Abstract
Chlamydia trachomatis (CT) is the most prevalent sexually transmitted bacterial pathogen worldwide and causes severe reproductive tract infections. Currently, nucleic acid amplification tests (NAATs) are the gold standard for clinical diagnosis but most NAATs are labor intensive and limited to specific CT serovars. We developed and validated a quantitative PCR assay that reproducibly detected CT serovars D, E, F, Ia and C. muridarum over a linear range of 2log10 to 10log10 genomes with low coefficients of variation (CV) from both experimental and human urine samples. CT DNA loads from human vaginal, endocervical and male urethral swabs correlated well with the BD ProbeTec ET assay run in parallel. In a preclinical microbicide evaluation, C. muridarum DNA loads in mouse swabs and tissues correlated well with an immunofluorescence assay. The optimized qPCR system provided enhanced sensitivity and facilitated the quantitative evaluation of clinical and experimental preclinical samples for anti-CT therapeutic and microbicide evaluation.
INTRODUCTION
Chlamydia trachomatis (CT) is a gram-negative, obligate intracellular bacterium implicated in ocular and genital tract infections of humans. CT (serovars D-K) is the most prevalent sexually transmitted bacterial pathogen worldwide (Gerbase et al., 1998) and is the leading cause of non-gonococcal urethritis and mucopurulent cervicitis in males and females, respectively (Shattock et al., 1998). Although most CT infections are subclinical, pelvic inflammatory disease (PID), endometritis and salpingitis can result from ascending infection of the female upper genital tract and may have significant deleterious effects on reproductive health (Eckert et al., 2002; Shattock, et al., 1998). Ultimately, tubal infertility and ectopic pregnancy are important sequelae and have intensified the need for more sensitive and cost-effective tools for clinical diagnostics and pre-clinical evaluation of prospective CT therapeutics.
Our intent was to develop an assay that would quantify the most prevalent genital serovars of CT and the C. muridarum Nigg II strain (formerly MoPn) commonly used in animal modeling of genital disease to provide a reproducible platform for CT detection and quantification. Until recently, cell culture and immunofluorescence have been the standards for clinical CT diagnosis but the sensitivity and reproducibility of these assays have been challenged by the development and acceptance of more specific nucleic acid amplification tests (NAATs) including the Gen-Probe APTIMA Combo 2 (Gen-Probe, San Diego, CA), Roche Cobas Amplicor (Roche Diagnostics, Pleasanton, CA) and BD ProbeTec ET (Becton Dickinson Diagnostic Systems, Franklin Lakes, NJ) (Gaydos et al., 2004; Martin et al., 2004; Templeton et al., 2001; Verkooyen et al., 2003). For laboratory studies, there are inherent problems with the use of fluorescent antibody labeling for quantitative detection of CT including subjective interpretation of results, retaining viability of the specimen following collection and an inconsistency in laboratory methods for CT infection in vitro. NAATs, however, can be highly specific and reproducible assays that detect an extended range of CT bacterial loads (Solomon et al., 2004). In general, conventional CT NAATs are purely qualitative primarily because small variations in the reaction conditions or components can produce large changes during amplification thus preventing quantitative end-point evaluation or reliable statistical analyses (Solomon, et al., 2004). In addition, conventional NAATs for laboratory use generally require post-run manipulation, such as gel electrophoresis, in order to evaluate the results and can introduce contaminating PCR products into the laboratory thereby providing an increased risk of false-positive results.
Quantification of CT in this manuscript studies was accomplished by targeting a highly conserved region within the single copy OmpA gene that encodes the major outer membrane protein (MOMP) for PCR amplification. By sequence comparisons of the OmpA gene from clinical CT serovars D, E, F, Ia, and C. muridarum we identified and targeted a region located at the 3’ end of variable domain IV (VDIV) (Baehr et al., 1988) that showed greater than 90% homology among all of the analyzed serovars. The medium to high-throughput 96-well format facilitates the simultaneous screening and quantitative analysis of samples in a closed-tube reaction that requires no post-run manipulation of the reaction products. The performance data from the quantitative PCR assay reported herein describe a highly specific and sensitive method of quantifying CT DNA from experimental preclinical samples and clinical specimens using a single primer pair and TaqMan probe.
MATERIALS AND METHODS
Bacterial isolates
Clinical isolates of CT serovars D, E, F and Ia, propagated in HeLa 229 cells, were kindly provided by Dr. Alison Quayle, Louisiana State University-Health Science Center, USA. The C. muridarum MoPn biovar strain Nigg II (ATCC VR-123), propagated similarly in McCoy-B cells (ATCC CRL-1696), were maintained in Minimum Essential Medium (MEM; Invitrogen) supplemented with 1x antibiotic-antimycotic, gentamicin (0.1mg/mL) and 10% fetal bovine serum. Infectious CT titers were determined by titration on McCoy-B cells using standard methods and expressed as inclusion forming units (IFU). Purified PCR-quality DNA from CT, normal and abnormal flora of the human urogenital tract including Lactobacillus spp., Prevotella bivia, Mobiluncus spp., group B β Streptococci spp., Mycoplasma genitalium and Atopobium vaginae were kindly provided by Dr. David H. Martin (Louisiana State University-Health Science Center, USA). Other tested species including Pseudomonas aeruginosa, Listeria monocytogenes, Escherichia coli K12, E. coli 083, E. coli 0157, E. coli HS, Klebsiella pneumoniae and Francisella tularensis LVS were kindly provided by Dr. Tonyia Eaves-Pyles (University of Texas Medical Branch, USA).
DNA Extraction
All specimens were processed using the DNeasy 96 Tissue Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol, eluted into 100uL of the kit-provided elution buffer and stored at −20° C until PCR reactions were assembled. The selected bacterial species used to test the cross-reactivity of the assay were extracted similarly, quantified using spectrophotometry (A260) and then diluted to 10ng of genomic DNA per reaction (∼106 genomic equivalents).
Real-time quantitative PCR
Using Beacon Designer v. 2.04 (Premier Biosoft, Intl., Palo Alto, CA, USA), candidate primer pairs and TaqMan probes (Roche Molecular Systems, Pleasanton, CA, USA) were designed to specifically amplify conserved regions of the major outer membrane protein gene in CT, OmpA. OmpA sequences for clinical CT Serovars D, E (2 isolates), F, Ia and C. muridarum strain Nigg II (formerly MoPn) (Genbank Accession no. AY535089, AY535110 and AY535111, AY535125, AY535150 and M64174, respectively) were aligned using the ClustalW algorithm in MacVector v7.0 (Accelrys, San Diego, CA, USA) to select a highly conserved region for primer and fluorogenic probe design (Fig. 1).
Figure 1.

Clustal 1W nucleotide alignment of the primer and TaqMan probe binding positions for C. trachomatis serovars D, E, F, Ia, and C. muridarum (MoPn) OmpA sequences. The presented OmpA regions are located at the 3’ end of variable domain IV and were used to design the forward and reverse primers and the TaqMan probe. Homology of the target DNA is >90% within the selected regions.
For each PCR reaction, 5uL of DNA template was added to 16.5uL of BioRad iQ Supermix (100mM KCl, 40mM Tris-HCl, 1.6mM dNTPs, 0.825U of iTaq DNA polymerase and 6mM MgCl2), 7.5 pMol of each primer (sense primer, 5’-GATACGTTGGACAAGAATTCCCTC-3’ and antisense primer, 5’-GTAAGGAGTGAACATATTCAGTCTGTAA-3’) and 11.25pMol of the TaqMan probe [5’-(6-FAM)CTTGCTTGCCACTCATGGTAATCGA(BHQ1A)-3’] that was then brought to a final volume of 25uL with sterile RNAse/DNAse-free water. Two-step cycling parameters were as follows: Initial denaturation at 95° C for 3 min followed by 50 cycles of 95° C for 20 sec and 63.5° C for 45 sec. A standard curve was generated using 10-fold serial dilutions of cloned OmpA that had been pre-quantified using spectrophotometry (260nm). The cloned OmpA standard dilutions were used as template in duplicate reactions for each PCR run and used to extrapolate unknown experimental CT DNA loads.
Experimental human clinical or murine specimens were normalized using the single-copy GAPDH gene target. Human GAPDH was quantified as described previously (Bourne et al., 2005). For each murine GAPDH PCR reaction, 5uL of DNA template was added to 16.5uL of BioRad iQ Supermix (100mM KCl, 40mM Tris-HCl, 1.6mM dNTPs, 0.825U of iTaq DNA polymerase, and 6mM MgCl2), 5pMol of each primer (sense primer, 5’-TGCTCCTCCCTGTTCCAGAGA-3’ and antisense primer, 5’-AATCCGTTCACACCGACCTTCA-3’) and 7.5pMol of the TaqMan probe [5’-(5-TX-RED)TCTTCTTGTGCAGTGCCAGCCTCGTCC(3BHQ2a)-3' in a final volume of 25uL. Real-time PCR was performed using the iQ5 multicolor real-time PCR detection system (Bio-Rad). The data was analyzed using the iQ5 Optical System software v1.0 to generate amplification plots and calculate PCR efficiencies. The results presented herein are representative of data generated by multiple operators (n=4) within the laboratory.
Detection of CT DNA in clinical specimens
The quantitative CT PCR assay was evaluated as a clinical diagnostic tool by testing human vaginal, endocervical and penile urethral swabs for the presence of CT DNA and comparing the quantitative PCR data to results obtained using the ProbeTec ET CT/NG detection system (BD Diagnostic Systems). Human specimens (n=40) were collected from clinical screening using the ProbeTec ET specimen collection and dry transport system (BD Diagnostic Systems). The samples were first fully processed by the UTMB clinical diagnostic laboratory for ProbeTec analysis. The remaining sample was stored sub-optimally at −20°C for 7–21d in a frost-free cycling freezer and then, with UTMB IRB approval, anonymized and provided for this study. The partially desiccated samples were resuspended in 0.5mL PBS and vortexed to enhance recovery of the collected material for DNA extraction. Two hundred microliters of each sample was removed and added to 180uL lysis buffer (Qiagen) for DNA purification as described above.
Detection of CT DNA in spiked human urine specimens
With UTMB Institutional Review Board approval, urine was collected from a healthy volunteer. Serial 5-fold dilutions of CT serovar E EBs (clinical isolate provided by Dr. Alison Quayle, LSUHSC, USA) were spiked into 400uL of urine to a final concentration of 1×106 to 1.6×103 IFU/mL (immunofluorescence assay, IFA; Chlamydiae Culture Conjugate Confirmation Reagent, Trinity Biotech, Carlsbad CA, USA). The spiked urine was centrifuged at 21,000 × g for 1h, re-suspended in 180uL Buffer ATL (Qiagen) and then processed for DNA extraction and PCR as described above. The influence of urine on the DNA extraction and the efficiency of the PCR was evaluated by comparing the intra-assay reproducibility of each CT dilution quantified in triplicate.
Detection of CT DNA in murine genital specimens
The described qPCR assay was employed in a candidate microbicide evaluation in order to determine whether the assay would be applicable for screening the efficacy of novel CT therapies in experimental animals. With full UTMB IACUC approval, 18–21g female Swiss Webster mice (Harlan, Indianapolis, IN, USA) were pre-treated either with the candidate microbicide or with the PBS vehicle and then challenged intravaginally with 15uL of an inoculum containing 104 IFU of C. muridarum strain Nigg II (Bourne et al., 2003). Vaginal swab samples were collected 3 and 6d post-inoculation (PI) into 1mL SPG medium and then stored at −80° C. CT ascending into the upper genital tract was detected from animals sacrificed 10d PI. Oviducts and ovaries collected from animal upper genital tracts were homogenized in 1mL SPG buffer and then stored at −80°C. Two hundred microliters of each collected swab or tissue homogenate was processed for DNA extraction and PCR quantification as described above. In parallel, the presence of viable CT was established by inoculating 200uL of clarified upper-tract supernatants or swab medium onto confluent McCoy cell monolayers. After 5d, infected cells were fixed with methanol (−20°C) for 3 min, stained using the IFA reagent as described above and then observed for the presence of fluorescent foci using fluorescence microscopy. C. muridarum DNA loads from mice treated with the prospective microbicide candidate or the PBS vehicle were compared to the results generated using the IFA.
RESULTS
CT detection and qPCR performance
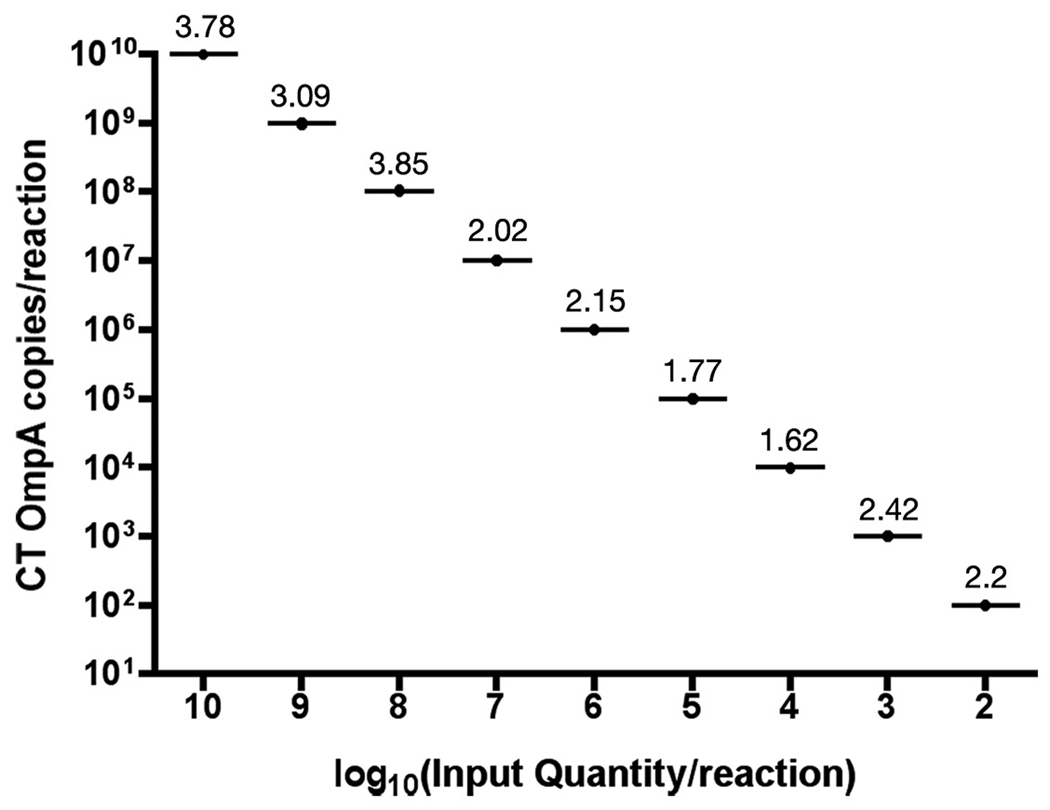
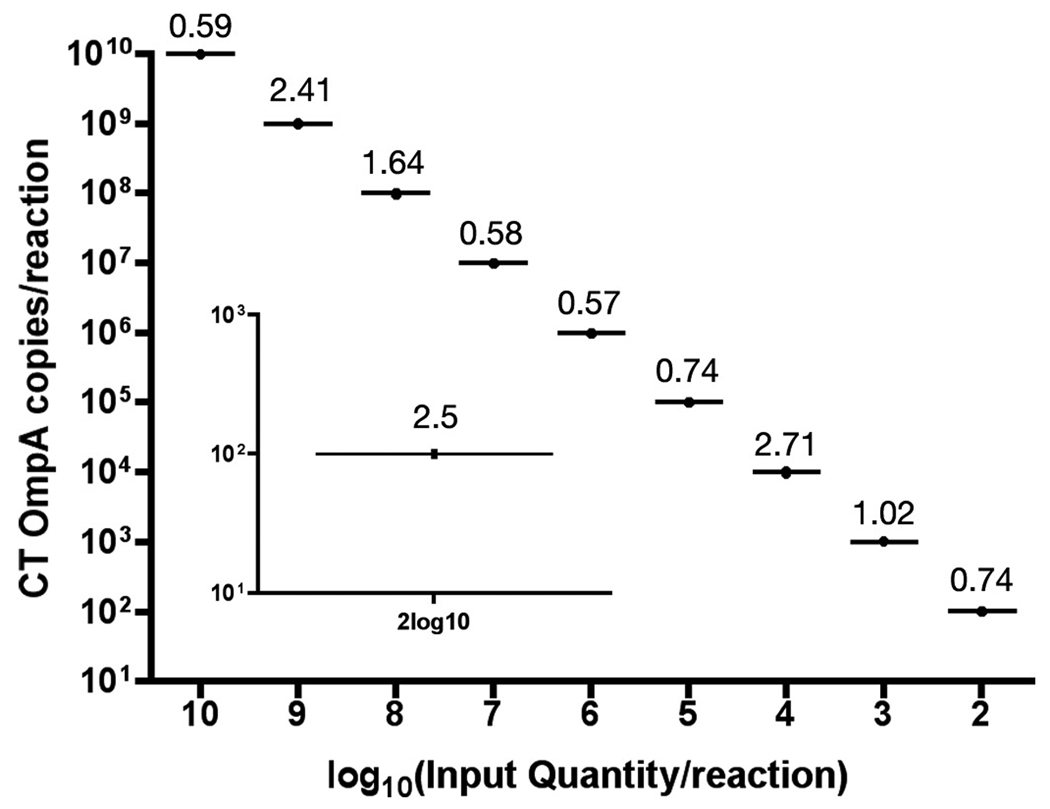
In order to determine the utility of the described qPCR assay we tested whether the assay would identify the most common genital serovars of CT and the widely used C. muridarum biovar for animal modeling of CT disease. All tested CT serovars including D, E, F, Ia and C. muridarum were reproducibly quantified from infected cell cultures (Table 1). The linear range of detection spanned from 2log10 to 10log10 copies per reaction (Fig. 2). Although the lower limit of linear detection was consistently 2log10 copies per reaction, 10 copies were detected in 50% of reactions (n=20, data not shown). The interassay reproducibility, completed by 2 technicians, showed that the complete linear range of detection was characterized by coefficient of variation (CV) values of less than 4% among six PCR runs (Fig. 2). Within a single reaction plate containing three standard curves of 10-fold OmpA serial dilutions, CV values were reduced to less than 2.7% (Fig. 3). The CV value at the lower limit of detection (2log10 copies per reaction) was 2.5 (n=8; Fig. 3, inset).
Table 1.
Microorganisms (n=20) and relevant human and murine specimens tested for specificity of the CT real-time qPCR assay.
| Genusa | Species, serovar, biovara | Result |
|---|---|---|
| Chlamydia | C. trachomatis Serovar D, E, F, Ia | Pos |
| Chlamydia | C. muridarum Nigg II | Pos |
| Lactobacillus | Lactobacillus spp. | Neg |
| Prevotella | P. bivia | Neg |
| Mobiluncus | Mobiluncus spp. | Neg |
| Streptococci | Group B β Streptococci spp. | Neg |
| Mycoplasma | M. genitalium | Neg |
| Atopobium | A. vaginae | Neg |
| Pseudomonas | P. aeruginosa | Neg |
| Listeria | L. monocytogenes | Neg |
| Escherichia | E. coli K12, O83, O157, HS | Neg |
| Klebsiella | K. pneumoniae | Neg |
| Francisella | F. tularensis (LVS) | Neg |
| uninfected human urine | Neg | |
| Human vaginal and cervical flora | Neg | |
| murine vaginal flora | Neg |
Selected organisms represent normal and abnormal flora isolated from the female genital tract and other mucosa.
Figure 2.
Inter-assay precision of the CT quantitative PCR assay. PCR reproducibility was determined by comparing standard curves of 10-fold serial dilutions of DNA plasmids that encoded CT OmpA from 2log10 to 10log10 copies per reaction among six independent PCR runs. Data is representative of 2 technicians. Values above each plot indicate % coefficient of variation.
Figure 3.
Intra-assay precision of the CT real-time PCR assay. Reproducibility within the same PCR run was evaluated by comparing three standard curves of 10-fold serially diluted OmpA DNA plasmids from 2log10 to 10log10 copies per reaction. Inset plot shows reproducibility (n=8 replicates) of the lower detection limit (2log10 copies per reaction). Values above each plot indicate % coefficient of variation.
PCR specificity was evaluated by testing selected bacterial species common to vaginal or urine samples as well as selected pathogens of other mucosa. Real-time PCR of approximately 6log10 genomic equivalents by SYBR-green detection showed no amplified products from any of the bacterial species tested for cross-reactivity. Importantly, no cross-reactivity was observed from any commensal flora present in samples from the normal human or mouse urogenital tract (Table 1). The lack of amplification was confirmed by agarose gel analyses (data not shown).
Evaluation of CT qPCR for clinical specimens
Patient specimens submitted for CT testing at the UTMB clinical laboratory (n=40) including vaginal, endocervical and male urethral swabs were tested in parallel for CT DNA content using our qPCR assay. The PCR results were compared to those generated using the ProbeTec CT/NG detection system. Twenty ProbeTec-positive and twenty ProbeTec-negative samples were blindly selected and randomized. Of 20 ProbeTec-positive samples the qPCR assay detected CT DNA in 18 samples (90%). The average CT DNA titer in each swab specimen was approximately 5000 genomic copies (100 copies per PCR reaction). Separate PCR amplification of human GAPDH was used to verify the presence of DNA and to normalize CT values based upon the integrity of the collected specimen whereby >108 GAPDH copies were observed in each sample (data not shown). Notably, the qPCR assay did identify CT DNA from a single specimen that was reported negative by the ProbeTec assay. The two samples reported positive by the ProbeTec assay that were negative by our qPCR assay were confirmed to be negative by 2 additional independent PCR runs. As a final confirmation of the status, using an independent CT-specific PCR assay (Solomon et al., 2003) the 2 samples again lacked amplifiable CT DNA (data not shown). The human GAPDH levels in both samples were concordant with the rest of the cohort.
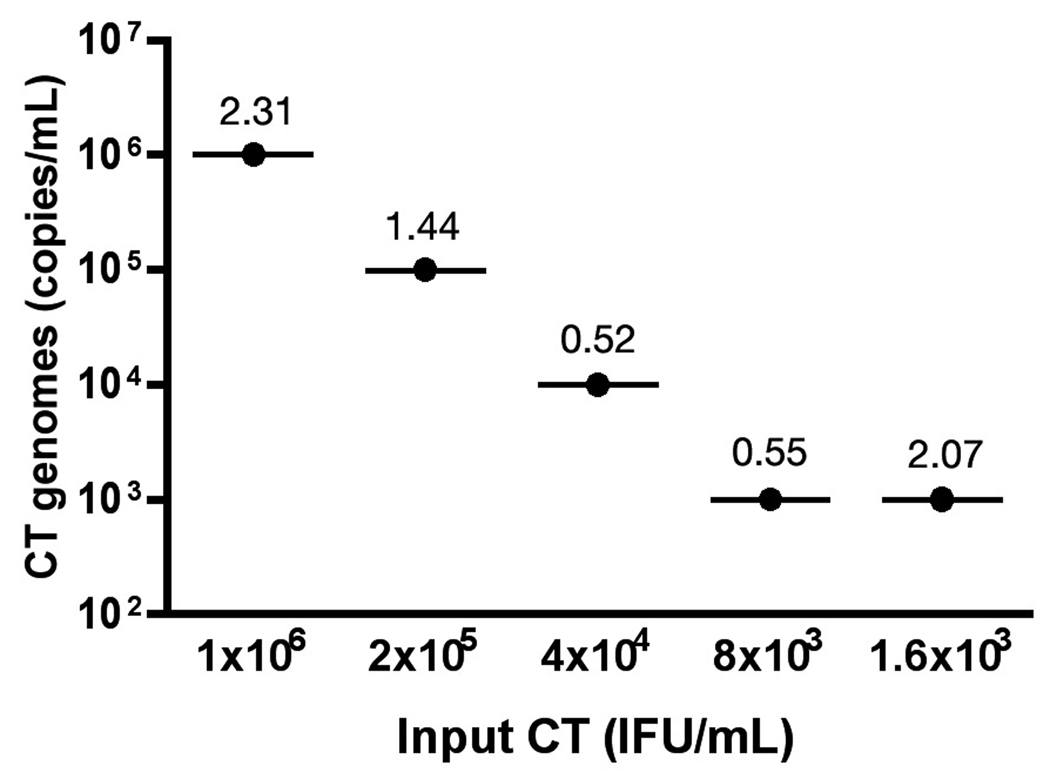
The utility of the qPCR assay for detection of CT DNA from human urine specimens was evaluated using human urine spiked with pre-quantified CT Serovar E IFU. Intra-assay CV values for spiked urine samples were less than 2.5% (n=3; Fig. 4) and qPCR copy numbers were similar to the IFU/mL input concentration at a 1:1 ratio of IFU to genomic copies for most dilutions. As expected, the lowest CT concentration deviated from the 1:1 ratio near the lower limit of detection for the PCR assay. The lowest quantity of DNA that was detected from urine using the described methods was approximately 10 copies per reaction (n=3, CV=3.8), a bacterial load equivalent to 500 IFU/mL (data not shown). Importantly, no negative impact on PCR efficiency or reproducibility was observed when processing human urine samples for CT quantification.
Figure 4.
Performance of the qPCR assay for CT detection from urine. Human urine was spiked with Chlamydia trachomatis serovar E elementary bodies (EB). Five-fold serial dilutions were spiked into urine at concentrations ranging from 3.2log10 to 6log10 IFU/mL and PCR-amplified in triplicate reactions. Infectious CT titers are shown in IFU/mL and compared to quantitative PCR titers presented as genomic copies per milliliter of urine. Values above each plot indicated % coefficient of variation from 3 replicate PCRs.
Quantification of C. muridarum DNA from murine genital specimens
In order to evaluate the utility of this assay for screening CT therapeutics in animals, we quantified bacterial loads in vaginal swabs and upper genital tract tissues from mice experimentally infected with C. muridarum after treatment with a candidate CT microbicide or its vehicle. The qPCR results correlated well with IFA data processed in parallel (Table 2). The predicted trends among microbicide-treated and untreated mice also showed strong correlations between the IFA and PCR results. Four samples were identified positive by IFA but negative by PCR. The PCR, however, did detect 3 samples identified as negative by IFA indicating some discordance between IFA testing and PCR quantification. The IFA-positive but PCR-negative samples were at the lowest detection limit (1 area of fluorescence in the undiluted specimen) and were not reproduced in a replicate IFA study suggesting false-positive IFA results. In a separate pre-clinical study using the CT PCR assay, more than 60% of the samples that were positive by PCR but negative by IFA had PCR titers greater than or equal to 3log10 copies per reaction (data not shown). Statistical differences (p<0.05; Student’s t-test) were observed in the microbicide trial study between the experimental compound and PBS-treated mice at each sampling time (Table 2), a quantitative comparison made possible by the real-time PCR assay. All CT values were normalized to the mean GAPDH value obtained in a separate PCR reaction.
Table 2.
Comparison of IFA and qPCR methods for preclinical evaluation of a candidate CT microbicide compound using a murine model of C. muridarum genital infection.
| Day 3 Swab | Vehicle | Compound |
| IFA-positivea | 11/13 | 1/10 |
| PCR-positiveb | 9/13 | 2/10 |
| Mean PCR Titer +/− SEMc | 2.2×103 +/− 1040 | 6.97 +/− 4.6 |
| Day 6 Swab | ||
| IFA-positive | 11/13 | 5/10 |
| PCR-positive | 11/13 | 3/10 |
| Mean PCR Titer +/− SEM | 2.4×104 +/− 7723 | 14.5 +/− 10.8 |
| Oviducts/Ovaries | ||
| IFA-positive | 10/13 | 1/10 |
| PCR-positive | 12/13 | 1/10 |
| Mean PCR Titer +/− SEM | 2.0×104 +/− 16551 | 4.81 |
Proportion of mice in treatment group identified positive by IFA (Vehicle n=13; Compound n=10)
Proportion of mice in treatment group identified positive by PCR
Average copies of CT genomes per 5uL reaction in positive samples
DISCUSSION
Genital CT infections in both men and women can result in significant reproductive sequelae that extend beyond the primary infection. It is now accepted that NAATs including PCR, ligase chain reaction (LCR), strand-displacement assays and transcription-mediated amplification of rRNA are more sensitive diagnostic tools than culture, IFA or non-amplified DNA probe assays (Black, 1997; Black et al., 2002). Fluorescent probe-based PCR assays, such as those utilizing a TaqMan probe, have advantages over other nucleic acid amplification tests (NAATs) or SYBR-green qPCR assays. The design and utilization of the probe greatly increases detection specificity by relying both on complimentarity of the primers and hybridization of a highly specific oligonucleotide probe. Importantly, real-time PCR can generate quantitative data extrapolated from a standard curve run in parallel thus permitting a more informative analysis of any specimen type in a high-throughput format.
The ideal CT detection system should reproducibly detect low levels of bacteria using minimally invasive collection methods and should recognize the most prevalent genital serovars in circulation. The genital serovars of CT are denoted D-K but recent analysis of CT genital infections in the United States indicated that serovars D, E, F and Ia currently are the most prevalent cause of urogenital infections (Millman et al., 2004). The assay described in this manuscript recognizes CT serovars D, E, F, Ia and C. muridarum for animal modeling of CT disease (Table 1). Based on nucleotide alignment of the primer and probe sequences, the assay should also efficiently recognize serovar G (data not shown) but this was not tested directly. Considering the other genital serovars, some nucleotide divergence was observed within the forward primer binding site of OmpA from Serovar J while OmpA sequence data spanning the target region is not yet available for serovars H and K. Finally, OmpA sequence from the closely related Chlamydia pneumoniae, a respiratory tract pathogen, has significant divergence in the target region suggesting it would not be recognized by this assay.
Among the tested serovars, quantitative results from the CT PCR assay showed highly reproducible inter-assay precision (Fig. 2) and intra-assay reproducibility among replicate samples within the same run (Fig. 3) indicating that the assay generates highly reproducible results within the same PCR run and among multiple technicians within the laboratory. The lower limit of detection for cloned OmpA plasmids was consistently 2log10 copies per reaction. However, the linear range of detection was broad and spanned from 2log10 to 10log10 copies per reaction (Fig. 2) providing increased utility both clinically and in the laboratory setting. Such quantitative outcomes would be especially useful for evaluations of antimicrobial responses in clinical trials of novel therapies. Robust and reproducible performance characteristics supported a more thorough evaluation of the assay utility for screening human clinical specimens.
Human vaginal, endocervical and penile urethral swabs were used to evaluate the utility of the described PCR assay for clinical CT detection. Results from the CT qPCR assay were compared to those generated by the ProbeTec ET CT detection system. The concordance rate among positive CT samples was very good (90%). The two clinical specimens that were identified positive by ProbeTec but not detected by the CT qPCR assay were confirmed negative in 2 additional PCR runs and had normal GAPDH copy numbers indicating sample collection, processing and DNA extraction were successful. Using an independent CT-specific PCR assay (Solomon et al., 2003), we verified the lack of detectable CT DNA in these two specimens (data not shown) suggesting false-positive ProbeTec results. However, ProbeTec-positive but PCR-negative results also could have been attributed to testing only a residual portion of the sample after the ProbeTec processing.
Ninety-five percent (19 of 20) of samples that were reported negative by ProbeTec also were reported negative by qPCR. However, CT DNA was detected from a single specimen that was reported negative by ProbeTec. These results were confirmed in a second PCR run with similar quantitative results for both CT and human GAPDH. Collectively, the preliminary evaluation of clinical utility indicated that, the CT PCR assay performance was similar to the ProbeTec system but may afford some enhanced sensitivity. Although a larger study would be required to fully validate the qPCR assay for clinical application, the concordance rate with the BD ProbeTec system and the quantitative nature of the PCR assay indicate the enhanced utility for quantifying CT loads in reproductive tract specimens and warrants continued investigation.
NAAT screening of urine for CT infection is an attractive diagnostic method because it is minimally invasive. The lower limit of detection reported for our qPCR system from human urine specimens is equivalent to 500 IFU/ml. Because patient urine was not available to us from the clinical laboratory, we tested urine spiked with CT serovar E EBs to construct clinical specimens whereby the total volume of urine for each tested specimen was less than 0.5mL. Clinically, a much larger volume of urine would be collected and therefore the lower limit of detection likely could be reduced by centrifugal concentration prior to DNA extraction. However, the dynamic range of the assay (2log10–10log10) largely reduces the need to dilute or further concentrate clinical or laboratory samples (Fig. 2) and facilitates use with cell culture-based and animal modeling studies in which higher titers would be predicted.
The presented results indicated that the CT PCR assay is a sensitive, specific and reproducible method to quantify CT from mouse lower genital tract swabs and upper-tract tissue specimens. In a candidate microbicide study, the qPCR assay enhanced the ability to evaluate efficacy with remarkably reproducible PCR titers at each sampling time that mirrored the qualitative IFA results generated in parallel (Table 2). Increased SEM values likely indicate the variability of CT disease manifestations in an outbred mouse population and provide a more informative quantitative measure of CT burden compared to IFA. Use of this qPCR assay will allow for more thorough quantitative analyses of candidate anti-CT compounds in laboratory animals thus resulting in reduced animal requirements to achieve analyzable outcomes. In addition, use of the qPCR assay in vitro will provide a quantitative measure of a candidate anti-CT compound’s efficacy and allow for prioritization of prospective therapies.
The qPCR system reported herein is a highly specific and sensitive method for the quantification of CT and C. muridarum DNA from various clinical and preclinical specimens using a single primer pair and TaqMan probe. The closed reaction 96-well format provides medium to high-throughput utility for sample screening and greatly reduces the possibility of exogenous contamination. Specific probe-based real-time PCR assays can reduce the clinical time to diagnosis, provide an accurate and informative monitor of therapy and provide quantitative outcome measures for in vitro and animal modeling of human CT disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Baehr W, Zhang YX, Joseph T, Su H, Nano FE, Everett KD, Caldwell HD. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci U S A. 1988;85:4000–4004. doi: 10.1073/pnas.85.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black CM. Current methods of laboratory diagnosis of Chlamydia trachomatis infections. Clin Microbiol Rev. 1997;10:160–184. doi: 10.1128/cmr.10.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black CM, Marrazzo J, Johnson RE, Hook EW, 3rd, Jones RB, Green TA, Schachter J, Stamm WE, Bolan G, St Louis ME, Martin DH. Head-to-head multicenter comparison of DNA probe and nucleic acid amplification tests for Chlamydia trachomatis infection in women performed with an improved reference standard. J Clin Microbiol. 2002;40:3757–3763. doi: 10.1128/JCM.40.10.3757-3763.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne N, Pyles RB, Yi M, Veselenak RL, Davis MM, Lemon SM. Screening for hepatitis C virus antiviral activity with a cell-based secreted alkaline phosphatase reporter replicon system. Antiviral Res. 2005;67:76–82. doi: 10.1016/j.antiviral.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Bourne N, Zaneveld LJ, Ward JA, Ireland JP, Stanberry LR. Poly(sodium 4-styrene sulfonate): evaluation of a topical microbicide gel against herpes simplex virus type 2 and Chlamydia trachomatis infections in mice. Clin Microbiol Infect. 2003;9:816–822. doi: 10.1046/j.1469-0691.2003.00659.x. [DOI] [PubMed] [Google Scholar]
- 6.Eckert LO, Hawes SE, Wolner-Hanssen PK, Kiviat NB, Wasserheit JN, Paavonen JA, Eschenbach DA, Holmes KK. Endometritis: the clinical-pathologic syndrome. Am J Obstet Gynecol. 2002;186:690–695. doi: 10.1067/mob.2002.121728. [DOI] [PubMed] [Google Scholar]
- 7.Gaydos CA, Theodore M, Dalesio N, Wood BJ, Quinn TC. Comparison of three nucleic acid amplification tests for detection of Chlamydia trachomatis in urine specimens. J Clin Microbiol. 2004;42:3041–3045. doi: 10.1128/JCM.42.7.3041-3045.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerbase AC, Rowley JT, Mertens TE. Global epidemiology of sexually transmitted diseases. Lancet. 1998;351(Suppl 3):2–4. doi: 10.1016/s0140-6736(98)90001-0. [DOI] [PubMed] [Google Scholar]
- 9.Martin DH, Nsuami M, Schachter J, Hook EW, 3rd, Ferrero D, Quinn TC, Gaydos C. Use of multiple nucleic acid amplification tests to define the infected-patient “gold standard” in clinical trials of new diagnostic tests for Chlamydia trachomatis infections. J Clin Microbiol. 2004;42:4749–4758. doi: 10.1128/JCM.42.10.4749-4758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millman K, Black CM, Johnson RE, Stamm WE, Jones RB, Hook EW, Martin DH, Bolan G, Tavare S, Dean D. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J Bacteriol. 2004;186:2457–2465. doi: 10.1128/JB.186.8.2457-2465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shattock RM, Patrizio C, Simmonds P, Sutherland S. Detection of Chlamydia trachomatis in genital swabs: comparison of commercial and in house amplification methods with culture. Sex Transm Infect. 1998;74:289–293. doi: 10.1136/sti.74.4.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon AW, Holland MJ, Burton MJ, West SK, Alexander ND, Aguirre A, Massae PA, Mkocha H, Munoz B, Johnson GJ, Peeling RW, Bailey RL, Foster A, Mabey DC. Strategies for control of trachoma: observational study with quantitative PCR. Lancet. 2003;362:198–204. doi: 10.1016/S0140-6736(03)13909-8. [DOI] [PubMed] [Google Scholar]
- 13.Solomon AW, Peeling RW, Foster A, Mabey DC. Diagnosis and assessment of trachoma. Clin Microbiol Rev. 2004;17:982–1011. doi: 10.1128/CMR.17.4.982-1011.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Templeton K, Roberts J, Jeffries D, Forster G, Aitken C. The detection of Chlamydia trachomatis by DNA amplification methods in urine samples from men with urethritis. Int J STD AIDS. 2001;12:793–796. doi: 10.1258/0956462011924416. [DOI] [PubMed] [Google Scholar]
- 15.Verkooyen RP, Noordhoek GT, Klapper PE, Reid J, Schirm J, Cleator GM, Ieven M, Hoddevik G. Reliability of nucleic acid amplification methods for detection of Chlamydia trachomatis in urine: results of the first international collaborative quality control study among 96 laboratories. J Clin Microbiol. 2003;41:3013–3016. doi: 10.1128/JCM.41.7.3013-3016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]