Abstract
Fanconi Anemia (FA) is a chromosome instability syndrome characterized by congenital abnormalities, cellular hypersensitivity to DNA crosslinking agents, and heightened cancer risk. Eight of the thirteen identified FA genes encode subunits of a nuclear FA core complex that monoubiquitinates FANCD2 and FANCI to maintain genomic stability in response to replication stress. The FA pathway has been implicated in the regulation of error-prone DNA damage tolerance via an undefined molecular mechanism. Here, we show that the FA core complex is required for efficient spontaneous and UVC-induced point mutagenesis, independently of FANCD2 and FANCI. Consistent with the observed hypomutability of cells deficient in the FA core complex, we also demonstrate that these cells are impaired in the assembly of the error-prone translesion DNA synthesis polymerase Rev1 into nuclear foci. Consistent with a role downstream of the FA core complex and like known FA proteins, Rev1 is required to prevent DNA crosslinker-induced chromosomal aberrations in human cells. Interestingly, PCNA monoubiquitination, known to contribute to Rev1 recruitment, does not require FA core complex function. Our results suggest a role for the FA core complex in regulating Rev1-dependent DNA damage tolerance independently of FANCD2, FANCI, and PCNA monoubiquitination.
1. INTRODUCTION
Fanconi anemia (FA) is a multigenic autosomal recessive and X-linked chromosome instability syndrome characterized by congenital abnormalities, progressive bone marrow failure, and elevated cancer susceptibility (1,2,3). At the cellular level, FA cells exhibit a marked hypersensitivity to DNA crosslinking agents such as cisplatin (CDDP) (4) and mitomycin C (MMC) (5), implicating the FA pathway in DNA repair. Somatic cell fusion studies and biochemical analyses have led to the identification of thirteen FA genes. The common cross-linker hypersensitivity of FA cells from distinct complementation groups and the characteristic clinical phenotype of FA patients suggest that FA proteins cooperate in a common DNA damage response pathway.
DNA damage detected during the synthesis (S) phase of the cell cycle triggers the activation of a nuclear complex of eight FA proteins (A, B, C, E, F, G, L and M) and two additional protein sub-units, FAAP100 and FAAP24, which have not been heretofore assigned to an FA complementation group (1,6,7). The activated FA core complex, which possesses E3 ubiquitin ligase activity, promotes the monoubiquitination of FANCD2 and FANCI (8,9). Monoubiquitinated FANCD2 and FANCI are subsequently localized into chromatin-associated foci containing DNA replication and repair factors such as BRCA2, Rad51, NBS1, and PCNA (10, 11). FA proteins, including the downstream components in the pathway, BRCA2/FANCD1 (12, 13), BRIP1/FANCJ (14) and PALB2/FANCN (15), are required for efficient homologous recombination (HR) repair (16,17).
Several lines of evidence also implicate FA proteins in the regulation of error-prone translesion synthesis (TLS), a DNA damage tolerance mechanism that allows the replicative bypass of damaged nucleotides (18). TLS is also an obligatory step in DNA inter-strand crosslink repair (19), and is believed to function analogous to replication-coupled damage bypass in this context (20). FA pathway-deficient patient-derived and chinese hamster (CHO) cells were found to be hypomutable in an HPRT mutagenesis assay, with a reduced frequency of point mutations and a concomitant increase in deletion and insertion mutagenesis (21,22). Furthermore, chicken DT40 cells deficient in FANCC are hypomutable at the endogenous IgM locus, (23) suggesting that the FA pathway is required for efficient spontaneous point mutagenesis. Whether all FA proteins, or only a subset, participate in TLS regulation remains unknown.
The capacity to bypass DNA damage during TLS resides in specialized DNA polymerases with unconstrained active sites that can accommodate damaged nucleotides as templates for DNA replication (24). Consequently, TLS polymerases can be error-prone, exhibiting low fidelity on undamaged and damaged DNA. Particularly important for mutagenic TLS is the Rev1 enzyme, a dCMP nucleotidyl transferase belonging to the Y family of TLS polymerases (25). Rev1 is recruited to sites of stalled DNA replication via its direct interaction with monoubiquitinated proliferating cell nuclear antigen (PCNA), the processivity clamp for DNA polymerases (26,27). Following DNA damage, PCNA is monoubiquitinated by the RAD6/RAD18 E2/E3 enzyme complex (28,29), and recruits Rev1 via the C-terminal ubiquitin binding motif (UBM) of Rev1 (27,30). The N-terminal BRCT domain of Rev1 is also required for its optimal interaction with monoubiquitinated PCNA and for mutagenesis in vivo (26,31). However, abrogation of PCNA monoubiquitination by genetic disruption of RAD18 results in a partial but not complete loss of Rev1-dependent DNA damage tolerance, suggesting that mechanisms independent of PCNA monoubiquitination could regulate Rev1 function (32,33).
How the FA pathway regulates error-prone TLS is unknown, but the mechanism appears to require downstream Rev1 activity. FANCC has been shown to be epistatic to Rev1 for cisplatin hypersensitivity in chicken DT40 cells and both FANCC and Rev1-deficient DT40 cells demonstrate hypomutability (23), suggesting that the FA pathway may regulate Rev1 function to promote efficient DNA crosslink repair and point mutagenesis.
To investigate the role of the FA pathway in error-prone TLS, we have deployed a shuttle-vector based mutagenesis assay to measure the mutation frequencies of FA patient-derived human cells of several genetic complementation groups. Our results demonstrate that the FA core complex subunits FANCA and FANCG are required for efficient point mutagenesis, whereas FANCD2, the downstream substrate for monoubiquitination, is not. Moreover, the recruitment of the error-prone TLS polymerase Rev1 into nuclear foci depends on an intact FA core complex. Interestingly, FA core complex-dependent Rev1 recruitment requires the BRCT domain of Rev1 and is independent of its UBM domain. Finally, we show that Rev1-depleted human cells demonstrate a characteristic FA phenotype-namely, crosslinker-induced DNA breaks and radial chromosomes. Our results indicate an important role for the FA core complex in regulating Rev1-dependent DNA damage tolerance.
2. MATERIALS AND METHODS
2.1. Cell lines and culture conditions
Fanconi Anemia-deficient and corrected patient-derived cell lines, PD326, GM6914, PD20 and Rad18-deficient mouse lung fibroblasts have been previously described (8,34). All cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 15% fetal calf serum, glutamine, and penicillin-streptomycin.
2.2. Antibodies and immunoblotting
Western blots were performed on whole-cell extracts separated on Nupage 3–8% Tris-Acetate or 4–12% Bis-Tris gradient gels (Invitrogen). Anti-GFP (JL-8) was obtained from BD Biosciences, anti-FANCD2 (FI-17), and anti-PCNA (PC10) from Santa Cruz Biotechnology.
2.3. siRNA and transfection
Synthetic siRNA oligonucleotides were used against the following target sequences:
human FANCA-5′-CAGCGTTGAGATATCAAAGAT-3′
human FANCD2-5′-TTGGAGGAGATTGATGGTCTA-3′
human RAD6-5′-AAGCGTGTTTCTGCAATAGTA
human RAD18-5′-CCCGAGGTTAATGTAGTTGTT-3′
SiRNAs were transfected using HiPerFect (Qiagen) and plasmid DNA using GeneJuice (Novagen), Fugene6 (Roche) or Effectene (Qiagen) according to the manufacturer’s protocol.
2.4. SupF Mutagenesis Assay
Fanconi Anemia-deficient and corrected patient-derived cell lines were transfected with either undamaged or UVC irradiated (254 nm radiation using a 2400 UV Stratalinker from Stratagene) pSP189 plasmid using GeneJuice (Novagen) or Effectene (Qiagen). Forty-eight hours post transfection, plasmid DNA was retrieved using a mini-prep kit (Promega), Dpn1 digested to remove unreplicated plasmid and ethanol precipitated. Recovered pSP189 plasmid was electroporated into the MBM7070 indicator bacterial strain containing an amber mutation in the β-galactosidase gene. Transformed bacteria were plated onto agar plates containing 100 μg/ml ampicillin, 1mM isopropyl-1-thio-b-D-galactopyranoside (IPTG) and 100 μg/ml X-gal. The ratio of white (mutant) to total (blue+white) colonies is the mutation frequency. The mutant colonies were re-streaked for verification, subjected to plasmid mini-prep (Qiagen) and sequenced using an automated DNA sequencer.
2.5. Fluoresence microscopy
Fluorescence microscopy on adherent cells was performed according to previously described protocols (27). Briefly, FA-deficient and corrected patient-derived cell lines or siRNA treated Hela cells were transfected with eGFP-mRev1 or eGFP-Poleta in 4-well chamber slides. Twenty hours post transfection, cells were exposed to 10J/m2 UVC radiation and 8–16 hours later, fixed with 4% paraformaldehyde.
2.6. Synthetic lethality and cell viability assay
FANCG-deficient patient-derived fibroblasts (PD326) and their cDNA corrected counterparts were treated with control, Rad6, or Rad18 specific siRNA as previously described (35). Seventy-two hours after transfection, cell viability was measured using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega) according to the manufacturer’s protocol. Mean cell viability and SEM were calculated from three independent experiments and plotted using GraphPad Prism version 3 (GraphPad Software).
3. RESULTS
3.1. The FA core complex, but not FANCD2, is required for point mutagenesis in the supF assay
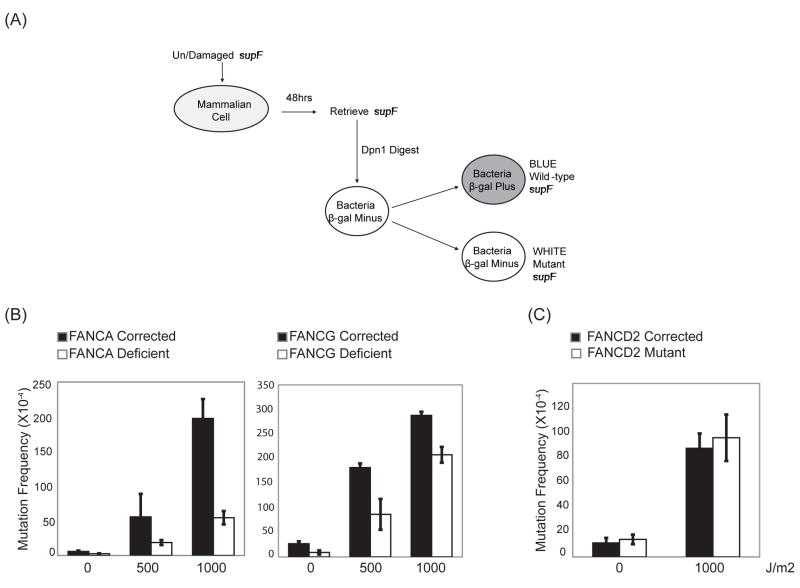
The FA core complex consists of eight protein subunits assigned to FA complementation groups: FANC-A, B, C, E, F, G, L, M, as well as FAAP24, and FAAP100 (1,36,7). Each subunit, with the exception of FAAP24, is required for the integrity of the entire complex (3). FANCA- (GM6914), FANCG- (PD326) and FANCD2- (PD20) deficient cell lines are patient-derived mutant cell lines that have previously been corrected with cDNA expression constructs thereby generating isogenic GM6914+FANCA, PD326+FANCG, and PD20+FANCD2 cell lines, respectively (8) (Table II). To investigate the involvement of the FA core complex in error-prone TLS, we subjected these FA patient-derived cells to the supF shuttle vector-based mutagenesis assay that allows SV40 T antigen-dependent replication of the supF plasmid in human cells (37,38,39) (Fig. 1A). As this assay favors point mutations for unirradiated as well as UVC-irradiated plasmids, it serves as a surrogate measure of error-prone TLS in vivo.
Table II.
Cell lines analyzed for supF mutagenesis and Rev1 foci assembly
| Cell Line | Genotype | supF Mutagenesis | Rev1 Foci |
|---|---|---|---|
| GM6914+FANCA | Wild-type | Normal | Normal |
| GM6914+Vector | FANCA Deficient | Hypomutable | Deficient |
| PD326+FANCG | Wild-type | Normal | Normal |
| PD326+Vector | FANCG Deficient | Hypomutable | Deficient |
| PD20+FANCD2 | Wild-type | Normal | N/A |
| PD20+Vector | FANCD2-Deficient | Normal | N/A |
| HEK293T+FANCD2 siRNA | FANCD2-Deficient | Normal | Normal |
| HEK293T+FANCI siRNA | FANCI-Deficient | Normal | Normal |
Figure 1. FANCA- and FANCG-, but not FANCD-deficient cells, are hypomutable in the supF assay.
(A) Schematic representation of the supF assay used to investigate the mutation frequency of various Fanconi Anemia cell lines. Undamaged or UVC-irradiated supF plasmid pSP189 is transfected into a cell line of interest, recovered 48 hrs post-transfection by alkaline lysis, Dpn1 digested, and electroporated into the MBM7070 indicator bacterial strain. The ratio of white to total number of colonies is the mutation frequency. (B) FANCA- (GM6914), FANCG- (PD326) and (C) FANCD2- (PD20) deficient patient-derived fibroblasts and their cDNA corrected counterparts were analyzed in the supF assay as described in (A). Error bars indicate the standard deviation of the mean between three independent experiments.
Patient-derived FANCA- and FANCG-deficient cells were hypomutable for both spontaneous and UVC-induced mutagenesis in comparison to their cDNA corrected counterparts (Fig. 1B). For instance, in the absence of UVC irradiation of the supF plasmid, FANCG-corrected cells demonstrated a mutation frequency of 27 X 10−4 versus 9 X 10−4 in FANCG-deficient cells (p=0.008). Upon 500 J/m2 of UVC irradiation of the supF plasmid, FANCG-corrected cells showed a mutation frequency of 181 X 10−4 compared to 87 X 10−4 in FANCG-deficient cells (p=0.02). This was not due to differences in DNA replication rates as FANCG-deficient and corrected cells demonstrate comparable rates of BrdU incorporation during S phase as previously described (35). Furthermore, similar amounts of plasmid DNA were retrieved from all cell lines analyzed, suggesting comparable replication rates of the supF plasmid.
The best described function of the FA core complex is the monoubiquitination of FANCD2 and FANCI (8,9). Next, we measured the mutation frequencies in FANCD2-mutant patient-derived cells compared to their cDNA corrected counterparts (Fig. 1C). Surprisingly, unlike FANCA and FANCG, FANCD2 did not appear to be required for mutagenesis in the supF assay. Also, 293T cells treated with FANCI-specific siRNA were proficient for point mutagenesis in this assay (data not shown). As FANCD2-mutant cells have been shown to be defective in FANCI ubiquitination (9), these data collectively suggest that the FA core complex is required for efficient point mutagenesis independently of FANCD2 and FANCI.
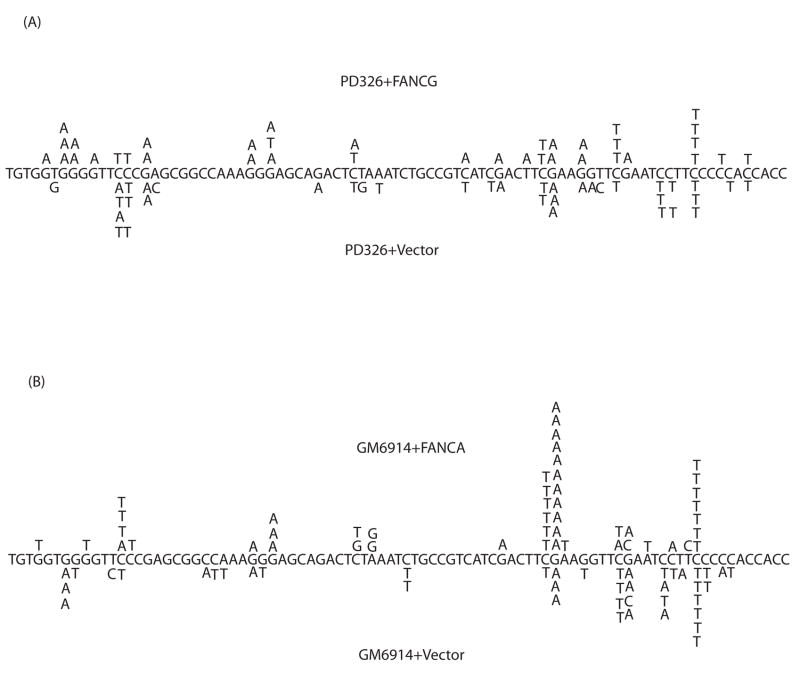
Next, UVC-irradiated supF plasmids that underwent replication in FA mutant and corrected cells were sequenced (56 clones each from FANCA-deficient and corrected cells, and 32 and 34 mutants from FANCG-deficient and corrected cells, respectively). Point mutations predominated the mutation spectrum with C/G to T/A transitions being the most prevalent (Table I and Fig. 2) in accordance with previous studies (38). The mutation spectrum between FA core complex-deficient and corrected cells was relatively unchanged (Table I and Fig. 2) suggesting that FA core complex-deficient cells show a general, rather than specific, impairment in point mutation frequency.
Table I.
UVC mutation spectrum of supF plasmids retrieved from Fanconi Anemia-deficient and cDNA corrected patient-derived cells
| GM6914 (FANCA-deficient) | PD326 (FANCG-deficient) | |||
|---|---|---|---|---|
| + FANCA | + Vector | + FANCG | + Vector | |
| C/G>T/A | 67.9% | 53.6% | 82.9% | 50% |
| Other Point Mutations | 23.2% | 17.9% | 5.7% | 10% |
| Multiple Point Mutations | 8.9% | 26.8% | 5.7% | 23.3% |
| Complex Mutations | 0% | 1.8% | 5.7% | 13.3% |
| Frameshifts | 0% | 0% | 0% | 3.3% |
Figure 2. Mutation spectra analysis of UVC-irradiated supF plasmids from FA-deficient and corrected cells.
UVC-irradiated mutant supF plasmids retrieved from (A) FANCA-deficient and corrected and (B) FANCG-deficient and corrected patient-derived fibroblasts were sequenced. Only mutations within the structural supF gene are shown.
3.2. The FA core complex is required for efficient Rev1 foci assembly
The majority of DNA damage-induced mutagenesis is a result of error-prone translesion DNA synthesis (24). Error-prone TLS, in turn, depends on specialized DNA polymerases: mainly, the deoxycytidyl (dCMP) transferase Rev1 and DNA polymerase ζ consisting of the Rev3/Rev7 heterodimer. As these TLS enzymes are mutagenic, their access to the primer-template junction is thought to be tightly regulated to prevent uncontrolled mutagenesis. In chicken DT40 cells, FANCC of the FA core complex was found to be epistatic to Rev1 and Rev3 for cisplatin sensitivity (23) although the mechanism by which FANCC might regulate TLS polymerase function remains unknown.
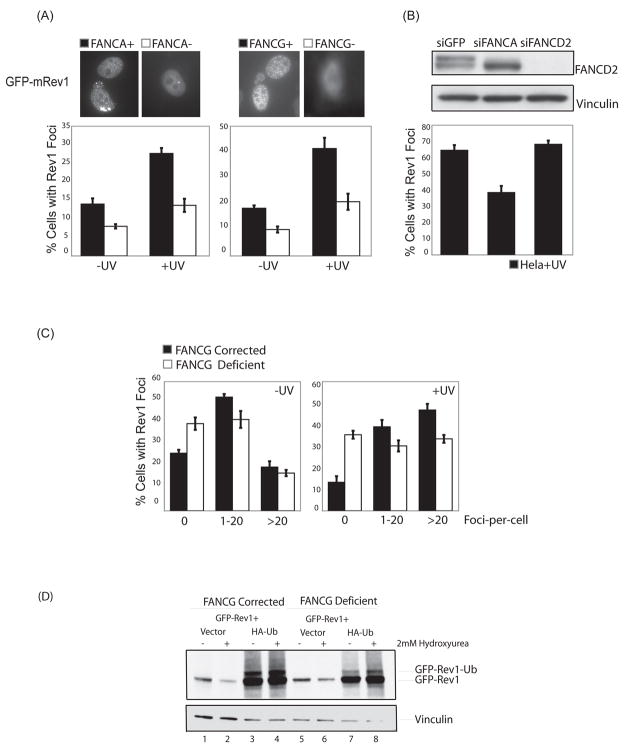
We tested whether FANCA and FANCG are required for the recruitment of Rev1 to spontaneous and DNA damage-induced nuclear foci (26,27) (Fig. 3 and Supplementary Fig. 1). FANCA- and FANCG-deficient patient-derived fibroblasts and their cDNA-complemented counterparts were transfected with murine eGFP-Rev1, damaged with 10J/m2 of UVC radiation and analyzed 8hrs later as previously described (26). Interestingly, in both FANCA- and FANCG-deficient cells, the efficiency of Rev1 foci formation was significantly reduced compared to their isogenic wild-type cellular counterparts (Fig. 3A). In the absence of exogenous DNA damage, ~20% of FANCG-corrected versus ~10% of FANCG-deficient cells demonstrated greater than five eGFP-Rev1 foci-per-cell (p=0.006) which increased to ~40% versus ~20% after UVC treatment respectively (p=0.01) (Fig. 3A). Similar results were obtained for FANCA-deficient and corrected cells. The efficiency of foci formation for the relatively error-free TLS polymerase, pol eta, on the other hand, was comparable between FA core complex-deficient and corrected cells (data not shown). Importantly, the expression of ectopically-expressed eGFP-Rev1 was comparable between FANCG-deficient and corrected cells (Fig. 3D, lanes 2 and 6). Also, consistent with the data in Figure 1C, FANCD2 siRNA treated Hela cells were proficient in eGFP-Rev1 foci formation whereas FANCA siRNA treated cells were not (Fig. 3B).
Figure 3. The FA core complex is required for efficient Rev1 foci assembly.
(A) FANCA- (GM6914) and FANCG- (PD326) deficient patient-derived fibroblasts and their cDNA corrected counterparts, and (B) siRNA-treated Hela cells were transiently transfected with murine eGFP-Rev1. 20 hours post-transfection, cells were treated with 10J/m2 of UVC radiation and 16 hours later, fixed with paraformaldehyde and analyzed via fluorescence microscopy. The ratio of cells with greater than five foci to cells with eGFP-Rev1 expression is indicated on the Y axis. Error-bars represent standard error between three independent transfections. The foci counts were conducted blind to reduce observer bias. (C) FANCG-deficient patient-derived cells (PD326) and their cDNA corrected counterparts were analyzed for eGFP-Rev1 distribution in terms of foci-per-cells as in (A). (D) FANCG-deficient and corrected cells were transiently transfected with murine eGFP-Rev1 with or without HA-Ub-encoding plasmid DNA, treated with 2mM hydroxyurea for 12 hrs, and analyzed by western blotting with an anti-GFP antibody.
Next, we investigated the distribution of eGFP-Rev1 in terms of foci-per-cell in FANCG-deficient versus corrected cells (Fig. 3C and Supplementary Fig. 1C). A significantly greater proportion of FANCG-deficient cells showed a complete absence of Rev1 foci (0 foci-per-cell) compared to FANCG-corrected wild-type cells before and after DNA damage (p=0.03 before UVC and 0.005 after UVC treatement). After UVC radiation, FANCG-deficient cells were impaired in the induction of eGFP-Rev1 foci across two distribution categories (1–20 and >20 foci-per-cell) compared to FANCG-corrected wild-type cells. For example, after UVC treatment, ~50% of FANCG-corrected versus ~30% of FANCG-deficient cells demonstrated >20 foci-per-cell (p=0.004). These data indicate that the FA core complex is required for the efficient loading of Rev1 into nuclear foci in response to replication fork arrest, thereby suggesting a possible molecular mechanism for FA core complex-mediated point mutagenesis.
Rev1 itself is monoubiquitinated (27), although the E3 ubiquitin ligase responsible for this modification and its functional relevance are unknown. Since Rev1 has a UBM domain, it may also undergo E3 ligase-independent monoubiquitination, as recently described (40). Since FANCG and FANCA were required for efficient Rev1 foci formation, we asked whether the FA core complex, given its E3 ubiquitin ligase activity, might regulate the monoubiquitination of Rev1. FANCG-deficient and corrected cells were transfected with either murine eGFP-Rev1 alone or in combination with a plasmid encoding HA-tagged ubiquitin (HA-Ub), which is essential to visualize clear monoubiquitination of Rev1 (27) (Fig. 3D). In the presence of exogenous ubiquitin, Rev1 was monoubiquitinated both in the absence and presence of DNA damage as previously observed (27). Moreover, the ubiquitination of Rev1 occurred independently of FANCG. These data suggest that the FA core complex regulates Rev1 localization into foci without affecting its ubiquitination status.
3.3. The FA core complex and Rad18 function in compensatory DNA damage response pathways
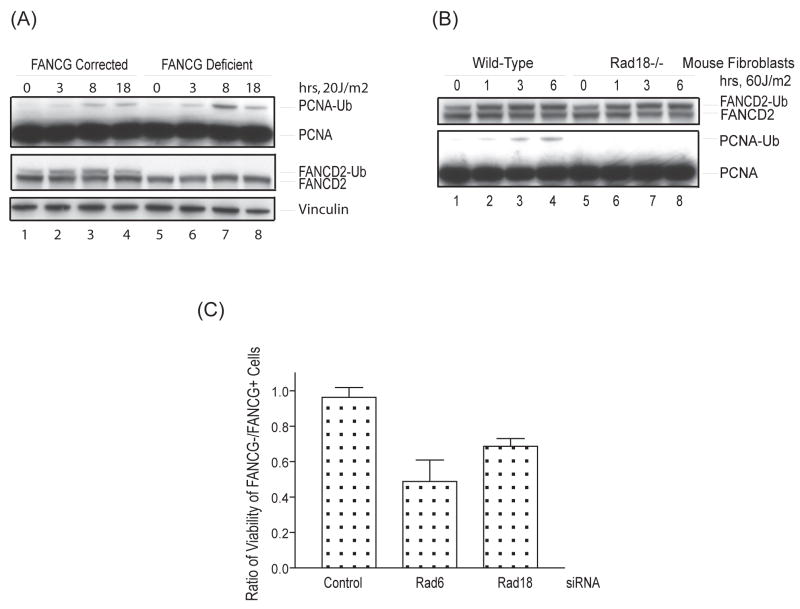
PCNA monoubiquitination by the Rad6/Rad18 E2/E3 enzyme complex is pivotal in regulating the access of Y family TLS polymerases, including Rev1, to sites of stalled DNA replication (28,29). Ubiquitin binding domains of the UBM or UBZ class in Y family polymerases engage monoubiquitinated PCNA at arrested replication forks, allowing for a transient bypass of DNA lesions (30). Therefore, we examined the state of monoubiquitinated PCNA in FANCG-deficient cells (Fig. 4). In response to UVC radiation, PCNA monoubiquitination occurred normally, and was even slightly elevated at certain time points, in FANCG-deficient versus corrected cells (Fig. 4A, lanes 3 and 7). Furthermore, Rad18, the E3 ubiquitin ligase for PCNA, did not appear to be required for the activation of the FA pathway. Rad18 mutant murine fibroblasts (34) were capable of supporting FANCD2 monoubiquitination in response UVC irradiation although PCNA ubiquitination was abolished, as previously described (41).
Figure 4. The FA core complex and Rad18 function in parallel DNA damage response pathways.
(A) FANCG-deficient patient-derived fibroblasts (PD326) and their cDNA corrected counterparts were treated with 20J/m2 of UVC and harvested at various time points, followed by western blotting with anti-PCNA, anti-FANCD2, and anti-vinculin antibodies. (B) Rad18-mutant versus wild-type murine fibroblasts were treated with 60J/m2 UVC and harvested at various time points, followed by western blotting with anti-PCNA and anti-mouse FANCD2 antibodies. (C) FANCG-deficient patient-derived fibroblasts (PD326) and their cDNA corrected counterparts were treated with control, Rad6-, or Rad18-specific siRNAs for 72 hours. Cell survival was assessed with a CellTiter-Glo Luminescent Cell Viability Assay kit (Promega). The ratio of cell survival in FANCG-deficient versus corrected cells is indicated. The error-bars indicate standard error between three independent experiments.
Rad18 and FANCC of the FA core complex have been shown to independently regulate cisplatin sensitivity and sister chromatid exchange levels in chicken DT40 cells (42). To test the hypothesis that PCNA monoubiquitination and the FA core complex function as compensatory DNA damage response pathways in human cells, we knocked down Rad6 or Rad18 in FANCG-deficient and corrected isogenic cells with siRNAs from a validated Qiagen siRNA library (Fig. 4C and Supplementary Fig. 2). As predicted, Rad6 and Rad18 siRNAs were significantly more toxic to FANCG-deficient compared to FANCG-corrected cells, specifically 0.96+/−0.06 for the control versus 0.49+/−0.12 and 0.69+/−0.04 for Rad6 and Rad18 siRNAs respectively. We surmise that Rad6/Rad18-mediated PCNA monoubiquitination and the FA core complex function in parallel compensatory pathways to promote endogenous DNA damage tolerance and cell survival.
3.4. FA core complex-mediated Rev1 recruitment requires the BRCT domain of Rev1
The Rev1 polypeptide has an N-terminal BRCT domain and a C-terminal ubiquitin binding UBM domain (Fig. 5A). Recent studies indicate that the UBM domain of Rev1 is required for its recruitment to monoubiquitinated PCNA at sites of DNA damage and for its functional interaction with monoubiquitinated PCNA in vitro (27,43). Although the BRCT domain is also required for damage-inducible Rev1 foci formation and PCNA binding, (26) a BRCT domain mutant of Rev1 is still stimulated by monoubiquitinated PCNA for TLS activity in vitro (43). How the BRCT domain of Rev1 interacts with PCNA remains unclear.
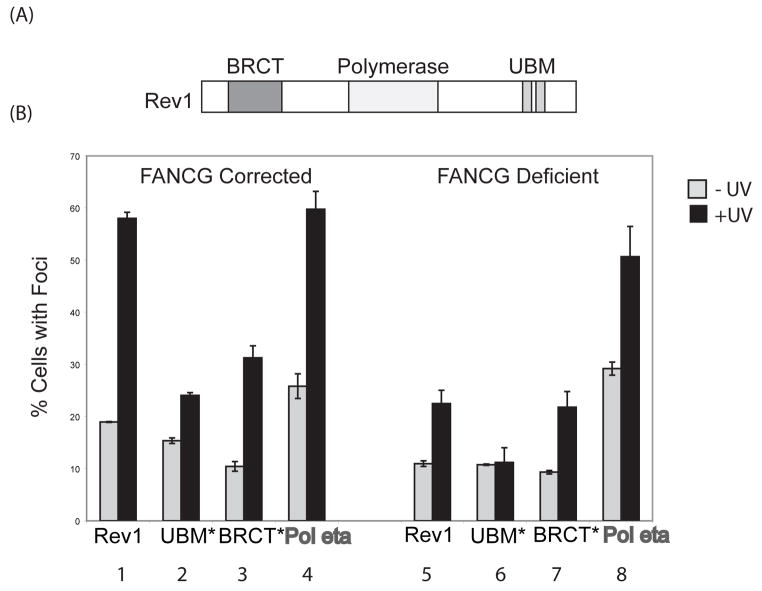
Figure 5. The FA core complex-mediated assembly of Rev1 nuclear foci requires the BRCT domain of Rev1.
(A) Schematic of the Rev1 polypeptide domain structure. (B) Bar graphs quantifying cells with eGFP-Rev1 foci. FANCG-deficient patient-derived fibroblasts (PD326) and corrected cells were transiently transfected with wild-type or mutant murine eGFP-Rev1 cDNAs. 20hrs post transfection, cells were treated with 10J/m2 UVC radiation and 8 hrs later, fixed with paraformaldehyde and analyzed by fluorescence microscopy. The ratio of cells with greater than five eGFP-Rev1 foci to cells with eGFP-Rev1 expression is indicated on the Y axis. At least 300 cells were counted for each data point. Error-bars represent the standard error between three independent experiments.
We transfected cDNAs encoding either the wildtype, BRCT mutant, or UBM mutant of murine Rev1 into FA-deficient and corrected cells for immunofluorescence analysis (Fig. 5B). FANCG-deficient cells displayed decreased UVC-induced Rev1 foci, as compared to FANCG corrected cells (Fig. 5B. Bars 1 and 5). A Rev1 UBM domain mutant was defective in UVC-induced foci formation in FANCG-corrected (wild-type) cells, as previously reported (27) (Fig. 5B. Bars 1 and 2). Disruption of the Rev1 UBM domain combined with a loss of the FA core complex resulted in a further decline in UVC-inducible Rev1 foci formation, suggesting that the FA core complex and the UBM domain of Rev1 independently regulate its recruitment (Fig. 5B. Bars 5 and 6). Disruption of the BRCT domain of Rev1 demonstrated impaired Rev1 foci in FANCG-corrected cells (Fig. 5B. Bars 1 and 3). Notably, disruption of the BRCT domain, combined with a loss of the FA core complex, resulted in no further loss of Rev1 foci, indicating that the BRCT domain of Rev1 is epistatic to FANCG for Rev1 foci formation (Fig. 5B. Bars 5 and 7). Taken together, these results suggest that the FA core complex promotes the assembly of Rev1 foci through an interaction, either direct or indirect, with the BRCT domain of Rev1.
3.5. Rev1 depleted cells demonstrate mitomycin C-induced chromosomal aberrations
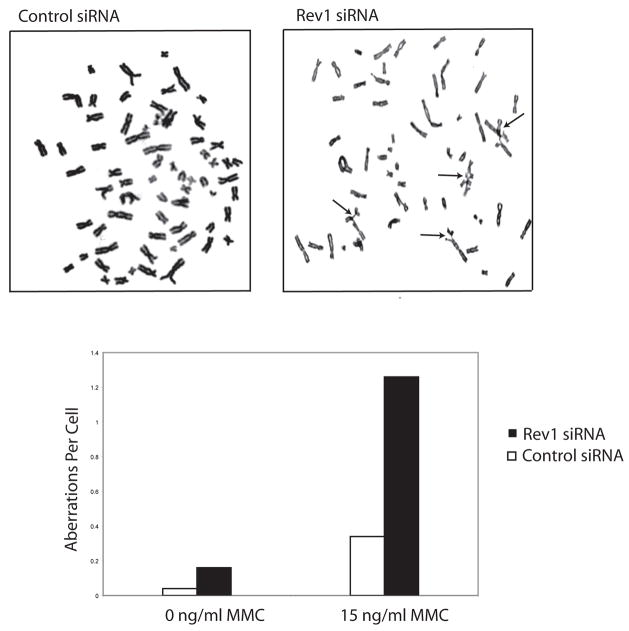
A characteristic feature of FA patient-derived cells that is utilized to diagnose the disease is chromosome breakage and radial formation upon exposure to the DNA interstrand crosslinking agent, mitomycin C (MMC) (3). Rev1-deficient chicken DT40 cells demonstrate cytotoxicity and chromosomal aberrations in response to the DNA crosslinking agent, cisplatin (23). We investigated the phenotype of Rev1 siRNA-treated human cells in a MMC-induced chromosome breakage test (Fig. 6). Analogous to FA patient-derived cells, Rev1 depleted HEK293T cells showed increased chromosome breakage and radial formation in response to 15 ng/ml of MMC compared to control siRNA treated cells. These results show that Rev1, like the known FA proteins, is required for genomic stability in response to DNA interstrand crosslinking agents in human cells.
Figure 6. Rev1 deficient cells display MMC-induced chromosomal aberrations.
HEK293T cells were treated with control or Rev1 specific siRNAs for 72 hrs, exposed to 15 ng/ml MMC for 48 hrs and analyzed for chromosomal aberrations.
4. DISCUSSION
The FA pathway is a DNA damage response pathway that has been implicated in the coordination of homologous recombination and translesion synthesis to maintain genomic stability in response to replication stress (23,44). Our results support a role for the FA pathway in TLS regulation. Interestingly, our data demonstrate that FA core complex-deficient (FANCG and FANCA), but not FANCD2-deficient cells, have reduced mutation efficiency and reduced Rev1 foci. This suggests that the FA core complex functions independently of its known ubiquitination substrates, FANCD2 and FANCI. The complex appears to regulate mutagenic TLS by channeling DNA lesions into damage bypass pathways and preventing more deleterious mutations, such as large insertions and deletions. Consistent with this model, FA cells are prone to an increased deletion frequency at the HPRT locus (45,22). Although FA core complex-deficient cells do not exhibit marked UVC hypersensitivity in terms of cytotoxicity (46), our results show that they are impaired in UVC-induced mutagenesis. The lack of UVC sensitivity in FA cells may be reflective of the presence of multiple specialized polymerases that can compensate for one another to allow DNA damage bypass, as well as intact nucleotide excision repair in FA core complex-deficient cells (47). Although Rev1-deficient chicken DT40 cells are sensitive to UVC in terms of cytotoxicity and chromosome breakage (48), human cells depleted for Rev1 are hypomutable, but not sensitive to the cytotoxic effects of UVC radiation (49,50). Whilst this may be a function of insufficient Rev1 knockdown by siRNA, it demonstrates that reduced levels of Rev1 can substantially impair mutagenesis without impacting UVC-induced cell survival, suggesting that these processes may be uncoupled.
Emerging evidence has demonstrated that the FA core complex functions in DNA repair independently of FANCD2 monoubiquitination (51). In accordance with this finding, we demonstrate that, in contrast to FANCA and FANCG disruption, FANCD2 deficiency results in normal levels of point mutagenesis, suggesting that for error-prone TLS regulation, the FA pathway is branched (Fig. 7).
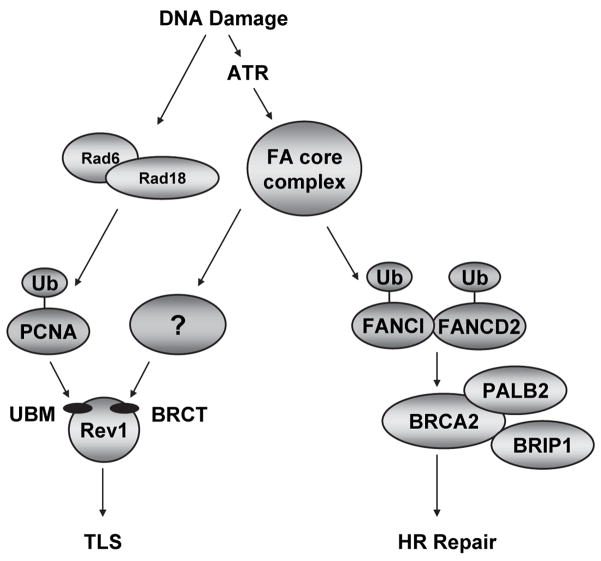
Figure 7. A model depicting the regulation of Rev1-dependent DNA damage tolerance by the FA core complex, independently of FANCD2 and FANCI.
Schematic representation of FA core complex-mediated regulation of Rev1. The FA core complex plays a well known role in monoubiquitinating FANCD2 and FANCI, leading to HR repair. The complex is also required for efficient Rev1 foci assembly. The mechanism underlying FA core complex-controlled regulation of Rev1 localization is unknown, but it appears to be independent of PCNA and Rev1 monoubiquitination as well as the UBM domain of Rev1 but requires the BRCT domain of Rev1.
To understand the molecular mechanism by which the FA core complex promotes point mutagenesis in response to replication fork arrest, we have investigated the role of FA proteins in the localization of error-prone Y family TLS polymerase, Rev1. Rev1 is required for spontaneous and UVC-induced point mutagenesis and is thought to play a structural rather than catalytic role in TLS (32,52). Our results show that Rev1 localization in spontaneous and DNA damage-induced foci requires an intact FA core complex, but not FANCD2. Therefore, there appears to be a correlation between point mutagenesis efficiency and Rev1 foci formation. Although FA core complex-deficient cells exhibit impaired Rev1 foci formation, they are proficient for the localization of Y family TLS polymerase eta into foci. As Rev1 was shown to interact with other TLS polymerases of the Y family (eta, iota and kappa) via its C-terminus (53), this suggests that Rev1 assembly into nuclear foci is not necessary for pol eta localization.
Although important for Rev1 regulation, PCNA monoubiquitination by Rad18 is not the sole determinant of Rev1 foci formation. The FA core complex, although required for Rev1 localization, does not regulate PCNA or Rev1 monoubiquitination. Furthermore, the combined deletion of the FA core complex subunit FANCG and Rad18 results in synthetic lethality, indicating that the FA core complex and Rad18 function in parallel, compensatory pathways (Fig. 7).
The N-terminal BRCT and C-terminal UBM domains of Rev1 participate in its localization at sites of stalled DNA replication. The BRCT domain has been studied extensively in yeast, and more recently in mammalian systems and has been shown to be required for Rev1-dependent DNA damage tolerance in vivo (31,26,54). The FA core complex is synergistic with the UBM domain but epistatic to the BRCT domain of Rev1 for efficient Rev1 foci formation. This suggests that the FA core complex, either directly or indirectly, regulates the BRCT domain-mediated loading of Rev1 into replication factories. Given that the BRCT domain of Rev1 promotes its interaction with PCNA, we have tested whether the FA core complex might promote the chromatin loading of unmodified and/or monoubiquitinated PCNA and thereby facilitate Rev1 binding. We observed normal or elevated PCNA chromatin localization in FA core complex-deficient cells (data not shown). Moreover, BRCT domains have been described as phospho-peptide binding modules (55,56). Several FA core complex proteins are phosphorylated in response to DNA damage and could serve as docking sites for the BRCT domain of Rev1 (3,57). We have not been able to detect interactions between FA core complex subunits and Rev1, although these interactions may be transient and weak. Alternatively, the recruitment of Rev1 by the FA core complex might be indirect. Besides possessing E3 ubiquitin ligase activity, some FA core complex sub-units, such as FANCM, have DNA processing activities (58,59). Consequently, the FA core complex may be involved in restructuring stalled replication forks to allow Rev1 loading into foci in a BRCT domain-dependent manner. Consistent with a role for Rev1 downstream of the FA core complex, we have shown that human cells depleted for Rev1 exhibit MMC-induced chromosome breakage and radial formation, a phenotype characteristic of FA pathway-deficient cells. Whether the role of Rev1 in preventing DNA crosslinker-induced chromosomal aberrations is linked to its function in error-prone DNA damage bypass remains unclear.
In conclusion, we present data demonstrating a role for the FA core complex in regulating point mutagenesis and Rev1 localization. Our results unveil a Rad18-independent mechanism to regulate the Rev1 enzyme. Understanding the molecular mechanism by which the FA core complex facilitates error-prone translesion synthesis and Rev1 function will shed light on the functions of this regulatory pathway in maintaining genomic integrity.
Supplementary Material
Acknowledgments
The authors would like to thank R. Kennedy, E. Deutsch, and J. More for technical assistance, and other members of the D’Andrea laboratory for critical reading of the manuscript. We thank M. Seidman for the supF reporter plasmid and for helpful discussions. We thank M. Bienko and I. Dikic for murine eGFP-Rev1 cDNA constructs, A. Lehmann for eGFP-pol eta cDNA constructs, and M. Yamaizumi for Rad18−/− mouse fibroblasts. This work was supported by NIH grants RO1-HL52725, RO1-DK43889 and PO1-HL54785 (A.D.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kennedy RD, D’Andrea AD. Genes Dev. 2005;19(24):2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 2.D’Andrea AD, Grompe M. Nat Rev Cancer. 2003;3(1):23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 3.Wang W. Nat Rev Genet. 2007;8(10):735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 4.Poll EH, Arwert F, Joenje H, Wanamarta AH. Hum Genet. 1985;71(3):206–210. doi: 10.1007/BF00284574. [DOI] [PubMed] [Google Scholar]
- 5.German J, Schonberg S, Caskie S, Warburton D, Falk C, Ray JH. Blood. 1987;69(6):1637–1641. [PubMed] [Google Scholar]
- 6.Gurtan AM, D’Andrea AD. DNA Repair (Amst) 2006;5(9–10):1119–1125. doi: 10.1016/j.dnarep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, Kalb R, Yan Z, Xue Y, Oostra AB, Auerbach AD, Hoatlin ME, Schindler D, Joenje H, de Winter JP, Takata M, Meetei AR, Wang W. Embo J. 2007;26(8):2104–2114. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. Mol Cell. 2001;7(2):249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 9.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, Elledge SJ. Cell. 2007;129(2):289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Andreassen PR, D’Andrea AD. Mol Cell Biol. 2004;24(13):5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. Hum Mol Genet. 2005;14(5):693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 12.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D’Andrea AD. Science. 2002;297(5581):606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 13.Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Mol Cell. 2001;7(2):273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 14.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. Nat Genet. 2005;37(9):953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 15.Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, Wang W, Livingston DM, Joenje H, de Winter JP. Nat Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 16.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D’Andrea AD, Wang ZQ, Jasin M. Proc Natl Acad Sci U S A. 2005;102(4):1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto K, Ishiai M, Matsushita N, Arakawa H, Lamerdin JE, Buerstedde JM, Tanimoto M, Harada M, Thompson LH, Takata M. Mol Cell Biol. 2003;23(15):5421–5430. doi: 10.1128/MCB.23.15.5421-5430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann AR. Experimental cell research. 2006;312(14):2673–2676. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Cell. 2005;123(7):1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Shen X, Jun S, O’Neal LE, Sonoda E, Bemark M, Sale JE, Li L. J Biol Chem. 2006;281(20):13869–13872. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- 21.Papadopoulo D, Guillouf C, Porfirio B, Moustacchi E. Prog Clin Biol Res. 1990;340A:241–248. [PubMed] [Google Scholar]
- 22.Hinz JM, Nham PB, Salazar EP, Thompson LH. DNA Repair (Amst) 2006;5(8):875–884. doi: 10.1016/j.dnarep.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 23.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. Mol Cell. 2004;15(4):607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Friedberg EC, Wagner R, Radman M. Science. 2002;296(5573):1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 25.Nelson JR, Lawrence CW, Hinkle DC. Nature. 1996;382(6593):729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 26.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. Mol Cell. 2006;23(2):265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 27.Guo C, Tang TS, Bienko M, Parker JL, Bielen AB, Sonoda E, Takeda S, Ulrich HD, Dikic I, Friedberg EC. Mol Cell Biol. 2006;26(23):8892–8900. doi: 10.1128/MCB.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 29.Kannouche PL, Wing J, Lehmann AR. Mol Cell. 2004;14(4):491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 30.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Science. 2005;310(5755):1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 31.Jansen JG, Tsaalbi-Shtylik A, Langerak P, Calleja F, Meijers CM, Jacobs H, de Wind N. Nucleic acids research. 2005;33(1):356–365. doi: 10.1093/nar/gki189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross AL, Simpson LJ, Sale JE. Nucleic acids research. 2005;33(4):1280–1289. doi: 10.1093/nar/gki279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM. PLoS biology. 2006;4(11):e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyase S, Tateishi S, Watanabe K, Tomita K, Suzuki K, Inoue H, Yamaizumi M. J Biol Chem. 2005;280(1):515–524. doi: 10.1074/jbc.M409219200. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, Moreau LA, Shimamura A, D’Andrea AD. The Journal of clinical investigation. 2007;117(5):1440–1449. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, Wang W, West SC. Mol Cell. 2007;25(3):331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Wang G, Levy DD, Seidman MM, Glazer PM. Mol Cell Biol. 1995;15(3):1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi JH, Pfeifer GP. DNA Repair (Amst) 2005;4(2):211–220. doi: 10.1016/j.dnarep.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Madzak C, Armier J, Stary A, Daya-Grosjean L, Sarasin A. Carcinogenesis. 1993;14(7):1255–1260. doi: 10.1093/carcin/14.7.1255. [DOI] [PubMed] [Google Scholar]
- 40.Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I. Mol Cell. 2007;26(6):891–898. doi: 10.1016/j.molcel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Embo J. 2004;23(19):3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano S, Yamamoto K, Ishiai M, Yamazoe M, Seki M, Matsushita N, Ohzeki M, Yamashita YM, Arakawa H, Buerstedde JM, Enomoto T, Takeda S, Thompson LH, Takata M. Embo J. 2005;24(2):418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood A, Garg P, Burgers PM. J Biol Chem. 2007;282(28):20256–20263. doi: 10.1074/jbc.M702366200. [DOI] [PubMed] [Google Scholar]
- 44.Thompson LH, Hinz JM, Yamada NA, Jones NJ. Environmental and molecular mutagenesis. 2005;45(2–3):128–142. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- 45.Papadopoulo D, Guillouf C, Mohrenweiser H, Moustacchi E. Proc Natl Acad Sci U S A. 1990;87(21):8383–8387. doi: 10.1073/pnas.87.21.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalb R, Duerr M, Wagner M, Herterich S, Gross M, Digweed M, Joenje H, Hoehn H, Schindler D. Radiat Res. 2004;161(3):318–325. doi: 10.1667/rr3138. [DOI] [PubMed] [Google Scholar]
- 47.Rothfuss A, Grompe M. Mol Cell Biol. 2004;24(1):123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson LJ, Sale JE. Embo J. 2003;22(7):1654–1664. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark DR, Zacharias W, Panaitescu L, McGregor WG. Nucleic acids research. 2003;31(17):4981–4988. doi: 10.1093/nar/gkg725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibbs PE, Wang XD, Li Z, McManus TP, McGregor WG, Lawrence CW, Maher VM. Proc Natl Acad Sci U S A. 2000;97(8):4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsushita N, Kitao H, Ishiai M, Nagashima N, Hirano S, Okawa K, Ohta T, Yu DS, McHugh PJ, Hickson ID, Venkitaraman AR, Kurumizaka H, Takata M. Mol Cell. 2005;19(6):841–847. doi: 10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Lawrence CW, Krauss BR, Christensen RB. Mutat Res. 1985;150(1–2):211–216. doi: 10.1016/0027-5107(85)90117-4. [DOI] [PubMed] [Google Scholar]
- 53.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Embo J. 2003;22(24):6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otsuka C, Kunitomi N, Iwai S, Loakes D, Negishi K. Mutat Res. 2005;578(1–2):79–87. doi: 10.1016/j.mrfmmm.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 55.Manke IA, Lowery DM, Nguyen A, Yaffe MB. Science. 2003;302(5645):636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 56.Yu X, Chini CC, He M, Mer G, Chen J. Science. 2003;302(5645):639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 57.Mi J, Qiao F, Wilson JB, High AA, Schroeder MJ, Stukenberg PT, Moss A, Shabanowitz J, Hunt DF, Jones NJ, Kupfer GM. Mol Cell Biol. 2004;24(19):8576–8585. doi: 10.1128/MCB.24.19.8576-8585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ. Nat Struct Mol Biol. 2005;12(9):763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 59.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. Mol Cell. 2008;29(1):141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.