Abstract
Activator protein 1 (AP-1) transcription factors are dimers of Jun, Fos, MAF and activating transcription factor (ATF) family proteins characterized by basic region and leucine zipper domains1. Many AP-1 proteins contain defined transcriptional activation domains (TADs), but Batf and the closely related Batf3 (refs 2, 3) contain only a basic region and leucine zipper and have been considered inhibitors of AP-1 activity3–8. Here we show that Batf is required for the differentiation of IL-17-producing T helper (TH17) cells9. TH17 cells comprise a CD4+ T cell subset that coordinates inflammatory responses in host defense but is pathogenic in autoimmunity10–13.Batf −/−mice have normal TH1 and TH2 differentiation, but show a defect in TH17 differentiation, and are resistant to experimental autoimmune encephalomyelitis (EAE).Batf −/−T cells fail to induce known factors required for TH17 differentiation, such as RORγt11 and the cytokine IL-21 (refs 14–17). Neither addition of IL-21 nor overexpression of RORγt fully restores IL-17 production in Batf−/− T cells. The IL-17 promoter is Batf-responsive, and upon TH17 differentiation, Batf binds conserved intergenic elements in the IL-17A/F locus and to the IL-17, IL-21 and IL-22 (ref 18) promoters. These results demonstrate that the AP-1 protein Batf plays a critical role in TH17 differentiation.
In a gene expression survey (Supplementary Fig. 1a), we identified the basic leucine zipper (bZIP) transcription factor ATF-like7 (Batf) to be highly expressed in TH1, TH2 and TH17 cells compared to naïve T cells and B cells. Batf and Batf3 (refs 2, 3) form heterodimers with Jun6,7 and are considered repressors of AP-1 activity3,5,6,8,19. To assess its role in T cell differentiation20, we generated Batf−/− mice (Supplementary Fig. 2a, b). Batf−/− mice lacked detectable Batf protein, were fertile and appeared healthy. Batf protein was low in naïve T cells, increased in TH2 cells, induced by activation (Supplementary Fig. 2), present in the nucleus and cytoplasm, but upon activation showed increased nuclear translocation (Fig. 1a and Supplementary Fig. 1b, c). Batf−/− mice had normal thymus, spleen and lymph node development and CD4+ and CD8+ T cell development (Supplementary Fig. 3, Supplementary Fig. 4a, b). Although Batf-transgenic mice had altered NKT cell development21, Batf−/− mice had normal development of NKT cells (Supplementary Fig. 4c), B cells (Supplementary Fig. 4d, e), conventional and plasmacytoid dendritic cells (Supplementary Fig. 5a, b).
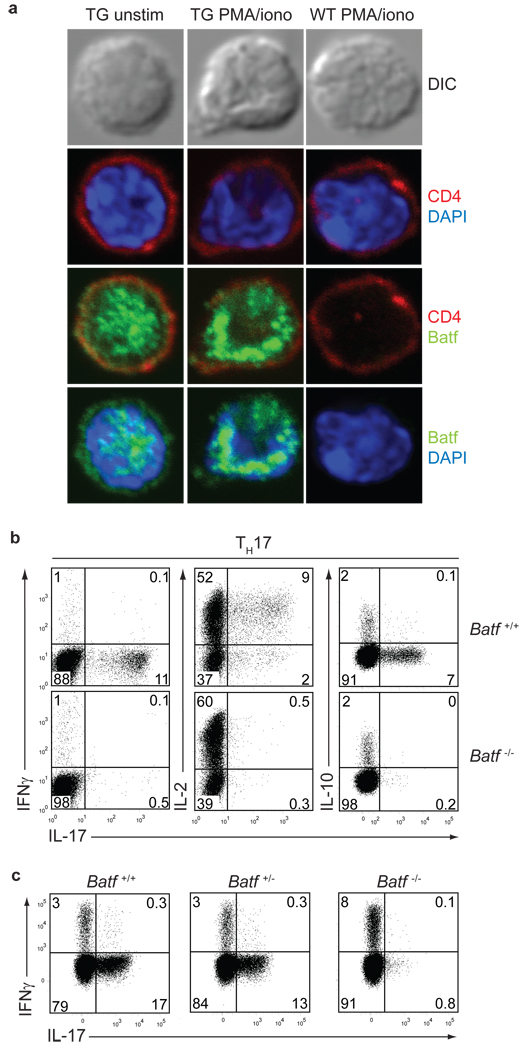
Figure 1. Loss of IL-17 production in Batf−/− T cells.
a, DO11.10+CD4+ T cells from CD2-N-FLAG-Batf transgenic mice or littermates were cultured with OVA/APCs under TH2 conditions for 7 days, and stained with antibodies to CD4 and FLAG. b,Batf+/+ and Batf−/− CD4+CD62L+CD25 T cells cultured under TH17 conditions were restimulated with PMA/ionomycin on days 7 (left panel) or 3 (middle and right panels) and stained for IL-17, IFN-γ, IL-2 and IL-10. c, IL-17 and IFN-γ expression in DO11.10+CD4+ T cells from Batf+/+Batf +/− and Batf−/− mice activated with OVA/APCs under TH17 conditions. Data are representative of at least 2 independent experiments.
Batf−/− T cells displayed normal TH1 and TH2 differentiation (Supplementary Fig. 6a). Under TH17 conditions, Batf−/− T cells, but not Batf+/− T cells, showed a dramatic reduction in IL-17 production, but had normal levels of IL-2, IFN-γ and IL-10 (Fig. 1b, c). Batf−/− DO11.10+ T cells showed loss of IL-17 even after several passages under TH17 conditions (Supplementary Fig. 6b). Batf−/− CD8+ T cells also failed to produce IL-17 (Supplementary Fig. 6c). We generated transgenic mice expressing FLAG-tagged Batf under the control of the CD2 promoter22. Batf-transgenic DO11.10+ CD4+ T cells and CD8+ T cells had increased IL-17 production under TH17 conditions compared to controls (Supplementary Fig. 6d, e). Lamina propria CD4+ T cells, which constitutively express IL-17 in wild type mice11, failed to produce IL-17 in Batf−/− mice (Supplementary Fig. 6f).
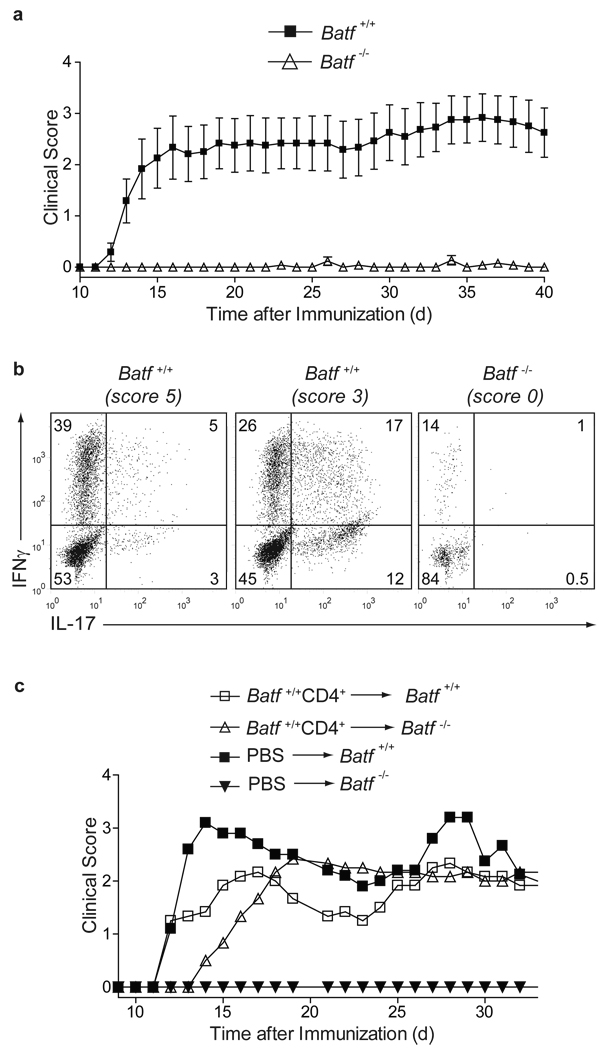
TH17 cells are the major pathogenic population in experimental autoimmune encephalomyelitis10 (EAE), although factors other than IL-17A and IL-17F can contribute to disease23. Batf +/+ mice immunized with myelin oligodendrocyte glycoprotein peptide 35–55 (MOG35–55) (Fig. 2) developed EAE, but Batf−/− mice were completely resistant (Fig. 2a). At peak disease, CNS-infiltrating and splenic CD4+ T cells from Batf +/+ mice produced abundant IL-17 and IFN-γ, while T cells from Batf−/− mice produced no IL-17 (Fig. 2b, Supplementary Fig. 7a). Since IL-6-deficient mice are resistant to EAE due to a compensatory increase in Foxp3+ T regulatory (Treg) cells14, we analyzed splenic Batf+/+ and Batf−/− CD4+ T cells for Foxp3 expression before and after MOG35–55 immunization (Supplementary Fig. 7b, c). Batf−/− mice had lower basal numbers of splenic Foxp3+ T cells compared to Batf+/+ mice, but showed no change in Foxp3+ expression after MOG35–55 immunization (Supplementary Fig. 7b, c), suggesting that their resistance to EAE is not due to an increase in Treg cells. To determine whether the resistance to EAE in Batf−/− mice resulted from a defect within T cells or other immune cells, we injected naïve Batf+/+ CD4+ T cells or PBS control buffer into mice before MOG35–55 immunization (Fig. 2c). Batf−/− mice receiving PBS remained resistant to EAE, but Batf−/− mice receiving naïve Batf+/+ CD4+ T cells developed severe EAE (Fig. 2c, Supplementary Table 1) with CNS-infiltrating IL-17-producing CD4+ T cells (Supplementary Fig. 7d). Thus, Batf−/− mice have a T cell-intrinsic defect preventing EAE.
Figure 2. Batf−/− mice are resistant to EAE.
a,Batf+/+ (n=12) and Batf−/− (n=13) mice were immunized with MOG33–35 peptide. (Mean clinical EAE scores +/− s.e.m, representative of two independent experiments). b, 13 days after EAE induction, CNS-infiltrating lymphocytes were stimulated with PMA/ionomycin, gated on CD4+ cells and stained for intracellular IL-17 and IFNγ (Clinical scores are in parentheses, data are representative of 2–3 mice per group). c,Batf+/+ and Batf−/− mice were injected with control PBS buffer (n=5) or 1×107Batf+/+ CD4+ T cells (n=6) four days prior to EAE induction. Mean clinical scores are shown.
Batf could control TH17 development by regulating IL-6 or TGF-β signaling. IL-6 receptor expression and IL-6-induced STAT3 phosphorylation were normal in Batf−/− T cells (Supplementary Fig. 8a and b). TGF-β induced normal levels of Foxp3 in Batf−/− CD4+ T cells (Supplementary Fig. 8d). While Batf−/− T cells failed to fully downregulate Foxp3 in response to IL-6 (ref 12), neutralization of IL-2 abrogated increased Foxp3 in Batf−/− T cells, without restoring IL-17 production (Supplementary Fig. 8d, e). Thus, Batf−/− T cells exhibit normal TGF-β signaling and proximal IL-6 signaling, implying Batf may regulate downstream target genes.
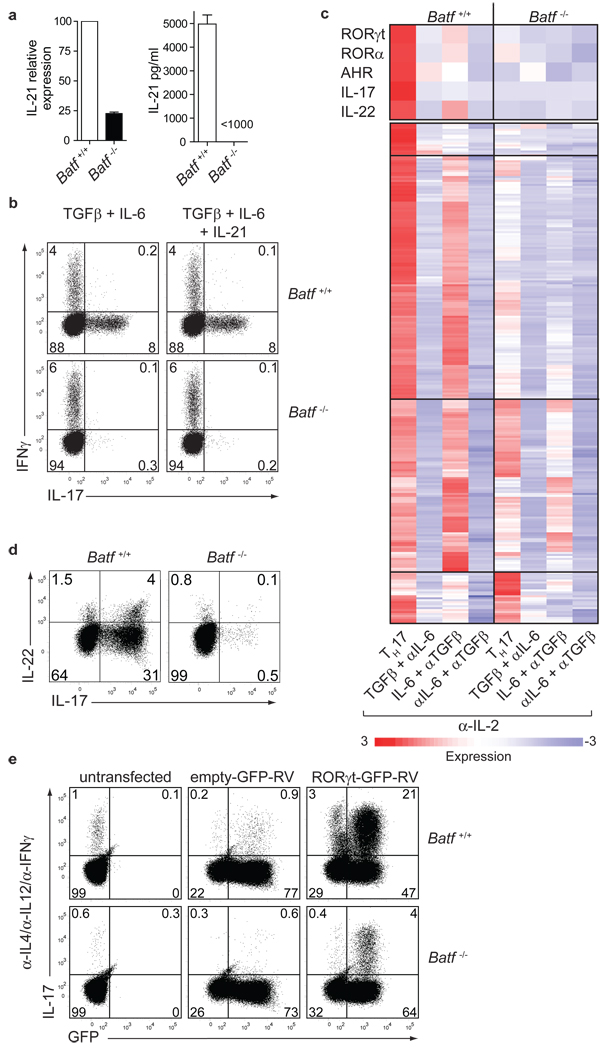
IL-21, an early target of IL-6 signaling in CD4+ T cells17, is required for TH17 development14–16. IL-21 was reduced in Batf−/− CD4+ T cells activated under TH17 conditions (Fig. 3a). Addition of IL-21 failed to rescue TH17 development in Batf−/− T cells (Fig. 3b) but IL-21-induced STAT3 phosphorylation was intact (Supplementary Fig. 8c), suggesting that Batf regulates other factors besides IL-21 during TH17 differentiation.
Figure 3. Batf controls multiple TH17-associated genes.
a, IL-21 expression in Batf+/+ or Batf−/− T cells cultured under TH17 conditions determined by qRT-PCR and ELISA. (mean + s.d. 3 mice). b, IL-17 and IFN-γ expression of CD4+CD62L+CD25− T cells cultured in a in the presence or absence of IL-21. c, Microarray analysis of anti-CD3/CD28-activated T cells at 72h, presented as heat maps of genes 5-fold-induced in Batf+/+ T cells under TH17 conditions. d, IL-17 and IL-22 expression in Batf+/+ or Batf−/− CD4+ T cells activated under TH17 conditions for 3 days. e, Anti−CD3/CD28-activated Batf+/+ or Batf−/− CD4+ T cells were left uninfected or infected with RORγt-GFP-RV or control-GFP-RV, and stained for IL-17.
We performed DNA microarrays and quantitative RT-PCR (qRT-PCR) of Batf+/+ and Batf−/− T cells activated with combinations of IL-6 and/or TGF-β (Fig. 3c, d and Supplementary Fig. 9). This analysis identified several genes known to regulate TH17 development as Batf-dependent (Fig. 3c, d, Supplementary Fig. 9c and Supplementary Table 2), including RORγt17, RORα24, the aryl hydrocarbon receptor25,26, IL-22 (ref 18) and IL-17. However, IRF-4 (ref 13) and SOCS gene expression were unchanged in Batf−/− T cells (Supplementary Fig. 9b and Supplementary Table 4). Early induction of RORγt was normal in Batf−/− T cells but was not maintained at 62h after stimulation (Supplementary Fig. 11a). Batf appeared necessary for expression of a subset of IL-6-induced genes, but was not required for expression of TGF-β-induced genes (Fig. 3c, Supplementary Fig. 9a and Supplementary Table 2, Supplementary Table 3). However, Batf did not globally affect IL-6-induced responses, since IL-6-induced liver acute phase responses appeared normal in Batf−/− mice (Supplementary Fig. 10).
Since RORγt acts directly on the IL-17 promoter27,28, we asked whether RORγt could rescue TH17 development in Batf−/− T cells. In Batf+/+ T cells, retroviral RORγt expression induced 38% IL-17 production, compared to only 1.6% IL-17 production induced by control retrovirus (Fig. 3e and Supplementary Fig. 11c)11,13. But in Batf−/− T cells, retroviral RORγt expression induced only 5.7% IL-17 production (Fig. 3e and Supplementary Fig. 11c). Even under TH17-inducing conditions, retroviral RORγt expression did not fully restore IL-17 production in Batf−/− T cells (Supplementary Fig. 11b, c). Retroviral expression of both Batf and RORγt in Batf−/− T cells induced 26% IL-17 production, compared to only 5% with RORγt alone, and 14% with Batf alone (Supplementary Fig. 11d), suggesting potential synergy between RORγt and Batf, and a possible direct action of Batf in transcription of IL-17 and other TH17-specific genes.
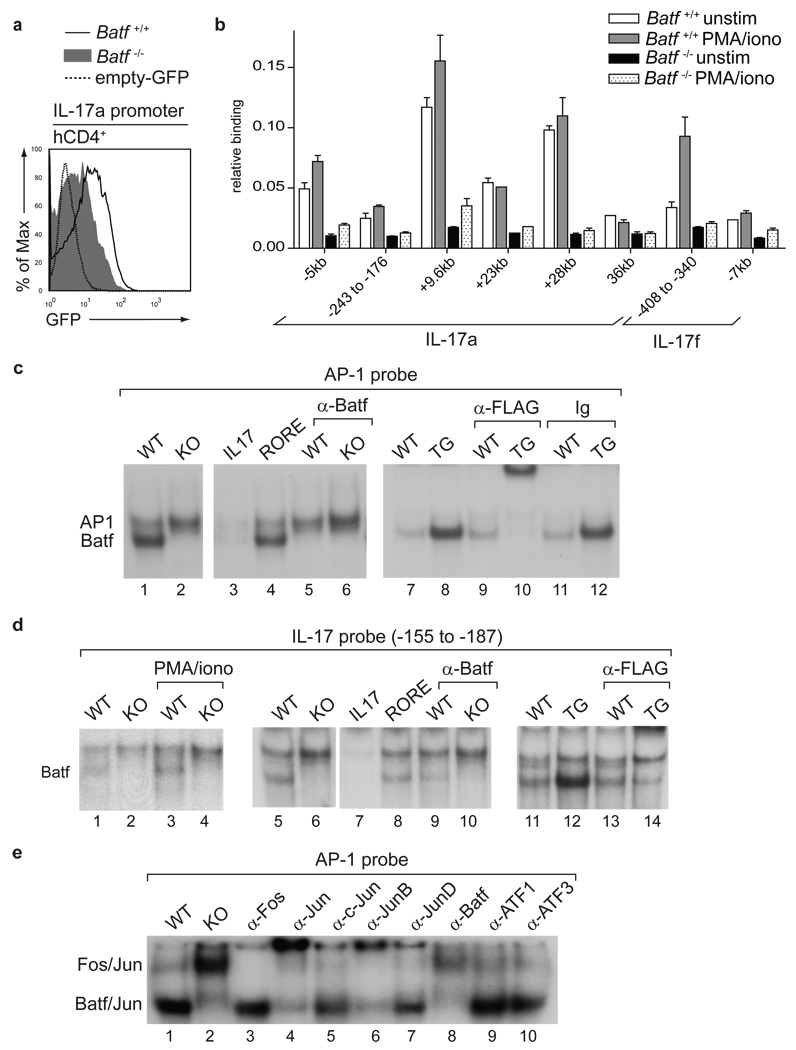
We used a reverse-strand retroviral reporter29 to examine IL-17 promoter activity in primary Batf+/+ and Batf−/− T cells (Fig. 4a). Three days after activation, Batf−/− CD4+ T cells showed considerably less reporter activity than Batf+/+ T cells, suggesting the proximal IL-17 promoter is Batf-responsive (Fig. 4a). Using chromatin immunoprecipitation (ChIP) analysis of several conserved regions within the IL-17a/IL-17f locus (Supplementary Fig. 12a), we found that Batf specifically bound to the +9.6kb and +28kb intergenic regions within 24h after activation (Fig. 4b, Supplementary Fig. 12b, c). By day 5 after stimulation, Batf bound specifically to several intergenic regions and to the proximal IL-17a and IL-17f promoters (Fig. 4b, Supplementary Fig. 12b, c), with distal elements showing more rapid and stronger binding than proximal elements.
Figure 4. Batf directly regulates IL-17 expression.
a,Batf+/+ and Batf−/− CD4+ T cells cultured under TH17 conditions were infected with hCD4-pA-GFP-RV-IL-17p reporter virus. GFP expression after PMA/ionomycin restimulation is shown. b, Batf+/+ and Batf−/− CD4+ T cells cultured under TH17 conditions for 5 days were subjected to ChIP analysis of the indicated regions using anti-Batf antibody (mean + s.d.). c, d, f, EMSA supershift analysis of TH17 whole cell extracts using a consensus AP-1 (c, f) or the IL-17(−155 to − 187) probe (d). (Batf+/+ (WT), Batf−/− (KO), CD2-N-FLAG-Batf transgenic (TG), IL-17(−155 to − 187) and RORE probes were used as competitors).
We next examined Batf binding to a consensus AP-1 probe6 by EMSA. This probe formed two complexes in Batf+/+ TH17 cell extracts (Fig. 4c) that were dependent on stimulation (Supplementary Fig. 13a). Only the upper complex formed in Batf−/− TH17 cells (Fig. 4c). An anti-Batf antibody inhibited the lower complex. In CD2-N-FLAG-Batf-transgenic TH17 cell extracts, the lower complex was specifically supershifted by an anti-FLAG antibody (Fig. 4c). Thus, only the lower complex binding the consensus AP-1 probe in TH17 cells contains Batf.
Several potential Batf binding sites were identified by EMSA in the IL-17, IL-21 and IL-22 proximal promoters, including the IL-17 promoter region (−188 to −210) that bound Batf in ChIP (Fig. 4b, Supplementary Fig. 13b–d). Another Batf-binding IL-17 promoter region (−155 to −187) overlapped with a reported RORγt-binding element27. As an EMSA probe, this region forms two complexes in TH17 cells (Fig. 4d), with the lower complex being selectively inhibited by anti-Batf antibody, absent in Batf−/− TH17 cells, and supershifted by an anti-FLAG antibody in Batf-transgenic TH17 extracts (Fig. 4d). We confirmed Batf binding to the IL-21 and IL-22 promoters by ChIP analysis (Supplementary Fig. 13e). The program CONSENSUS30 determined that the Batf-binding element in the IL-17, IL-21 and IL-22 promoters resembles canonical AP-1 elements at positions 1 through 3, with variation at remaining nucleotides (Supplementary Fig. 13f). CONSENSUS did not identify other transcription factor binding sites enriched near Batf binding elements. We determined the composition of the Batf-containing complex using supershift analysis (Fig. 4e). The upper complex supershifted with pan-anti-Fos antibody, whereas the lower complex supershifted with a pan-anti-Jun and anti-Batf antibodies. Anti-JunB supershifted the majority of the lower complex, but antibodies to c-Jun, JunD, ATF1 or ATF3 did not. Thus, Batf forms heterodimers preferentially with JunB during TH17 differentiation.
Although Batf and Batf3 were considered AP-1 inhibitors3–8, we have shown that they are required for the development of specific immune lineages2. Batf is selectively required for TH17 development, but unlike Irf4 (Ref 13), is not required for TH2 development. Since Batf is also expressed in TH1 and TH2 cells, it likely cooperates with other TH17-specific factors to regulate target genes. Future work will determine whether the actions of Batf involve distinct DNA binding specificity or unique protein-protein interactions with TH17 specific factors.
Methods Summary
Mice
Batf−/− mice were generated by homologous recombination, deleting exons 1 and 2 of the Batf gene on the pure 129SvEv genetic background. The neomycin resistance cassette was removed from the targeted Batf allele in ES cells before generation of mice.
T cell differentiation assays
Naïve CD4+CD62L+CD25− T cells were isolated by cell sorting and activated with plate-bound anti-CD3 and soluble anti-CD28 antibodies. Cultures were supplemented with anti-IL-4 (11B11; hybridoma supernatant), IFN-γ (Peprotech; 0.1ng/ml) and IL-12 (Genetics Institute; 10U/ml) for TH1; anti-IFN-γ (H22; BioXcell; 10µg/ml), anti-IL-12 (Tosh; BioXcell; 10µg/ml) and IL-4 (Peprotech; 10ng/ml) for TH2; anti-IL-4, anti-IL-12, anti-IFN-γ, IL-6 (Peprotech 20ng/ml) and TGF-β (Peprotech; 0.5ng/ml) for TH17 differentiation. Unless otherwise indicated, three days after activation cells were restimulated with PMA/ionomycin for 4h for intracellular cytokine analysis by flow cytometry.
Intracellular Staining
For intracellular cytokine staining, cells were stained for surface markers followed by fixation with 2% formaldehyde for 15 minutes at room temperature. Cells were then washed once in 0.05% saponin and stained with anti-cytokine antibodies in 0.5% saponin. Anti-phospho-STAT3 antibody (BD Pharmingen) was used according to the manufacturer’s recommendations. Briefly, cells were stained for surface markers followed by fixation with 90% methanol at −20°C overnight. Cells were then washed and stained for phospho-Stat3 in PBS containing 3% FCS. Foxp3 staining was performed according to the manufacturer’s recommendations using Foxp3 staining buffers (eBioscience).
Induction of EAE
Mice (7–10 weeks old) were immunized subcutaneously with 100µg MOG35–55 peptide (Sigma) emulsified in CFA (IFA supplemented with 500µg Mycobacterium tuberculosis). One and three days later mice were given 300ng Pertussis Toxin (List Biological Laboratories) intraperitonally (i.p.). Clinical scores were assessed as described in methods. For T cell transfer experiments mice were injected with either PBS or 107 Batf+/+ CD4+ T cells 4 days prior to MOG35–55 immunization13.
Supplementary Material
Acknowledgements
We thank Dr. Roger Lallone (Brookwood Biomedical) for anti-Batf antibody preparation, and Dr. Barry Sleckman for Cre-expressing adenovirus. This work was supported by the Howard Hughes Medical Institute (KMM), and grants from the NIH HG00249 and training grant GM07200 (GDS), AI035783 (CTW), AR049293 (RDH) and from Daiichi-Sankyo Co. Ltd. (CTW).
Footnotes
Microarray data are available at Array Express, E-MEXP-1518, E-MEXP-2152 and E-MEXP-2153. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 2.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iacobelli M, Wachsman W, McGuire KL. Repression of IL-2 promoter activity by the novel basic leucine zipper p21SNFT protein. J Immunol. 2000;165:860–868. doi: 10.4049/jimmunol.165.2.860. [DOI] [PubMed] [Google Scholar]
- 4.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 5.Williams KL, et al. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur. J Immunol. 2001;31:1620–1627. doi: 10.1002/1521-4141(200105)31:5<1620::aid-immu1620>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Echlin DR, Tae HJ, Mitin N, Taparowsky EJ. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene. 2000;19:1752–1763. doi: 10.1038/sj.onc.1203491. [DOI] [PubMed] [Google Scholar]
- 7.Dorsey MJ, et al. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene. 1995;11:2255–2265. [PubMed] [Google Scholar]
- 8.Thornton TM, Zullo AJ, Williams KL, Taparowsky EJ. Direct manipulation of activator protein-1 controls thymocyte proliferation in vitro. Eur. J Immunol. 2006;36:160–169. doi: 10.1002/eji.200535215. [DOI] [PubMed] [Google Scholar]
- 9.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 10.Langrish CL, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp. Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 13.Brustle A, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 14.Korn T, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurieva R, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 16.Wei L, Laurence A, Elias KM, O'Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 18.Liang SC, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp. Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bower KE, Fritz JM, McGuire KL. Transcriptional repression of MMP-1 by p21SNFT and reduced in vitro invasiveness of hepatocarcinoma cells. Oncogene. 2004;23:8805–8814. doi: 10.1038/sj.onc.1208109. [DOI] [PubMed] [Google Scholar]
- 20.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 21.Williams KL, et al. BATF transgenic mice reveal a role for activator protein-1 in NKT cell development. J Immunol. 2003;170:2417–2426. doi: 10.4049/jimmunol.170.5.2417. [DOI] [PubMed] [Google Scholar]
- 22.Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
- 23.Haak S, et al. IL-17A and IL-17F do not contribute vitally to autoimmune neuroinflammation in mice. J Clin. Invest. 2009;119:61–69. doi: 10.1172/JCI35997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veldhoen M, et al. The aryl hydrocarbon receptor links T(H)17-cell-mediated autoimmunity to environmental toxins. Nature. 2000 doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 26.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 27.Ichiyama K, et al. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J Biol Chem. 2008;283:17003–17008. doi: 10.1074/jbc.M801286200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H, et al. Unexpected characteristics of the IFN-gamma reporters in nontransformed T cells. J. Immunol. 2001;167:855–865. doi: 10.4049/jimmunol.167.2.855. [DOI] [PubMed] [Google Scholar]
- 30.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.