Abstract
Sleep is universal, strictly regulated, and necessary for cognition. Why this is so remains a mystery, though recent work suggests a link between sleep, memory, and plasticity. However, little is known about how wakefulness and sleep affect synapses. Using Western blots and confocal microscopy in Drosophila, we found that protein levels of key components of central synapses were high after waking and low after sleep. These changes were related to behavioral state rather than time of day and occurred in all major areas of the Drosophila brain. The decrease of synaptic markers during sleep was progressive and sleep was necessary for their decline. Thus, sleep may be involved in maintaining synaptic homeostasis altered by waking activities.
There is increasing evidence for a link between sleep need and neuronal plasticity (1). Sleep need increases after learning, and learning tasks that involve local brain regions lead to local changes in sleep intensity. Neural activity during sleep can reactivate neural circuits involved in learning. Moreover, sleep consolidates memories, whereas sleep deprivation interferes with memory acquisition. Presumably, sleep exerts these effects by modifying synapses (1). In rats, the number of cortical and hippocampal GluR1-containing AMPA receptors (AMPARs) is high after waking and low after sleep, and the slope of cortical evoked responses is steeper after waking relative to sleep (2). These data suggest that periods of wakefulness are associated with a net increase in synaptic strength, and periods of sleep with a net decrease. Sleep, therefore, could play an important role in renormalizing synaptic changes caused by learning during wakefulness (3). But how general is this finding, and does it apply to species with brains very different from mammals? Both plasticity and sleep are universal across animal species. If the opposing effects of wakefulness and sleep on synaptic markers reflect a fundamental function of sleep, they should also occur in non-mammalian species.
To address this issue, we turned to Drosophila melanogaster, whose sleep shares most of the features of mammalian sleep: it consists of long periods of behavioral immobility with increased arousal threshold (4, 5), and is associated with changes in brain electrical activity and gene expression (1). In flies like in mammals, the duration and intensity of sleep increase with the duration of prior waking, sleep deprivation reduces performance (6), and learning and enriched experience increase sleep need and sleep intensity (7, 8). Fly brains are different from mammalian brains, but Drosophila central synapses share with mammals key pre- and postsynaptic components, and use similar mechanisms of synaptic plasticity (9, 10).
Here we tested whether waking and sleep affect synaptic markers in the Drosophila CNS by measuring changes in protein levels of several pre- and postsynaptic components (Fig. 1A). Bruchpilot (BRP) is an essential constituent of the active zone of all synapses (Fig. 1A) and excitatory synapses lacking BRP show reduced evoked release of glutamate (11). In vivo, a strong BRP signal is associated with fully formed synapses (11), and BRP staining is used to quantify synapse number ((12) and Methods).
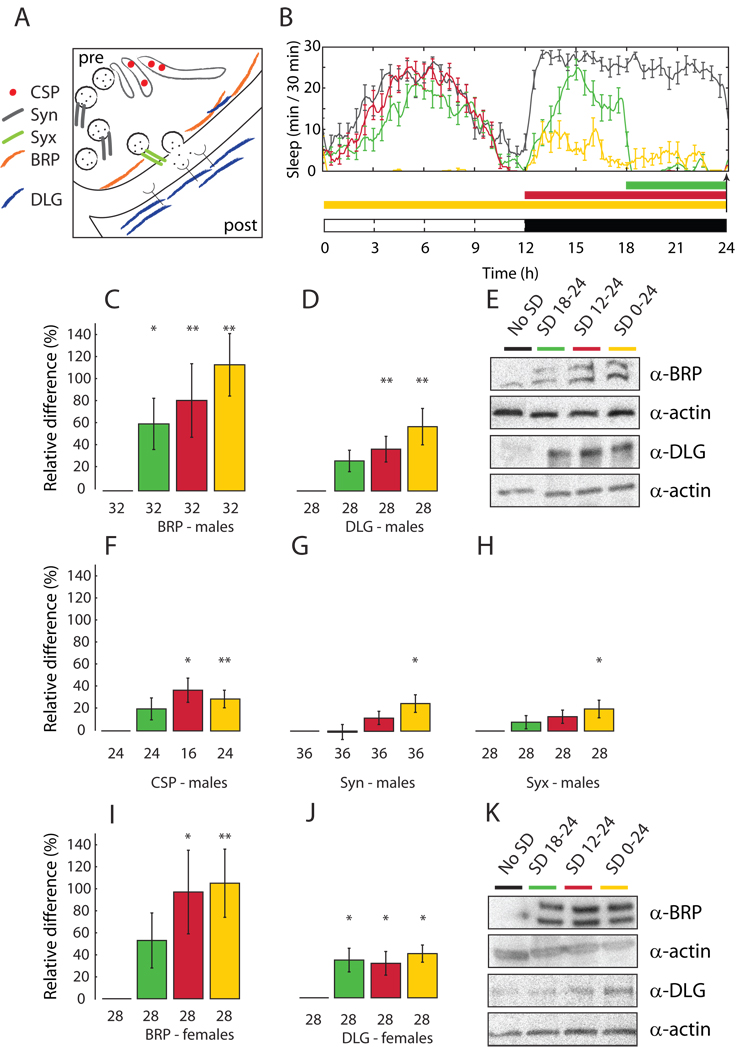
Fig. 1. Levels of synaptic proteins are high after sleep deprivation and low after sleep.
(A) Presynaptic (pre), postsynaptic (post), Bruchpilot (BRP), Cysteine string protein (CSP), Discs-large (DLG), Synapsin (Syn), Syntaxin (Syx). (B) Daily pattern of sleep in Canton-S males sleep deprived (SD) for the last 6h of the night (green), 12h at night (red), or 24h (yellow) and control siblings left undisturbed (no SD, black line). Arrow shows when flies were collected. White and black bars: light and dark periods. Each group includes 12–16 flies and represents one of the 2–3 experiments used for panels C–H. (C–H) Representative immunoblots (E) and gel quantification (mean ± standard deviation; n of flies below each bar). SD values (color-coded as in B) expressed as % change relative to sleep (= 0%). (I–K) Canton-S females. *, p < 0.05; **, p < 0.01 (one-way ANOVA followed by Tukey’s HSD Post Hoc test).
Wild type Canton-S (CS) males were kept awake using an established mechanical method of sleep deprivation that causes a post-deprivation increase in sleep duration and intensity that is proportional to the amount of sleep lost (6). Sleep deprivation was performed for 6h, 12h, and 24h and was highly effective (flies lost at least 80% of their baseline sleep (Fig. 1B). At the end of sleep deprivation fly brains were dissected and BRP expression measured by Western blots. Sleep deprived flies showed higher BRP levels relative to controls allowed to sleep ad libitum, and the increase in BRP expression was smaller after 6h of forced waking and larger when sleep was prevented for 12–24h (Fig. 1C,E). Similar BRP increases were seen after sleep deprivation in males of another strain, white1118 (Fig. S1A,C), and in CS females (Fig. 1I,K). In the latter, BRP levels increased similarly after 12 and 24h of sleep deprivation, consistent with the fact that females concentrate most of their sleep at night (5, 6), whereas males take a nap in the middle of the day (Fig. 1B and Fig. S2).
We then asked whether sleep deprivation also increases the expression of postsynaptic proteins, and focused on Discs-large (DLG), the Drosophila homologue of the postsynaptic density protein PSD-95/SAP-90 (13). DLG is mainly expressed postsynaptically (Fig. 1A) (14) and is abundant at the neuromuscular junction and in the CNS neuropil, where its pattern of expression overlaps with that of BRP (13, 15). DLG regulates glutamate release and postsynaptic structure (16), and controls the postsynaptic clustering of glutamatergic receptors (12). At the Drosophila neuromuscular junction, postsynaptic activation of DLG increases the number and size of the presynaptic active zones (17). In mammals, the synaptic delivery and incorporation of AMPARs partly depend on PSD-95 (18), and overexpression of PSD-95 enhances the activity of postsynaptic glutamatergic receptors and the number and size of dendritic spines (19). In CS male flies, sleep deprivation increased DLG levels in CNS, and more so after 24h of sleep loss (Fig. 1D,E). Similar results were observed in CS females (Fig. 1J,K) and in 2 other strains (Fig. S1B,D).
We then tested whether sleep loss affects the expression not only of structural proteins such as BRP and DLG, but also of components of the secretory machinery (Fig. 1A): synapsin (Syn; (20)), syntaxin (Syx, (21)) and cystein string protein (CSP; (22)). After 24h of sleep deprivation all three proteins increased their expression levels by ~ 30% (Fig. 1F–H), with CSP showing a significant increase already after 12h of continuous waking.
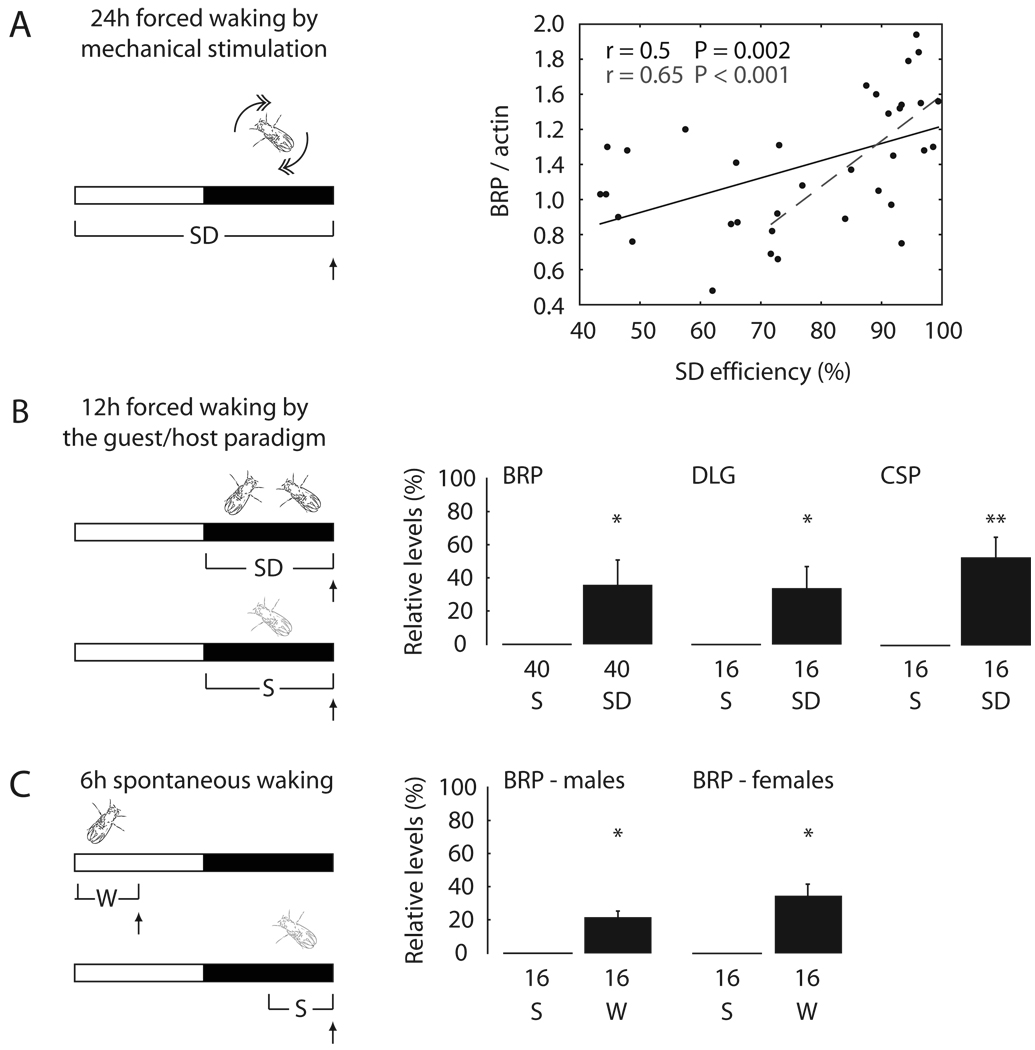
Because both sleeping and sleep deprived flies were collected at light onset (Fig. 1B), these results cannot be ascribed to circadian differences in the time of collection. However, because flies were stimulated mechanically to keep them awake, it was important to demonstrate that lack of sleep, and not stimulation per se, was responsible for the observed results. To do so, we carried out 3 sets of experiments. First, sleep loss was enforced for 24h using mechanical stimulation as before, but flies were grouped according to the efficiency of sleep deprivation. Despite the fact that all flies received the same amount of mechanical stimuli across the 24h, the more effective the sleep deprivation, the higher were the levels of BRP, and the correlation was stronger for highly effective sleep deprivation (> 70%, corresponding to ¾ of all flies; Fig. 2A). In a second set of experiments we used a different method to keep flies awake. A wild type (red-eyed) CS male was first kept in standard conditions for at least 2 days, then a “guest” white-eyed male fly was introduced in the recording tube at dark onset. As confirmed by continuous video recording, both flies were awake most of the night (Fig. S3A–C). As soon as the guest fly was removed the next morning, the host fly showed a sleep rebound, and the increase in sleep duration over the first 3h of recovery sleep was similar to that observed after 12h of sleep loss by mechanical stimulation (Fig. S3D). Relative to flies that slept as usual in single tubes, flies kept awake with this method showed increased levels of BRP, DLG and CSP (Fig. 2B). Thus, nonspecific factors due to the sleep deprivation methods cannot account for the effects of behavioral state on synaptic markers. Finally, we investigated whether the expression of synaptic proteins also increases after spontaneous waking. BRP levels were measured in flies collected in the middle of the day, after they had been spontaneously awake almost continuously for 6h, and at the end of the night, after 6h spent mainly asleep (Fig. 2C). In both female and male CS flies, BRP levels were again higher after waking than after sleep (Fig. 2C). The increase after 6h of spontaneous wakefulness was not as pronounced as after 12–24h of sleep deprivation.
Fig. 2. The expression of synaptic proteins increases due to sleep loss.
(A–C, left) SD, sleep deprivation; S, sleep; W, spontaneous waking. Vertical arrows show when flies were collected. (A, right) Correlation between BRP levels and sleep deprivation efficiency during the last 24h in Canton-S males (Pearson correlation). Each dot represents 4 flies with similar SD efficiency. (B–C, right) Levels of synaptic proteins after SD or W, expressed as % change relative to sleep (= 0%). (mean ± standard deviation; n of flies below each bar). *, p < 0.05; **, p < 0.01 (Student's t-test).
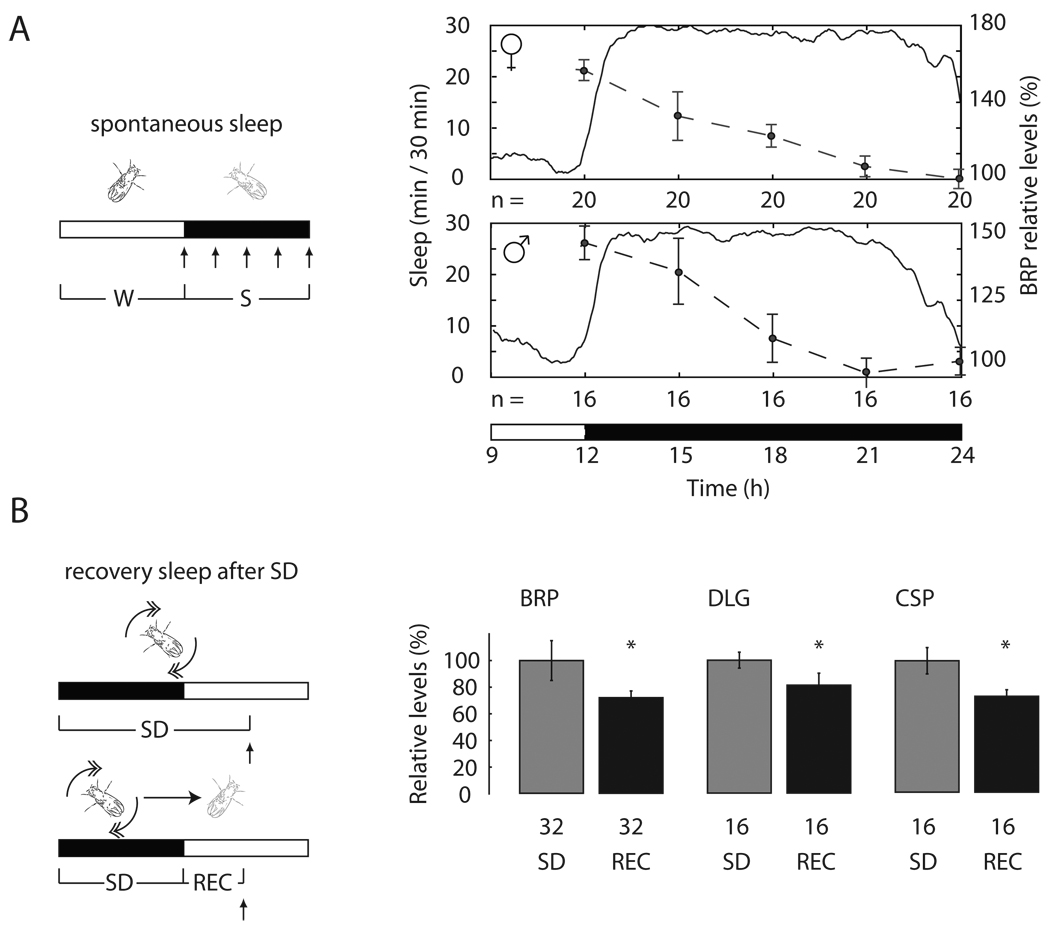
To determine the time course of the decline in BRP expression during sleep, we collected flies at the end of the light phase, after they had been mostly awake for several hours, and every 3h across the entire dark phase, when they slept. In both males and females BRP levels declined progressively in the course of sleep, reaching the lowest level towards the end of the night (Fig. 3A). Is sleep per se responsible for the decline in BRP expression, or merely the passage of time? To answer this question, two groups of flies were sleep deprived by mechanical stimulation for 12h during the night; one group was then allowed to recover sleep for the first 6h of the next day, while the other group was kept awake for the same period of time. BRP, DLG and CSP levels were ~ 20–40% lower in flies that could sleep relative to those that could not (Fig. 3B). Because BRP levels do not decrease, but rather increase, when flies remain awake spontaneously for 6h (Fig. 2C), these results are consistent with the idea that sleep is necessary for downregulating the expression of synaptic proteins.
Fig. 3. Synaptic markers decrease during sleep.
(A–B, left) Rec, recovery sleep after SD. Vertical arrows show when flies were collected. (A, right) BRP levels expressed as % change relative to sleep at the end of the night (= 100%). (B, right) Levels of synaptic proteins after recovery sleep, expressed as % change relative to SD (=100%; mean ± standard deviation; n of flies is below each bar). *, p < 0.05 (Student's t-test).
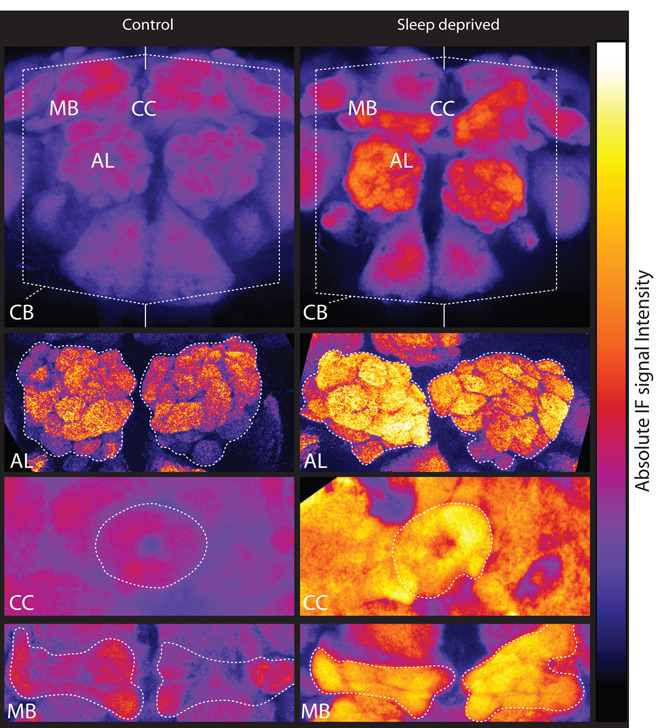
All changes described so far were detected using whole brain homogenates, and thus it remained unclear whether they were restricted to select brain areas. We therefore used confocal microscopy to measure BRP levels in dissected brains of 2 groups of flies, both collected immediately after light onset. Controls slept undisturbed, while the other group was sleep deprived for the preceding 16h. An increase in the level of BRP protein could be observed across the whole central brain of sleep deprived flies compared to controls (Fig. 4), and was due to sleep loss (two-way ANOVA, “sleep loss”, F = 16.27, P < 0.0001; “brain tissue”, F = 0.68, P = 0.56; “sleep * tissue” interaction”, F = 0.24, P = 0.86). This increase was similar in 4 major structures of the fly brain, including antennal lobes (mean of % ± SEM, 69 ± 11), β lobes of the mushroom bodies (40 ± 11), ellipsoid body of the central complex (67 ± 18), and entire central brain (76 ± 15). In flies as in mammals, learning and memory, and activity-dependent structural plasticity in general, involve complex and widespread circuits that span most of the brain (10, 23). Thus, it is perhaps not surprising that forcing flies to stay awake through a complex range of tactile, auditory, and visual stimuli affects synaptic activity in most brain regions. Finally, we examined whether prolonged sleep loss is also associated with overall changes in the volume of antennal lobes, because activity-dependent plasticity leads to volumetric increases in this structure (24). In flies that were sleep deprived for 16h, antennal lobes were slightly but significantly bigger than in flies that had slept (+15%, p = 0.01; N of flies, S = 27; SD = 19), a result that is compatible with a diffuse increase in either the number or the volume of synaptic connections.
Fig. 4. Widespread BRP increase after sleep loss.
Representative examples of BRP immunofluorescence (IF) in controls and flies sleep deprived for 16h ending at light onset (sum of selected optical stacks, false colored using a quantitative scale). Immunoreactivity levels were measured in antennal lobes (AL), beta lobes of the mushroom bodies (MB), ellipsoid body of the central complex (CC) and central cerebrum (excluding the optic lobes, CB).
The main finding of this study is that, in Drosophila, the expression of several bona fide synaptic markers increases after wakefulness and decreases after sleep, independent of time of day. There are 3 documented examples of daily structural changes in Drosophila neuronal circuits. First, neuronal branches at the neuromuscular junction are thicker and carry larger synaptic boutons during the day, when flies are active, than during the night (25). Second, optic lobe interneurons have larger axons at the beginning of the night than at the beginning of the day, and their dendritic trees also undergo daily morphological changes (26). Third, the dorsal axonal projections of the small ventral lateral neurons, which are involved in rhythmic behavior, show a higher degree of arborisation in the morning than at night (27). It is thought that these forms of structural plasticity are controlled by the circadian clock, because they are largely maintained in constant darkness and are abolished in flies carrying mutations of circadian genes (25, 27, 28). However, the sleep/waking cycle is an endogenous circadian rhythm, it persists in constant darkness, and is disrupted by mutations of circadian genes. Thus, structural changes in neural circuits occurring between day and night may be due, at least in part, to changes in behavioral state. Consistent with this, the number of synaptic terminals in ventral lateral neurons is reduced during sleep, and this decline is prevented by sleep deprivation (29).
What could be the functional significance of the large, widespread decrease in synaptic markers that takes place during sleep? Processes of synaptic potentiation and depression, which are often associated with structural changes, are usually assumed to occur concurrently so as to maintain an overall synaptic balance (3). However, the increase in synaptic markers observed after periods of wakefulness in rats (2) and now in flies suggests that this balance may not be fully preserved. An increase of synaptic strength by the end of the waking day would result in higher energy consumption, larger synapses that take up precious space, and saturation of the capacity to learn (3). Also, a net strengthening of synapses likely represents a major source of cellular stress, due to the need to synthesize and deliver cellular constituents ranging from mitochondria to synaptic vesicles. Sleep may thus play an important role in renormalizing synapses to a baseline level that is sustainable and ensures cellular homeostasis. Because similar changes appear to occur in phylogenetically distant species with different brain organizations, synaptic homeostasis may represent a cellular correlate of wakefulness and sleep that is conserved across evolution.
Supplementary Material
References and Notes
- 1.Cirelli C, Tononi G. Plos Biology. 2008;6:e216. doi: 10.1371/journal.pbio.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U, Tononi G. Nat Neurosci. 2008 Feb;11:200. doi: 10.1038/nn2035. [DOI] [PubMed] [Google Scholar]
- 3.Tononi G, Cirelli C. Sleep Med Rev. 2006 Feb;10:49. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Hendricks JC, et al. Neuron. 2000 Jan;25:129. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 5.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Science. 2000 Mar 10;287:1834. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 6.Huber R, et al. Sleep. 2004 Jun 15;27:628. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 7.Huber R, Ghilardi MF, Massimini M, Tononi G. Nature. 2004 Jul 1;430:78. doi: 10.1038/nature02663. [DOI] [PubMed] [Google Scholar]
- 8.Ganguly-Fitzgerald I, Donlea J, Shaw PJ. Science. 2006 Sep 22;313:1775. doi: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- 9.Guan Z, Saraswati S, Adolfsen B, Littleton JT. Neuron. 2005 Oct 6;48:91. doi: 10.1016/j.neuron.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 10.Margulies C, Tully T, Dubnau J. Curr Biol. 2005 Sep 6;15:R700. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagh DA, et al. Neuron. 2006 Mar 16;49:833. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Chen K, Featherstone DE. BMC Biol. 2005;3:1. doi: 10.1186/1741-7007-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woods DF, Bryant PJ. Cell. 1991 Aug 9;66:451. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 14.Lahey T, Gorczyca M, Jia XX, Budnik V. Neuron. 1994 Oct;13:823. doi: 10.1016/0896-6273(94)90249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albornoz V, et al. Gene Expr Patterns. 2008 Jul;8:443. doi: 10.1016/j.gep.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Budnik V, et al. Neuron. 1996 Oct;17:627. doi: 10.1016/s0896-6273(00)80196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazama H, Nose A, Morimoto-Tanifuji T. Neuroscience. 2007 Mar 30;145:1007. doi: 10.1016/j.neuroscience.2006.12.066. [DOI] [PubMed] [Google Scholar]
- 18.Ehrlich I, Malinow R. J Neurosci. 2004 Jan 28;24:916. doi: 10.1523/JNEUROSCI.4733-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. Science. 2000 Nov 17;290:1364. [PubMed] [Google Scholar]
- 20.Klagges BR, et al. J Neurosci. 1996 May 15;16:3154. doi: 10.1523/JNEUROSCI.16-10-03154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulze KL, Broadie K, Perin MS, Bellen HJ. Cell. 1995 Jan 27;80:311. doi: 10.1016/0092-8674(95)90414-x. [DOI] [PubMed] [Google Scholar]
- 22.Zinsmaier KE, Eberle KK, Buchner E, Walter N, Benzer S. Science. 1994 Feb 18;263:977. doi: 10.1126/science.8310297. [DOI] [PubMed] [Google Scholar]
- 23.Heisenberg M, Heusipp M, Wanke C. J Neurosci. 1995 Mar;15:1951. doi: 10.1523/JNEUROSCI.15-03-01951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sachse S, et al. Neuron. 2007 Dec 6;56:838. doi: 10.1016/j.neuron.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 25.Mehnert KI, et al. Dev Neurobiol. 2007 Mar;67:415. doi: 10.1002/dneu.20332. [DOI] [PubMed] [Google Scholar]
- 26.Gorska-Andrzejak J, Keller A, Raabe T, Kilianek L, Pyza E. Photochem Photobiol Sci. 2005 Sep;4:721. doi: 10.1039/b417023g. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez MP, Berni J, Ceriani MF. PLoS Biol. 2008 Mar 25;6:e69. doi: 10.1371/journal.pbio.0060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyza E, Meinertzhagen IA. J Neurobiol. 1999 Jul;40:77. doi: 10.1002/(sici)1097-4695(199907)40:1<77::aid-neu7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Donlea Science. in press. [Google Scholar]
- 30.We thank D. Paasch, D. Denucci and M. Uy for technical assistance with fly work, Martha Pfister-Genskow for help with Western blot analysis, Lance Rodenkirch and the UW Keck Laboratory for Biological Imaging for assistance in confocal microscopy, and Dr. D. Bushey for discussion and comments to the manuscript. The study was supported by NIGMS (R01 GM075315 to CC) and NIH Director’s Pioneer award to GT.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.