Abstract
DLC1, which encodes a Rho GTPase activating protein (Rho-GAP), is a potent tumor suppressor gene that is frequently inactivated in several human cancers. DLC-1 is a multidomain protein that has been shown previously to bind members of the tensin gene family. Here we show that p120Ras-GAP (Ras-GAP; also known as RASA1) interacts and extensively colocalizes with DLC1 in focal adhesions. The binding was mapped to the SH3 domain located in the N-terminus of Ras-GAP and to the RhoGAP catalytic domain, located in the C-terminus of the DLC1. In vitro analyses with purified proteins determined that the isolated Ras-GAP SH3 domain inhibits DLC1 RhoGAP activity, suggesting that Ras-GAP is a negative regulator of DLC1 RhoGAP activity. Consistent with this possibility, we found that ectopic overexpression of Ras-GAP in a Ras-GAP-insensitive tumor line impaired the growth suppressing activity of DLC1 and increased RhoA activity in vivo. Our observations expand the complexity of proteins that regulate DLC1 function and define a novel mechanism of the cross-talk between Ras and Rho GTPases.
Keywords: Deleted in liver cancer, Ras-GAP, protein interaction, modulation of tumor cell proliferation, Rho-GAP, SH3, H-ras mutation
Introduction
In the past several years, the DLC1 (Deleted in Liver Cancer 1) gene, which encodes a ubiquitously expressed Rho GTPase-activating protein (Rho-GAP), has attracted considerable interest because it is frequently down-regulated in a variety of tumor types and possesses potent oncosuppressive activity in cell lines derived from several major cancers (Yuan et al., 1998; Durkin et al., 2007a). Consistent with these observations, DLC1 has recently been confirmed as a bona fide tumor suppressor gene that regulates RhoA in an in vitro/in vivo reconstitution mouse model of hepatocellular carcinogenesis (Xue et al., 2008). Recent genome-wide sequencing analyses of prostate, colon and pancreatic cancers have also identified missense mutation of DLC1 (Liao et al., 2008; Jones et al., 2008). DLC1 is the prototype of a multigene family that includes two other closely related genes, DLC2 and DLC3, which have been studied less extensively than DLC1. DLC2 and DLC3 are widely expressed in normal cells and, as is true of DLC1, are also inactivated in several different cancers. Limited comparative analysis between the three DLC genes has suggested that at least one of the DLC genes is expressed in many tumors (Durkin et al., 2007b; Ullmannova and Popescu, 2006).
The full inhibitory effects on cell growth and tumorigenicity induced by wild type DLC genes require their RhoGAP activity, which can negatively regulate RhoA, RhoB, RhoC, and, to a lesser degree, CDC42 (Leung et al., 2005; Wong et al., 2005; Healy et al., 2007;Kawai et al 2007; Kim et al., 2007;Guan et al 2008). Inactivation of DLC1, via genetic and epigenetic changes, may represent the most frequent mechanism for aberrant activation of Rho GTPases in human oncogenesis. However, the function of DLC1 appears to be complex, as analysis of DLC1 mutants in which the RhoGAP function has been inactivated indicates that RhoGAP-independent mechanisms may also contribute to the anti-oncogenic activity of DLC1 (Kim et al., 2007; Healy et al., 2007; Qian et al., 2007; Wong et al., 2005). Consistent with this hypothesis, DLC1 has a multidomain structure that includes near its N-terminus a sterile alpha motif (SAM), which in other proteins has been implicated in protein-protein interactions, and at its C-terminus a steroidogenic acute regulatory–related lipid transfer (START) domain, in addition to the RhoGAP domain, which is located upstream from the START domain (Yuan et al., 1998). The presence of these various motifs, together with the RhoGAP-independent activities, has suggested that DLC1 might interact with other proteins in addition to Rho GTPases. Identification of such interacting partners could enhance our understanding of the mechanisms that regulate the function of DLC1. Thus far, the only additional binding partners that have been clearly identified are members of tensin family, which interact with DLC1 sequences located between the SAM and Rho-GAP domains (Liao et al., 2007; Yam et al., 2006; Qian et al., 2007). Structure-function analysis of DLC1 demonstrated that suppression of cell migration and anchorage-independent growth by DLC1 in a lung cancer cell line required cooperation between the RhoGAP activity and tensin binding activity, although the two functions did not depend on each other (Qian et al., 2007).
To identify other binding partners, a yeast two-hybrid screen with DLC1 was carried out. Here, we report that p120Ras-GAP (Ras-GAP), which is a ubiquitously expressed negative regulator of Ras that is encoded by the Ras-GAP gene (also know as RASA1), was identified by this screen as a novel binding partner for DLC1. We have confirmed that this interaction occurs in mammalian cells, mapped the regions of DLC1 and Ras-GAP that mediate binding, and provided data suggesting that Ras-GAP can serve as a negative regulator of the RhoGAP and tumor suppressor activity of DLC1.
Materials and methods
Cell culture and transfection
The human SK-Hep1 (hepatocellular carcinoma), PC-3 (prostate carcinoma), HCT15 (colon carcinoma), and HEK293 (embryonic kidney) cell lines were obtained from ATCC (Manassas, VA). SK-Hep1 cells were cultured in DMEM medium, and the others in RPMI 1640 medium containing 10% (v/v) fetal calf serum at 37°C in a humidified 5% CO2 atmosphere. Transient transfection was carried out with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Stable HCT15 clones expressing Ras-GAP were generated by transfection with Lipofectamine2000 followed by selection with neomycin.
Yeast two-hybrid assay
The yeast two-hybrid screen was performed by Myriad Genetics (Salt Lake City, UT). A DLC-1 cDNA fragment encoding a 291 amino acid polypeptide (residues 590−880) was subcloned in-frame into pGBT.super B containing the GAL4 DNA-binding domain. A human lung cDNA library was screened with the PNY 200 yeast strain according to the instructions of the manufacturer. The bait and prey plasmids were co-transformed into naive yeast strain to confirm the interaction, and analyzed by DNA sequencing.
Vector construction and adenovirus production
An adenovirus encoding the DLC1 cDNA was prepared as previously described (Guan et al., 2008; Zhou et al., 2008). The Ras-GAP encoding plasmid was kindly provided by Alex Papageorge (NCI, Bethesda, MD). Bacterial expression vectors encoding glutathione S-transferase (GST) fusion proteins containing the Ras-GAP SH3 domain were provided by Jeffrey Settleman (MGH, Charlestown, MA). To narrow the domain of DLC1 that interacts with Ras-GAP, a series of vectors for expressing truncated fragments of DLC1 (residues 590−641, 641−847, 798−880, 847−880,590−880) with an N-terminal V5 tag were constructed. Briefly, cDNAs corresponding to these truncated fragments were subcloned into the pENTER/D-TOPO vector (Invitrogen, Carlsbad, CA), and the cDNA inserts were then transferred into the pcDNA3.1/nV5-DEST vector by means of the Gateway system using LR Clonase (Invitrogen, Carlsbad, CA). All constructs were verified by DNA sequencing.
Immunoprecipitation and immunoblotting
DLC1-positive SK-Hep1 cells or HEK293 cells transiently transfected with truncated DLC1 fragments with the V5 tag were collected and lysed in immunoprecipitation lysis buffer. Anti-DLC1 (H260) polyclonal antibodies, anti-Ras-GAP (B4F8) monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-V5 monoclonal antibodies (Invitrogen, Carlsbad, CA), and protein G-Sepharose slurry (Zymed Inc, San Francisco, CA) were added into pre-cleared cell lysates. Immunoprecipitates were washed and subjected to immunoblotting analyses using the antibodies to DLC1 or Ras-GAP.
Pull down assay
Agarose-conjugated Ras-GAP N-SH2 (175−274), SH3 (277−346) or C-SH2 (343−442) domain proteins with the GST tag were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). SK-Hep1 cells and truncated fragments of DLC1 (residues 641−847) with an N-terminal V5 tag transiently transfected into HEK293 cells were lysed, and cell lystates were pre-cleared. Ten micrograms of each agarose-conjugated recombinant protein were added to 500 μg of total cell lysate, respectively, and rotated at 4°C for 4 h. The pellets were then washed with lysis buffer and collected for immunoblotting analysis using the anti-DLC1 polyclonal antibodies, anti-V5 antibody or anti-GST antibody (Pierce, Rockford, IL).
Confocal microscopy
PC-3 cells infected with Ad-DLC1 at 50 M.O.I and SK-Hep1 were grown on a glass chamber and incubated at 37 °C for 48 h. Cells were fixed and permeabilized in ice-cold methanol and then blocked with 1% BSA at room temperature. Coverslips were stained with the anti-DLC1 (H260), anti-Vinculin (H300) or anti-Ras-GAP (B4F8) antibody (Santa Cruz Biotechnology, Santa Cruz, CA) followed by anti-Rabbit Alexa488 or anti-mouse Alexa 568-conjugated secondary antibody (Molecular Probes Inc. Eugene, OR) and examined by a Zeiss 510 confocal microscopy.
In Vitro RhoGAP Assay
The in vitro GAP activity of DLC1 was measured with a fluorescence-based technique as previously described (Healy et al., 2007). This assay uses a fluorescent labeled, phosphate-binding protein (PBP; provided by Martin Webb, National Institute for Medical Research, Mill Hill, London) sensor. A change of PBP conformation, caused by binding to a single Pi, is coupled to a measurable increase in fluorescence of the fluorophore (Shutes and Der, 2005). Briefly, RhoA GST-fusion protein, the Rho-GAP domain of DLC1 (609−878), and the SH3 and SH2-SH3-SH2 domains of Ras-GAP were purified from BL-21 E. coli cells by glutathione Sepharose 4B chromatography. The RhoA-GST protein was preloaded with GTP. Hydrolysis assays were initiated by adding 30 nM DLC1 RhoGAP domain in an assay buffer. A molar excess (1 μM) of SH3 or SH2-SH3-SH2 domains of Ras-GAP were also added to the DLC1 RhoGAP domain. The increases in Pi production from GTP hydrolysis were measured with a SpectraMAX Gemini (Molecular Devices) spectrofluorimeter by checking increases in fluorescence (λex = 425 nm and λem = 465 nm). Hydrolysis curves were fitted to a single-exponential function with GraphPad Prism software to determine the times to 50% hydrolysis.
RhoA activity analysis
Active RhoA was measured using an ELISA-based RhoA activation assay kit (Cytoskeleton, Denver, CO) according to the manufacturer's protocol. Briefly, after 24 h of serum-starvation and 30 min of calpeptin stimulation, cells were lysed, and the protein concentration was determined. Thirty micrograms of cell lysate from each sample were incubated in micro-wells coated with the isolated Rhotekin Rho-binding domain, and active RhoA was measured using indirect immunodetection followed by a colorimetric reaction measured by absorbance at 490 nm.
MTT assay
Cells were seeded in 96-well plates and infected with Ad-DLC1 or Ad-LacZ at a multiplicity of infection of 50. The MTT [3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide] (Roche, Indianapolis, IN) assay was used to test cell proliferation as previously described (Guan et al., 2008).
Ras Activity assay
Ras activity was determined using an EZ-Detect™ Ras Activation Kit (Pierce, Rockford, IL) according to the manufacturer's protocol. Briefly, 500 μg of cell lysate from each sample were incubated with a SwellGel Immobilized Glutathione Disc and GST-Raf-RBD for 1 h at 4°C, and pellets were washed with lysis buffer and boiled in sample buffer. Samples were then subjected to western blotting with anti-Ras antibody (Pierce, Rockford, IL).
Results
Identification of Ras-GAP as a binding partner of DLC1
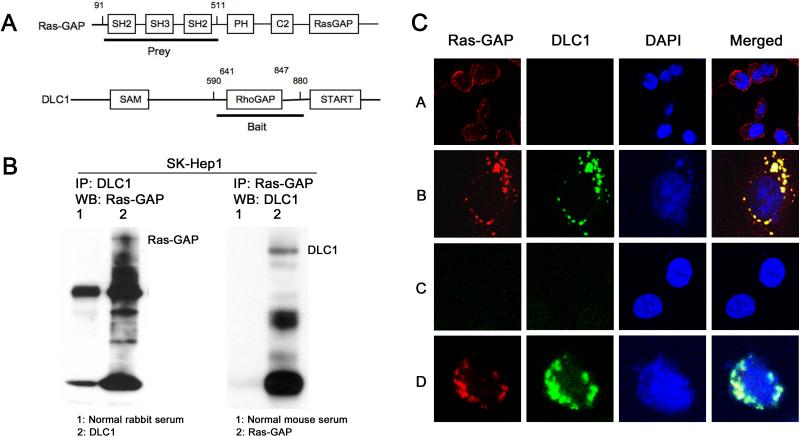
In a yeast two-hybrid assay, an interaction was identified between DLC1 and Ras-GAP. This positive result occurred when amino acids 590−880 of human DLC1, which is composed mainly of the RhoGAP domain, were used as bait to trap N-terminal residues 91−511 of Ras-GAP as prey (Fig. 1A). To confirm that this interaction also occurs with the endogenous proteins in mammalian cells, we carried out immunoprecipitation and Western blotting with extracts from a human hepatocellular carcinoma (HCC) cell line, SK-Hep1, and a human prostate cancer cell line, DU145 that express high levels of endogenous Ras-GAP and DLC1 proteins. Consistent with the yeast two-hybrid results, complex formation between Ras-GAP and DLC1 was detected in the cell extracts (Fig. 1B; data shown for SK-Hep1).
Figure 1. Identification of Ras-GAP as a DLC1 binding partner.
A: The interaction regions of Ras-GAP (upper) and DLC1 (lower) identified by yeast two-hybrid assay. The numbers refer to the amino acids of the DLC1 bait and p120Ras-GAP prey that gave the positive yeast two-hybrid result.
B: DLC1 forms a complex with Ras-GAP. Extracts from SK-HEP1 cells were immunoprecipitated (IP) using Ras-GAP (left panel, lane 2) or DLC1 antibody (right panel, lane 2) and analyzed by immunoblotting (WB; left panel, Ras-GAP antibody; right panel, DLC1 antibody). The arrowhead in the left panel indicates coprecipitated Ras-GAP, and the arrow in the right panel shows coprecipitated DLC1. Normal rabbit or mouse serum (lane 1 in both panels) was used as IP control.
C: Colocalization of DLC1 with Ras-GAP in human cells. DLC1-negative PC-3 cells were infected with Ad-LacZ (A) or Ad-DLC1 (B), and Sk-HEP1 (C, D) cells, which contain endogenous DLC1, were stained with DLC1 and Ras-GAP antibodies. Ectopic (B) and endogenous (D) DLC1 protein as well as endogenous Ras-GAP protein were detected primarily in peripheral punctate areas in the cytoplasm, with extensive colocalization of the two proteins (B, D). Arrow indicates one area of colocalization of Ras-GAP with ectopic (B) endogenous (D) DLC1 in the cytoplasm.
Ras-GAP colocalizes with DLC1
To determine the subcellular compartment(s) in which DLC1 interacts with Ras-GAP in mammalian cells, double immunofluorescent staining was performed in SK-Hep1 cells, which as noted above contain endogenous DLC1 and Ras-GAP, and in the DLC1-negative prostate carcinoma PC-3 line that had been infected with Ad-DLC1, a recombinant adenovirus that expresses DLC1. Using anti-DLC1 or anti-Ras-GAP antibodies and staining with Alexa 488 (green) or Alexa 568 (orange-red)-conjugated secondary antibody, there was extensive co-localization of DLC1 and Ras-GAP to peripheral punctuate areas in the cytoplasm (Fig. 1C). By using vinculin as a marker for focal adhesions, it was confirmed that Ras-GAP and DLC1 are co-localized predominantly at focal adhesions (Supplementary Fig.1).
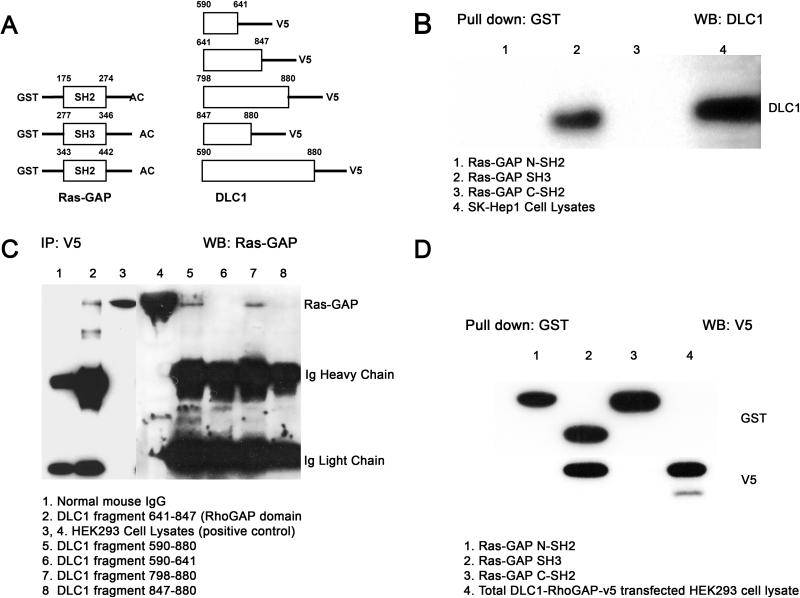
DLC1 RhoGAP catalytic domain binds to Ras-GAP SH3 domain
Ras-GAP contains multiple domains, including an amino-terminal SH3 domain flanked by two SH2 domains, a pleckstrin homology domain and a carboxy-terminal GAP domain (Fig. 1A) (Trahey et al., 1988; Vogel et al., 1988). The binding region of Ras-GAP identified in the yeast two-hybrid, amino acids 91−511, contains both of the SH2 domains and the SH3 domain. In other proteins, these domains are common docking sites for protein-protein interaction (Bradshaw and Wassman, 2002; Musacchio, 2002; Mayer and Eck, 1995). Deletion mapping indicated that the SH3 domain was sufficient to bind full-length endogenous DLC1 in SK-Hep1 cells, and that the SH2 domains did not interact with DLC1 (Fig. 2A, B). For DLC1, the yeast two-hybrid bait spanned amino acids 590−880, which, as noted above, is composed primarily of the catalytic RhoGAP domain of DLC1 (amino acids 641−867). Deletion mapping of interaction by this fragment indicated that the DLC1 RhoGAP catalytic domain was sufficient for binding Ras-GAP in HEK293 cells, and some of this binding occurs via amino acids 798−847 (Fig. 2A, C, D).
Figure 2. DLC1 Rho-GAP domain binds to Ras-GAP SH3 domain.
A: Illustration of fragments of agarose-conjugated (AC) Ras-GAP N-SH2, SH3, and CSH2 with GST tag (left) and constructs of DLC1 with V5 tag (right). The numbers refer to the amino acids for the respective domains of each protein.
B: SK-Hep1 cell lysates were incubated with agarose-conjugated Ras-GAP N-SH2, SH3 and C-SH2 GST fusion proteins and bound proteins were subjected to Western Blotting analysis using DLC1 antibody. Total cell lysate was used as positive control. Lane 1, Ras-GAP N-SH2; Lane 2, Ras-GAP SH3; Lane 3, Ras-GAP C-SH2; Lane 4, cell lysate.
C: HEK293 cells were transiently transfected with V5-tagged DLC1 constructs, cell lysates were immunoprecipitated with V5 antibody, and pellets were analyzed by immunoblotting with Ras-GAP antibody. Total cell lysate was used as positive control, and normal mouse serum as control. The arrow indicates Ras-GAP. Lane 1, negative control; Lane 2, DLC1 fragment 641−847 (Rho-GAP domain); Lanes 3,4 positive control; Lane 5, fragment 590−880; Lane 6, fragment 590−641; Lane 7, fragment 798−880; Lane 8, fragment 847−880.
D: Lysates from HEK293 cells transfected with DLC1 Rho-GAP-V5 (641−847) were incubated with Ras-GAP agarose-conjugated (AC) N-SH2, SH3, and C-SH2 GST fusion proteins, and bound proteins were analyzed by immunoblotting with anti-GST (upper) or anti-V5 antibody (lower). Total cell lysate was used as positive control. Lane 1, Ras-GAP N-SH2; Lane 2, Ras-GAP SH3; Lane 3, Ras-GAP C-SH2; Lane4, cell lysate.
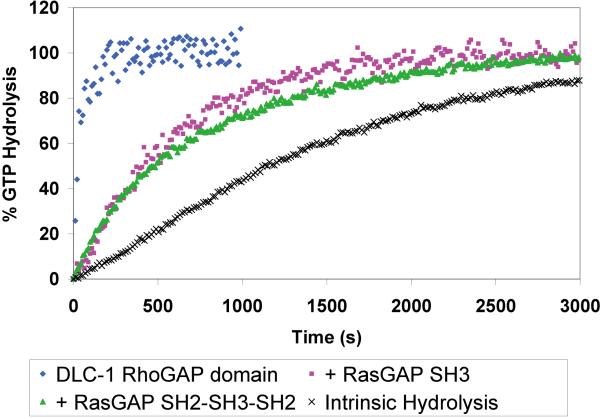
SH3 domain of Ras-GAP inhibits DLC1 Rho-GAP activity in vitro
To assess the functional relevance of Ras-GAP binding to DLC1, we investigated whether the isolated SH3 or tandem SH2-SH3-SH2 domains of Ras-GAP could affect the GAP activity of the DLC1 RhoGAP catalytic domain towards RhoA. We used a single-turnover fluorescent assay to measure in real-time the hydrolysis of GTP, using purified bacterially expressed proteins. As shown in Figure 3, the RhoGAP domain of DLC1 induced a massive increase in the rate of RhoA-GTP hydrolysis. Interestingly, both the isolated SH3 domain and the tandem SH2-SH3-SH2 domains strongly inhibited the RhoGAP activity, by 15- and 18-fold, respectively (Table 1). These observations serve as further confirmation of direct binding between the Ras-GAP SH3 domain and the DLC1 RhoGAP domain and, more importantly, show that Ras-GAP binding has the functional effect of inhibiting the RhoGAP activity of DLC1.
Figure 3.
Effect of Ras-GAP SH3 domain on the catalytic activity of DLC-1 Rho-GAP. Bacterially expressed DLC-1 Rho-GAP domain and the SH3 and tandem SH2-SH3-SH2 domains of Ras-GAP were purified for in vitro analysis of Rho-GAP activity. Purified RhoA was preloaded with GTP, and GTP hydrolysis was monitored with a single-turnover assay in which a fluorescent PBP (phosphate binding protein) undergoes an increase in fluorescence upon binding free inorganic phosphate.
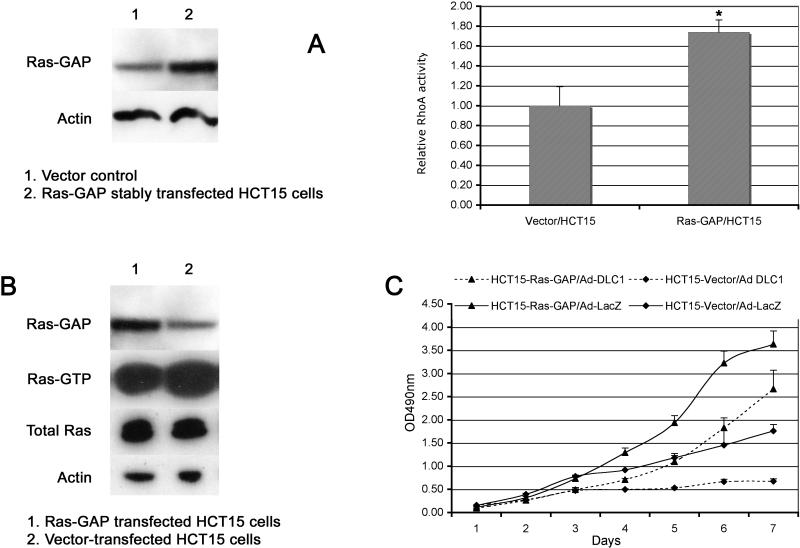
Ras-GAP increases RhoA activity and reduces growth suppression
We also wished to examine the effects of Ras-GAP on RhoA-GTP (active Rho) and cell growth in a relevant cell line. The above biochemical results predict that the Ras-GAP-mediated inhibition of the DLC1 RhoGAP activity would be expected to increase the level of active RhoA, which would therefore attenuate the growth-suppressive activity of DLC1. However, this putative biological effect of Ras-GAP on DLC1 could be obscured by the well know growth inhibitory effects of Ras-GAP that are mediated by its negative regulation of wild type Ras (Tocque et al., 1997; Zhang et al., 1990). We therefore studied the effects of Ras-GAP in a cell line that contains mutationally activated Ras, because mutant Ras is resistant to negative regulation by Ras-GAP, which means that the effects of Ras-GAP in such a line should be Ras-independent. To this end, HCT15 was identified as a colon carcinoma cell line that contains mutant Ras (Gly12-H-Ras-Ser; supplementary Fig. 2), as well as low levels of endogenous Ras-GAP (Shankavaram et al., 2007), high levels of DLC2, and undetectable levels of DLC1 (data not shown).
To determine whether the interaction of DLC1 with Ras-GAP would influence the regulation of RhoA, we examined the effect of Ras-GAP on RhoA activity in HCT15 cells, as determined by a pull down assay using the isolated Rho-GTP binding domain of Rhotekin, which associates preferentially with active RhoA-GTP. Stable transfection of these cells with Ras-GAP significantly increased its expression and RhoA activity compared with the vector control (p=0.017, Fig.4A).
Figure 4. Ras-GAP increases RhoA activity and antagonizes growth suppression by DLC1.
A: Ras-GAP promotes RhoA activity. Left panel: Ras-GAP expression increased 2-fold in Ras-GAP stably transfected HCT15 cells (lane2) as compared with vector control (lane1). Actin was used as loading control. Right panel: Column 1, vector control. Column 2, Ras-GAP stably transfected cells. Data shown represent the mean ± SD of duplicate wells from two independent experiments. The relative level of RhoA activity was normalized relative to vector control. *Statistical significance (t-test) was set at P<0.05.
B: Ras (Ras-GTP) HCT15 cells, which contain Gly12Ser mutation of H-Ras, is resistant to negative regulation by Ras-GAP. 1, Ras-GAP transfected HCT15 cells; 2, vector transfected HCT15 cells.
C: Ras-GAP attenuates growth suppression induced by DLC1. Ras-GAP stably transfected HCT15 cells were infected with Ad-DLC1 as indicated. Cell proliferation was measured with MTT assay. Data shown were the mean absorbance 490nm ± SD from three independent experiments.
We also verified that the level of active Ras was not altered by the Ras-GAP in HCT15 cells (Fig. 4B). The effect of Ras-GAP on the growth suppressing activity of DLC1 was also examined. Previous studies observed an antiproliferative effect of DLC1 in other cancers, and recent data showed in addition that suppression of DLC1 by RNA interference promoted cell growth and migration in a human colon carcinoma cell line (Jin et al., 2008). Consistent with these observations, we found that adenovirus-mediated overexpression of DLC1 induced a substantial reduction in growth compared with that of parental colon carcinoma cells infected with the control Adenovirus that expresses LacZ (Fig. 4C). By contrast, adenovirus-mediated expression of DLC1 in cells stably transfected with Ras-GAP resulted in approximately four times as many cells after one week in culture, compared with the cells expressing only DLC1, indicating that Ras-GAP can antagonize the growth-suppressive effects induced by DLC1. The cells stably expressing Ras-GAP and the control adenovirus-mediated LacZ accumulated approximately twice as many cells as the parental cells during one week in culture.
Discussion
In this study, we have generated evidence that leads us to conclude that Ras-GAP attenuates the tumor suppressor activity of DLC1 by interfering with the Rho-GAP activity of DLC1. We found that endogenous levels of the DLC1 tumor suppressor protein form a stable complex with endogenous Ras-GAP, and that there is extensive co-localization of the two proteins by immunomicroscopy. The interaction was mapped to the Rho-GAP catalytic domain of DLC1 and the SH3 domain of Ras-GAP. We showed that this interaction can inhibit the RhoGAP activity of DLC1 in vitro. Furthermore, overexpression of Ras-GAP, in a cell line that harbored mutant Ras and thus was resistant to the negative regulation of Ras by Ras-GAP, increased the level of endogenous active Rho and antagonized the growth suppressive effects of DLC1. Although the current analysis has been confined to the interaction between the Ras-GAP and the RhoGAP domain of DLC1, we speculate that a similar interaction may occur between the other two DLC family members, DLC2 and DLC3, whose Rho-GAP domains share 80% and 71%, respectively, with this region of DLC1.
The regulation of DLC1 by Ras-GAP has several distinct features. A prior study from one of our laboratories suggested that DLC1 has an intramolecular mechanism responsible for limiting its catalytic activity, as the SAM domain of DLC1 appears to function as an inhibitor of its RhoGAP activity (Kim et al., 2008). The current observations with DLC1 and Ras-GAP show that an intermolecular mechanism can also regulate this activity. In addition, the functional consequences that result from the binding of DLC1 with Ras-GAP differ from those of DLC1 binding to members of the tensin family (Fig.5). That interaction, although it contributes to the physiologic activity of DLC1, is not associated with regulation of the Rho-GAP activity of DLC1 (Qian et al., 2007). Thus, at least two different types of protein interactions with DLC1 exist. It seems possible that DLC1 tumor suppressor function may ultimately be found to depend on its interaction with other proteins in a complex network.
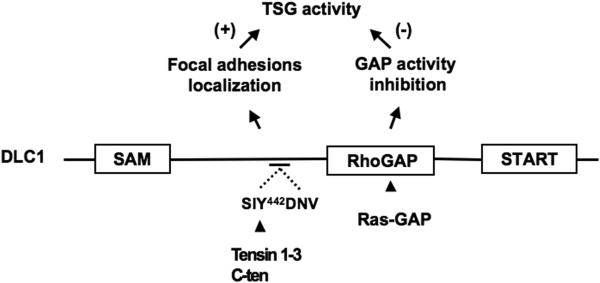
Figure 5.
Regulation of DLC1 activity. The schematic model illustrates the interacting partners of DLC1, tensin and Ras-GAP, and their functional effects on the tumor suppressor activity of DLC1. Members of tensin family recruit DLC1 to focal adhesions via the consensus sequence SIY442DNV, shared by all three DLC proteins, thus contributing to the physiologic activity of DLC1. The SH3 domain of Ras-GAP binds to RhoGAP domain of DLC1 and attenuates the tumor suppressor activity of DLC1 by interfering with the RhoGAP activity of DLC1. TSG, tumor suppressor gene. (+), increases, (−), inhibits.
The DLC1 amino acids that mediate the interaction with the Ras-GAP SH3 domain may be unusual. SH3 domains contain well-conserved aromatic amino acids that most commonly recognize proline-rich sequences, typically PxxP, in its interacting proteins (Li, 2005). However, none of the 5 PxxP motifs identified by the SH3-Hunter software (Ferraro et al., 2007) in DLC1 were located in the binding region (aa 590−880; supplementary Fig. 3). This observation indicates that the binding of the DLC1 RhoGAP catalytic domain to the Ras-GAP SH3 domain is not mediated through such a proline-rich motif, thus suggesting atypical peptide recognition. This feature which may be attributable to structural differences between the Ras-GAP SH3 domain and other SH3 domains (Ross et al., 2007), as three important residues (Leu 288, Val 332, and Leu335) that participate in the docking of proline-rich motifs (PxxP) are not conserved in Ras-GAP (Ross et al., 2007). Indeed, Ras-GTPase-activating protein SH3-domain-binding protein (G3BP) does not use its several proline-rich motifs to bind Ras-GAP (Kennedy et al., 2001).
Ras-GAP has been implicated previously in cross-talk between Ras and Rho. p190Rho-GAP, which is another major regulator of cellular Rho protein, binds simultaneously to the two SH2 domains of Ras-GAP via its two closely linked tyrosine-containing peptides (Bryant et al., 1995). Although this binding does not appear to alter the enzymatic RhoGAP activity of p190Rho-GAP, complex formation between p190Rho-GAP and Ras-GAP is believed to increase the negative regulation of Rho by mediating the Ras-GAP-dependent translocation of complexed p190Rho-GAP to the plasma membrane (Bradley et al., 2006). Thus, the Ras-GAP-induced effects on Rho that are mediated through its association with p190Rho-GAP are the opposite of the Ras-GAP effects on Rho that are mediated by its interaction with DLC1. As DLC1 is localized primarily to focal adhesions, while p190Rho-GAP may be diffusely cytoplasmic or associated with the plasma membrane, the opposite activities of Ras-GAP towards these two Rho regulators might be related to their distinct sub-cellular localization and imply that p190Rho-GAP and DLC1 mediate different physiologic functions. The latter possibility is supported by the observations that genetic disruption of p190Rho-GAP or DLC1 in the mouse results in distinct phenotypes associated with post-natal and embryonic lethality, respectively (Brouns et al., 2000; Durkin et al., 2005).
Other studies have also implicated the SH3 domain of Ras-GAP in cross-talk with Rho, where it could be relevant that p190Rho-GAP-binding induces Ras-GAP to undergo a conformational change that results in a 100-fold increase in the accessibility of the SH3 domain (Hu and Settleman, 1997). A brain-specific Rho-GAP with various names, including p200Rho-GAP, Grit, Rics, and p250 GAP, has been shown to interact with Ras-GAP by binding the SH3 domain of Ras-GAP (Shang et al., 2007). In addition, a monoclonal antibody that binds the Ras-GAP SH3 domain can block neurite outgrowth of PC12 cells stimulated with lysophosphatidic acid and prevent the formation of actin stress fibers in Swiss 3T3 cells stimulated with growth factors (Leblanc et al., 1998). Importantly, the inhibitory effects of the antibody could be overcome by activated Rho, but not by activated Ras, implying that the antibody had induced a reduction in Rho activity. Although it is unknown whether these effects depend on DLC, they are compatible with the scenario that the monoclonal antibody had increased the RhoGAP activity of DLC by blocking the interaction with Ras-GAP. As suggested from the above studies, the function of Ras-GAP is complicated. The role of its C-terminus, which contains the Ras-GAP catalytic domain, as a negative regulator of wild type Ras is well established, but the N-terminus of Ras-GAP, which contains the SH2 and SH3 domains, has been implicated in effectors functions, which could be Ras-independent (Abdellatif et al. 1998; Clark et al., 1997; Tocque et al., 1997).
Our studies suggesting that Ras-GAP can promote the growth of cells containing mutant Ras add to a growing body of evidence suggesting that Ras-GAP might represent a valid therapeutic target. Approximately 30% of all tumors carry a mutant Ras (Forbes et al., 2006), and there is a particular need for improved therapeutic approaches to tumors with mutant Ras, which are often associated with treatment resistance and poor prognosis (Baselga and Rosen, 2008). Several activities that may depend on the SH3 domain of Ras-GAP could promote growth and/or be anti-apoptotic, including the activation of survivin and Akt and the inactivation of DLC (Gigoux et al., 2002; Yue et al., 2004; Zhou et al., 2004). The monoclonal antibody that binds the SH3 domain of Ras-GAP can be pro-apoptotic (Lelanc et al., 1999). Similar results have also been seen, in tumor lines with mutant Ras, and even one with wild type Ras, with an aptamer that specifically binds the Ras-GAP SH3 domain (Pamonsinlapatham, et al., 2008). We speculate that the observed pro-apoptotic activity may be result from interfering with the multiple pathways that appear to be inhibited by blocking the binding activity of the SH3 domain, which therefore make it an attractive molecular target for therapeutic intervention.
Supplementary Material
Fig. 1: Colocalization of endogenous p120Ras-GAP with endogenous focal adhesion marker vinculin. Human prostate cancer cells PC3 were grown on coverslips. p120Ras-GAP protein as well as endogenous vinculin protein were detected primarily in peripheral punctate areas in the cytoplasm, with colocalization of the two proteins.
Fig. 2: Identification of Gly12Ser mutation in H-Ras coding sequence in HCT15 cell line. Arrow indicates a G→A mutation that resulted in a Gly→Ser shift.
Fig. 3: Location of 5 PxxP proline rich motifs in DLC1 protein identified by SH3 hunter software. http://cbm.bio.uniroma2.it/SH3-Hunter/
Acknowledgements
The Intramural Research Program of the National Cancer Institute, Center for Cancer Research, supported this work, NIH. CJD is also supported by an NIH grant (1R01CA129610), and DV was supported by a fellowship from the American Cancer Society.
References
- Abdellatif M, Packer SE, Michael LH, Zhang D, Charng MJ, Schneider MD. A Ras-dependent pathway regulates RNA polymerase II phosphorylation in cardiac myocytes: implications for cardiac hypertrophy. Mol Cell Biol. 1998;18:6729–36. doi: 10.1128/mcb.18.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Rosen N. Determinants of RASistance to anti-epidermal growth factor receptor agents. J Clin Oncol. 2008;26:1582–4. doi: 10.1200/JCO.2007.15.3700. [DOI] [PubMed] [Google Scholar]
- Bradley WD, Hernandez SE, Settleman JS, Koleske AJ. Intergrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol Bio Cell. 2006;17:4827–4836. doi: 10.1091/mbc.E06-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw JM, Waksman G. Molecular recognition by SH2 domains. Adv Protein Chem. 2002;61:161–210. doi: 10.1016/s0065-3233(02)61005-8. [DOI] [PubMed] [Google Scholar]
- Brouns MR, Matheson SF, Hu KQ, Delalle I, Caviness VS, Silver J, et al. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 2000;127:4891–4903. doi: 10.1242/dev.127.22.4891. [DOI] [PubMed] [Google Scholar]
- Bryant SS, Briggs S, Smithgall TE, Martin GA, McCormick F, Chang JH, et al. Two SH2 domains of p120 Ras GTPase-activating protein bind synergistically to tyrosine phosphorylated p190 Rho GTPase-activating protein. J Biol Chem. 1995;270:17947–17952. doi: 10.1074/jbc.270.30.17947. [DOI] [PubMed] [Google Scholar]
- Clark GJ, Westwick JK, Der CJ. p120 GAP modulates Ras activation of Jun kinases and transformation. J Biol Chem. 1997;272:1677–1681. doi: 10.1074/jbc.272.3.1677. [DOI] [PubMed] [Google Scholar]
- Durkin ME, Avner MR, Huh CG, Yuan BZ, Thorgeirsson SS, Popescu NC. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 2005;579:1191–1196. doi: 10.1016/j.febslet.2004.12.090. [DOI] [PubMed] [Google Scholar]
- Durkin ME, Yuan BZ, Zhou X, Zimonjic DB, Lowy DR, Thorgeirsson SS, et al. DLC-1: a Rho GTPase-activating protein and tumor suppressor. J Cell Mol Med. 2007a;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin ME, Ullmannova V, Guan M, Popescu NC. Deleted in liver cancer 3(DLC-3), a novel RhoGTPase-activating protein,is downregulated in cancer and inhibits tumor cell growth. Oncogene. 2007b;26:4580–4589. doi: 10.1038/sj.onc.1210244. [DOI] [PubMed] [Google Scholar]
- Ferraro E, Peluso D, Via A, Ausiello G, Helmer-Citterich M. SH3-Hunter: discovery of SH3 domain interaction sites in proteins. Nucleic Acids Res. 2007;35:W451–4. doi: 10.1093/nar/gkm296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes S, Clements J, Dawson E, Bamford S, Webb T, Dogan A, et al. COSMIC 2005. Br J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigoux V, L'Hoste S, Raynaud F, Camonis J, Garbay C. Identification of Aurora kinases as RasGAP Src homology 3 domain-binding proteins. J Biol Chem. 2002;277:23742–6. doi: 10.1074/jbc.C200121200. [DOI] [PubMed] [Google Scholar]
- Guan M, Tripathi V, Zhou X, Popescu NC. Adenovirus-mediated restoration of the expression of the tumor suppressor gene DLC1 inhibits the proliferation and tumorigenicity of aggressive, androgen-independent human prostate cancer cell lines: prospects for gene therapy. Cancer Gene Therapy. 2008;15:371–378. doi: 10.1038/cgt.2008.13. [DOI] [PubMed] [Google Scholar]
- Healy KD, Hodgson L, Kim TY, Shutes AT, Maddileti S, Juliano RL. DLC1 suppresses non-small lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol Carcinog. 2007;47:326–337. doi: 10.1002/mc.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KQ, Settleman J. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 1997;16:473–483. doi: 10.1093/emboj/16.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Tian X, Shang Y, Huang P. Inhibition of DLC-1 gene expression by RNA interference in the colon cancer LoVo cell line. Oncol Rep. 2008;19:669–674. [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K, Kiyota M, Seike J, Deki Y, Yagisawa H. START-GAP3/DLC3 is a GAP for RhoA and Cdc42 and is localized in focal adhesions regulating cell morphology. Biochem Biophys Res Commun. 2007;364:783–789. doi: 10.1016/j.bbrc.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Kennedy D, French J, Guitard E, Ru K, Tocque B, Mattick J. Characterization of G3BPs: tissue specific expression, chromosomal localisation and rasGAP(120) binding studies. J Cell Biochem. 2001;84:173–187. doi: 10.1002/jcb.1277. [DOI] [PubMed] [Google Scholar]
- Kim TY, Lee JW, Kim HP, Jong HS, Kim TY, Jung M, et al. DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology, and migration in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2007;355:72–77. doi: 10.1016/j.bbrc.2007.01.121. [DOI] [PubMed] [Google Scholar]
- Kim TY, Healy KD, Der CJ, Sciaky N, Bang Y, Juliano R. Effects of Rho GTPase-acivating protein DLC-1 on cell morphology and migration.Sept11. J Biol Chem. 2008 doi: 10.1074/jbc.M800617200. doi/10.1074/jbc/M8006117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc V, Delumeau I, Tocqué B. Ras-GTPase activating protein inhibition specifically induces apoptosis of tumour cells. Oncogene. 1999;18:4884–9. doi: 10.1038/sj.onc.1202855. [DOI] [PubMed] [Google Scholar]
- Leblanc V, Tocque B, Delumeau I. RasGAP Controls Rho-Mediated Cytoskeletal Reorganization through Its SH3 Domain. Mol Cell Biol. 1998;18:5567–5578. doi: 10.1128/mcb.18.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung TH, Ching YP, Yam JW, Wong CM, Yau TO, Jin DY, et al. Deleted in liver cancer 2 (DLC2) suppresses cell transformation by means of inhibition of RhoA activity. Proc Natl Acad Sci USA. 2005;102:15207–15212. doi: 10.1073/pnas.0504501102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YC, Si L, deVere White RW, Lo SH. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J Cell Biol. 2007;176:43–49. doi: 10.1083/jcb.200608015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YC, Shih YP, Lo SH. Mutations in the focal adhesion targeting region of deleted in liver cancer-1 attenuate their expression and function. Cancer Res. 2008;68:7718–7722. doi: 10.1158/0008-5472.CAN-08-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer BJ, Eck MJ. SH3 domains. Minding your p's and q's. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- Musacchio A. How SH3 domains recognize proline. Adv Protein Chem. 2002;61:211–268. doi: 10.1016/s0065-3233(02)61006-x. [DOI] [PubMed] [Google Scholar]
- Pamonsinlapatham P, Hadj-Slimane R, Raynaud F, Bickle M, Corneloup C, Barthelaix A, et al. A RasGAP SH3 peptide aptamer inhibits RasGAP-Aurora interaction and induces caspase-independent tumor cell death. PLoS ONE. 2008;3:e2902. doi: 10.1371/journal.pone.0002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Li G, Asmussen HK, Asnaghi L, Vass WC, Braverman R, et al. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc Natl Acad Sci U S A. 2007;104:9012–9017. doi: 10.1073/pnas.0703033104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross B, Kristensen O, Favre D, Walicki J, Kastrup JS, Widmann C, et al. High resolution crystal structures of the p120 RasGAP SH3 domain. Biochem Biophys Res Commun. 2007;353:463–468. doi: 10.1016/j.bbrc.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Shang X, Moon SY, Zheng Y. p200 RhoGAP promotes cell proliferation by mediating cross-talk between Ras and Rho signaling pathways. J Biol Chem. 2007;282:8801–8811. doi: 10.1074/jbc.M609375200. [DOI] [PubMed] [Google Scholar]
- Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- Shutes A, Der CJ. Real-time in vitro measurement of GTP hydrolysis. Methods. 2005;37:183–189. doi: 10.1016/j.ymeth.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Tocque B, Delumeau I, Parker F, Maurier F, Multon MC, Schweighoffer F. Ras-GTPase activating protein (GAP): a putative effector for Ras. Cell Signal. 1997;9:153–158. doi: 10.1016/s0898-6568(96)00135-0. [DOI] [PubMed] [Google Scholar]
- Trahey M, Wong G, Halenbeck R, Rubinfeld B, Martin GA, Ladner M, et al. Molecular Cloning of Two Types of GAP Complementary DNA from Human Placenta. Science. 1988;242:1697–1700. doi: 10.1126/science.3201259. [DOI] [PubMed] [Google Scholar]
- Ullmanova V, Popescu NC. Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. Int. J. Oncol. 2006;29:1127–1132. [PubMed] [Google Scholar]
- Vogel US, Dixon RA, Schaber MD, Diehl RE, Marshall MS, Scolnick EM, et al. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988;335:90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- Wong CM, Yam JW, Ching YP, Yau TO, Leung TH, Jin DY, et al. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- Xue W, Krasnitz A, Lucito R, Sordella R, VanAelst L, Cordon-Cardo C, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam JW, Ko FC, Chan CY, Jin DY, Ng IO. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 2006;66:8367–8372. doi: 10.1158/0008-5472.CAN-05-2850. [DOI] [PubMed] [Google Scholar]
- Yuan BZ, Miller MJ, Keck C, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- Yue Y, Lypowy J, Hedhli N, Abdellatif M. Ras GTPase-activating protein binds to Akt and is required for its activation. J Biol Chem. 2004;279:12883–12889. doi: 10.1074/jbc.M312308200. [DOI] [PubMed] [Google Scholar]
- Zhang K, DeClue JE, Vass WC, Papageorge AG, McCormick F, Lowy. DR. Suppression of c-ras transformation by GTPase-activating protein. Nature. 1990;346:754–756. doi: 10.1038/346754a0. [DOI] [PubMed] [Google Scholar]
- Zhou X, Zimonjic DB, Park SW, Yang XY, Durkin ME, Popescu. NC. DLC-1 suppresses distant dissemination of human hepatocellular carcinoma cells in nude mice through reduction of RhoA GTPase activity, actin cytoskeletal disruption and down regulation of gene involved in metastasis. Int J Oncol. 2008;32:1285–1291. doi: 10.3892/ijo_32_6_1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Thorgeirsson SS, Popescu NC. Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 2004;23:1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. 1: Colocalization of endogenous p120Ras-GAP with endogenous focal adhesion marker vinculin. Human prostate cancer cells PC3 were grown on coverslips. p120Ras-GAP protein as well as endogenous vinculin protein were detected primarily in peripheral punctate areas in the cytoplasm, with colocalization of the two proteins.
Fig. 2: Identification of Gly12Ser mutation in H-Ras coding sequence in HCT15 cell line. Arrow indicates a G→A mutation that resulted in a Gly→Ser shift.
Fig. 3: Location of 5 PxxP proline rich motifs in DLC1 protein identified by SH3 hunter software. http://cbm.bio.uniroma2.it/SH3-Hunter/