Abstract
Mouse knockout technology provides a powerful means of elucidating gene function in vivo, and a publicly available genome-wide collection of mouse knockouts would be significantly enabling for biomedical discovery. To date, published knockouts exist for only about 10% of mouse genes. Furthermore, many of these are limited in utility because they have not been made or phenotyped in standardized ways, and many are not freely available to researchers. It is time to harness new technologies and efficiencies of production to mount a high-throughput international effort to produce and phenotype knockouts for all mouse genes, and place these resources into the public domain.
Now that the human and mouse genome sequences are known1-3, attention has turned to elucidating gene function and identifying gene products that might have therapeutic value. The laboratory mouse (Mus musculus) has had a prominent role in the study of human disease mechanisms throughout the rich, 100-year history of classical mouse genetics, exemplified by the lessons learned from naturally occurring mutants such as agouti4, reeler5 and obese6. The large-scale production and analysis of induced genetic mutations in worms, flies, zebrafish and mice have greatly accelerated the understanding of gene function in these organisms. Among the model organisms, the mouse offers particular advantages for the study of human biology and disease: (i) the mouse is a mammal, and its development, body plan, physiology, behavior and diseases have much in common with those of humans; (ii) almost all (99%) mouse genes have homologs in humans; and (iii) the mouse genome supports targeted mutagenesis in specific genes by homologous recombination in embryonic stem (ES) cells, allowing genes to be altered efficiently and precisely.
The ability to disrupt, or knock out, a specific gene in ES cells and mice was developed in the late 1980s (ref. 7), and the use of knockout mice has led to many insights into human biology and disease8-11. Current technology also permits insertion of ‘reporter’ genes into the knocked-out gene, which can then be used to determine the temporal and spatial expression pattern of the knocked-out gene in mouse tissues. Such marking of cells by a reporter gene facilitates the identification of new cell types according to their gene expression patterns and allows further characterization of marked tissues and single cells.
Appreciation of the power of mouse genetics to inform the study of mammalian physiology and disease, coupled with the advent of the mouse genome sequence and the ease of producing mutated alleles, has catalyzed public and private sector initiatives to produce mouse mutants on a large scale, with the goal of eventually knocking out a substantial portion of the mouse genome12,13. Large-scale, publicly funded gene-trap programs have been initiated in several countries, with the International Gene Trap Consortium coordinating certain efforts and resources14-17.
Despite these efforts, the total number of knockout mice described in the literature is relatively modest, corresponding to only ~10% of the ~25,000 mouse genes. The curated Mouse Knockout & Mutation Database lists 2,669 unique genes (C. Rathbone, personal communication), the curated Mouse Genome Database lists 2,847 unique genes, and an analysis at Lexicon Genetics identified 2,492 unique genes (B.Z., unpublished data). Most of these knockouts are not readily available to scientists who may want to use them in their research; for example, only 415 unique genes are represented as targeted mutations in the Jackson Laboratory’s Induced Mutant Resource database (S. Rockwood, personal communication).
The converging interests of multiple members of the genomics community led to a meeting to discuss the advisability and feasibility of a dedicated project to produce knockout alleles for all mouse genes and place them into the public domain. The meeting took place from 30 September to 1 October 2003 at the Banbury Conference Center at Cold Spring Harbor Laboratory. The attendees of the meeting are the authors of this paper.
Is a systematic project warranted?
A coordinated project to systematically knock out all mouse genes is likely to be of enormous benefit to the research community, given the demonstrated power of knockout mice to elucidate gene function, the frequency of unpredicted phenotypes in knockout mice, the potential economies of scale in an organized and carefully planned project, and the high cost and lack of availability of knockout mice being made in current efforts. Moreover, implementing such a systematic and comprehensive plan will greatly accelerate the translation of genome sequences into biological insights. Knockout ES cells and mice currently available from the public and private sectors should be incorporated into the genome-wide initiative as much as possible, although some may be need to be produced again if they were made with suboptimal methods (e.g., not including a marker) or if their use is restricted by intellectual property or other constraints. The advantages of such a systematic and coordinated effort include efficient production with reduced costs; uniform use of knockout methods, allowing for more comparability between knockout mice; and ready access to mice, their derivatives and data to all researchers without encumbrance. Solutions to the logistical, organizational and informatics issues associated with producing, characterizing and distributing such a large number of mice will draw from the experience of related projects in the private sector and in academia, which have made or phenotyped hundreds of knockout mice using a variety of techniques. Lessons learned from these projects include the need for redundancy at each step to mitigate pipeline bottlenecks and the need for robust informatics systems to track the production, analysis, maintenance and distribution of thousands of targeting constructs, ES cells and mice.
Null-reporter alleles should be created
The project should generate alleles that are as uniform as possible, to allow efficient production and comparison of mouse phenotypes. The alleles should achieve a balance of utility, flexibility, throughput and cost. A null allele is an indispensable starting point for studying the function of every gene. Inserting a reporter gene (e.g., β-galactosidase or green fluorescent protein) allows a rapid assessment of which cell types normally support the expression of that gene. Therefore, we propose to produce a nullreporter allele for each gene. Making each mutation conditional in nature by adding cis-elements (e.g., loxP or FRT sites) would be desirable, but we do not advocate this as part of the mutagenesis strategy unless the technological limitations currently associated with generating conditional targeted mutations on a large scale and in a cost-effective manner can be overcome.
A combination of methods should be used
Various methods can be used to create mutated alleles, including gene targeting, gene trapping and RNA interference. Advantages of conventional gene targeting include flexibility in design of alleles, lack of limitation to integration hot spots, reliability for producing complete loss-of-function alleles, ability to produce reporter knock-ins and conditional alleles, and ability to target splice variants and alternative promoters. BAC-based targeting has the potential advantages of higher recombination efficiencies and flexibility for producing complex mutated alleles18. Gene trapping is rapid, is cost-effective and produces a large variety of insertional mutations throughout the genome but can be somewhat less flexible17,19-21. There is uncertainty regarding the percentage of gene traps that produce a true null allele and the fraction of the genome that can ultimately be covered by gene-trap mutations. Trapping is not entirely random but shows preference for larger transcription units and genes more highly expressed in ES cells. In recent studies, gene trapping was estimated to potentially produce null alleles for 50-60% of all genes, perhaps more if a variety of gene-trap vectors with different insertion characteristics is used17,21. RNA interference offers enormous promise for analysis of gene function in mice22 but is not yet sufficiently developed for large-scale production of gene modifications capable of reliably producing true null alleles. Both gene-targeting and gene-trapping methods are suitable for producing large numbers of knockout alleles, and, given their complementary advantages, a combination of these methods should be used to produce the genome-wide collection of null-reporter alleles most efficiently.
What should the deliverables be?
A genome-wide knockout mouse project could deliver to the research community a trove of valuable reagents and data, including targeting and trapping constructs and vectors, mutant ES cell lines, live mice, frozen sperm, frozen embryos, phenotypic data at a variety of levels and detail, and a database with data visualization and mining tools. At a minimum, we believe that a comprehensive genome-wide resource of mutant ES cell lines from an inbred strain, each with a different gene knocked out, should be produced and made available to the community. Choosing an inbred line (129/SvEvTac or C57BL/6J), and evaluating the alternative of using F1 ES cells and tetraploid aggregation to provide potential time savings, merits additional scientific review and discussion23,24. ES cells should be converted into mice at a rate consistent with project funding and the ability of the worldwide scientific community to analyze them. Although the value and cost-effectiveness of systematically characterizing the mice is a matter of debate, a limited set of broad and cost-effective screens, probably including assessment of developmental lethality, physical examination, basic blood tests, and histochemical analysis of reporter gene expression, would be useful. More detailed phenotyping, based on findings from the initial screen or existing knowledge of the gene’s function, could be done at specialized centers. All ES cell clones and mice (as frozen embryos or sperm) should be available to any researcher at minimal cost, and all mouse phenotyping and reporter expression data should be deposited into a public database.
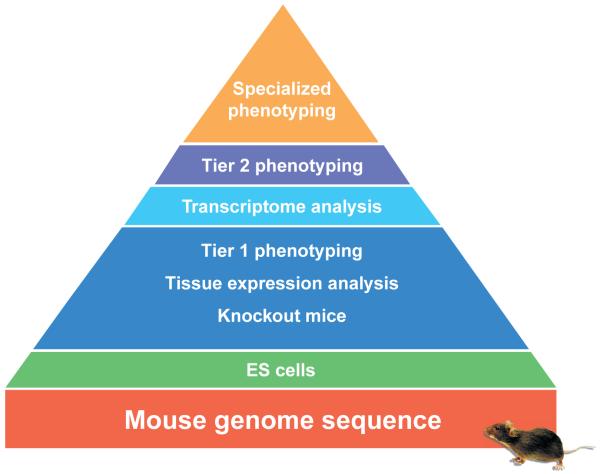
In determining how to implement the project, utility to the research community should be the standard for judging value. Each step after ES cell generation (e.g., mouse creation, breeding, expression analysis, phenotyping) will make the resource useful to more researchers but will also increase costs and scientific complexity. We therefore advocate a ‘pyramid’ structure for the project (Fig. 1). At the base of the pyramid is the genome-wide collection of mutant ES cells for every mouse gene. Over time, a subset of these mutant ES cells should be made into mice and characterized with an initial phenotype screen (Tier 1; Fig. 1) and analysis of tissue reporter-gene expression. A subset of these lines should be profiled by microarray analysis, and a subset of these profiled by system-specific (Tier 2) phenotyping, based on the results of the Tier 1 phenotyping, array studies, existing knowledge of the gene’s function and the gene’s tissue expression pattern. With time, the upper tiers of the pyramid will be filled out, eventually transforming the pyramid into a cube, with information of all types available for all genes.
Figure 1.
Structure of resource production in the proposed KOMP. Using the mouse genome sequence as a foundation, knockout alleles in ES cells will be produced for all genes. A subset of ES cell knockouts will be used each year to produce knockout mice, determine the expression pattern of the targeted gene in a variety of tissues and carry out screening-level (Tier 1) phenotyping. In a subset of mouse lines, transcriptome analysis and more detailed system-specific (Tier 2) phenotyping will be done. Finally, specialized phenotyping will be done on a smaller number of mouse lines with particularly interesting phenotypes. All stages will occur within the purview of the KOMP except for the specialized phenotyping, which will occur in individual laboratories with particular expertise.
This project will require the resolution of numerous intellectual property claims involving the production and use of knockout mice. To deal with the existing patents that cover the technologies and processes involved in the production of mutant mice, we suggest that a ‘patent pool’, such as that used in the semiconductor industry25, should be generated. Several individuals who represent entities that control patents on mouse knockout technologies are authors on this paper, and they agree with this approach. We also agree that any mutant ES cells or mice produced should be placed immediately in the public domain.
Mechanisms and costs
ES cell production
Automated knockout construct and ES cell production should be carried out in coordinated centers to ensure efficiency and uniformity. We estimate that most known mouse genes could be knocked out in ES cells within 5 years, using a combination of gene-trapping and gene-targeting techniques. Gene trapping can produce a large number of mutated alleles quickly, but its progress should be monitored closely to determine when its yield of new genes diminishes17 and, therefore, when targeting should be increasingly relied on. As large-scale trapping projects have already defined gene classes that probably cannot be knocked out by trapping (e.g., single-exon GPCRs, genes that are not expressed in ES cells), we propose that targeting begin on those classes immediately. All ES cells should be made available to the research community, because this collection itself would be a valuable resource. Efforts in the public and private sectors have already knocked out many genes in ES cells, and, to the degree that the alleles produced fit the prescribed characteristics (i.e., null alleles with a reporter) and are available, every effort should be made to incorporate these into the planned public resource. Costs for generating this part of the resource were estimated at between $9-11 million/year for five years (these and all subsequent figures are direct costs).
Mouse production
The subset of ES cells made into mice each year should be chosen by a peer-review process. Central facilities for high-efficiency mouse production, genotyping, breeding, maintenance and archiving should be funded, to take advantage of efficiencies of scale in mouse creation and distribution. Researchers could apply to produce groups of mice outside the centers, as long as they meet the cost specifications of the program. All mice should be made available immediately to researchers as frozen embryos or sperm, for nominal distribution cost. An initial target of 500 new mouse lines per year would double the current rate at which new genes are knocked out in the public sector; we feel that this rate is within the capacity of the biomedical research community worldwide to absorb and analyze. We estimated the initial cost of this level of mouse production to be $12.5-15 million per year.
Reporter tissue expression analysis
Approximately 30 tissues from adult and developmental stages should be sampled to cover the main organ systems. Analysis methods should be customized to the organ system and marker, and a searchable database of the sites of gene expression, and the images showing them, should be produced. Centers to carry out these analyses and data curation should be selected by peer review. We estimated the cost of this component for 500 mouse lines to be $2.5-5 million per year, depending on how much tissue sectioning and cell-level analysis is done.
Phenotyping
Tier 1 phenotyping should be a low-cost screen for clear phenotypes and should be done on all mouse lines produced. Tier 1 should include home-cage observation, physical examination, blood hematological and chemistry profiles, and skeletal radiographs. The centers producing the mice should carry out the Tier 1 analyses, at an estimated cost of $2.5 million per year for 500 lines. Selected lines, chosen on the basis of findings from Tier 1 phenotyping, tissue expression patterns, microarray data and the scientific literature, should undergo more detailed and system-focused Tier 2 phenotyping. Tier 2 phenotyping should be done in specialized phenotyping centers, akin to those already in operation for phenotyping of mice produced by ENU mutagenesis. All Tier 1 and Tier 2 phenotyping should be done on a uniform genetic background by dedicated groups of individuals in single locations, to facilitate consistency and cross-comparison of results among different mouse lines. All Tier 1 and Tier 2 phenotyping results should be deposited into a central project database freely accessible to the research community. More detailed and specialized phenotyping could be done by individual researchers in their own laboratories; deposition of this more detailed phenotype data would be encouraged.
Transcriptome analysis
Transcriptome profiling of tissues from each knockout line, collected in a uniform way across all mice and tissues and placed into a searchable relational database, would add substantially to the scientific value of the project, though it would also add considerably to its cost. Transcriptome analysis should therefore be done on a subset of mice, chosen by peer review. We estimate that, with the best currently available array technology, an analysis of ten tissues would cost ~$18,000 per line.
Conclusions
This project, tentatively named the Knockout Mouse Project (KOMP), will be a crucial step in harnessing the power of the genome to drive biomedical discovery. By creating a publicly available resource of knockout mice and phenotypic data, KOMP will knock down barriers for biologists to use mouse genetics in their research. The scientific consensus that we achieved—that a dedicated project should be undertaken to produce mutant mice for all genes and place them into the public domain—is important but is only the beginning. Implementation of these recommendations will require additional input from the greater scientific community, including those responsible for programmatic direction and financial support of biomedical research in the public and private sectors. This ambitious and historic initiative must be carried out as a collaborative effort of the worldwide scientific community, so that all can contribute their skills, and all can benefit. International discussions among scientific and programmatic staffs since the Banbury meeting at Cold Spring Harbor, in both the public and private sectors, have shown that there is great enthusiasm and commitment to this vision. The next step for KOMP will be to move this visionary plan from conceptualization to implementation, with an urgency befitting the benefits it will bring to science and medicine.
Footnotes
URLs. The curated Mouse Knockout & Mutation Database is available at http://research.bmn.com/mkmd/. The curated Mouse Genome Database is available at http://www.informatics.jax.org/. Patent pools: A solution to the problem of access in biotechnology patents? is available at http://www.uspto.gov/web/offices/pac/dapp/opla/patentpool.pdf.
References
- 1.International Human Genome Sequencing Consortium Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Mouse Genome Sequencing Consortium Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Bultman SJ, Michaud EJ, Woychik RP. Cell. 1992;71:1195–1204. doi: 10.1016/s0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- 5.D’Arcangelo G, et al. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein JL. Nat. Med. 2001;7:1079–1080. doi: 10.1038/nm1001-1079. [DOI] [PubMed] [Google Scholar]
- 8.D’Orleans-Juste P, Honore JC, Carrier E, Labonte J. Curr. Opin. Pharmacol. 2003;3:181–185. doi: 10.1016/s1471-4892(03)00017-1. [DOI] [PubMed] [Google Scholar]
- 9.Horton WA. Lancet. 2003;362:560–569. doi: 10.1016/S0140-6736(03)14119-0. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DC. Am. J. Med. Genet. 2001;106:71–93. doi: 10.1002/ajmg.1393. [DOI] [PubMed] [Google Scholar]
- 11.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 12.Zambrowicz BP, et al. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 13.Nadeau JH, et al. Science. 2001;291:1251–1255. doi: 10.1126/science.1058244. [DOI] [PubMed] [Google Scholar]
- 14.Wiles MV, et al. Nat. Genet. 2000;24:13–14. doi: 10.1038/71622. [DOI] [PubMed] [Google Scholar]
- 15.Stryke D, et al. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen J, et al. Proc. Natl. Acad. Sci. USA. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skarnes WC, et al. Nat. Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valenzuela DM, et al. Nat. Biotechnol. 2003;21:652–629. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 19.Chen WV, Delrow J, Corrin PD, Frazier JP, Soriano P. Nat. Genet. 2004;36:304–312. doi: 10.1038/ng1306. [DOI] [PubMed] [Google Scholar]
- 20.Stanford WL, Cohn JB, Cordes SP. Nat. Rev. Genet. 2001;2:756–768. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- 21.Zambrowicz BP, et al. Proc. Natl. Acad. Sci. USA. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunath T, et al. Nat. Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- 23.Seong E, Saunders TL, Stewart CL, Burmeister M. Trends Genet. 2004;20:59–62. doi: 10.1016/j.tig.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Eggan K, et al. Nat. Biotechnol. 2002;20:455–459. doi: 10.1038/nbt0502-455. [DOI] [PubMed] [Google Scholar]
- 25.Clark J, Piccolo J, Stanton B, Tyson K. Patent pools: A solution to the problem of access in biotechnology patents? US Patent and Trademark Office; 2000. [Google Scholar]