Abstract
Objective
Human adiposity is highly heritable, but few of the genes that predispose to obesity in most humans are known. We tested candidate genes in pathways related to food intake and energy expenditure for association with measures of adiposity.
Methods
We studied 355 genetic variants in 30 candidate genes in 7 molecular pathways related to obesity in two groups of adult subjects: 1,982 unrelated European Americans living in the New York metropolitan area drawn from the extremes of their body mass index (BMI) distribution and 593 related Yup'ik Eskimos living in rural Alaska characterized for BMI, body composition, waist circumference, and skin fold thicknesses. Data were analyzed by using a mixed model in conjunction with a false discovery rate (FDR) procedure to correct for multiple testing.
Results
After correcting for multiple testing, two single nucleotide polymorphisms (SNPs) in Ghrelin (GHRL) (rs35682 and rs35683) were associated with BMI in the New York European Americans. This association was not replicated in the Yup'ik participants. There was no evidence for gene × gene interactions among genes within the same molecular pathway after adjusting for multiple testing via FDR control procedure.
Conclusion
Genetic variation in GHRL may have a modest impact on BMI in European Americans.
Key Words: Obesity, Body composition, Body mass index, Candidate gene, Ghrelin
Introduction
Human adiposity resolves complex interactions among genetic, developmental, behavioral, and environmental influences. Obesity is primarily the result of a net imbalance of caloric intake over energy expenditure. Even small differences resulting in positive energy balance – when integrated over long periods of time – can produce increased adiposity. In the United States, 65% of adults are overweight (BMI 25.0–29.9) and more than 30% of the adult population is obese (BMI ≥30) [1]. The problem also affects children in whom the percentage with BMI >95 percentile between the ages of 6–19 defined by the CDC growth charts is now 16% [2]. Increasing in parallel with these trends in obesity are the frequently associated comorbidities of diabetes, hypertension, and cardiovascular disease [3].
Evidence for potent genetic contributions to human obesity is provided by familial clustering of increased adiposity, including a 3–7 fold increased relative risk (λs) among siblings [4], estimates of heritability for fat mass between 40 and 70% in twin studies [5, 6], and rare monogenic causes of syndromic and nonsyndromic obesity [7]. Severe loss-of-function mutations in genes causing monogenic forms of obesity do not account for the heritable component of adiposity in the majority of individuals, although mutations in the Melanocortin 4 Receptor (MC4R) may account for obesity in 1–5% of the extremely obese [8]. The phenotypic differences among individuals at the extremes of adiposity presumably reflects – to a substantial degree – allelic variation at genes that affect energy intake, expenditure and the chemical form in which excess calories are stored (‘partitioning’). The nature of these genes and their interactions is of obvious interest.
Both linkage and association studies have been undertaken to identify obesity candidate genes. The 2005 Human Obesity Gene Map update reports over 1,100 studies including 500 genes, markers, and chromosomal regions apparently linked to, or associated with, human obesity phenotypes in specific cohorts [9]. Yet only 22 genes have been reproducibly implicated in ≥5 studies as statistically significant contributors to common obesity. For monogenic obesity, 176 mutations have been identified in 11 genes, and 50 loci have been mapped corresponding to Mendelian syndromes related to obesity [9]. Genome-wide association studies have and will likely continue to identify additional genes associated with obesity, but often identify genetic variants that are not themselves functionally significant and genes which were not previously known to be implicated in energy homeostasis. In most individuals, the genetic basis for obesity is complex and likely to involve the interaction of multiple genes as well as gene-by-environment interactions.
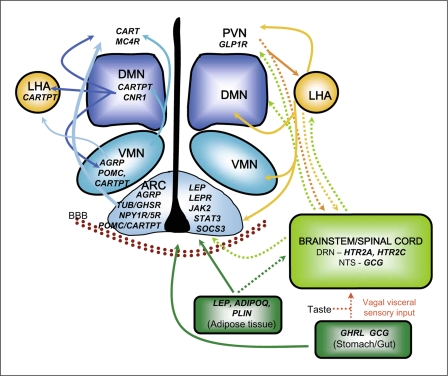
In most candidate-based genetic analyses of complex disease phenotypes, fewer than five candidate genes are interrogated in a population. Moreover, the coverage of coding and non-coding polymorphisms genotyped within a given candidate gene may be sparse, and pathways of functionally-related genes are rarely simultaneously investigated. In the present study, we evaluated seven groups of functionally-related obesity candidate genes (30 total genes) in two racially divergent population groups (European Americans and Alaskan Yup'ik Eskimos). Many of the candidate genes were selected from hypothalamic arcuate pathways since the hypothalamus integrates complex neuroendocrine, autonomic, and behavioral signaling pathways that determine food intake, energy expenditure, and nutrient partitioning (fig. 1).
Fig. 1.
The molecular pathways affecting energy homeostasis. Depicted are candidate genes focused on hypothalamic control of energy homeostasis. The location of neurons in the hypothalamus and brainstem, and cells in the stomach and adipose tissue expressing various genes involved in energy homeostasis are demonstrated with interactions between cells shown with arrows. Nuclei in the hypothalamus include the arcuate nucleus (ARC), ventral medial nucleus (VMN), dormal medial nucleus (DMN), paraventricular nucleus (PVN) and lateral hypothalamic area (LHA). Brainstem regions shown include the dorsal raphe nucleus (DRN) and nucleus tractus solitarius (NTS). BBB = blood brain barrier. The figure is modified from [10,11,12,13].
The seven groups of genes were included amongst the candidate genes:
(1) The leptin pathway included the hormone leptin (LEP), its receptor (LEPR), and downstream signaling molecules (JAK2, SOCS3, and STAT3).
(2) The melanocortin pathway included agouti related peptide (AGRP), melanocortin 4 receptor (MC4R), proopiomelanocortin (POMC), and carboxypeptidase E (CPE) that processes preprohormones.
(3) The ghrelin pathway included ghrelin (GHRL) and its receptor, GHSR.
(4) The glucagon-like peptide 1 pathway included GCG and its receptor GLP1R.
(5) The neuropeptide Y pathway included the receptors NPY1R and NPY5R.
(6) The serotonin pathway included the serotonin receptors HTR2A and HTR2C.
(7) The Bardet-Biedl group included the genes identified for BBS1, BBS2, ARL6, BBS4, BBS5, MKKS, BBS7, and TTC8.
Five genes that encode proteins involved in the leptin signaling pathway (LEP, LEPR, JAK2, STAT3, SOCS3) were selected for investigation since leptin is a key hormone secreted in proportion to peripheral fat mass as a signal to the hypothalamus regarding the state of long-term energy stores [14]. Central regulation of energy homeostasis begins as leptin binds receptors on (at least) two sets of neurons in the arcuate nucleus to decrease transcription of Agouti Related Peptide (AgRP) and Neuropeptide Y (NPY) or to increase transcription of Pro-Opiomelanocortin (POMC) and Cocaine and Amphetamine Related Transcript (CARTPT) that act reciprocally to increase and decrease food intake, respectively [14]. AgRP is a naturally occurring inverse agonist of MC3R and MC4R that stimulates food intake. Agouti Related Peptide (AGRP), Melanocortin 4 Receptor (MC4R), POMC and Carboxypeptidase E (CPE) represent orexigenic (AGRP) and anorexigenic (POMC) signaling pathways and downstream effector molecules (MC4R). CPE processes prohormones in this pathway including POMC. Neuropeptide Y receptors (NPY1R and NPY5R) are expressed on downstream effector neurons responding to the orexigenic NPY signaling events originating in the arcuate nucleus. The orexigenic peptide ghrelin (GHRL) is secreted from the stomach and duodenum and binds to the ghrelin receptor (GHSR) in the arcuate nucleus activating AGRP/NPY neurons to stimulate food intake. Glucagon-like peptide 1 (GCG) is produced by the small intestine and neurons in the nucleus of tractus solitarius post-prandially and decreases food intake and slows gastric emptying upon binding to the glucagon-like peptide 1 receptor (GLP1R) in the brain. Serotonin is involved in central signaling of satiety in hypothalamic downstream effector neurons and acts through the serotonin receptor genes (HTR2A and HTR2C). Lastly, included in the list of candidate genes are eight genes for Bardet-Biedl syndrome (BBS1, BBS2, ARL6, BBS4, BBS5, MKKS, BBS7, TTC8), a syndromic, oligogenic form of obesity resulting from mutations in at least twelve genes, some of which are apparent components of a ciliary motor that can individually, or in some cases in combination, produce a dysmorphic phenotype that includes obesity, polydactyly, and retinopathy [15].
In summary, our candidate gene list (table 1) includes genes encoding signaling peptides, receptors and downstream signaling molecules expressed in central nervous system and/or peripheral tissues, including adipocytes and gastrointestinal tract, involved in food intake, energy expenditure or energy storage. Bardet-Biedl syndrome is also caused by mutations within a common molecular pathway (ciliary structure), and hence also provides a paradigm for genetic variants in multiple candidate genes within the same pathway interacting to affect adiposity.
Table 1.
Candidate genes analyzed
| Gene symbol | Gene name | Chromosomal location | Size of gene bp | Number of polymorphic SNPs genotyped | Average interval between SNPs, kb | Number of nsSNPs |
|---|---|---|---|---|---|---|
| AGRP | agouti related peptide | 16q22 | 1,242 | 3 (2) | 110 bp | 1 |
| ADIPOQ | adiponectin, C1Q and collagen domain containing | 3q27 | 15,789 | 18 (17) | 14.0 | 1 |
| BBS1 | Bardet-Biedl syndrome 1 | 11q13 | 22,965 | 10 (9) | 5.3 | 0 |
| BBS2 | Bardet-Biedl syndrome 2 | 16q21 | 33,895 | 9 (8) | 5.1 | 0 |
| ARL6 | BBS3, ADP-ribosylation factor-like 6 | 3q11.2 | 33,778 | 7 (7) | 26.4 | 0 |
| BBS4 | Bardet-Biedl syndrome 4 | 15q22.3-q23 | 52,281 | 8 (7) | 12.0 | 0 |
| BBS5 | Bardet-Biedl syndrome 5 | 2q31 | 27,172 | 6 (6) | 76.0 | 0 |
| MKKS | BBS6, McKusick-Kaufman syndrome | 20p12 | 29,029 | 6 (5) | 2.6 | 0 |
| BBS7 | Bardet-Biedl syndrome 7 | 4q27 | 46,005 | 6 (4) | 14.0 | 0 |
| TTC8 | BBS8, tetratricopeptide repeat domain 8 | 14q31.3 | 53,354 | 4 (4) | 8.4 | 0 |
| CARTPT | cocaine- and amphetamine-regulated transcript | 5q13.2 | 1,878 | 6 (6) | 11.1 | 0 |
| CNR1 | cannabinoid receptor 1 | 6q14-q15 | 5,469 | 10 (10) | 3.0 | 0 |
| CPE | carboxypeptidase E | 4q32.3 | 119,388 | 24 (23) | 5.3 | 0 |
| GHRL | ghrelin/obestatin preprohormone | 3p26-p25 | 5,111 | 11 (8) | 1.1 | 1 |
| GHSR | ghrelin receptor, growth hormone secretagogue receptor | 3q26.31 | 3,252 | 10 (9) | 2.6 | 0 |
| GCG | glucagon | |||||
| glucagon-like peptide 1 (glucagon/GLP1/GLP2/GRPP) | 2q24.2 | 9,369 | 6 (6) | 4.0 | 1 | |
| GLP1R | glucagon-like peptide 1 receptor | 6p21 | 38,903 | 24 (22) | 2.6 | 2 |
| HTR2A | 5-hydroxytryptamine (serotonin) receptor 2A | 13q14-q21 | 62,662 | 31 (31) | 2.7 | 2 |
| HTR2C | 5-hydroxytryptamine (serotonin) receptor 2C | Xq23 | 326,073 | 20 (19) | 18.0 | 1 |
| JAK2 | Janus kinase 2 (a protein tyrosine kinase) | 9p24 | 142,750 | 18 (17) | 8.8 | 1 |
| LEP | leptin | 7q32 | 5,637 | 23 (21) | 3.7 | 0 |
| LEPR | leptin receptor | 1p31 | 212,190 | 35 (34) | 15.6 | 3 |
| MC4R | melanocortin 4 receptor | 18q22 | 998 | 10 (10) | 19.7 | 1 |
| NPY1R | neuropeptide Y receptor Y1 | 4q31.3-q32 | 8,631 | 4 (4) | 1.5 | 1 |
| NPY5R | neuropeptide Y receptor Y5 | 4q31-q32 | 7,995 | 7 (7) | 3.9 | 1 |
| PLIN | perilipin | 15q26 | 14,987 | 11 (8) | 12.8 | 3 |
| POMC | proopiomelanocortin | 2p23 | 7,664 | 13 (9) | 10.2 | 0 |
| SOCS3 | suppressor of cytokine signaling 3 | 17q25.3 | 3,294 | 6 (6) | 2.2 | 1 |
| STAT3 | signal transducer and activator of transcription 3 | 17q21 | 75,170 | 19 (18) | 4.9 | 0 |
| TUB | tubby homolog | 11p15.4 | 67,472 | 19 (18) | 4.7 | 0 |
For number of polymorphic SNPs genotyped, the number in parentheses indicates number of SNPs that were determined to be polymorphic in this study. nsSNPs are non-synonymous SNPs.
This study was designed to prioritize the relative contributions of individual sequence variants, as well as to examine interactions among functionally related candidate genes and pathways. In an effort to identify more universally relevant genes/alleles, two racially divergent populations were genotyped in this study: a population of European American adults living in the New York City tri-state region, and a population of Yup'ik Eskimos from southwest Alaska. GHRL was associated with BMI in the NY European Americans, even after correction for multiple testing, suggesting that genetic variation in GHRL may contribute to relative adiposity in this population.
Materials and Methods
Subjects
European American participants were recruited from the New York Health Project (NYHP), a prospective cohort study undertaken under the auspices of the Academic Medicine Development Corporation (AMDeC) of New York to analyze general health and cancer-related epidemiology [16]. Approximately 18,000 subjects were enrolled between January, 2000 and December, 2002 at fourteen sites across the five boroughs of New York City. The institutional review boards at each of the fourteen sites and Columbia University approved the protocol. Enrollees had to be 30 years of age or older, reside in New York, New Jersey, or Connecticut, and have a literacy level sufficient to complete a consent and simple follow-up questionnaire. All subjects signed a written informed consent. A trained interviewer administered a questionnaire regarding health status and behaviors, drew blood, and measured standing height on a stadiometer and weight on a scale to the nearest pound with light clothing. Written informed consent was obtained from all subjects. Race/ethnicity was assessed by self report using the categories of the 2000 Census (http://www.census.gov/main/www/cen2000.html). A total of 17,709 subjects with valid data were recruited. Of these, 10,260 (57.9%) were European Americans (non-Hispanic white). Individuals at the extremes of the BMI distribution are potentially the most informative in this context and were, therefore, selected for initial analysis. Cancer history was by self-report and confirmed in state cancer registries when possible. All subjects with a history of cancer were eliminated from the study groups to eliminate the possible effect of cancer and its treatment on body weight. European American subjects were sorted by ascending BMI. Subjects below the 10th percentile for BMI were not included to avoid the confounding effects of medical conditions that themselves might cause weight loss. Gender-and age-matched 1,003 pairs of European Americans with BMI between the 10th and 30th percentile or above the 80th percentile were selected for genotyping. In the European American subjects living in New York, with the sampling method we used we had 80% power at α level of 5% to detect a genetic variant accounting for 0.27% of the variance in BMI. This sampling method involved identifying the 1,003 lean subjects between 10th and 30th percentiles for BMI and the 1,003 obese above the 80th percentile of the total sample BMI distribution. A random selection of 2,006 subjects from the same 10,260 individuals would have 80% power to detect a genetic variant accounting for 0.39% of the phenotypic variance. Using all of the available (10,260) subjects would have provided 80% power to detect a gene accounting for 0.077% of the phenotypic variance. Therefore, this strategy of selective genotyping of subjects at the extremes of phenotype can be used to reduce genotyping costs without substantially diminishing power [17].
Recruitment of Yup'ik Eskimo study participants started in December, 2003 and continued through May, 2005. All residents 14 years of age and older from seven communities were invited to participate [18]. All seven communities were located in rural southwest Alaska. On average, approximately 43% of eligible community members participated in the research project. The age distribution of our study population reflects the age distribution among eligible participants. All participants were consented using protocols approved by the University of Alaska Institutional Review Board, the National and Alaska Area Indian Health Service Institutional Review Boards, and the Yukon Kuskokwim Human Studies Committee.
593 Yup'ik Eskimo participant samples were included in this study. The analysis for this report is based upon participants who identified themselves as non-pregnant Alaska Native Yup'ik Eskimos, ≥18 years, and as having fasted at least 8 h. Anthropometric measurements included height, weight, four circumferences (triceps, waist, hip, and thigh), four skinfolds (triceps, subscapular, abdominal and thigh), and were obtained by trained observers using previously published protocols implemented in the NHANES III survey [19]. Percent body fat was estimated by electrical bioimpedance using a Tanita TBF-300A body fat analyzer. Participants were selected from pedigrees based on availability of DNA for individuals within the family, individuals phenotyped for at least five obesity related traits, and a composite obesity score that is a weighted average of the primary phenotype (BMI) and secondary phenotypes (percent body fat, waist circumference, and four skinfold thicknesses) and weights were a function of the correlations between primary and secondary phenotypes (See the details of derivation of composite score in the Appendix A). The Yup'ik data set consists of 157 families with 731 individuals. 593 individuals were included in the present study based on the following selection strategy. First we selected families with available DNA and at least five phenotypes within a family. We ordered the families with respect to the smallest amount of missing data. The family highest in this ranking is the family with the smallest amount of missing data among all individuals in the family. Then, we selected a minimum number of families that would enable us to achieve at least 80% power at significance level of 5% for traits with a broad-sense heritability of 50% and 5% locus heritability. Using these criteria, we identified 593 individuals belonging to the top 53 families (see Appendix A, table A1).
Table A1.
Power of association test for given locus and polygenic heritability when only 593 individuals have phenotype and genotype data available
| h2 total | h2 locus | h2 poly | Power | |
|---|---|---|---|---|
| α = 0.05 | α = 0.01 | |||
| 0.2 | 0.05 | 0.15 | 0.766 | 0.618 |
| 0.1 | 0.1 | 0.940 | 0.923 | |
| 0.5 | 0.05 | 0.45 | 0.827 | 0.673 |
| 0.1 | 0.4 | 1.000 | 0.951 | |
| 0.7 | 0.05 | 0.65 | 0.837 | 0.719 |
| 0.1 | 0.6 | 1.000 | 1.000 | |
Candidate Gene Selection
Candidate genes were selected according to their physiological function and/or reported association/linkage with obesity or a related endophenotype. The 30 candidate genes are listed in table 1. The relationships between genes in the respective molecular pathways is depicted in figure 1.
SNP Selection
Single nucleotide polymorphisms (SNPs) were gathered from multiple public databases, NCBI (http://www.ncbi.nlm.nih.gov/projects/SNP/), Celera (ABI) (now publicly available through dbSNP), HapMap (http://www.hapmap.org), the literature, and our local resequencing data. For each locus, SNPs were selected based upon the following criteria: (1) All SNPs with a minor allele frequency >0.05 between 10,000 base pair (bp) upstream and 10,000 bp downstream of the gene's coding sequence were considered; (2) only one SNP from a group of SNPs in linkage disequilibrium with r2 > 0.80 using the aggressive multimarker tagging method of the Haploview software (http://www.broad.mit.edu/mpg/haploview/download.php); (3) coding SNPs and non-coding SNPs with previous evidence of linkage or association with obesity in the literature or from our local data with a minor allele frequency >0.05 were included; (4) non-coding SNPs within putative transcription factor binding sites or sequences conserved across species with a minor allele frequency >0.05 were included; (5) for large genes with large numbers of non-coding SNPs such as LEPR, we limited SNPs to within 5,000 bp upstream and downstream of the coding sequence, and (6) all nonsynonymous coding SNPs (nsSNPs) with an allele frequency of at least 0.01 were included.
Genomic DNA was extracted from whole blood and stored at 4°C. SNPs were genotyped on an Illumina BeadArray® assay according to the manufacturer's directions. Due to technical characteristics of the Illumina BeadArray® assay [20], we could not use SNPs within 60 bp of another SNP or SNPs with more than 3 alleles or insertion/deletion polymorphisms. 384 SNPs in 30 genes were selected for a custom-designed BeadArray (supplemental table S1; for online supplementary material, see www.karger.com/doi/10.1159/000181158).
Eighteen subjects from the NYHP and 14 Yup'iks were blindly genotyped in duplicate to assess assay reproducibility. In addition, 28 Illumina controls (4/plate) were included as positive and negative controls to assess bead assay performance. Twenty-five additional subjects for whom genotypes had been independently determined for a subset of the candidate genes were also included to independently determine accuracy of the bead array genotyping method.
Statistical Analysis
For the NYHP BMI data, the BMI distribution for 1,982 individuals with valid genotypic data was evaluated, and were log-transformed to normalize the residual distribution. The sample selected from 2 extremes of population distribution was highly skewed. The log transformation made it a distribution of 2 extremes of comparatively normal distribution. The Ordinary Least Squares (OLS) regression method is quite robust to the distribution. Past research has shown that when OLS regression is used, extreme sampling not only provides more power than random sampling on a per subject basis, but also that the tests are robust to the marked non-normality induced by the extreme sampling [21,22,23]. Similarly, for the Yup'ik data, BMI, subscapular skinfold thickness, and waist circumference phenotypes were log transformed; thigh-skinfold phenotype was square root transformed to render the residuals approximately normally distributed after adjusting for age, age2, and sex. The phenotypes were then tested for univariate and multivariate outliers. Multivariate outliers were identified by chi-square testing using Mahalanobis distance [24]. For the NYHP data, the bivariate distribution of BMI and age was used to calculate Mahalanobis distance; for the Yup'ik data, multivariate distributions of all 8 phenotypes were used to identify possible outliers. None of the individuals in the NYHP study was found to be an outlier. Thirteen individuals were identified as outliers in the Yup'ik data, and their phenotypic data were not included in our final analyses. We did, however, perform an association analysis of all phenotypes in Yup'ik data including the 13 outlier individuals. The results did not vary significantly in comparison to those excluding the outliers. In single SNP, gene, and gene-group association analysis, Pearson's correlation between the resulting p values corresponding to the significance of the test of the association for all phenotypes, including and excluding outliers were found to be between 0.65 and 0.95. The phenotypes in both populations were adjusted for the covariates sex, age, age2, and their respective interactions.
The 355 SNP markers were tested for Hardy-Weinberg equilibrium in both population data sets. For 17 SNPs in the NYHP data and 21 SNPs in the Yup'ik data, all subjects were homozygous for the major allele and were thus not polymorphic. These SNPs were excluded from further analysis. 43 SNPs in the NYHP data and 73 SNPs in the Yup'ik data were found to be in Hardy-Weinberg disequilibrium. In NYHP data there are 17 SNPs that are not in HWE among the entire sample but are in HWE among lean subjects. There would be 11 more SNPs in HWE if the threshold were moved from 0.05 to 0.01. We performed the association analyses including and excluding the SNPs that were in HW disequilibrium. None of the SNPs in HW disequilibrium demonstrated statistical significance at the 0.05 significance level.
For the NYHP data, associations between BMI and both additive and dominance effects of each of 355 SNPs were tested using the Ordinary Least Squares (OLS) regression method. The Yup'ik data, comprised of families, could not be analyzed using the OLS regression method because of correlations present among related individuals. Before analyzing the Yup'ik data set, we compared Within Cluster Resampling (WCR) and Mixed Model procedures to determine the validity of these methods suited for Yup'ik familial data set (see Appendix B for more details). We found that the WCR method gave more false positive results than expected. The false positive rate for the WCR method increased at lower significance thresholds. The mixed model always gave valid type I error rates. Thus, association analysis on these data was performed using a mixed model approach in which we used family membership as a random effect by fitting a mixed model with phenotype as a dependent variable and family ID as a random effect, and SNPs and covariates as fixed effects using the PROC MIXED procedure in the Statistical Analysis System (SAS 9.1, SAS Institute Inc. Cary, N.C., USA). Because of multiple testing (for 355 SNPs) in both data sets, association results were tested at threshold significance level of 0.05 using the false discovery rate method of corrected tests [25].
To assess the overall effect of multiple genetic variants in a particular gene on relevant phenotypes, each gene was tested for association with each phenotype by linear regression of each phenotype on additive and dominance effects of all SNPs in the gene. For the Yup'ik family data, we used a similar method of regression using a mixed model in which family membership was treated as a random effect. As noted, the candidate genes were included (non-exclusively) in 7 groups based upon their membership in molecular/physiological pathways mediating energy homeostasis. As was done for the individual genes, the effects of these groups on relevant phenotypes were tested using linear regression modeling all SNPs in the group.
The haplotype analysis was performed using a sliding window of size 2, 3, and greater until all of the markers within each gene were analyzed [26]. For each haplotype analysis, the score test, was used in the R package Haplo Stats [27, 28]. In the haplotype association analyses, age and gender were included as covariates. Multiple testing adjustment was performed following the strategy proposed in [18].
We also searched for interactions among SNPs by fitting a logistic regression model with at most nine parameters: intercept, additive and dominance effects at each locus, and four interaction effects. We used the log-likelihood ratio test statistic (LRT) computed as χ2df = 2(Lfull – Lreduced) where Lfull is the log-likelihood of the data computed under a fully specified model and Lreduced is the log-likelihood of the data computed under the constraint that one or more parameters in the model are equal to zero, to explore presence of epistatic interactions. Since testing for interaction among several SNPs tends to increase multiple testing error rates, which are largely responsible for false positive results and failure to replicate numerous phenotype-genotype associations [29], we chose to minimize the impact of multiple testing adjustment by estimating the number of truly null hypotheses from observed p values by a finite mixture model approach [30]. Posterior probabilities of true positives (PTP), true negatives (PTN) and false negatives (PFN) were also estimated. Mix-o-matic approach uses maximum likelihood (ML) to fit a mixture model to observed p values, which represent two hypotheses groups: the null (modeled as uniformly distributed) and alternative (modeled as beta distributed).
Results
Phenotypic characteristics of the 1,982 New York European American NYHP subjects with valid genotype data are summarized in table 2a. Obese and lean subjects were matched for sex (56.7% female) and age. The mean age ± standard deviation of obese patients was 47.7 ± 10.2 years, and the lean subjects were 46.5 ± 10.7 years. The BMI of the obese subjects was 36.3 ± 4.6 kg/m2; the lean subjects had a BMI of 22.7 ± 1.4 kg/m2.
Table 2.
Characteristics of the participants
| Obese >80th perc. | Lean 10th to 30th perc. | |
|---|---|---|
| a NYHP European Americans | ||
| Number (F/M) | 990 (561/429) | 992 (563/429) |
| Subjects with diabetes (F/M) | 61 (37/24) | 9 (3/6) |
| Age, years | 47.66 ± 10.24 | 46.48 ± 10.68 |
| BMI, kg/m2 | 36.28 ± 4.64 | 22.68 ± 1.40 |
| b Yup'ik | ||
| Number (F/M) | 593 (333/260) | |
| Age, years | 38.33 ± 17.42 | |
| BMI, kg/m2 | 27.58 ± 5.95 | |
| Triceps skinfold, mm | 21.03 ± 10.81 | |
| Subscapular skinfold, mm | 21.24 ± 11.80 | |
| Abdominal skinfold, mm | 27.53 ± 14.29 | |
| Thigh skinfold, mm | 22.86 ± 13.04 | |
| Body fat, % | 29.44 ± 10.84 | |
| Waist circumference, cm | 90.17 ± 14.18 | |
Data are the means ± standard deviations.
The phenotypes of the 593 Yup'ik participants (333 females and 260 males) are summarized in table 2b. They had a mean age of 38.3 years, mean BMI of 27.6 kg/m2, mean percent body fat of 29.4%, and mean waist circumference of 90.2 cm (table 2b). The mean triceps, subscapular, abdominal and thigh skinfold thicknesses were 21.0, 21.2, 27.5, and 22.9 mm, respectively.
355 of the original 384 SNPs were used in these analyses (suppl. table S1). The 29 SNPs that were not used were not polymorphic in European Americans and/or Yup'ik Eskimos. For the 18 replicate European American subjects and 14 replicate Yup'ik Eskimo participants, all of the genotypes were 100% concordant for the 355 SNPs genotyped. There was 100% concordance of genotypes determined by the bead array and other methods of genotyping including dideoxy sequencing and TaqMan.
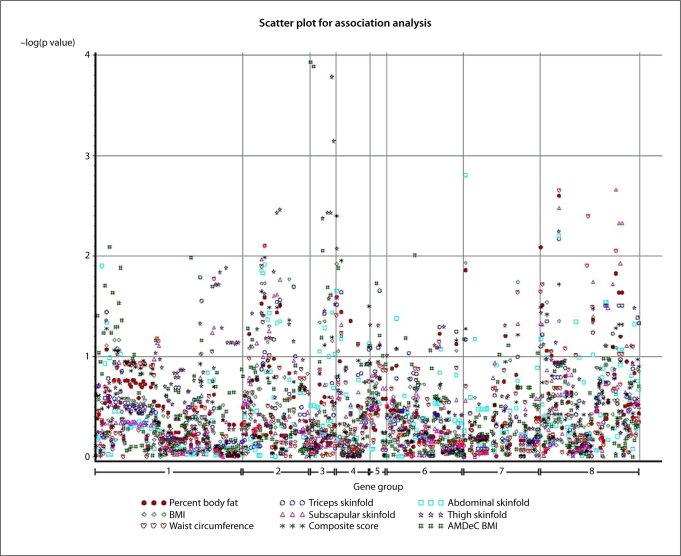
When each of the SNPs was analyzed individually, 17 SNPs in 9 genes (GLP1R, GHRL, JAK2, GHSR, HTR2A, LEP, LEPR, NPY5R, and SOCS3) demonstrated association with BMI in the NYHP European Americans sample at significance level of 0.05 (table 3). In the Yup'ik participants, 142 SNPs in 21 genes (ADIPOQ, ARL6, BBS4, BBS5, CNR1, CPE, GHRL, GCG, GHSR, GLP1R, HTR2A, HTR2C, JAK2, LEPR, MC4R, NPY1R, NPY5R, PLIN, SOCS3, STAT3, and TUB) were associated with measures of adiposity at significance level of 0.05 (suppl. table 2). The complete distribution of observed p values in both data sets is given in figure 2. After correction for multiple testing by False Discovery Rate (FDR) controlling procedure, two SNPs (rs35682 and rs35683) in GHRL within 600 base pairs of each other had FDR values below 0.05 in the NYHP European American sample. However, after such correction, none of the associations in the Yup'ik participants remained significant.
Table 3.
SNPs associated with BMI in NYHP European Americans
| Gene | SNP name | Map location | BMI (NYHP) |
|
|---|---|---|---|---|
| raw p value | FDR | |||
| GHRL | rs35683 | 10303250 | 0.0001 | 0.04 |
| GHRL | rs35682 | 10303782 | 0.0001 | 0.02 |
| GHSR | rs560994 | 173636722 | 0.009 | 0.75 |
| GHSR | rs4144707 | 173641907 | 0.02 | 0.57 |
| GHSR | rs495225 | 173648735 | 0.02 | 0.59 |
| GLP1R | rs9380825 | 39118941 | 0.01 | 0.63 |
| HTR2A | rs912127 | 46349287 | 0.01 | 0.66 |
| JAK2 | rs3780365 | 5058520 | 0.008 | 0.91 |
| JAK2 | rs966871 | 5111070 | 0.01 | 0.55 |
| JAK2 | rs2230722 | 5040706 | 0.02 | 0.66 |
| JAK2 | rs1410779 | 5073173 | 0.02 | 0.60 |
| JAK2 | rs3780379 | 5102519 | 0.03 | 0.66 |
| JAK2 | rs3808850 | 4973311 | 0.04 | 0.82 |
| LEP | rs791601 | 127460064 | 0.04 | 0.86 |
| LEPR | rs3790419 | 65779130 | 0.01 | 0.58 |
| NPY5R | Y5R1c52 | 164410779 | 0.02 | 0.70 |
| SOCS3 | rs4969170 | 73872133 | 0.02 | 0.62 |
Raw p value (p values without correction for multiple testing) standard deviations. and false discovery rate (FDR) estimates.
Fig. 2.
The distribution of −log(p value) distribution corresponding to single SNP association tests for both data sets given location of each SNP within gene groups as shown in table 6.
When all SNPs within a gene for each of the 30 candidate genes were evaluated jointly, polymorphisms in GHRL, GLP1R, and JAK2 were significantly associated with BMI in the NYHP European American sample, with each gene accounting for approximately 1% of the variance. Table 4 provides the p values associated with genes significant at α level of ≤5%, R2, and adjusted R2 values. Note that none of the genes were significant after correcting for multiple testing procedures by FDR. R2 is the proportion of variability in the phenotypic data that is accounted for by a statistical model, which is a combined additive and dominant effect of one SNP or group of SNPs on a gene. Adjusted R2 is a modification of R2 that adjusts for the number of predictors (SNPs in this context) in a model. The adjusted R2 value increases only if the newly added predictor improves the model more than would be expected by chance. In the Yup'ik participants, four genes were associated with obesity phenotypes (suppl. table 3), but when corrected for multiple testing, none of the associations with all the SNPs in any of the genes remained significant.
Table 4.
Gene association p values for BMI at α level of 5% and corresponding R2 accounted for by the genes in NYHP European Americans using all SNPs genotyped within the gene
| Target locus | BMI |
||
|---|---|---|---|
| raw p value | R2 | adjusted R2 | |
| GHRL | 0.03 | 0.018 | 0.010 |
| GLP1R | 0.02 | 0.033 | 0.011 |
| JAK2 | 0.04 | 0.026 | 0.0084 |
For none of the genes were the FDR values less than 0.05.
Haplotype analysis demonstrated an association of BMI with AGRP (p = 0.03), GHRL (p = 0.04), and NPY1R-5R (0.008), in the NYHP European Americans after correction for multiple testing (table 5). No haplotypes were associated with any measures of adiposity in the Yup'ik participants after multiple testing correction.
Table 5.
Haplotype analysis of genes associated with adiposity in the NYHP European Americans
| AGRP | GHRL | NPY1R-5R | |
|---|---|---|---|
| Number of SNPs | 2 | 8 | 2 |
| Permutation p value | 0.03 | 0.04 | 0.008 |
p value is the adjusted p value using 2,000 simulations corresponding to permutation test.
As noted, the candidate genes were selected in part based upon the molecular pathways in which they interact (fig. 1). Genes were analyzed in the groups shown in table 6 by modeling all SNPs within the genes in that pathway. The regression analyses of SNPs for genes within a pathway showed significant associations with BMI in the NYHP European American subjects, in particular, the GHRL/GHSR pathway (p = 0.03) and the GLP/GLP1R pathway (p = 0.01) (table 6). However, after correcting for multiple testing by the FDR procedure, neither of these associations had FDR values below 0.05. GHRL/GHSR and GLP1R pathways both account for approximately 1% of the attributable variance in BMI. Similar analyses revealed that none of the gene pathways in the Yup'ik participants had FDR values below 0.05.
Table 6.
Results of regression analysis of gene groups [‘pathway’ components] in NYHP European Americans
| Gene group | Genes in the group | BMI |
||
|---|---|---|---|---|
| raw p value | R2 | adjusted R2 | ||
| 1 | LEP, LEPR, SOCS3, JAK2, STAT3 | 0.22 | 0.099 | 0.009 |
| 2 | AGRP, CPE, MC4R, POMC | 0.64 | 0.040 | −0.002 |
| 3 | GHRL, GHSR | 0.03 | 0.027 | 0.010 |
| 4 | GCG, GLP1R | 0.01 | 0.033 | 0.011 |
| 5 | NPY1R, NPY5R | 0.12 | 0.012 | 0.002 |
| 6 | HTR2A, HTR2C | 0.16 | 0.056 | 0.007 |
| 7 | BBS1, BBS2, BBS3, BBS4, BBS5, BBS6, BBS7, BBS8 | 0.40 | 0.036 | 0.002 |
For none of the genes were the FDR values less than 0.05.
The results of interaction analyses are presented in suppl. fig. 1 and suppl. table 4. Of the over 50,000 interaction hypotheses tested, we estimate that for 16.4% (95% confidence interval, 13.5–17.5%) the null hypotheses were estimated by finite mixture models [30] to be false, which implies that a model including epistatic interactions will better characterize the available data. The ten pair-wise interactions with the lowest observed p values i.e., ≤10–3 and the highest posterior probabilities of being true positives (>30%) are presented in suppl. table 4.
The SNP with the most significant association with sex-, age- and diabetes-adjusted BMI in NYHP European Americans was rs35683 in GHRL with A/A genotypes having an adjusted BMI of 29.47 ± 0.01 kg/m2, A/C genotypes having an adjusted BMI of 29.50 ± 0.008 kg/m2, and C/C genotypes having the lowest adjusted BMI of 29.44 ± 0.01 kg/m2.
Discussion
We studied two independent groups of subjects for association with 30 candidate genes for obesity involved in 7 molecular pathways regulating energy homeostasis. Of the 30 genes analyzed, two SNPs within GHRL (rs35682 and rs35683 within 600 base pairs of each other) are associated with BMI in the New York European American population. Polymorphisms in GHRL have previously been associated with obesity, impaired glucose tolerance, and the metabolic syndrome in several previous studies in European American children and adults [31,32,33,34,35,36,37,38,39,40]. Both rs35682 and rs35683, the two SNPs in GHRL most highly associated with BMI, are intronic and are not located in regions that are conserved across species. The two SNPs themselves are not predicted to result in changes in splicing or expression. Therefore, the biological effect of these SNPs is likely through another variant in linkage disequilibrium with rs35682 and rs35683. Notably, the GHRL SNPs rs696217 (Leu72Met) and rs27647 (5′UTR polymorphism) previously associated with adiposity in children and adults from Japan, Finland, Italy, and North America [33,34,35,36, 38, 40] were not the SNPs most highly associated with BMI in our study. The phenotypic effect of the GHRL variants is modest, and account for a difference in BMI of only 0.06 kg/m2, with the heterozygotes having a higher BMI than either of the homozygous genotypes. This is an example of heterosis at a single locus conferring an increased BMI for the heterozygotes.
Our studies include two very different racial groups in two very different environments, European Americans living in the New York metropolitan area and Yup'ik Eskimos living in rural Alaska. Only one SNP in one gene, rs560994 in GHSR, was found to be associated with BMI in both populations, but was not significant in the Yup'iks or European Americans after correction for multiple testing. The lack of replication may be in part based upon genetic and environmental differences between the groups, and is not surprising. Our study underscores the need to study large numbers of subjects in multiple populations.
We analyzed the family data (Yup'ik participants) using a mixed model with correction for multiple testing using the FDR procedure. This methodology is conservative and protects against false positives. Our results that did not meet rigorous FDR cutoffs might still be the basis for testing these genes in other populations.
The genetics of complex traits such as obesity are likely to result from interactions among genes and between genes and the environment. We used the molecular physiology of the control of energy homeostasis to cluster genes within molecular pathways and to examine interactions among these genes. Bardet Biedel syndrome represents an oligogenic example of interacting genes [15] and provides a rationale for this approach. By using prior knowledge of genes within networks/pathways, the number of tests for interactions that must be performed to test hypotheses of gene × gene interactions can be reduced. Given the number of genes we studied, testing for all possible combinations of gene × gene interactions would have resulted in a prohibitive penalty for multiple testing. Even using our strategy of testing only for interactions of genes within common pathways, we were unable to demonstrate significance for the interactions within the ghrelin/ghrelin receptor and GLIP1R pathways in the European Americans after correcting using the false discovery method. Our experience highlights the need for extremely large studies to adequately power analyses of complex traits for gene × gene and gene × environment interactions.
Supplementary Material
Supplementary Materials
Acknowledgements
We would like to thank Josue Martinez for help with manuscript preparation. These studies were supported by NIH DK 52431, and NIHDKP30-26687 (NYORC). The Center for Alaska Native Health Research is funded by a grant from the National Center for Research Resources at the National Institutes of Health (RR16430).
Appendix
Appendix A: Derivation of Composite Score
Often we are required to select the individuals from large data sets with a multivariate phenotype such that the sub-sample is best representative of the sample and preserves the correlation structure among the multivariate phenotype as well as provides optimal chance of finding putative genetic loci. In the Yup'ik sample, we have adiposity related multivariate phenotypic data consisting of BMI, skinfolds (subscapular, suprailiac, thigh, triceps), waist circumference, and percent body fat. However, it is not clear which phenotype or combination of phenotypes should be used in determining the sub-sample. In most situations investigators use a primary phenotype of interest to create a sub-sample by using an extreme sampling design (i.e. selecting only individuals from both extremes of the distribution using fixed threshold). This sampling design does provide optimal power for the primary phenotype, but not necessarily for other correlated phenotypes which may also be of interest. We used the selection strategy using all correlated phenotypes and incorporating the correlation structure in the pedigrees. Here, we provide a short description of a potentially useful measure to estimate individual phenotypic value based on multivariate data following Allison and Franklin [41].
Consider k correlated phenotypes Yi measured on individuals in a sample. Then the minimum variance estimator of combined phenotypes Yi for j-th individual is given by
where k = the number of observed variables for individual j, Yij = the standardized i-th variable for the j-th individual, Wj = the weight for the i-th variable.
Without loss of generality, we can assume Y1 is the primary trait. Let ρ1i = genetic correlation between i-th secondary variable and primary variable Y1 (for example, BMI can be considered as a primary variable and percent body fat (PBF) or skin fold measurements can be considered as secondary variables). Then following Allison and Franklin [41], we can define and the weight for the correlated secondary n-th variable is defined as Note that weights as function of heritability of primary trait and/or genetic correlations between primary and secondary traits will allow us to capture the genetic variance of each trait in the composite score. We can further refine the combined score by using variance shrinkage method, and the new updated combined score is given by . Note that this score can easily accommodate missing data and also preserves the correlation between primary trait and secondary traits.
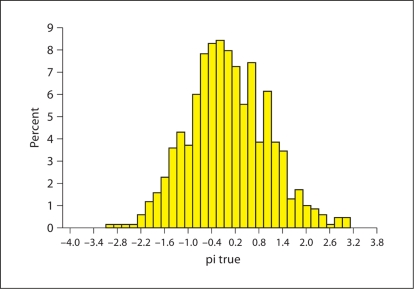
After calculating a composite score for each individual in the pedigrees, we calculated the total variance of the composite score for each family. The distribution of the composite score is given in figure A1 and the power estimates using selected pedigrees are given in table A1. The power was calculated using simulated data set those mimic the Alaska pedigrees. We simulated 1,000 replicates of pedigrees keeping structure of the pedigrees exactly same as Alaska pedigrees and also only simulating phenotypes and genotypes of individuals with available DNA and obesity related traits as in real data. The analysis was performed using ASSOC of SAGE suite of programs. We have sufficient power to detect putative loci with locus heritability of 0.05 at 5% α level.
Fig. A1.
The distribution of composite scores for obesity for the non-pregnant Alaska Native Yup'ik Eskimos, ≥ 18 years. The composite measure score is a weighted average of BMI and percent body fat, waist circumference, and four skinfold thicknesses.
Appendix
Appendix B
Before analyzing the Yup'ik data set that consists of genotypes and multivariate phenotypes in pedigrees, we compared the Within Cluster Resampling and Mixed-Model procedure to determine the validity of these methods suited for Yup'ik familial data set. We briefly describe both methods.
Within-Cluster Resampling (WCR)
WCR was first introduced by Hoffman et al. [42] and subsequently expanded upon by Follmann et al. [43]. WCR is an elegant method for analyzing correlated clustered data and is a simple but computationally intensive estimation method. The steps of the methods are described below.
Step 1: Select one individual from each cluster with replacement. Repeat this m times to create m data sets.
Step 2: Analyze each of the m data sets using standard complete-data methods such as SAS or SPSS.
Step 3: Integrate or combine m analyses to get a single estimate of parameters and their variances. This involves averaging the values of the parameter estimates across the m samples to produce a single point estimate and variance. Formally, we can describe it as follows:
Let m = the number of data sets analyzed, = Estimate of the parameter of interest from the i-th set, = Variance Estimate of the from the i-th set.
The point estimate from the WCR method is the average of the estimates from m analyses and is given by
and the total variance estimate of the point estimate is the difference of within replicate variance
and between replicate variance
and is given by
The statistic
is approximately distributed as t with νm degrees of freedom, where
The original method was used for binary correlated clusters where cluster size is informative, but it could easily be extended to continuous phenotypes.
Mixed-Model Procedure
The linear mixed effects model may be viewed as a generalization of the variance component and regression analysis models in which SNPs are modeled as fixed effects and family ID as random effect. In particular, we can write the model as
where ϒ is the outcome variable (phenotype) such as BMI, α is overall mean of the phenotype, β1 is treated as fixed effect, β2 is treated as random effect, and ε as non-shared random effect.
Simulation Procedure
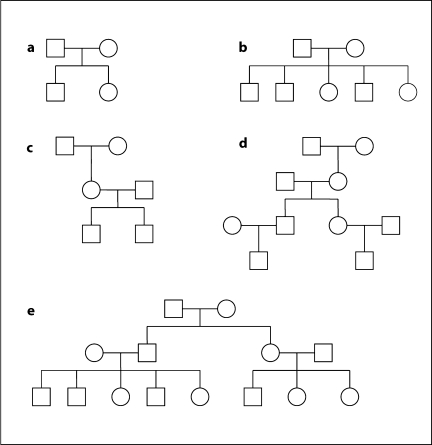
We simulated 100,000 replicates each with 50 unequal size families shown in figure B1.
Fig. B1.
Pedigrees of unequal size used for simulation.
Quantitative Trait Simulation
We simulated a mixed model quantitative trait for each individual in the family structures described above assuming no major locus effect to determine the behavior of the null distribution. The polygenic component was assumed to be normally distributed with mean 0 and variance 0.30 and the random environmental component was assumed to be normally distributed with mean 0 and variance 0.70.
Marker Simulation
Marker with 2 alleles was simulated for testing the null hypothesis of no association with allele frequency of 0.2. Genotypes of founders were simulated by two random draws from binomial distribution with p = 0.2 and n = 2 to get both alleles in the genotype. The non-founders genotypes were randomly assigned following Mendelian inheritance.
The Type I error rate of both WCR and Mixed Model is given in the table B1 at nominal α levels at 0.05, 0.01, 0.001, and 0.0001, respectively. Clearly, WCR produces inflated type I error rates compared to mixed model. Mixed model always gave valid type I error rates. Thus, we have shown that mixed model is an attractive choice as a method to test association in pedigree data since it represents a good balance between quality of results and ease of use.
Table B1.
Comparison of WCR and mixed model methods with respect to Type I error rates
| Method | Nominal α level |
|||
|---|---|---|---|---|
| 0.05 | 0.01 | 0.001 | 0.001 | |
| WCR | 0.07935 | 0.03476 | 0.01673 | 0.01027 |
| Mixed model | 0.05036 | 0.00996 | 0.00091 | 0.00010 |
The results are shown for 100,000 replicates of 50 unequal size pedigrees. We used 10,000 bootstrap samples in WCR.
References
- 1.Flegal KM, Carroll MD, Ogden CL, Johnson CL. Prevalence and trends in obesity among US adults, 1999–2000. JAMA. 2002;288:1723–1727. doi: 10.1001/jama.288.14.1723. [DOI] [PubMed] [Google Scholar]
- 2.Hedley AA, Ogden CL, Johnson CL, Carroll MD, Curtin LR, Flegal KM. Prevalence of overweight and obesity among US children, adolescents, and adults, 1999–2002. JAMA. 2004;291:2847–2850. doi: 10.1001/jama.291.23.2847. [DOI] [PubMed] [Google Scholar]
- 3.Calle EE, Thun MJ, Petrelli JM, Rodriguez C, Heath CW., Jr Body-mass index and mortality in a prospective cohort of U.S. adults. N Engl J Med. 1999;341:1097–1105. doi: 10.1056/NEJM199910073411501. [DOI] [PubMed] [Google Scholar]
- 4.Allison DB, Faith MS, Nathan JS. Risch's lambda values for human obesity. Int J Obes Relat Metab Disord. 1996;20:990–999. [PubMed] [Google Scholar]
- 5.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256:51–54. [PubMed] [Google Scholar]
- 6.Stunkard AJ, Harris JR, Pedersen NL, McClearn GE. The body-mass index of twins who have been reared apart. N Engl J Med. 1990;322:1483–1487. doi: 10.1056/NEJM199005243222102. [DOI] [PubMed] [Google Scholar]
- 7.Chung WK, Leibel RL. Molecular physiology of syndromic obesities in humans. Trends Endocrinol Metab. 2005;16:267–272. doi: 10.1016/j.tem.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Alharbi KK, Spanakis E, Tan K, Smith MJ, Aldahmesh MA, O'Dell SD, Sayer AA, Lawlor DA, Ebrahim S, Davey Smith G, O'Rahilly S, Farooqi S, Cooper C, Phillips DI, Day IN. Prevalence and functionality of paucimorphic and private MC4R mutations in a large, unselected European British population, scanned by meltmadge. Hum Mutat. 2007;28:294–302. doi: 10.1002/humu.20404. [DOI] [PubMed] [Google Scholar]
- 9.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: The 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 10.Ahima RS, Osei SY. Molecular regulation of eating behavior: New insights and prospects for therapeutic strategies. Trends Mol Med. 2001;7:205–213. doi: 10.1016/s1471-4914(01)01989-x. [DOI] [PubMed] [Google Scholar]
- 11.Berthoud HR. Multiple neural systems controlling food intake and body weight. Neurosci Biobehav Rev. 2002;26:393–428. doi: 10.1016/s0149-7634(02)00014-3. [DOI] [PubMed] [Google Scholar]
- 12.Kalra SP, Dube MG, Pu S, Xu B, Horvath TL, Kalra PS. Interacting appetite-regulating pathways in the hypothalamic regulation of body weight. Endocr Rev. 1999;20:68–100. doi: 10.1210/edrv.20.1.0357. [DOI] [PubMed] [Google Scholar]
- 13.Saper CB, Chou TC, Elmquist JK. The need to feed: Homeostatic and hedonic control of eating. Neuron. 2002;36:199–211. doi: 10.1016/s0896-6273(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 14.Chung WK, Leibel RL. Molecular physiology of monogenic and syndromic obesities in human. In: Donohue P, editor. Energy and Metabolism and Obesity: Research and Clinical Applications. New Jersey: Humana Press; 2008. pp. 1–22. [Google Scholar]
- 15.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: An emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell MK, Gregersen PK, Johnson S, Parsons R, Vlahov D. The New York cancer project: Rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81:301–310. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allison DB, Thiel B, St Jean P, Elston RC, Infante MC, Schork NJ. Multiple phenotype modeling in gene-mapping studies of quantitative traits: Power advantages. Am J Hum Genet. 1998;63:1190–1201. doi: 10.1086/302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyer BB, Mohatt GV, Lardon C, Plaetke R, Luick BR, Hutchison SH, de Mayolo GA, Ruppert E, Bersamin A. Building a community-based participatory research center to investigate obesity and diabetes in Alaska Natives. Int J Circumpolar Health. 2005;64:281–290. doi: 10.3402/ijch.v64i3.18002. [DOI] [PubMed] [Google Scholar]
- 19.Lohman T, Roche A. Anthropometric standardization reference manual. Champaign, IL: Human Kinetics Books; 1988. [Google Scholar]
- 20.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. Beadarray technology: Enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;(Suppl):56–58. 60–51. [PubMed] [Google Scholar]
- 21.Alf EF, Abrahams NM. The use of extreme groups in assessing relationships. Psychometrika. 1975;40:563–572. [Google Scholar]
- 22.Feldt LS. The use of extreme groups to test for the presence of a relationship. Psychometrika. 1961;26:307–316. [Google Scholar]
- 23.Fowler RL. Using the extreme groups strategy when measures are not normally distributed. Appl Psychol Meas. 1992;16:249–259. [Google Scholar]
- 24.Filzmoser P, Garrett RG, Reimann C. Multivariate outlier detection in exploration geochemistry. Comp Geosci. 2005;31:579–587. [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 26.Becker T, Knapp M. A powerful strategy to account for multiple testing in the context of haplotype analysis. Am J Hum Genet. 2004;75:561–570. doi: 10.1086/424390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered. 2003;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 28.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Allison DB, Gadbury GL, Heo M, Fernandez JR, Leed C-K, Prolla TA, Weindruch R. A mixture model approach for the analysis of microarray gene expression data. Comp Stat Data Anal. 2002;39:1–20. [Google Scholar]
- 31.Ando T, Ichimaru Y, Konjiki F, Shoji M, Komaki G. Variations in the preproghrelin gene correlate with higher body mass index, fat mass, and body dissatisfaction in young Japanese women. Am J Clin Nutr. 2007;86:25–32. doi: 10.1093/ajcn/86.1.25. [DOI] [PubMed] [Google Scholar]
- 32.Hinney A, Hoch A, Geller F, Schafer H, Siegfried W, Goldschmidt H, Remschmidt H, Hebebrand J. Ghrelin gene: Identification of missense variants and a frameshift mutation in extremely obese children and adolescents and healthy normal weight students. J Clin Endocrinol Metab. 2002;87:2716. doi: 10.1210/jcem.87.6.8672. [DOI] [PubMed] [Google Scholar]
- 33.Jo DS, Kim SL, Kim SY, Hwang PH, Lee KH, Lee DY. Preproghrelin leu72met polymorphism in obese Korean children. J Pediatr Endocrinol Metab. 2005;18:1083–1086. doi: 10.1515/jpem.2005.18.11.1083. [DOI] [PubMed] [Google Scholar]
- 34.Kuzuya M, Ando F, Iguchi A, Shimokata H. Preproghrelin leu72met variant contributes to overweight in middle-aged men of a Japanese large cohort. Int J Obes (Lond) 2006;30:1609–1614. doi: 10.1038/sj.ijo.0803296. [DOI] [PubMed] [Google Scholar]
- 35.Mager U, Lindi V, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Tuomilehto J, Laakso M, Pulkkinen L, Uusitupa M. Association of the leu72met polymorphism of the ghrelin gene with the risk of type 2 diabetes in subjects with impaired glucose tolerance in the Finnish diabetes prevention study. Diabet Med. 2006;23:685–689. doi: 10.1111/j.1464-5491.2006.01870.x. [DOI] [PubMed] [Google Scholar]
- 36.Miraglia del Giudice E, Santoro N, Cirillo G, Raimondo P, Grandone A, D'Aniello A, Di Nardo M, Perrone L. Molecular screening of the ghrelin gene in Italian obese children: The leu72met variant is associated with an earlier onset of obesity. Int J Obes Relat Metab Disord. 2004;28:447–450. doi: 10.1038/sj.ijo.0802572. [DOI] [PubMed] [Google Scholar]
- 37.Steinle NI, Pollin TI, O'Connell JR, Mitchell BD, Shuldiner AR. Variants in the ghrelin gene are associated with metabolic syndrome in the Old Order Amish. J Clin Endocrinol Metab. 2005;90:6672–6677. doi: 10.1210/jc.2005-0549. [DOI] [PubMed] [Google Scholar]
- 38.Ukkola O, Ravussin E, Jacobson P, Perusse L, Rankinen T, Tschop M, Heiman ML, Leon AS, Rao DC, Skinner JS, Wilmore JH, Sjostrom L, Bouchard C. Role of ghrelin polymorphisms in obesity based on three different studies. Obes Res. 2002;10:782–791. doi: 10.1038/oby.2002.106. [DOI] [PubMed] [Google Scholar]
- 39.Vartiainen J, Kesaniemi YA, Ukkola O. Sequencing analysis of ghrelin gene 5′ flanking region: Relations between the sequence variants, fasting plasma total ghrelin concentrations, and body mass index. Metabolism. 2006;55:1420–1425. doi: 10.1016/j.metabol.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 40.Vivenza D, Rapa A, Castellino N, Bellone S, Petri A, Vacca G, Aimaretti G, Broglio F, Bona G. Ghrelin gene polymorphisms and ghrelin, insulin, IGF-1, leptin and anthropometric data in children and adolescents. Eur J Endocrinol. 2004;151:127–133. doi: 10.1530/eje.0.1510127. [DOI] [PubMed] [Google Scholar]
- 41.Franklin RD, Allison DB. The rev.: An IBM BASIC program for Bayesian test interpretation. Behav Res Methods Instruments Computers. 1992;24:491–492. [Google Scholar]
- 42.Hoffman EB, Sen PK, Weinberg CR. Within-cluster resampling. Biometrika. 2001;88:1121–1134. [Google Scholar]
- 43.Follmann D, Proschan M, Leifer E. Multiple outputation: Inference for complex clustered data by averaging analyses from independent data. Biometrics. 2003;59:420–429. doi: 10.1111/1541-0420.00049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials