Abstract
Understanding inter-individual differences in stress response requires the explanation of genetic influences at multiple phenotypic levels, including complex behaviours and the metabolic responses of brain regions to emotional stimuli. Neuropeptide Y (NPY) is anxiolytic1,2 and its release is induced by stress3. NPY is abundantly expressed in regions of the limbic system that are implicated in arousal and in the assignment of emotional valences to stimuli and memories4–6. Here we show that haplotype-driven NPY expression predicts brain responses to emotional and stress challenges and also inversely correlates with trait anxiety. NPY haplotypes predicted levels of NPY messenger RNA in postmortem brain and lymphoblasts, and levels of plasma NPY. Lower haplotype-driven NPY expression predicted higher emotion-induced activation of the amygdala, as well as diminished resiliency as assessed by pain/stress-induced activations of endogenous opioid neurotransmission in various brain regions. A single nucleotide polymorphism (SNP rs16147) located in the promoter region alters NPY expression in vitro and seems to account for more than half of the variation in expression in vivo. These convergent findings are consistent with the function of NPY as an anxiolytic peptide and help to explain inter-individual variation in resiliency to stress, a risk factor for many diseases.
Variation in stress resiliency influences many human characteristics, including both normal and pathological behaviour7. Maladaptive responses to stress are critical in the development of many psychiatric disorders, including mood and anxiety disorders8,9. Anxiety and emotionality (neuroticism) are moderately to highly heritable traits (40–60%) but are also strongly influenced by exposures to stress in a pattern consistent with gene–environment interaction10. These observations point to the importance of genes that modulate the effects of stress. Genes—such as the serotonin transporter—that have so far been implicated in emotional responses have small effects on complex behavioural traits11, but larger effects on the metabolic responses of the brain to emotional stimuli accessed by brain imaging12–14.
We evaluated effects of neuropeptide NPY on emotion and stress resiliency using a haplotype-based approach intended to capture effects of unknown loci or locus combinations. We analysed functionally grouped NPY haplotypes against a complex behaviour, trait anxiety, and also on intermediate phenotypes accessed by two different brain imaging modalities in which gene effects might be more strongly manifested. An NPY seven-marker panel (Supplementary Fig. 1a) genotyped in 516 Finnish Caucasians captured the major haplotypes and linkage disequilibrium features observed in the International HapMap Project (http://www.hapmap.org). A block of strong pairwise linkage disequilibrium encompasses 70% of the gene and extends from the 5′ region to exon 3 (Supplementary Fig. 1b). Five haplotypes (H1–H5) account for 93.8% of chromosomes in this block (Fig. 1a).
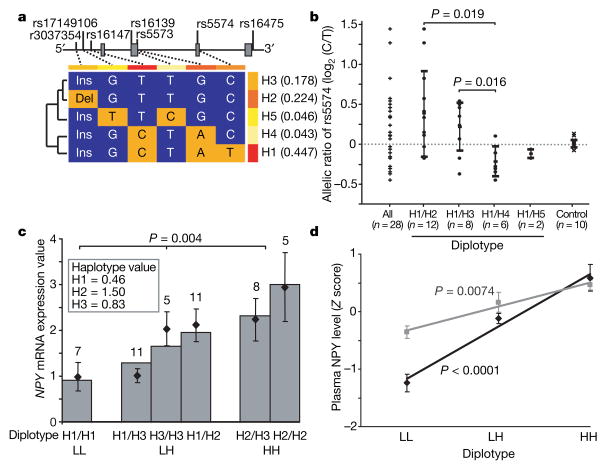
Figure 1. Haplotype-predicted NPY expression in brain, lymphoblasts and plasma.
aConfiguration, frequencies (right) and cladistic clustering (left) of major NPY haplotypes (H1, H2, H3, H4 and H5; frequency more than 1%) in Finnish Caucasians (n = 516). b, Differential allele expression of NPY mRNA showing allelic ratio (log2(C/T)) of rs5574 in 28 heterozygous human post-mortem cerebella (Miami sample) grouped according to diplotypes (columns 2–5, means ± s.d.) to infer expression levels of H2, H3, H4 and H5 relative to H1 (the T allele is found only within H1, as shown in a). Diplotypes are compared using a two-tailed t-test. Column 1 presents all samples; column 6 shows controls for allelic amplification efficiency.
c, Expression levels of the six common diplotypes in 47 lymphoblastoid cell lines derived from healthy Finnish Caucasians. Expression values of the three major haplotypes (H1, H2 and H3) were calculated from observed diplotype values (points and error bars show means ± s.e.m.) by multiple regression and were used to predict diplotype values (bars) under a co-dominant model (see Supplementary Fig. 2 for details, including fit to model). Diplotypes were grouped as low (LL), intermediate (LH) or high (HH) in expression. Numbers above columns are n. d, Diplotype-predicted plasma NPY levels (means ± s.e.m., standardized with Z scores; New Haven sample) in controls (black diamonds; LL, n = 16; LH, n = 113; HH, n = 39) and alcoholic patients (grey squares; LL, n = 29; LH, n = 72; HH, n = 27). P values were calculated by regression analysis.
We observed haplotype-driven NPY mRNA expression in postmortem brain (US Caucasians, Miami sample) by detecting the differential expression of alleles at single nucleotide polymorphism (SNP) rs5574 C/T, selected because of its high frequency and location in the transcript. Of these 28 samples, chosen because all were heterozygous for rs5574, 16 (57%) showed differential allele expression at an allele ratio of more than 1.2, in either direction. H1 and H4 were low-expression haplotypes, H2 was high, H3 was intermediate and H5 was unclassified because only two H1/H5 heterozygous brains were available (Fig. 1b). This expression-based functional classification is consistent with a cladistically based clustering of haplotypes, indicating that expression variation is linked to gene ancestry (Fig. 1a). The effects on expression of the more common H1, H2 and H3 haplotypes were verified in 47 lymphoblastoid cell lines derived from healthy Finnish men (Fig. 1c) representing the six common diplotypes (72% of all diplotypes). On the basis of lymphoblast NPY mRNA levels, the expression value for each haplotype was calculated by regression analysis. Expression values for the six common diplotypes were well predicted under a co-dominant model and had a threefold range (see Supplementary Fig. 2 for details). Diplotypes were clustered into three expression groups: low (LL: H1/H1), intermediate (LH: H1/H3, H3/H3 and H1/H2) and high (HH: H2/H3 and H2/H2) (Fig. 1c). We applied this grouping in subsequent analyses. Two loci, rs3037354 and rs16147, which differentiate the three common haplotypes (see Fig. 1a), correlated with expression (Supplementary Fig. 3). However, NPY haplotypes accounted for more variation. Finally, haplotype-predicted NPY expression was correlated with plasma NPY peptide levels available in a US sample (New Haven) in both controls (n = 24) and alcoholic patients (n = 18) sampled during a no-stress condition. Individuals with the low-expression LL diplotype had lower NPY levels than those with high-expression HH diplotypes; individuals with LH diplotypes were intermediate (P < 0.0001 in controls and P = 0.0074 in alcoholic patients; Fig. 1d).
These common, functionally significant NPY haplotypes were evaluated for their effect on brain responses to emotion and stress. Amygdala activation in response to threat-related facial expressions and other provocative stimuli predicts affective arousal, including anxiety responses15,16. We employed a widely used functional magnetic resonance (fMRI) probe12,17 to assess whether NPY diplotypes predicted amygdala reactivity to threat-related facial expressions in 71 healthy volunteers (Pittsburgh sample). This model has been used to identify greater amygdala reactivity in individuals possessing the lower-transcribing allele of the serotonin-transporter-linked polymorphic region (5-HTTLPR)12,18. As shown in Fig. 2, amygdala activation in individuals with the low-NPY-expression (LL) diplotype was higher than in those with the high-expression (HH) diplotype (P = 0.003). NPY diplotype predicted amygdala reactivity in an allele-dosage fashion, and it accounted for 9% of the variance in the fMRI amygdala response to emotional challenge. Task-related hippocampal activation was similarly predicted in an allele-dosage fashion (P = 0.006; Fig. 2). Functional interactions of the amygdala and hippocampus are crucial for emotional memories, and long-lasting changes in hippocampal architecture are induced by stress19.
Figure 2. Effect of diplotype-predicted NPY mRNA expression on fMRI-measured amygdala and hippocampal activation in response to threat-related facial expressions.
Top: statistical parametric maps representing NPY diplotype-biased (LL > LH > HH) mean right dorsal amygdala (x = 16, y = −8, z = −14, 64 voxels, t = 2.82, P = 0.003) and right hippocampal (x = 24, y = −20, z = −12, 132 voxels, t = 2.56, P = 0.006) activation is shown overlaid on an average sagittal and coronal MRI. Bottom: right amygdala (black diamonds) and hippocampal (grey squares) activities (means and s.e.m.) from clusters grouped by NPY diplotypes.
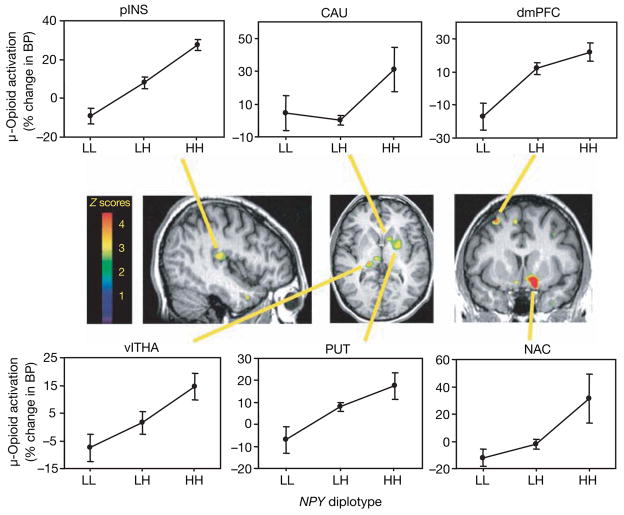
We also tested the ability of NPY haplotypes in a model of physical and emotional stress involving moderate levels of sustained muscular pain. This physical and emotional stress activates endogenous opioid neurotransmission in regions of the brain that regulate pain, stress and emotion20,21. Endogenous opioid release suppresses pain, stress and anxiety-like responses in animal models22,23 and humans21. The behavioural effects of NPY are mediated, at least in part, through interactions with the endogenous opioid system24,25. We measured endogenous opioid release by decreases in the availability of μ-opioid receptors in vivo during the painful stressor, quantified by means of positron emission tomography (PET) with the selective μ-opioid receptor radiotracer [11C]carfentanil20,21 (see Supplementary Methods for details). In 35 healthy volunteers (Ann Arbor sample), we found that highly expressed NPY diplotypes predicted significantly higher levels of stress-induced μ-opioid system activation in several brain regions (Fig. 3 and Supplementary Table 1) including prefrontal cortex, posterior insula, medial and lateral thalamus, ventral basal ganglia (ventral caudate, ventral putamen and nucleus accumbens) and amygdala (analysis of variance (ANOVA), P < 0.05 after correction for multiple comparisons). NPY diplotype accounted for 13% of the variance in activation of μ-opioid neurotransmission in the amygdala, 18–35% of the variance in prefrontal cortex, thalamus and nucleus accumbens, and 37% of the variance in posterior insular cortex. In comparison with its effects on the activation of endogenous opioid neurotransmission by painful stress, NPY explained less of the variance in the more complex, self-rated pain and affective response phenotypes. NPY diplotypes accounted for 3% of the variance in subjective pain (McGill Pain Questionnaire sensory subscale) and 5% of the variance in emotional experience (Positive and Negative Affectivity Scale negative affect) (Supplementary Fig. 4).
Figure 3. Effect of diplotype-predicted NPY mRNA expression on pain/stress-induced μ-opioid system activation.
Activation (means ± s.e.m.) measured by the percentage change in the μ-opioid receptor binding potential (BP) among the NPY diplotypes (LL, n = 8; LH, n = 21; HH, n = 6) are shown in the dorsomedial prefrontal (dmPFC) and posterior insular (pINS) cortices, ventrolateral thalamus (vlTHA), ventral putamen (PUT) and caudate (CAU) nuclei, nucleus accumbens (NAC) and ventral amygdala (AMY; Supplementary Table 1). Z scores represented by the pseudocolour scale (left) are superimposed on an anatomically standardized MRI image. See also Supplementary Table 1 for details of the localization and statistics.
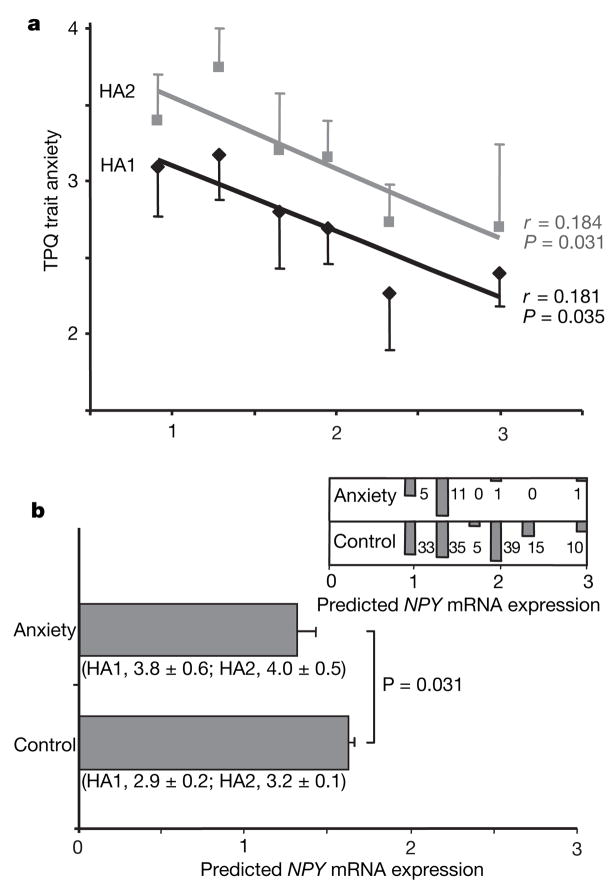
In comparison with gene-influenced brain imaging responses in which allele action has been evident even in small data sets12–14, trait anxiety is a complex behaviour for which gene effects are small11, and it is the type of gene-influenced behaviour that is perturbed by external factors such as exposure to stress10,26. However, it is important to understand the role of NPY in complex behaviours and in different contexts. In a relatively modest sample of 137 healthy Finnish Caucasian controls, expression predicted by NPY diplotype was inversely correlated with trait anxiety (Fig. 4a), measured with the Tridimensional Personality Questionnaire (TPQ) Harm Avoidance subscales HA1 (Fear of Uncertainty; P = 0.035) and HA2 (Anticipatory Worry; P = 0.031). There was no correlation with the HA3 (Shyness with Strangers) or HA4 (Fatigability and Asthenia) subscales. Diplotype-predicted NPY mRNA expression was lower in the small number of Finnish participants with clinical anxiety disorders (n = 18) diagnosed with the Structured Clinical Interview for the Diagnostic and Statistical Manual of Mental Disorders, 3rd edition revised (SCID), in comparison with the same 137 healthy Finnish controls (Fig. 4b). Within Finnish participants with SCID-diagnosed alcoholism (n = 138), drug addiction (n = 38), anxiety disorders (n = 18) and major depression (n = 22), we observed no correlation of NPY-diplotype-predicted expression with any of the HA subscale scores. This may indicate dysregulation of the stress axis in these patients; in addition, the sample sizes were not large. None of the seven individual markers (Supplementary Fig. 1a) were associated with HA subscales. In contrast with the larger effects of NPY diplotype on brain functional responses, the effects of NPY on trait anxiety were modest, accounting for 3.3% of the variance in HA1 and 3.4% of the variance in HA2. Very large study samples are required for the consistent detection of gene effects on a complex behaviour such as trait anxiety. We have presented converging results in which modest effects of genes at the level of complex behaviour are supported, and mechanistically clarified, by larger effects on brain-imaging phenotypes reflecting response to emotion and stress.
Figure 4. Correlations of diplotype-predicted NPY mRNA expression with TPQ trait anxiety and anxiety disorders in Finnish Caucasians.
a, Bivariate regression of HA1 and HA2 scores (means and s.e.m.) and diplotype-predicted expression values in 137 healthy subjects (left to right: H1/H1, n = 33; H1/H3, n = 35; H3/H3, n = 5; H1/H2, n = 39; H2/H3, n = 15; H2/H2, n = 10). b, Diplotype-predicted NPY mRNA levels (means and s.e.m.) of n = 137 healthy controls and n = 18 anxiety patients by a two-tailed t-test (means ± s.e.m. for HA1 and HA2 are also shown). At the top right is a histogram comparing the numbers of controls and anxiety patients within each of the six diplotypes.
Population stratification, a potential confounding factor, was addressed with the use of ancestry informative markers (AIMs). As detailed (Supplementary Fig. 5), 186 highly informative AIMs were genotyped; this process was followed by a factor analysis anchored against a panel of 1,017 Centre d’Étude du Polymorphisme Humain (CEPH) worldwide diversity samples representing 52 populations, and yielding ethnic factor scores for each individual. These 186 AIMs yielded a similar seven-factor solution to that observed27 on the basis of short tandem repeat markers genotyped in the same populations. Our analyses of PET pain/stress response, plasma NPY levels and TPQ Harm Avoidance subscales were not confounded by ethnicity, as revealed by comparisons of individuals above and below medians. For the emotional fMRI imaging sample we had available a set of 15 AIMs, again with no difference between low and high responders (data not shown).
We identified a locus accounting for part of the NPY haplotype effect by testing the four moderately common variants found in the NPY promoter region (−1016 base pairs (bp) to 63 bp; see Supplementary Fig. 6a for details) for their ability to influence mRNA expression. The five naturally occurring allele combinations were inserted into a promoterless reporter gene vector (pDsRed2-1) and NPY promoter haplotype-driven expression was analysed by transient transfection into the raphe neuronal cell line RN46A. As shown in Supplementary Fig. 6b and Supplementary Table 2, the −399C allele (rs16147) accounted for a 30% decrease in basal expression determined by comparing the allelic variations between the promoter haplotypes. In addition, the TGins allele located at −883 bp (rs3037354) may decrease expression, although to an extent not reaching statistical significance. The combination of −399C and −883TGins reduced expression 47%. These results are consistent with the effect of −399C and the smaller effect of −883TGins on mRNA expression in post-mortem brain and lymphoblastoid cell lines, and with the greater predictive value of haplotypes both in vitro and in vivo (Fig. 1b, c, and Supplementary Fig. 3), and underlies our choice to emphasize haplotype effects of NPY.
Haplotype-based association analysis maximizes the ability to capture information, haplotypes serving as proxies for unknown alleles. However, the existence of multiple haplotypes can lead to a loss of analytic power without some mode of clustering, for example on a cladistic basis28. In this study we functionally grouped most NPY haplotypes and diplotypes according to levels of expression in vivo. There were two less common haplotypes (H4, with a frequency of 4.3%, and H5, with a frequency of 4.6%) that were excluded from association analyses because of a lack of definitive in vivo expression data. However, on the basis of in vitro data for both (Supplementary Fig. 6b), and limited brain expression data for H4 (Fig. 1b), it is likely that H4 is a low-expression haplotype and H5 is a high-expression haplotype. In addition, the H5 haplotype uniquely contains the mis-sense variant Leu 7 → Pro (rs16139C). Several studies have associated Pro 7 with disordered glucose and lipid metabolism29,30 but not with anxiety, and Pro 7 was not associated with anxiety in our data set (data not shown).
We observed effects of haplotype-predicted NPY expression on human trait anxiety and on neurobiological circuits and neurotransmitter systems implicated in the regulation of emotional and stress responses. Although the effect of haplotype-predicted NPY expression was modest for trait anxiety, consistent effects were observed across related measures and were more evident for brain metabolic responses to emotional images as measured with fMRI and molecular imaging measures of the activation of the endogenous opioid system after a stressful challenge. These findings indicate the important role of NPY in modulating inter-individual variation in emotion and stress resiliency, and reflect the value of a multilevel approach to the genetic analysis of behaviour.
METHODS SUMMARY
Participants from five independent samples were analysed. A sample of 516 Finnish Caucasians was used to identify NPY haplotype configuration and linkage disequilibrium structure, and subsamples were analysed for lymphoblast NPY mRNA, trait anxiety, and anxiety disorders. The New Haven sample (n = 42) was measured for plasma NPY. The Pittsburgh sample (n = 71) received fMRI after an emotional probe. The Ann Arbor sample (n = 35) underwent [11C]carfentanil PET during a pain/stress challenge. A collection of postmortem brains from the Miami sample was genotyped and 28 heterozygous cerebella were used for differential allele expression. A full description of participants’ demographic data and psychiatric diagnoses is provided in Supplementary Methods. Informed consent was obtained under the auspices of human research protocols approved by institutional review boards of the National Institutes of Health, the University of Helsinki, the University of Pittsburgh, the University of Michigan, and Yale University. NPY genotyping was performed with 5′-nuclease assays. Genotyping with 186 AIMs was performed with an Illumina 1536-SNP array. NPY mRNA quantification in lymphoblastoid cell lines was performed with reverse transcription (RT) followed by TaqMan real-time PCR. Detection of allele-specific NPY expression in postmortem brain was performed with an RT-coupled 5′ nuclease assay. Plasma concentrations of NPY were measured by radioimmunoassay. DNAs containing NPY promoter haplotypes were inserted into a reporter vector, pDsRed2-1, and transfected to the RN46A cell line for the analysis of promoter variants. Details of the fMRI emotional model, the blood-oxygenation-level-dependent (BOLD) fMRI acquisition parameters, the experimental design for pain stressor challenge, the PET scan processes, image reconstruction, and the data analysis are provided in Supplementary Methods.
Supplementary Material
Acknowledgments
This work was supported by grants R01 DA 016423 to J.-K.Z., K01 MH072837 and a NARSAD Young Investigator Award to A.R.H., R01-AA13892, P50-DA16556 and K02-DA17232 to R.S., PO1 HL040962 and RO1 HL065137 to S.B.M., and R01 DE 15396 to C.S.S.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Z.Z. and G.Z. performed molecular biology and genetic analyses. A.R.H., R.E.F., S.B.M. and S.M.B. generated and analysed fMRI data. J.-K.Z., D.S. and C.S.S. generated and analysed PET data. R.S. and R.L.H. generated plasma NPY data. J.-K.Z., A.R.H., Z.Z., G.Z., D.S., M.V. and D.G. all contributed to conceptual issues. M.V. and D.G. were involved in the collection of the Finnish data set. D.C.M. provided post-mortem tissue samples. M.-A.E., R.H.L, X.-Z.H., C.H., K.X., B.B., Q.Y. and P.-H.S. contributed to data analysis. D.G. conceived and planned the study. Z.Z. and D.G. wrote the paper.
References
- 1.Broqua P, Wettstein JG, Rocher MN, Gauthier-Martin B, Junien JL. Behavioral effects of neuropeptide Y receptor agonists in the elevated plus-maze and fear-potentiated startle procedures. Behav Pharmacol. 1995;6:215–222. [PubMed] [Google Scholar]
- 2.Heilig M, Soderpalm B, Engel JA, Widerlov E. Centrally administered neuropeptide Y (NPY) produces anxiolytic-like effects in animal anxiety models. Psychopharmacology (Berl) 1989;98:524–529. doi: 10.1007/BF00441953. [DOI] [PubMed] [Google Scholar]
- 3.Thorsell A, Carlsson K, Ekman R, Heilig M. Behavioral and endocrine adaptation, and up-regulation of NPY expression in rat amygdala following repeated restraint stress. Neuroreport. 1999;10:3003–3007. doi: 10.1097/00001756-199909290-00024. [DOI] [PubMed] [Google Scholar]
- 4.Adrian TE, et al. Neuropeptide Y distribution in human brain. Nature. 1983;306:584–586. doi: 10.1038/306584a0. [DOI] [PubMed] [Google Scholar]
- 5.Allen YS, et al. Neuropeptide Y distribution in the rat brain. Science. 1983;221:877–879. doi: 10.1126/science.6136091. [DOI] [PubMed] [Google Scholar]
- 6.Tatemoto K, Carlquist M, Mutt V. Neuropeptide Y—a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature. 1982;296:659–660. doi: 10.1038/296659a0. [DOI] [PubMed] [Google Scholar]
- 7.McEwen BS, Stellar E. Stress and the individual. Mechanisms leading to disease. Arch Intern Med. 1993;153:2093–2101. [PubMed] [Google Scholar]
- 8.McEwen BS. Allostasis and allostatic load: implications for neuropsychopharmacology. Neuropsychopharmacology. 2000;22:108–124. doi: 10.1016/S0893-133X(99)00129-3. [DOI] [PubMed] [Google Scholar]
- 9.Sinha R, Garcia M, Paliwal P, Kreek MJ, Rounsaville BJ. Stress-induced cocaine craving and hypothalamic–pituitary–adrenal responses are predictive of cocaine relapse outcomes. Arch Gen Psychiatry. 2006;63:324–331. doi: 10.1001/archpsyc.63.3.324. [DOI] [PubMed] [Google Scholar]
- 10.Caspi A, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 11.Sen S, Burmeister M, Ghosh D. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet B Neuropsychiatr Genet. 2004;127:85–89. doi: 10.1002/ajmg.b.20158. [DOI] [PubMed] [Google Scholar]
- 12.Hariri AR, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 13.Heinz A, et al. Amygdala–prefrontal coupling depends on a genetic variation of the serotonin transporter. Nature Neurosci. 2005;8:20–21. doi: 10.1038/nn1366. [DOI] [PubMed] [Google Scholar]
- 14.Pezawas L, et al. 5-HTTLPR polymorphism impacts human cingulate–amygdala interactions: a genetic susceptibility mechanism for depression. Nature Neurosci. 2005;8:828–834. doi: 10.1038/nn1463. [DOI] [PubMed] [Google Scholar]
- 15.Haas BW, Omura K, Constable RT, Canli T. Emotional conflict and neuroticism: personality-dependent activation in the amygdala and subgenual anterior cingulate. Behav Neurosci. 2007;121:249–256. doi: 10.1037/0735-7044.121.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Somerville LH, Kim H, Johnstone T, Alexander AL, Whalen PJ. Human amygdala responses during presentation of happy and neutral faces: correlations with state anxiety. Biol Psychiatry. 2004;55:897–903. doi: 10.1016/j.biopsych.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Tessitore A, et al. Dopamine modulates the response of the human amygdala: a study in Parkinson’s disease. J Neurosci. 2002;22:9099–9103. doi: 10.1523/JNEUROSCI.22-20-09099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hariri AR, et al. A susceptibility gene for affective disorders and the response of the human amygdala. Arch Gen Psychiatry. 2005;62:146–152. doi: 10.1001/archpsyc.62.2.146. [DOI] [PubMed] [Google Scholar]
- 19.McEwen BS. Stress and hippocampal plasticity. Annu Rev Neurosci. 1999;22:105–122. doi: 10.1146/annurev.neuro.22.1.105. [DOI] [PubMed] [Google Scholar]
- 20.Zubieta JK, et al. COMT val158met genotype affects μ-opioid neurotransmitter responses to a pain stressor. Science. 2003;299:1240–1243. doi: 10.1126/science.1078546. [DOI] [PubMed] [Google Scholar]
- 21.Zubieta JK, et al. Regional mu opioid receptor regulation of sensory and affective dimensions of pain. Science. 2001;293:311–315. doi: 10.1126/science.1060952. [DOI] [PubMed] [Google Scholar]
- 22.Moles A, Kieffer BL, D’Amato FR. Deficit in attachment behavior in mice lacking the μ-opioid receptor gene. Science. 2004;304:1983–1986. doi: 10.1126/science.1095943. [DOI] [PubMed] [Google Scholar]
- 23.Watkins LR, Mayer DJ. Organization of endogenous opiate and nonopiate pain control systems. Science. 1982;216:1185–1192. doi: 10.1126/science.6281891. [DOI] [PubMed] [Google Scholar]
- 24.Britton KT, Southerland S. Naloxone blocks ‘anxiolytic’ effects of neuropeptide Y. Peptides. 2001;22:607–612. doi: 10.1016/s0196-9781(01)00371-0. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Li JJ, Yu LC. Anti-nociceptive effect of neuropeptide Y in the nucleus accumbens of rats: an involvement of opioid receptors in the effect. Brain Res. 2002;940:69–78. doi: 10.1016/s0006-8993(02)02594-5. [DOI] [PubMed] [Google Scholar]
- 26.Caspi A, et al. Role of genotype in the cycle of violence in maltreated children. Science. 2002;297:851–854. doi: 10.1126/science.1072290. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Templeton AR. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping or DNA sequencing. V Analysis of case/control sampling designs: Alzheimer’s disease and the apoprotein E locus. Genetics. 1995;140:403–409. doi: 10.1093/genetics/140.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karvonen MK, et al. Association of a leucine(7)-to-proline(7) polymorphism in the signal peptide of neuropeptide Y with high serum cholesterol and LDL cholesterol levels. Nature Med. 1998;4:1434–1437. doi: 10.1038/4027. [DOI] [PubMed] [Google Scholar]
- 30.Niskanen L, et al. Leucine 7 to proline 7 polymorphism in the neuropeptide Y gene is associated with retinopathy in type 2 diabetes. Exp Clin Endocrinol Diabetes. 2000;108:235–236. doi: 10.1055/s-2000-7748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.