged tandem repeat. Here, we observe a very large intergenic region at the location corresponding to the tandem pair in K. waltii, not found in S. kluyveri. (C) Syntenic region of K. thermotolerans KLTH0F12276g HGT candidate in K. waltii and S. kluyveri. Same legend as (A). Note that this region is syntenic with the four other protoploid species (Figure 4B). Remarkably, we observe a synteny breakpoint in K. waltii. This region involves a tRNA proline gene in K. thermotolerans and S. kluyveri, and another tRNA glycine in K. waltii, identified using tRNAscan (Material and Methods). (D) Syntenic region of K. thermotolerans KLTH0H12914g HGT candidate in K. waltii and S. kluyveri. Same legend as (A). Here again, we observe large intergenic regions in K. thermotolerans and K. waltii. Interestingly, this region is even larger in S. kluyveri, and contains two tRNA genes and remnants from transposable elements (Long-Terminal Repeats, LTRs).
(0.13 MB PPT)
Part of conserved synteny regions surrounding the putative serine recombinase genes in S. kluyveri (SAKL0H06314g and SAKL0H06600g are represented in Figure 5). (A) SAKL0B05940g was not initially identified as an intervening gene due to the synteny breakpoint occurring in Ergo0F/Ergo0B precisely at the locus corresponding to the serine recombinase insertion. (B) SAKL0G04686g was detected as an intervening gene despite the fact that the region is highly rearranged. A large sequence inversion is present in both Klth0G and Klwa.s47 (black hatched rectangle), compared to Sakl0G, ZYRO0D, ERGO0A and KLLA0A (the flanking genes have been inverted). The orange pentagon at the inversion border represents a conserved tRNA gene identified using tRNAscan (Materials and Methods). The yellow pentagon (ERGO0A07744r) represents a non-coding tRNA gene whose localization is specific to E. gossypii. Black arrows in rectangles symbolize LTR or relics of LTR. (C) SAKL0H03674g was not initially identified as an intervening gene due to the synteny breakpoint in Klla0E that separated the syntenic region into two distant regions of this chromosome. The blue pentagon represents a tRNA gene conserved in all species. It is interesting to note that the synteny breakpoint region is also highly rearranged as in (B) with the presence of LTRs. (D) SAKL0B01782g was detected as an intervening gene in a synteny block made of 9 anchor points, five of them being represented on this figure. White arrows correspond to distinct intervening genes.
(0.18 MB PPT)
Set of 244 species-specific intervening genes which have no significant hit in NR database.
(0.21 MB DOC)
Measures of dN/dS ratio for duplicated HGT genes.
(0.12 MB DOC)
Yeast strains used in this work.
(0.12 MB DOC)
Results of PCR amplification of the six serine recombinase genes in S. kluyveri strains.
(0.12 MB DOC)
Primers used to amplify the serine recombinase genes.
(0.11 MB DOC)
Acknowledgments
TR is very thankful to Benno Schwikowski and Institut Pasteur Systems Biology lab members for useful discussions and strong technical support. CN thanks Marie-Agnès Petit for stimulating discussions on serine recombinase. BD is a member of Institut Universitaire de France.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by funding from ANR (ANR-05-BLAN-0331, GENARISE) and from CNRS (GDR 2354, Genolevures). TR is supported by a doctoral fellowship from the French Ministere de l'Enseignement Superieur et de la Recherche. BD is a member of Institut Universitaire de France. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Campbell A. Phage integration and chromosome structure. A personal history. Annu Rev Genet. 2007;41:1–11. doi: 10.1146/annurev.genet.41.110306.130240. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Wolf YI. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acid Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohan FM, Koeppel AF. The origins of ecological diversity in prokaryotes. Curr Biol. 2008;18:R1024–34. doi: 10.1016/j.cub.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nature Reviews Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 5.Andersson JO. Lateral gene transfer in eukaryotes. Cell Mol Life Sci. 2005;62:1182–97. doi: 10.1007/s00018-005-4539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Temporini ED, VanEtten HD. An analysis of the phylogenetic distribution of the pea pathogenicity genes of Nectria haematococca MPVI supports the hypothesis of their origin by horizontal transfer and uncovers a potentially new pathogen of garden pea: Neocosmospora boniensis. Curr Genet. 2004;46:29–36. doi: 10.1007/s00294-004-0506-8. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Vallve S, Romeu A, Palau J. Horizontal gene transfer in bacterial and archaeal genomes. Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosewich UL, Kistler HC. Role of horizontal gene transfer in the evolution of fungi. Annu Rev Physiopathol. 2000;38:325–63. doi: 10.1146/annurev.phyto.38.1.325. [DOI] [PubMed] [Google Scholar]
- 9.Walton JD. Horizontal gene transfer and the evolution of secondary metabolite gene cluster in fungi: An hypothesis. Fungal Genetics and Biology. 2000;30:167–71. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- 10.Khaldi N, Wolfe KH. Elusive origins of the extra genes in Aspergillus oryzae. PloS One. 2008;3:e3036. doi: 10.1371/journal.pone.0003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–87. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Scannell DR, Butler G, Wolfe KH. Yeast genome evolution-the origin of the species. Yeast. 2007;24:929–42. doi: 10.1002/yea.1515. [DOI] [PubMed] [Google Scholar]
- 13.The Génolevures Consortium. Comparative genomics of protoploid Saccharomycetaceae. 2009. Genome Res published online June 12, 2009 doi:10.1101/gr.091546.109. [DOI] [PMC free article] [PubMed]
- 14.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, et al. Life with 6000 genes. Science. 1996;274: 546, 563–7. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 15.Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci U S A. 2007;104:12825–30. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borneman AR, Forgan AH, Pretorius IS, Chambers PJ. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 2008;8:1185–95. doi: 10.1111/j.1567-1364.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 17.Doniger SW, Kim HS, Swain D, Corcuera D, Williams M, et al. A catalog of neutral and deleterious polymorphism in yeast. PloS Genet. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liti G, Carter DM, Moses AM, Warringer J, Parts L, et al. Population genomics of domestic and wild yeasts. Nature Epub. 2009 doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall C, Brachat S, Dietrich FS. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryotic Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 21.Andersson JO, Roger AJ. Evolution of glutamate dehydrogenase genes: evidence for lateral gene transfer within and between prokaryotes and eukaryotes. BMC Evol Biol. 2003;3:14. doi: 10.1186/1471-2148-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gojkovic Z, Knecht W, Zameitat E, Warneboldt J, Coutelis JB, et al. Horizontal gene promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol Gen Genomics. 2004;271:387–393. doi: 10.1007/s00438-004-0995-7. [DOI] [PubMed] [Google Scholar]
- 23.Hall C, Dietrich FS. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics. 2007;177:2293–2307. doi: 10.1534/genetics.107.074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woolfit M, Rozpedowska E, Piskur J, Wolfe KH. Genome survey sequencing of the wine spoilage yeast Dekkera (Brettanomyces) bruxellensis. Eukaryot Cell. 2007;6:721–33. doi: 10.1128/EC.00338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzpatrick DA, Logue ME, Butler G. Evidence of recent interkingdom horizontal gene transfer between Bacteria and Candida parapsilosis. BMC Evolutionary Biology. 2008;8:181. doi: 10.1186/1471-2148-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietrich FS, Voegeli S, Brachat S, Lerch A, Gates K, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- 27.Payen C, Fischer G, Marck C, Proux C, Sherman DJ, et al. Unusual composition of a yeast chromosome arm is associated with its delayed replication. Submitted to Genome Res. 2009 doi: 10.1101/gr.090605.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durand F, Dagkessamanskaia A, Martin-Yken H, Graille M, Van Tilbeurgh H, et al. Structure-function analysis of Knr4/Smi1, a newly member of intrinsically disordered proteins family, indispensable in the absence of a functional PKC1-SLT2 pathway in Saccharomyces cerevisiae. Yeast. 2008;25:563–76. doi: 10.1002/yea.1608. [DOI] [PubMed] [Google Scholar]
- 29.Vandamme P, Holmes B, Coenye T, Goris J, Mahenthiralingam E, et al. Burkholderia cenocepacia sp. nov. — a new twist to an old story. Res in Microbiology. 2003;154:91–96. doi: 10.1016/S0923-2508(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 30.Loftus B, Anderson A, Davies R, Alsmark UC, Samuelson J, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 31.Kersulyte D, Mukhopadhyay AK, Shirai M, Nakazawa T, Berg DE. Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori. J Bacteriol. 2000;182:5300–5308. doi: 10.1128/jb.182.19.5300-5308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith MC, Thorpe HM. Diversity in the serine recombinases. Mol Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 33.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 34.Kondrashov FA, Koonin EV, Morgunov IG, Finogenova TV, Kondrashova MN. Evolution of glyoxalate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol Direct. 2006:1–31. doi: 10.1186/1745-6150-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakashima K, Yamada L, Satou Y, Azuma J, Satoh N. The evolutionary origin of animal cellulose synthase. Dev Genes Evol. 2004;214:81–8. doi: 10.1007/s00427-003-0379-8. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, Mullapudi N, Lancto CA, Scott M, Abrahamsen MS, et al. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal transfer in Cryptosporidium parvum. Genome Biol. 2004;5:R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, et al. The genome of the social Amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stachel SE, Zambryski PC. Bacteria-yeast conjugation. Generic trans-kingdom sex? Nature. 1989;340:190–1. doi: 10.1038/340190a0. No abstract available. [DOI] [PubMed] [Google Scholar]
- 39.Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 40.Sacerdot C, Casaregola S, Lafontaine I, Tekaia F, Dujon B, et al. Promiscuous DNA in the nuclear genomes of hemiascomycetous yeasts. FEMS Yeast Res. 2008;8:846–57. doi: 10.1111/j.1567-1364.2008.00409.x. [DOI] [PubMed] [Google Scholar]
- 41.Naumova ES, Naumov GI, Masneuf-Pomarède I, Aigle M, Dubourdieu D. Molecular genetic study of introgression between Saccharomyces bayanus and S. cerevisiae. Yeast. 2005;22:1099–115. doi: 10.1002/yea.1298. [DOI] [PubMed] [Google Scholar]
- 42.Muller LAH, McCusker JH. A multispecies-based taxonomic microarray reveals interspecies hybridization and introgression in Saccharomyces cerevisiae. FEMS Yeast Res. 2009;9:143–152. doi: 10.1111/j.1567-1364.2008.00464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novo M, Bigey F, Beyne E, Galeote V, Gavory F, et al. The genome sequence of Saccharomyces cerevisiae EC1118 reveals multiple gene transfer events that have shaped the genome of wine yeasts. Submitted. 2009 doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González SS, Barrio E, Querol A. Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl Environ Microbiol. 2008;74(8):2314–20. doi: 10.1128/AEM.01867-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belloch C, Perez-Torrado R, Gonzalez SS, Perze-Ortin JE, Garcia-Martinez J, et al. Chimeric genomes of natural hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Appl Environ Microbiol. 2009;75(8):2534–44. doi: 10.1128/AEM.02282-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hellborg L, Woolfit M, Arthursson-Hellborg M, Piskur J. Complex evolution of the DAL5 transporter family. BMC Genomics. 2008;9:164. doi: 10.1186/1471-2164-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci USA. 1997;94:5213–8. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PloS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003;4(3):233–45. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 50.Kellis M, Birren BW, Landert ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–24. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 51.Sherman DJ, Martin T, Nikolski M, Cayla C, Souciet JL, Durrens P for the Génolevures Consortium. Génolevures: Protein families and synteny among complete hemiascomycetous yeasts proteomes and genomes. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkn859. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seret ML. Comparative genomics of yeasts: Identification of orthologs by studying chromosomal environment. MSc thesis, Université de Louvain. 2007 [Google Scholar]
- 53.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Misawa K, Kuma KI, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 57.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 58.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–52. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 59.Chevenet F, Brun C, Banuls AL, Jacq B, Chisten R. Treedyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perrière G, Gouy M. WWW-Query: An on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 61.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Computer Applications in Biosciences. 1997;13:555–6. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignments of single HGT candidates in K. lactis, K. thermotolerans and S. kluyveri. These alignments have been produced using ClustalW tool (Materials and Methods). (A) Sequence alignment of KLLA0B05269p with bacterial proteins. Aeromonas salmonicida protein is annotated as hypothetical protein, and Bacillus cereus protein as member of the SMI1/KNR4 family. (B) Sequence alignment of KLLA0C09218p with bacterial proteins. Xanthomonas axonopodis and Oceanicaulis alexandrii proteins are annotated as hypothetical, Xanthomonas campestris protein as putative glyoxalase/bleomycin resistance protein/dioxygenase, and Sphingomonas wittichii protein as glyoxalase/bleomycin resistance protein/dioxygenase. The N-terminal extension in our candidate sequence with respect to bacterial sequences goes beyond a conserved in-frame methionine, and could be explained by an alternative upstream start codon or an incorrect annotation. (C) Sequence alignment of KLTH0E10032p with the Sorangium cellulosum protein, annotated as hypothetical. The sequences share 36% identity, as measured on total length of query protein. (D) Sequence alignment of SAKL0H22528p with bacterial and moss proteins. Physcomitrella patens protein is predicted, and Burkholderia cenocepacia strain MC0-3 protein is annotated as MoeA domain protein.
(0.54 MB PPT)
Part of conserved syntenic regions surrounding HGT candidates in K. thermotolerans and K. waltii. Gaps in syntenic chromosomal regions supports a possible loss of genes. (A) Syntenic region of K. waltii HGT candidate (KLWA_20732) in K. thermotolerans and S. kluyveri. Orthologous genes, inferred from sequence similarity for K. waltii, are colored, intervening genes are white, and tRNA genes are indicated by short hatched arrows. Arrows represent gene orientation. Note that the scale is not respected. We observe a well conserved synteny between all three species of the Lachancea clade, with a large intergenic region at the location corresponding to KLWA_20732 in K. thermotolerans, but not in S. kluyveri. (B) Syntenic region of K. thermotolerans tandem pair HGT candidates in K. waltii and S. kluyveri. Same legend as (A). Grey gene KLWA_23011 has an ectopic homolog. Note that KLTH0C07744g shows poor similarity with the genes of the tandem pair, and thus may also represent a diverged tandem repeat. Here, we observe a very large intergenic region at the location corresponding to the tandem pair in K. waltii, not found in S. kluyveri. (C) Syntenic region of K. thermotolerans KLTH0F12276g HGT candidate in K. waltii and S. kluyveri. Same legend as (A). Note that this region is syntenic with the four other protoploid species (Figure 4B). Remarkably, we observe a synteny breakpoint in K. waltii. This region involves a tRNA proline gene in K. thermotolerans and S. kluyveri, and another tRNA glycine in K. waltii, identified using tRNAscan (Material and Methods). (D) Syntenic region of K. thermotolerans KLTH0H12914g HGT candidate in K. waltii and S. kluyveri. Same legend as (A). Here again, we observe large intergenic regions in K. thermotolerans and K. waltii. Interestingly, this region is even larger in S. kluyveri, and contains two tRNA genes and remnants from transposable elements (Long-Terminal Repeats, LTRs).
(0.13 MB PPT)
Part of conserved synteny regions surrounding the putative serine recombinase genes in S. kluyveri (SAKL0H06314g and SAKL0H06600g are represented in Figure 5). (A) SAKL0B05940g was not initially identified as an intervening gene due to the synteny breakpoint occurring in Ergo0F/Ergo0B precisely at the locus corresponding to the serine recombinase insertion. (B) SAKL0G04686g was detected as an intervening gene despite the fact that the region is highly rearranged. A large sequence inversion is present in both Klth0G and Klwa.s47 (black hatched rectangle), compared to Sakl0G, ZYRO0D, ERGO0A and KLLA0A (the flanking genes have been inverted). The orange pentagon at the inversion border represents a conserved tRNA gene identified using tRNAscan (Materials and Methods). The yellow pentagon (ERGO0A07744r) represents a non-coding tRNA gene whose localization is specific to E. gossypii. Black arrows in rectangles symbolize LTR or relics of LTR. (C) SAKL0H03674g was not initially identified as an intervening gene due to the synteny breakpoint in Klla0E that separated the syntenic region into two distant regions of this chromosome. The blue pentagon represents a tRNA gene conserved in all species. It is interesting to note that the synteny breakpoint region is also highly rearranged as in (B) with the presence of LTRs. (D) SAKL0B01782g was detected as an intervening gene in a synteny block made of 9 anchor points, five of them being represented on this figure. White arrows correspond to distinct intervening genes.
(0.18 MB PPT)
Set of 244 species-specific intervening genes which have no significant hit in NR database.
(0.21 MB DOC)
Measures of dN/dS ratio for duplicated HGT genes.
(0.12 MB DOC)
Yeast strains used in this work.
(0.12 MB DOC)
Results of PCR amplification of the six serine recombinase genes in S. kluyveri strains.
(0.12 MB DOC)
Primers used to amplify the serine recombinase genes.
(0.11 MB DOC)