Summary
The Ca2+-ATPase of cardiac muscle cells transports Ca2+ ions against a concentration gradient into the sarcoplasmic reticulum and is regulated by phospholamban, a 52-residue integral membrane protein. It is known that phospholamban inhibits the Ca2+ pump during muscle contraction and that inhibition is removed by phosphorylation of the protein during muscle relaxation. Phospholamban forms a pentameric complex with a central pore. The solid-state magic angle spinning (MAS) NMR measurements presented here address the structure of the phospholamban pentamer in the region of Gln22-Gln29. Rotational echo double resonance (REDOR) NMR measurements show that the side chain amide groups of Gln29 are in close proximity, consistent with a hydrogen-bonded network within the central pore. 13C MAS NMR measurements are also presented on phospholamban that is 1-13C-labeled at Leu52, the last residue of the protein. pH titration of the C-terminal carboxyl group suggests that it forms a ring of negative charge on the lumenal side of the sarcoplasmic reticulum membrane. The structural constraints on the phospholamban pentamer described in this study are discussed in the context of a multifaceted mechanism for Ca2+ regulation that may involve phospholamban as both an inhibitor of the Ca2+ ATPase and as an ion channel.
Introduction
Phospholamban is essential for the rapid regulation of Ca2+ levels across the sarcoplasmic reticulum (SR) membrane of muscle fibers during muscle contraction and relaxation [1–3]. During muscle contraction, when Ca2+-release channels are open in the SR membrane, the Ca2+ ATPase is inhibited by phospholamban. During muscle relaxation, inhibition of the pump is removed by phosphorylation of phospholamban at Ser16 and Thr17 in the hydrophilic N-terminus by cAMP-dependent and calmodulin-dependent protein kinases.
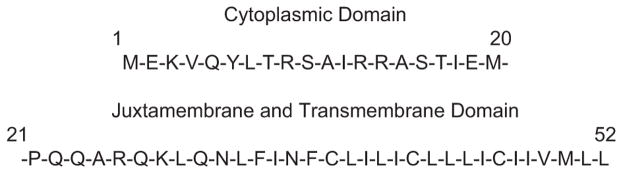
The phospholamban sequence is typically divided into three regions: an N-terminal cytoplasmic region, a central hinge region containing a proline at position 21, and a transmembrane region from Leu28 to Leu52 (Figure 1). The structure of the N-terminal region is important since arginine residues at positions 9, 13 and 14 interact with Asp398 and Asp399 in the nucleotide binding domain of the Ca2+ ATPase [4, 5]. There is a general agreement for helical secondary structure in the N-terminal region of phospholamban derived from NMR and CD studies of both the full length protein and N-terminal fragments [6–10]. The orientation of the N-terminal region is less certain. We [11] and others [6, 12] have previously proposed that the N-terminal helix extends into the cytoplasm, while other groups have used NMR and ESR data to argue that the N-terminal helix interacts with and lies parallel to the membrane surface [13, 14]. There is general agreement that the N-terminal region is more dynamic than the transmembrane region and adopts multiple conformations. The ability of Ser 16 and Thr17 to become phosphorylated by protein kinase C strongly argues that this region may adopt, at least transiently, an extended conformation.
Figure 1.
Phospholamban sequence highlighting the residues in the cytoplasmic, juxtamembrane and transmembrane regions of the protein.
The transmembrane region from Leu28 to Leu52 is important in the structure and regulatory function of phospholamban. The transmembrane residues are largely hydrophobic, fold into helical secondary structure and anchor the protein in the SR membrane. Moreover, specific non-covalent interactions between transmembrane helices stabilize a pentameric protein complex [15, 16]. The transmembrane region is involved in the association of phospholamban with the Ca2+ ATPase leading to inhibition [17, 18]. The strongest inhibition is exhibited by a monomeric form of phospholamban produced by mutation of specific transmembrane residues [19, 20]. Interestingly, the phospholamban pentamer exhibits Ca2+-selective ion conductance [21] and has also been suggested to function as a Cl− ion channel [22]. However, such channel activity is generally considered secondary to phospholamban’s role in regulating the activity of the Ca2+ ATPase.
The monomeric subunit within the pentameric complex of phospholamaban is largely α-helical. The rotational orientation of the helices that form the phospholamban pentamer is known only for the transmembrane domain [6, 23]. Mutational studies have shown that the most sensitive residues for pentamer stability are Leu37, Ile40, Leu44, and Ile47, suggesting that these sites are in the helix-helix interfaces [15, 16]. This packing arrangement is in agreement with cysteine reactivity studies of Thomas and coworkers showing that Cys41 is unreactive to sulfhydryl reagents [24] and deuterium NMR lineshape studies of Leu42, 43 and 44 showing that Leu42 is oriented toward the surrounding lipids [25].
In order to establish the rotational orientation of the phospholamban helix as it emerges from the membrane, we have focused on Gln22, Gln26 and Gln29 at the transition between the transmembrane and juxtamembrane regions of the protein. If the transmembrane and juxtamembrane residues form a continuous α-helix, the charged groups Arg25 and Lys27, would be oriented away from the center of the pore at the level of the membrane interface [26]. This orientation places Gln22, Gln26 and Gln29 roughly in the middle of the helical bundle where they would be in a position to form interhelical hydrogen bonds.
Solid-state NMR using magic angle spinning (MAS) is well-suited for obtaining high-resolution distance constraints of proteins embedded in membrane bilayers. Short internuclear distances (< 5 Å), which are characteristic of hydrogen bonding interactions or residues in van der Waals contact, can be measured with high resolution using specific 13C and 15N-labeling [27]. We have previously targeted specific regions of the phospholamban sequence in the N-terminal cytoplasmic region, juxtamembrane region and transmembrane regions of the protein and shown that they exist as helix in the membrane bound pentamer [26]. Here we use rotational echo double resonance (REDOR) NMR methods [28] to measure the distances between 5-15N-labeled Gln and 5-13C-labeled Gln side chains in the juxtamembrane region of the protein. These distances establish the rotational orientation of the transmembrane helices of phospholamban as they emerge from the membrane bilayer. In addition, we present 13C MAS measurements on Leu52, the last residue in the phospholamban sequence, addressing the protonation state of the C-terminal Leu52 carboxyl group. The C-terminal carboxyl group is not protected and can exist as a negatively charged carboxylate or as a protonated carboxylic acid. The charge on the C-terminal Leu52 carboxyl group will influence the location of the peptide in the bilayer; a charged C-terminus will likely extend into the SR lumen, while a protonated C-terminus is able to partition into the headgroup region of the bilayer. A C-terminal charge will also regulate phospholamban’s ability to function as an ion channel; a negative charge is complementary for functioning as a Ca2+ channel, but inhibitory for functioning as a Cl− channel. The pentameric structure suggested by these measurements is discussed in terms of phospholamban’s regulatory function.
Results
Hydrogen bonding of Gln26 and Gln29 in the phospholamban pentamer
The REDOR methods for measuring heteronuclear dipolar couplings require the acquisition of two spectra, a full echo spectrum, S(full), with no dephasing pulses on the 15N channel, and a dephased spectrum, S(reduced), resulting from the application of a train of dephasing pulses on the 15N channel. Analysis of the normalized echo difference, ΔS/S(full), yields the internuclear distance. Figure 2 presents experimental REDOR spectra of phospholamban containing 13C- and 15N-labeled Gln29 at dipolar evolution times of 9.6 msec (A), 12.8 msec (B) and 17.6 msec (C). The 13C-observe full echo spectrum (solid line) is overlayed with the dephased echo spectrum (dotted line). Parallel data was obtained for phospholamban labeled at Gln22 and Gln26. In these experiments, phospholamban peptides containing a single 5-13C Gln at either position 22, 26 or 29 were reconstituted into POPC:POPS bilayers at a 1:1 molar ratio with phospholamban peptides containing a 5-15N-Gln at the same position. The lipid:protein molar ratio was 50:1 and the ratio of POPC:POPS was 5:1. The intense resonance at ~180 ppm is due to the 13C=O amide signal from 13C-labeled glutamine, and the signal at ~173 ppm is due to the natural abundance 13C signal from the lipid. The experiments were all performed at −10°C, below the phase transition temperature of the lipids, but where there is still a significant amount of mobile water as determined from the narrow 1H linewidth of the water resonance (data not shown). The lower temperature reduces rotational motion of the pentameric complex in the membranes and allows for the measurement of the full dipolar couplings. We have previously measured 13C…13C and 13C…15N dipolar couplings in solid-state MAS experiments of membrane-embedded peptides and determined that a temperature near −10°C is sufficient to retain the full dipolar couplings, but allows for residual motion to narrow the observed linewidths and improve spectral resolution [29].
Figure 2.
Intermolecular 13C-15N REDOR spectra and simulations. Panels A-C present the full (solid line) and dephased (dashed line) spectra of 5-15N-Gln29 phospholamban complexed with 5-13C-Gln29 phospholamban. Spectra are shown for dipolar evolution times of 9.6 msec (A), 12.8 msec (B) and 17.6 msec (C) at a MAS frequency of 5.0 kHz. (D) REDOR data and simulated dephasing curves are presented for 5-15N Gln29-labeled phospholamban in association with 5-13C Gln29-labeled phospholamban (filled squares). The experimental REDOR data are shown for experiments obtained under the same conditions for 5-15N Gln26 phospholamban complexed with 5-13C Gln26 phospholamban (filled circles) and for 5-15N Gln22 phospholamban complexed with 5-13C Gln22 phospholamban (open squares). The experimental data points correspond to the intensity of the 5-13C carbonyl resonance in the REDOR measurement with 15N-dephasing pulses (Sreduced) normalized to the full echo spectrum obtained without 15N-dephasing pulses. The reduced signal is corrected for reconstitution of the 15N- and 13C-labeled peptides in a 1:1 molar ratio. For all three phospholamban samples, the results are shown for dipolar evolution times of 9.6 msec, 12.8 msec and 17.6 msec with a MAS frequency of 5.0 kHz. Simulated REDOR curves are shown for different internuclear 13C…15N distances from 3.0 Å to 6.0 Å. The spectra were obtained with 60,000 –100,000 transients for each spectrum with the larger number of transients needed for the longer evolution times to obtain similar signal:noise ratios.
Comparison of the REDOR data for Gln22, Gln26 and Gln29 shows that the largest changes between the full and dephased echo spectra occur for Gln29 (Figure 2D). Integration of the 13C signal shows a reduction of the 5-13C resonance by ~25% after a 17.6 msec dipolar evolution period with 15N dephasing pulses (Figure 2C). The reduced echo intensity observed in the REDOR experiment can be related to the dipolar coupling and internuclear distance by simulating the REDOR dephasing curve. A series of simulated dephasing curves are shown in Figure 2D based on isolated spin pairs having internuclear 13C…15N distances from 3.0 Å to 6.0 Å. The reduced intensities presented in this graph are corrected to account for reconstitution of the 15N- and 13C-labeled peptides in a 1:1 molar ratio. The correction for the reconstitution in a 1:1 ratio of 15N-labeled and 13C-labeled peptides is to multiply the reduced intensity by a factor of 2, based on the assumption that the peptides associate randomly and that the observed 13C=O…H-15N hydrogen bonded pairs are only 50% of the total, the remaining hydrogen bonding pairs being 13C=O…H-14N (25%) and 12C=O…H-15N (25%). The strongest dipolar coupling is for the interaction between Gln29 side chains and corresponds to an internuclear distance of 4.1 Å ± 0.2 Å. The close proximity is consistent with hydrogen bonding of the Gln29 side chain amide functional groups. The distance may be larger if more than a single 15N site is contributing to the dephasing of each 13C=O site (i.e. the simulations assume an isolated 13C…15N pair).
Weaker dipolar couplings are observed for Gln26 suggesting that while the Gln26 side chains in the pentamer are still in close proximity to one another (~5.5 Å), they do not form a tightly hydrogen bonded network. The data for Gln22 indicate that the intermolecular distance between Gln22 side chain labels is greater than ~5.5 Å.
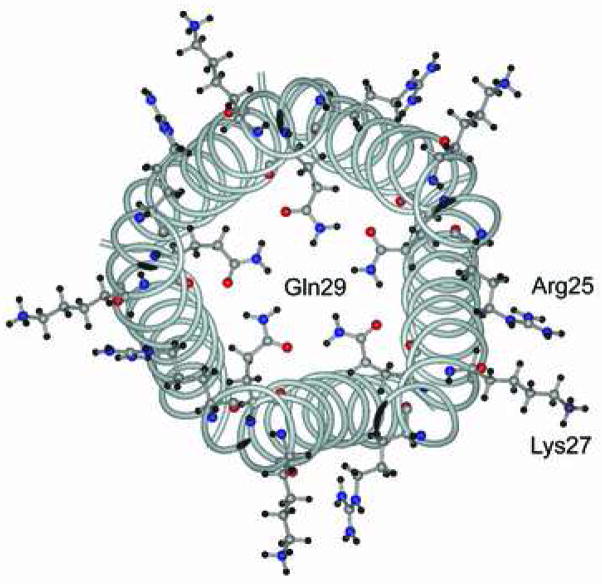
Figure 3 presents a model of the phospholamban pentamer in the region of Gln29 based on our previous NMR data indicating a helical geometry from Pro21 to Leu52 [26]. In a pentameric arrangement, interhelical hydrogen bonding of Gln29 results in a structure similar to that observed in the COMP protein [30]. The fact that Gln26 and Gln29 point toward the pore of the pentamer defines the rotational orientation of the juxtamembrane helix and is consistent with an orientation of Arg25 and Lys27 toward the membrane. Fujii et al. [31] mutated Gln22-Gln23 to Ala-Ala, Gln26-Asn27 to Glu-Asp and Gln29-Asn30 to Glu-Asp. These mutations did not disrupt pentamer formation indicating that the dominant force for oligomerization is within the transmembrane helices.
Figure 3.
Molecular model of the phospholamban structure in the region of Gln29. The helical secondary structure in the region between Pro21 and Leu52 is based on NMR [26] and IR spectroscopy [11]. The position of Gln29 is based on the strong REDOR signal observed in Figure 2. Arg25 and Lys27 are shown oriented away from the central pore in a position where they can interact with the negatively charged lipid headgroups.
Leu52 forms a ring of negative charge on the lumenal side of the SR membrane
In order to address the position of the C-terminal residue of phospholamban relative to the membrane surface, we performed MAS NMR measurements on 1-13C-Leu52 phospholamban reconstituted into POPC:POPS bilayers. Leu52 is the last residue in the phospholamban sequence and the 1-13C position of Leu52 corresponds to the C-terminal carboxyl group. Figure 4 shows the region of the 13C MAS spectrum containing the resonances from Leu52 (1-13C) and the lipid acyl chain carbonyls as a function of pH. The 1-13C Leu52 resonance is sensitive to the protonation state of the C-terminus. Deprotonated carboxyl groups are observed at an average chemical shift of 177 ± 6 ppm, while protonation characteristically shifts the 13COOH resonance upfield, with an average observed chemical shift of 175 ± 6 ppm [32, 33]. Titration of vesicles containing 1-13C Leu52 labeled phospholamban exhibited a decrease in intensity of the resonance corresponding to the unprotonated carboxyl group at 179.3 ppm and a shift of intensity to ~173 ppm. At low pH, the 1-13C Leu52 resonance was not resolved from the natural abundance resonance of the lipid acyl chain carbonyls. The intensity changes show that the C-terminal carboxyl group is charged at neutral pH (with a pKa of ~6.0) and exposed to the bulk aqueous medium.
Figure 4.
13C-MAS spectra of 1-13C Leu52-labeled phospholamban as a function of pH. The spectra were obtained at pH 7.5 (A) pH 6.5 (B) pH 6.0 (C), pH 5.5 (D) and pH 4.5 (E). The 1-13C Leu52-labeled phospholamban was reconstituted into POPC:POPS bilayers. The lipid:protein molar ratio was 50:1 and the ratio of POPC:POPS was 5:1.
Figure 5 presents an electrostatic potential map of the phospholamban pentamer showing the region from Gln22 to Leu52. The observation of a negatively charged ring surrounding the central pore on the C-terminal side of the pentameric bundle of helices has a number of consequences. First, the charged nature of the C-terminal side of the bundle indicates that the pentamer crosses the bilayer. Second, a C-terminal charge will also regulate phospholamban’s ability to function as an ion channel; a negative charge is complementary for functioning as a Ca2+ channel, but inhibitory for functioning as a Cl− channel [22]. Computational studies have previously shown that a negative ring of charge at the C-terminus makes the energetically selective for Ca++ over Cl− ions [34].
Figure 5.

Electrostatic potential of the C-terminus of the phospholamban pentamer from Gln22 to Leu52. The figure was made using PyMol based on the pentamer structure of phospholamban.
The ring of negative charge is strikingly similar to the structures of membrane channels that are selective for cations (e.g. the K+ channel from Streptomyces lividans [35], the acetylcholine receptor [36], and the CorA cation channel [37, 38]). In particular, the highest resolution crystal structure of CorA at 2.9 Å [37] reveals an assembly of five core transmembrane helices that resembles the phospholamban structure we have previously proposed [11, 26]. In CorA, the helices define a hydrophobic pore that is lined with only a few weakly polar serine and threonine residues. The pore diameter is restricted by three rings of hydrophobic residues, and importantly, there is a highly conserved Asp residue (Asp277) at the entrance of the pore that can be compared to the ring of carboxylate groups formed by Leu52 in phospholamban.
Discussion
In the current study, NMR measurements address the structure of the phospholamban pentamer between Pro21 and Leu52 in membrane bilayers. We show that the side chain amide groups of Gln29 are in close proximity, consistent with a hydrogen-bonded network within the central pore. In addition, pH titration of the 13C-labeled C-terminal carboxyl group shows that it forms a ring of negative charge near the surface of POPC:POPS membranes. In order to evaluate how the Gln29 interaction and Leu52 charge constrain the structure of the pentameric complex in membranes, we review the structural basis for our current model of phospholamban and discuss the similarities and differences with models that have been proposed to date.
Our original model of the pentamer was based on studies using polarized infrared (IR) spectroscopy to establish the secondary structure and orientation of wild-type phospholamban reconstituted into membrane bilayers [11]. One of the challenges for structural studies on hydrophobic membrane peptides that lack intrinsic activity is to reconstitute the peptides into membrane bilayers in a well-defined transmembrane orientation. We have found that polarized IR spectroscopy provides a rapid method to assess different reconstitution methods based on the assumption that the membrane spanning region of long hydrophobic peptides has helical secondary structure oriented roughly parallel to the bilayer normal.
In IR spectroscopy, protein secondary structure is characterized by the frequency of the amide I vibration. Helical structure yields an amide I vibration in the range of 1650 – 1660 cm−1. The orientation relative to the bilayer normal of the helical elements in membrane protein structures can be determined in polarized IR experiments by the dichroic ratio (R), which is defined by the intensity of the amide I vibration obtained using light polarized parallel and perpendicular to the bilayer normal in oriented membranes [29]. For example, using the parameters described previously [29, 39], a dichroic ratio of R = 3.7 corresponds to a helix that is parallel to the membrane normal, while a dichroic ratio of R = 1.5 corresponds to a helix that is perpendicular to the membrane normal.
In our original studies, we compared the ability of different reconstitution protocols to incorporate phospholamban into membrane vesicles. Detergent dialysis proved to be the most robust approach for reconstituting phospholamban as a transmembrane α-helix. In contrast, we found that the most widely used reconstitution protocol in the literature based on the co-solubilization of lipid and peptide in organic solvent, rehydration in buffer and sonication, yielded a high fraction of non-helical secondary structure and poorly oriented helices. The results on phospholamban, which were not published, are similar to the results found in a detailed comparison of different reconstitution approaches we have reported for the transmembrane domain of glycophorin A [29].
For full-length phospholamban, we established that ~40 of the 52 residues are in helical secondary structure. We reported a maximum experimental dichroic ratio for the isolated phospholamban transmembrane domain (residues Arg25 – Leu52) of R = 3.43, and for the full-length sequence (Met1 - Leu52) of R = 3.31. A dichroic ratio of 3.3 – 3.4 corresponds to a helix tilt of ~16–19° using a transition moment angle for amide I vibration of 41.8° (see [29] for a discussion). To obtain the maximum dichroic ratio, we carried out several independent reconstitutions since the homogeneity of the sample can vary between reconstitutions. We selected the reconstitution yielding the highest dichroic ratio (i.e. corresponding to an orientation of the transmembrane helix closest to the bilayer normal) as the most homogeneous sample. The homogeneity can be verified by comparing dichroic ratios obtained from samples before and after running sucrose gradients [11, 29].
Tamm and coworkers [12] also proposed a model for the phospholamban pentamer based on polarized IR measurements. They obtained a dichroic ratio of R = 3.06 for the isolated phospholamban transmembrane domain by a variation of detergent dialysis (i.e. solubilization of protein and lipid using octyl-β-glucoside, followed by the removal of the detergent with biobeads) [12]. Their use of a detergent-based reconstitution method yielded a high dichroic ratio for the isolated transmembrane domain that was comparable to our value of R = 3.31 for full-length phospholamban. Tamm and co-workers also measured the dichroic ratio of full-length phospholamban. For these reconstitutions, they started with detergent dialysis, but subsequently lyophilized and solubilized their sample in organic solvent (chloroform and trifluoroethanol). They then followed the standard protocol of removing the solvent with nitrogen gas, followed by rehydration in buffer, vortexing and sonication. With this reconstitution method, they obtained a dichroic ratio of 2.28. This value is typical of the dichroic ratios we obtained for full-length phospholamban by reconstituting directly from organic solvents without detergent dialysis (unpublished results). On the basis of their polarized IR measurements, they proposed a model of phospholamban with the transmembrane sequence folding into an α-helix and the juxtamembrane region folding into antiparallel β-strands. Importantly, with an overall dichroic ratio of 2.3 they concluded that the N-terminal helix was tilted from the membrane normal by 61 +/− 13°, or alternatively could assume a dynamic range of orientations (e.g. including orientations parallel to the membrane surface).
Polarized IR measurements have not generally been reported for other structural studies on phospholamban. However, we can predict the dichroic ratios for two of the additional models proposed in the literature on the basis of the orientations proposed for the transmembrane and cytoplasmic helices, and using a value of 41.8° for the orientation of the amide I transition moment relative to the helix axis (see [29] for a discussion). For the model of monomeric and pentameric phospholamban proposed by Veglia and Thomas [13, 40] with 18 cytoplasmic and 34 transmembrane amino acids, we calculate a dichroic ratio of R = 2.66 (R = 3.27 for a transmembrane helix with a 21° tilt and R = 1.52 for a cytoplasmic helix with a 93° tilt). For the model of the phospholamban pentamer proposed by Oxenoid and Chou [6], we calculate a dichroic ratio of R = 3.46 (R = 3.34 for a transmembrane helix with 19° tilt and R = 3.61 for a cytoplasmic helix with a 10° tilt). The dichroic ratio that is predicted for the pentamer structure of Oxenoid and Chou is similar to that experimentally observed.
These studies have led us to conclude that the structure of the phospholamban pentamer depends on the method of reconstitution. There is a general consensus for helical secondary structure in the N-terminal region of phospholamban from NMR and CD studies of both the full-length protein and N-terminal fragments [6–10]. One exception is the structure of the AFA monomer determined by Baldus and co-workers in DOPC:DOPE bilayers that has an unstructured N-terminus [41]. A common element among the structural studies undertaken to date is that the N-terminal region may adopt multiple conformations or orientations. The polarized IR data discussed above can be interpreted in this light. In our original studies [11], the average dichroic ratio for the full-length protein was 2.9, and in our subsequent NMR analysis of several full-length phospholamban peptides [26], the average dichroic ratio was 2.8. As in the analysis of Tamm and co-workers, these data are consistent with a range of orientations that can include orientations parallel to the membrane surface. The differences between proposed models of the N-terminal region of phospholamban may simply reflect how different reconstitution protocols favor conformations with membrane associated helices or helices extending into the cytoplasm.
The focus of the current study is on the structure of the phospholamban sequence between Pro21 and Leu52, which includes the transmembrane domain of the protein. As presented in the introduction, there is general agreement that the transmembrane domain forms a bundle of helices that pack together with left-handed crossing angles. There are differences among structural models at the cytoplasmic and luminal ends of the helices as they emerge from the hydrophobic core of the bilayer.
In the structure of the phospholamban pentamer we have previously proposed [26], the basic residues (Arg25 and Lys27) are roughly 45 Å from the C-terminal carboxyl group, sufficient to span a membrane bilayer with a thickness of 45 Å – 50 Å. The transmembrane helices extend through the juxtamembrane region to Pro21. The proposed structure suggested that interactions between the juxtamembrane helices are mediated by hydrogen bonding interactions involving Gln22, Gln26 and Gln29. In the solution NMR structure solved in dodecylphosphocholine (DPC) micelles by Oxenoid and Chou [6], the transmembrane helix within the hydrophobic core has roughly the same helix tilt angle as in membrane bilayers. However, the juxtamembrane helix in DPC micelles becomes more significantly tilted (relative to the central pore axis) in the region between Pro21 and Asn34, and the distance is only ~35 Å between the C-terminal end of the transmembrane helix and the charged residues, Arg25 and Lys27. As a result, the detergent structure would predict that in membranes Leu52 (and the C-terminal carboxyl group) and/or the basic residues (Arg25 and Lys27) at the cytoplasmic boundary of the transmembrane domain are located within the headgroup region of the bilayer. In addition, although the glutamine residues at positions 22, 26 and 29 are oriented roughly toward the central pore of the pentamer, they are not in a position to form interhelical contacts because the helices splay away from one another in this region of the detergent structure.
Our current results on the protonation state of the Leu52 carboxyl group and hydrogen bonding interactions between Gln29 side chains argue that the membrane and detergent structures of the phospholamban pentamer are different. The differences are reminiscent of the differences between the membrane [42] and detergent [43] structures of the transmembrane dimer of glycophorin A. These structures are subtly, but distinctly different in at least two ways [29]. The interior of detergent micelles is more aqueous than the interior of membrane bilayers. As a result, in the detergent structure of glycophorin A, Thr87 is solvated and does not form interhelical hydrogen bonds that stabilize the dimer structure as in the membrane bound dimer. A similar situation may influence the interhelical hydrogen bonding of Gln29. Second, detergent micelles and membrane bilayers differ in their geometry. Spherical micelles can favor dimer structures with larger crossing angles than in planar bilayers. In the glycophorin A dimer, the crossing angle of the helices in membranes is less than in detergent. In the detergent structure of phospholamban, the large helix tilt in the region between Pro21 and Asn34 may reflect the spherical geometry of the micelle.
Together the structural constraints involving the glutamine residues in the juxtamembrane region of phospholamban and the observation of a C-terminal charge on the protein more tightly define the structure of the phospholamban pentamer in membranes, and set the stage for detailed structural studies on the N-terminal region of the peptide.
Methods
Lipids (Avanti Polar Lipids, Alabaster, AL) used were 1-palmitoyl, 2-oleoyl phosphoserine (POPS) and 1-palmitoyl, 2-oleoyl phosphocholine (POPC). 13C- and 15N-labeled amino acids were obtained from Cambridge Isotope Labs (Andover, MA) or Mass Trace (Woburn, MA) as t-Boc derivatives or derivatized using standard methods.
The 52 residue sequence of phospholamban was synthesized by solid-phase methods using t-butoxycarbonyl (t-Boc) chemistry at the Institute for Protein Research (Osaka, Japan) or the Keck Facility for Peptide Synthesis (Yale University). The peptide was purified by high performance liquid chromatography (HPLC) on an ion exchange TSK-gel CM 3SW (7.5 × 75 mm) column using trichloromethane:methanol:water in a 4:4:1 ratio and checked for purity by mass spectroscopy and reverse phase HPLC on a Zorbax 300SB-CN (9.4 × 250 mm) column. The protein was reconstituted into POPC:POPS bilayers by first cosolubilizing lipid, peptide (lyophilized) and detergent (octyl β-glucoside) in trifluoroethanol. The lipid:protein molar ratio was 50:1 and the ratio of POPC:POPS was 5:1. The trifluoroethanol was then removed by evaporation using argon gas and then with vacuum. The dry lipid/peptide/detergent mixture was rehydrated with buffer such that the final concentration of octyl β-glucoside was 5% (w/v). A series of different buffers containing 50 mM NaCl were used depending on the final pH (HEPES, pH 7–7.5; MES, pH 5.5–6.5, malate, pH 4.5–5.5). The octyl β-glucoside was removed by dialysis using Spectra-Por dialysis tubing (3500 MW cutoff) at ~37°C. Sucrose gradient (10%–40% w/v) ultracentrifugation using a Beckman ultracentrifuge at 150,000 × g for 12 h at 20°C was used to isolate homogeneous lipid vesicles.
The sample is then typically spun in an MAS rotor at 3–4 kHz for 30 min to further pellet the membranes and remove excess water. This step helps balance the rotor for the MAS experiments. The level of hydration can be measured based on the intensity of the water 1H resonances relative to those of the lipid and peptide. The hydration levels after this procedure are typically in the range of 80–100% (w/w) water. At this level of hydration, lipid phase transition temperatures are not changed.
Magic angle spinning NMR experiments were performed on a Bruker Avance spectrometer operating at 359.6 MHz for 1H and using double and triple resonance 5 mm MAS probes from Doty Scientific (Columbia, SC). The 1H, 13C and 15N pulse lengths were typically ~3.5 μsec, 4 μsec, and 5 μsec, respectively. Ramped amplitude cross polarization (Metz et al., 1994) was used to improve quantification of the measured NMR intensities. The total contact time for cross polarization was 4 ms and the recycle delay was typically 2.5 s. TPPM proton decoupling [44] was used to decrease linewidths and improve sensitivity. Proton decoupling field strengths were generally higher than ~85 MHz. The REDOR pulse sequence uses a train of 15N π pulses with two pulses per rotor cycle. XY4 phase cycling was used to suppress resonance offset effects [45]. A single 13C π pulse was used to refocus the chemical shift interaction. REDOR experiments were carried out with dipolar evolution times 9.6 msec, 12.8 msec and 17.6 msec. The temperature was maintained at −10°C
Acknowledgments
This work was supported by NIH-NSF instrumentation grants (S10 RR13889 and DBI-9977553), a grant from the National Institutes of Health (GM-46732) to S.O.S. We gratefully acknowledge the W.M. Keck Foundation for support of the NMR facilities in the Center of Structural Biology at Stony Brook. We thank Terry Gullion for the REDOR simulation program, and Martine Ziliox for technical assistance with the NMR measurements and critical reading of the manuscript.
Abbreviations
- DPC
dodecylphosphocholine
- MAS
magic angle spinning
- NMR
nuclear magnetic resonance
- POPC
1-palmitoyl, 2-oleoyl phosphocholine
- POPS
1-palmitoyl, 2-oleoyl phosphoserine
- REDOR
rotational echo double resonance
- SR
sarcoplasmic reticulum
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tada M, Kirchberger MA, Katz AM. Phosphorylation of a 22,000-dalton component of cardiac sarcoplasmic-reticulum by adenosine 3′–5′-monophosphate-dependent protein kinase. J Biol Chem. 1975;250:2640–2647. [PubMed] [Google Scholar]
- 2.Katz AM, Tada M, Kirchberger MA. Control of calcium transport in the myocardium by the cyclic AMP-protein kinase system. Adv Cyclic Nucleotide Res. 1975;5:453–472. [PubMed] [Google Scholar]
- 3.Simmerman HK, Jones LR. Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 4.Toyofuku T, Kurzydlowski K, Tada M, MacLennan DH. Amino acids Lys-Asp-Asp-Lys-Pro-Val402 in the Ca2+-ATPase of cardiac sarcoplasmic reticulum are critical for functional association with phospholamban. J Biol Chem. 1994;269:22929–22932. [PubMed] [Google Scholar]
- 5.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 6.Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Natl Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamberth S, Schmid H, Muenchbach M, Vorherr T, Krebs J, Carafoli E, Griesinger C. NMR solution structure of phospholamban. Helv Chim Acta. 2000;83:2141–2152. [Google Scholar]
- 8.Mortishire-Smith RJ, Pitzenberger SM, Burke CJ, Middaugh CR, Garsky VM, Johnson RG. Solution structure of the cytoplasmic domain of phopholamban: phosphorylation leads to a local perturbation in secondary structure. Biochemistry. 1995;34:7603–7613. doi: 10.1021/bi00023a006. [DOI] [PubMed] [Google Scholar]
- 9.Zamoon J, Mascioni A, Thomas DD, Veglia G. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys J. 2003;85:2589–2598. doi: 10.1016/s0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollesello P, Annila A. Structure of the 1–36 N-terminal fragment of human phospholamban phosphorylated at Ser-16 and Thr-17. Biophys J. 2002;83:484–490. doi: 10.1016/S0006-3495(02)75184-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arkin IT, Rothman M, Ludlam CF, Aimoto S, Engelman DM, Rothschild KJ, Smith SO. Structural model of the phospholamban ion channel complex in phospholipid membranes. J Mol Biol. 1995;248:824–834. doi: 10.1006/jmbi.1995.0263. [DOI] [PubMed] [Google Scholar]
- 12.Tatulian SA, Jones LR, Reddy LG, Stokes DL, Tamm LK. Secondary structure and orientation of phospholamban reconstituted in supported bilayers from polarized attenuated total reflection FTIR spectroscopy. Biochemistry. 1995;34:4448–4456. doi: 10.1021/bi00013a038. [DOI] [PubMed] [Google Scholar]
- 13.Traaseth NJ, Verardi R, Torgersen KD, Karim CB, Thomas DD, Veglia G. Spectroscopic validation of the pentameric structure of phospholamban. Proc Natl Acad Sci USA. 2007;104:14676–81. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abu-Baker S, Lorigan GA. Phospholamban and its phosphorylated form interact differently with lipid bilayers: A P-31, H-2, and C-13 solid-state NMR spectroscopic study. Biochemistry. 2006;45:13312–13322. doi: 10.1021/bi0614028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arkin IT, Adams PD, MacKenzie KR, Lemmon MA, Brünger AT, Engelman DM. Structural organization of the pentameric transmembrane α-helices of phospholamban, a cardiac ion channel. EMBO J. 1994;13:4757–4764. doi: 10.1002/j.1460-2075.1994.tb06801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmerman HK, Kobayashi YM, Autry JM, Jones LR. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J Biol Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 17.Kimura Y, Kurzydlowski K, Tada M, MacLennan DH. Phospholamban regulates the Ca2+-ATPase through intramembrane interactions. J Biol Chem. 1996;271:21726–21731. doi: 10.1074/jbc.271.36.21726. [DOI] [PubMed] [Google Scholar]
- 18.Asahi M, Kimura Y, Kurzydlowski K, Tada M, MacLennan DH. Transmembrane helix M6 in sarco(endo)plasmic reticulum Ca2+-ATPase forms a functional interaction site with phospholamban: Evidence for physical interactions at other sites. J Biol Chem. 1999;274:32855–32862. doi: 10.1074/jbc.274.46.32855. [DOI] [PubMed] [Google Scholar]
- 19.Kimura Y, Kurzydlowski K, Tada M, MacLennan DH. Phospholamban inhibitory function is activated by depolymerization. J Biol Chem. 1997;272:15061–15064. doi: 10.1074/jbc.272.24.15061. [DOI] [PubMed] [Google Scholar]
- 20.Reddy LG, Autry JM, Jones LR, Thomas DD. Co-reconstitution of phospholamban mutants with the Ca-ATPase reveals dependence of inhibitory function on phospholamban structure. J Biol Chem. 1999;274:7649–7655. doi: 10.1074/jbc.274.12.7649. [DOI] [PubMed] [Google Scholar]
- 21.Kovacs RJ, Nelson MT, Simmerman HK, Jones LR. Phospholamban forms Ca2+-selective channels in lipid bilayers. J Biol Chem. 1988;263:18364–18368. [PubMed] [Google Scholar]
- 22.Decrouy A, Juteau M, Rousseau E. Examination of the Role of Phosphorylation and Phospholamban in the Regulation of the Cardiac Sarcoplasmic-Reticulum Cl- Channel. J Membr Biol. 1995;146:315–326. doi: 10.1007/BF00233951. [DOI] [PubMed] [Google Scholar]
- 23.Torres J, Adams PD, Arkin IT. Use of a new label 13C=18O in the determination of a structural model of phospholamban in a lipid bilayer. Spatial restraints resolve the ambiguity arising from interpretations of mutagenesis data. J Mol Biol. 2000;300:677–685. doi: 10.1006/jmbi.2000.3885. [DOI] [PubMed] [Google Scholar]
- 24.Karim CB, Stamm JD, Karim J, Jones LR, Thomas DD. Cysteine reactivity and oligomeric structures of phospholamban and its mutants. Biochemistry. 1998;37:12074–12081. doi: 10.1021/bi980642n. [DOI] [PubMed] [Google Scholar]
- 25.Ying WW, Irvine SE, Beekman RA, Siminovitch DJ, Smith SO. Deuterium NMR reveals helix packing interactions in phospholamban. J Am Chem Soc. 2000;122:11125–11128. [Google Scholar]
- 26.Smith SO, Kawakami T, Liu W, Ziliox M, Aimoto S. Helical structure of phospholamban in membrane bilayers. J Mol Biol. 2001;313:1139–1148. doi: 10.1006/jmbi.2001.5101. [DOI] [PubMed] [Google Scholar]
- 27.Smith SO, Aschheim K, Groesbeek M. Magic angle spinning NMR spectroscopy of membrane proteins. Q Rev Biophys. 1996;29:395–449. doi: 10.1017/s0033583500005898. [DOI] [PubMed] [Google Scholar]
- 28.Gullion T, Schaefer J. Rotation-echo double-resonance NMR. J Magn Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Smith SO, Eilers M, Song D, Crocker E, Ying WW, Groesbeek M, Metz G, Ziliox M, Aimoto S. Implications of threonine hydrogen bonding in the glycophorin A transmembrane helix dimer. Biophys J. 2002;82:2476–2486. doi: 10.1016/S0006-3495(02)75590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malashkevich VN, Kammerer RA, Efimov VP, Schulthess T, Engel J. The crystal structure of a five-stranded coiled coil in COMP: a prototype ion channel? Science. 1996;274:761–765. doi: 10.1126/science.274.5288.761. [DOI] [PubMed] [Google Scholar]
- 31.Fujii J, Maruyama K, Tada M, MacLennan DH. Expression and site-specific mutagenesis of phospholamban. Studies of residues involved in phosphorylation and pentamer formation. J Biol Chem. 1989;264:12950–12955. [PubMed] [Google Scholar]
- 32.Gu ZT, McDermott A. Chemical shielding anisotropy of protonated and deprotonated carboxylates in amino-acids. J Am Chem Soc. 1993;115:4282–4285. [Google Scholar]
- 33.Gu ZT, Zambrano R, McDermott A. Hydrogen-bonding of carboxyl groups in solid state amino acids and peptides: comparison of carbon chemical shielding, infrared frequencies, and structures. J Am Chem Soc. 1994;116:6368–6372. [Google Scholar]
- 34.Sansom MSP, Smith GR, Smart OS, Smith SO. Channels formed by the transmembrane helix of phospholamban: a simulation study. Biophys Chem. 1997;69:269–281. doi: 10.1016/s0301-4622(97)00109-9. [DOI] [PubMed] [Google Scholar]
- 35.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, Mackinnon R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 36.Unwin N. Nicotinic acetylcholine receptor at 9 Å resolution. J Mol Biol. 1993;229:1101–1124. doi: 10.1006/jmbi.1993.1107. [DOI] [PubMed] [Google Scholar]
- 37.Eshaghi S, Niegowski D, Kohl A, Molina DM, Lesley SA, Nordlund P. Crystal structure of a divalent metal ion transporter CorA at 2.9 Å resolution. Science. 2006;313:354–357. doi: 10.1126/science.1127121. [DOI] [PubMed] [Google Scholar]
- 38.Lunin VV, Dobrovetsky E, Khutoreskaya G, Zhang RG, Joachimiak A, Doyle DA, Bochkarev A, Maguire ME, Edwards AM, Koth CM. Crystal structure of the CorA Mg2+ transporter. Nature. 2006;440:833–837. doi: 10.1038/nature04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bechinger B, Ruysschaert JM, Goormaghtigh E. Membrane helix orientation from linear dichroism of infrared attenuated total reflection spectra. Biophys J. 1999;76:552–563. doi: 10.1016/S0006-3495(99)77223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traaseth NJ, Buffy JJ, Zamoon J, Veglia G. Structural dynamics and topology of phospholamban in oriented lipid bilayers using multidimensional solid-state NMR. Biochemistry. 2006;45:13827–13834. doi: 10.1021/bi0607610. [DOI] [PubMed] [Google Scholar]
- 41.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 42.Smith SO, Song D, Shekar S, Groesbeek M, Ziliox M, Aimoto S. Structure of the transmembrane dimer interface of glycophorin A in membrane bilayers. Biochemistry. 2001;40:6553–6558. doi: 10.1021/bi010357v. [DOI] [PubMed] [Google Scholar]
- 43.MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 44.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 45.Gullion T, Schaefer J. Elimination of resonance offset effects in rotational-echo, double-resonance NMR. J Magn Reson. 1991;92:439–442. [Google Scholar]