Abstract
Genomics provides a comprehensive view of the complete genetic makeup of an organism. Individual sequence variations, as manifested by single nucleotide polymorphisms (SNPs), can provide insight into the basis for a large number of phenotypes and diseases including cancer. The ability rapidly screen for SNPs will have a profound impact on a number of applications, most notably personalized medicine. Here we demonstrate a new approach to SNP detection through the application of surface-enhanced Raman scattering (SERS) to the ligase detection reaction (LDR). The reaction uses two LDR primers, one of which contains a Raman enhancer and the other a reporter dye. In LDR, one of the primers is designed to interrogate the SNP. When the SNP being interrogated matches the discriminating primer sequence, the primers are ligated and the enhancer and dye are brought into close proximity enabling the dye’s Raman signature to be detected. By detecting the Raman signature of the dye rather than its fluorescence emission, our technique avoids the problem of spectral overlap which limits number of reactions which can be carried out in parallel by existing systems. We demonstrate the LDR-SERS reaction for the detection of point mutations in the human K-ras oncogene. The reaction is implemented in an electrokinetically active microfluidic device that enables physical concentration of the reaction products for enhanced detection sensitivity and quantization. We report a limit of detection of 20 pM of target DNA with the anticipated specificity engendered by the LDR platform.
Introduction
Single nucleotide polymorphisms (SNPs) are single base pair differences in DNA among individuals where the less common variant occurs in at least 1% of the total population.1,2 The decoding of the human genome has revealed more than 3 million SNPs (roughly 1 every 100–300 bases) and opened up exciting new capabilities for associating individual SNPs, haplotypes, and linkage disequilibrium with disease states and pharmacological responses.2,3 Owing to large-scale SNP discovery, genetic variation in the human genome is now an emerging resource for the study of cancer-related genes.4–6 SNPs represent the most common variations across a genome, and they can be used to directly detect alleles responsible for a trait of interest.2,7 Since cancer is at least in part caused by the accumulation of inherited and/or somatic mutations, SNPs are also emerging as an invaluable tool for cancer association studies.5,8 In some cases single base pair mutations are the direct cause of the cancer,9 while in others they represent well-defined molecular markers indicative of an increased risk of cancer. In either case, numerous SNPs have been shown to be good biomarkers for many classes of cancer and have further been shown to correlate with various clinicopathological features of different cancer subtypes.8,10–13 For example, point mutations in the proto-oncogene K-ras have been identified which induce its oncogenic function at codons 12, 13, and 61.14 Mutations in the BRCA genes associated with breast cancer also serve as a model. The p53 tumor suppressor gene and its negative regulator MdM2, have also been associated with oncogenic activation after various point mutations, which has been extensively analyzed and reviewed.15,16 Successive SNP–SNP interactions which may increase risk or severity of cancers have also been described.17 Analysis from SNP arrays have also shown that determination of copy number from specific SNP populations is also a useful indicator for cancer progression.18 Polymorphisms themselves have additionally been linked as statistically significant indicators of cancer progression.19 Recently, SNPs have been identified as key markers in pharmacogenomics, the study of inheritable drug metabolism and reactivity, directly related to cancer treatment.20
To make good use of these discoveries, faster and cheaper technological methods are needed to discover new SNPs, for genotyping them in many individuals, and ultimately for clinical diagnostics.21,22 All allele-specific SNP discrimination techniques suitable for high-throughput genetic analysis can be categorized as primer extension, oligonucleotide ligation, invasive cleavage, or hybridization based.22 There are numerous variants on the primer extension technique; however, all are based on the ability of DNA polymerase to incorporate specific deoxyribonucleotides that are complementary to the sequence of the template DNA.23 A detailed review of all the implementations is beyond the scope of what can be covered here (see recent reviews by Perkel24 for details) but of the various methods Real-Time PCR or RT-PCR25–27 is likely of the simplest. The primary reason for this is that the reaction is carried out in a homogeneous format and requires no post-PCR processing. This reduces the time and labor required for analysis while minimizing the number of potential sources of error and contamination. To increase the specificity of the RT-PCR format for more stringent SNP detection, a number of researchers have moved toward oligonucleotide ligation-based methods such as the ligase detection reaction (LDR).28–30 In LDR, two primers anneal onto the DNA template at the site of a SNP. A discriminating base complimentary to the wild template (WT) or mutant (MT) allele is present at the 3′ end of the upstream primer. A downstream primer common to both alleles is also present in the reaction. If the primers match the template perfectly, ligation occurs. Ligation will not occur if the primer and template are mismatched at the discriminating base. The ligation, which permanently links the two primers together, can be detected with a variety of different including FRET31 and autoradiography.32,33 Most existing LDR and RT-PCR protocols use fluorescence tags34 as reporters, and thus, all have the same fundamental limitation in that spectral overlap between the reporter dyes limits the degree to which the reactions can be multiplexed. Typical florescent dyes have an emission spectrum with a full width-half-maximum on the order of 50 nm. Thus, over the useful detection range of the spectrum (about 500–750 nm) one can at maximum expect to be able to discriminate on the order of six different dyes. This extends then to single tube reactions in that a maximum of five different SNPs can be screened for at once (one color is used as an internal calibration). To increase the number of molecular markers which can be screened for in a single step, one would like to develop a homogeneous detection system as simple as these but without the spectral overlap limitation.
One method by which this could be accomplished is through the use of a surface-enhanced Raman spectroscopy (SERS). A number of authors have developed SERS-based techniques for sequence specific DNA detection. Cao et al.35 for example demonstrated a three-component sandwich assay using Ag staining on Au seed particles to enhance SERS signals in DNA microarray format. In another example, Fabris et al.36 developed a peptide nucleic acid (PNA)-based SERS DNA assay which enabled more rapid hybridization rates since the neutrally charged PNA were not limited by the typical electrostatic repulsion between cDNA strands. A number of so-called “molecular beacon”-based SERS probe techniques have also been developed.37–39 These methods typically use a DNA hairpin structure with a Raman-active molecule at one end and a metallic nanoparticle at the other which become separated following a hybridization, thereby reducing the strength of the emitted SERS signal. Qian et al.33 and Graham et al.40 have also recently presented dye-coded DNA-functionalized metal nanoparticles-based SERS methods which enabled greater sensitivity and repeatability in obtaining the SERS spectrum.
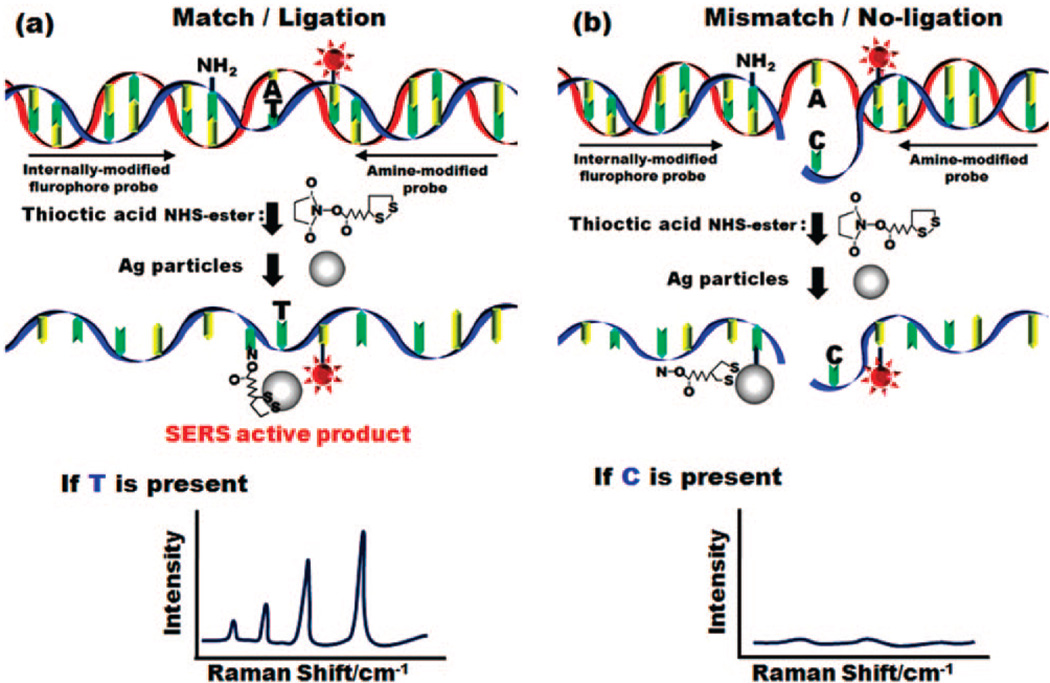
Here we present a new technique for SNPs detection that combines the selectivity and ease of use of the LDR reaction with the potential for large bandwidth and sensitivity of SERS.1,36,41 The LDR for discrimination of alleles is more accurate than the common hybridization reaction and provides high sensitivity and parallel analysis of several loci directly on genomic DNA in order to distinguish point mutations.42 In this technique (illustrated in Figure 1) the upstream primer contains a SERS active dye and the discriminating 3′ base, while the downstream primer contains an amine to which a silver nanoparticle (which acts as the SERS enhancer) is attached. When the two primers are ligated together (which occurs in the case of a perfect match with the template DNA) the dye is brought into close proximity to the nanoparticle and its Raman signature is detectable. In the case where ligation is not successful, the dye and nanoparticle remain separated and SERS enhancement will not occur. In this paper we demonstrate the LDR-SERS detection reaction and apply it to the detection of point mutations in the K-ras oncogene. We implement the reaction in a microfluidic chip containing electrokinetically active microwell devices43 that enable us to enhance SERS detection by concentrating the reaction products from bulk phase into a confined volume for enhanced optical interrogation. In addition to SNP detection, we also demonstrate the quantitative nature of the reaction and characterize the limit of detection.
Figure 1.
Overview of SERS enhanced PCR/LDR detection reaction.
Experimental Section
Materials
All chemicals and solvents were purchased at the highest purity grade available. For the SERS enhancers, 50 nm diameter silver colloid solutions were purchased from Nanocs (New York, NY). The buffer solution used was 10 mM phosphate-buffered saline (PBS) buffer solution (0.6 M NaCl, pH 7.6) and stored in a freezer until use. Poly(dimethylsiloxane) (PDMS) microfluidics were made using a Sylgard 184 silicon elastomer kit (Dow Corning, Midland, MI). The thermostable ligase 9°N DNA Ligase was purchased from New England Biolabs Inc. which included a buffer for the LDR reaction. All reactions were carried out at room temperature unless otherwise specified.
LDR Reaction
The oligonucleotide sequences of all the probes and templates used in these experiments are shown in Table 1. All DNA primers and templates were synthesized by Integrated DNA technologies and adapted by previous work done by Khanna et al.38 During primer design, the fluorophore and the amine by which nanoparticles are attached were placed 14 bp apart. To maximize the Raman signal, the enhancer and fluorophore must be as close together as possible. The LDR reaction contained the following in 10 µL reaction: 25 pM of template, 100 pM of each primer, 1 µL of ligase, 1 µL of 10 × Buffer, and water to 10 µL. The LDR reactions used the following thermocycler program: (1) at 90 °C for 2 min (2) at 90 °C for 30 s (3) at 50 °C for 4 min (4) repeat steps 2–4 100 times.
Table 1.
Synthetic DNA Template and Primers Used in LDR-SERS Experiments
| template/primer | sequence for LDR (5′ 3′) |
|---|---|
| WT K-Ras template | TCC ACA AAA TGA TTC TGA ATT AGC TGT ATC GTC AAG |
| GCA CTC TTG CCT ACG CCA CCA GCT CCA ACT ACC ACA AGT | |
| TTA TAT TCA GTC ATC | |
| MT K-Ras template | TCC ACA AAA TGA TTC TGA ATT AGC TGT ATC GTC AAG |
| GCA CTC TTG CCT ACG CCA TCA GCT CCA ACT ACC ACA AGT | |
| TTA TAT TCA GTC ATC | |
| common LDR primer | 5Phosa/TGG CG/AmTb/ AGG CAA GAG TGC CTT GAC |
| mutant LDR primer | GAA TAT AAA CTT GTG GTA G/FlurTc/T GGA GCT GA |
5Phos denotes a 5′ phosphorylation.
AmT denotes an aminated thymine.
FlurT denotes a fluorescein dT.
The italicized bases of the template sequences are the complementary nucleotides to both primers. The 3′ base in mutant LDR primer (bold) allows for specific discrimination of the two templates.
LDR Functionalization and Purification
The LDR reaction mixture was treated with 2 µL of DMSO to lower the melting temperature of the single-stranded template and primers and reduce postligation binding to each other. The NHS ester of thioctic acid (100 pmol) was added to the treated LDR reaction and allowed to react for 1 h. Thioctic acid was chosen as a linker due to its high affinity for Ag and greater stability than thiols when coupling oligonucleotides to nanoparticles.44 The amine modification present in the primer is an amine-modified deoxythymidine with a c6 spacer. The reaction mixture was then added to 200 µL of 50 nm Ag nanoparticles and allowed to react for 1 h. After the reaction, the solution was put onto a 37 mL size exclusion column using Superdex 200 resin and flowed at 2 mL/min using DW eluent solution. The first elution peak by Abs280 was collected and determined to be the particles. Further evidence of purification is seen by the negative control where fluorophore conjugated DNA is exposed to the silver nanoparticles then purified. Very little signal is seen as compared to the experimental samples. The particles were then concentrated using a 30 kD spin filter, spinning at 2500g for 1.5 min intervals and resuspending completely between spins. It is important to note that if the particles are not resuspended or are spun too hard, they will irreversibly stick to the filter membrane.
Gel Shift Assay
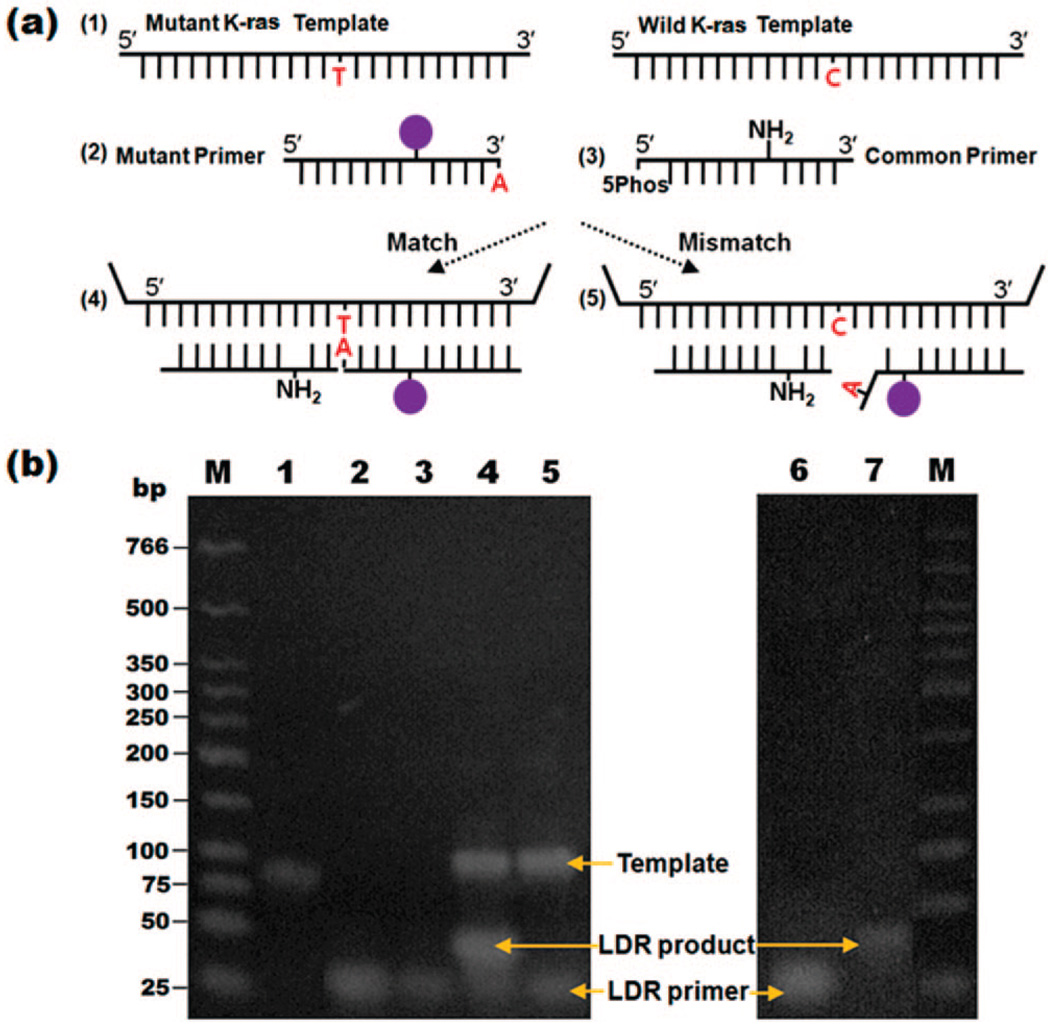
An expected positive and negative LDR reaction were run on a 3% Low-Melt temp TAE agarose gel for 50 min at 200 V in a cold room and subsequently stained with ethidium bromide (Figure 2, Lanes 1–5) or directly (Figure 2, Lanes 6–7). LDR-SERS reactants and products were characterized via fluorescent imaging after ethidium bromide staining as compared to DNA fragments of known size (Lanes 1–5, Figure 2). Gaffney et al.12 reported that the allelic probes can be designed to have unique lengths, so that the wild type and variant ligation products can be separated and detected on the basis of size. In this study, the allelic probes were labeled with fluorophores, enabling the ligation products to be additionally discriminated by the presence of fluorescence without being stained by ethidium bromide (lanes 6 and 7, Figure 2.
Figure 2.
Ethidium bromide-stained agarose gel showing the results of LDR-SERS reaction. Standard marker (lane 1), templates (lane 2), mutant LDR primer (lane 3), LDR product by MT template (lane 4), LDR product by WT template (lane 5), final WT and MT LDR products illuminated without EtBr staining (lanes 6 and 7).
Microfluidic Device
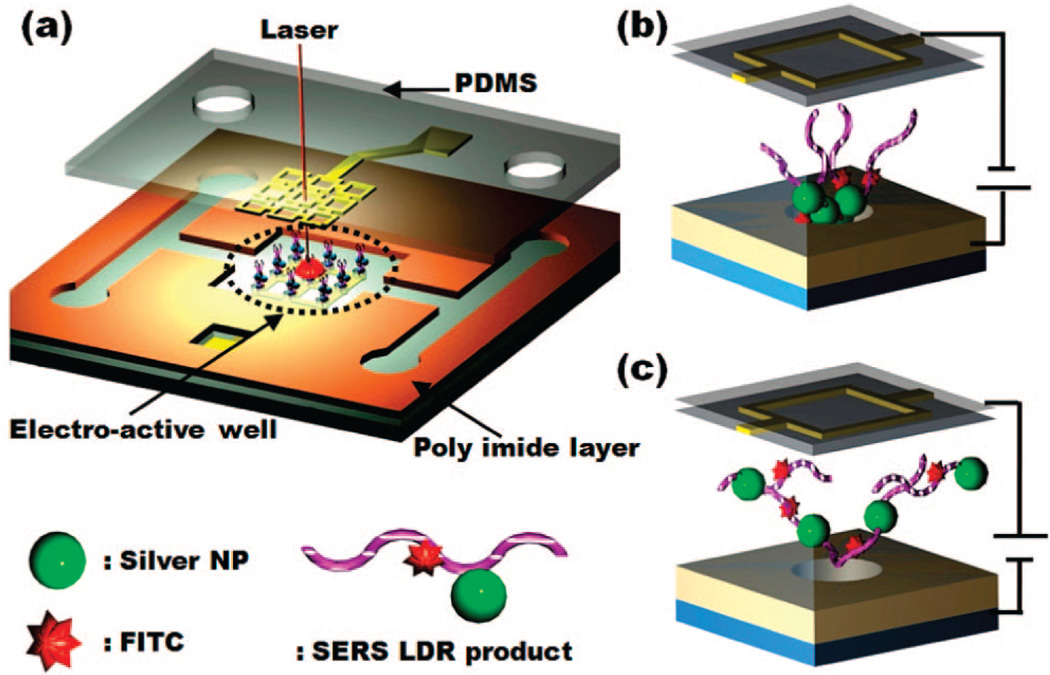
Figure 3 shows a schematic of the microfluidic device used here. As mentioned above, in addition to the sample delivery channels, the active element in the device are the electrokinetically active microwells which serve to rapidly concentrate the reaction products from the bulk solution phase into a confined volume for optical probing. Details of the fabrication are outlined in our previous work;44 however, briefly the overall structure consists of three functional layers, namely, a lower substrate which contains the attraction electrode, a polyimide (PI) dielectric layer into which the channels and microwell were defined, and an upper electrode. The device was manufactured by first spinning positive and lithographically patterning photoresist S1813 (Shipley, Marlborough, MA) onto a Pyrex substrate to define the lower electrode pattern. Following this, 5 nm Ti/ 50 nm gold was evaporated and a lift-off process carried out with 1165 photoresist stripper (Shipley Microposit) overnight, leaving the lower electrode on the glass surface. A two-layer photoactive PI process was used to pattern the microchannels and wells as shown in Figure 3. The upper gold electrode was patterned on PDMS using a similar technique to that described by Lee et al.45 To bond the upper and lower surfaces, both layers were activated in oxygen plasma, and the two halves were aligned and pressed together using our homemade aligner.
Figure 3.
(a) Illustration electroactive microwell device for LDR-SERS-based SNPs detection. Schematic representation of the system showing the lower electrode on the Pyrex glass substrate, the microwell array (diameters of 10 µm and height of 8 µm), and the upper electrically functionalized PDMS gold electrode. Applying the polarity shown in (b) attracts particles and (c) rejects them.
To record the SERS emission spectrum, the LDR-SERS products were concentrated in the microwell by applying the attraction potential. As described in our previous work,44 the approach uses electrokinetically active microwells to physically concentrate the bulk phase reaction product into a well-defined volume for optical interrogation. For each of the measurements reported here, we take spectra from three different points in the 10 µm well (using a 2 µm laser spot size) and report the average measurement (with error bars to indicate the standard deviation). Further details on this concentration approach (including detailed comparison with other approaches) are available in this earlier work.44 The excitation laser was focused at the microwell through the upper gold electrode patterned PDMS layer. In all cases a Hewlett-Packard 6234A dual-output power supply and a Keithley 236 were used to apply and measure the electrical potentials.
Raman Spectroscopy Measurements
Raman measurements were made using an in Via Raman microscope spectrometer coupled to a Leica microscope. The experiments were conducted by focusing the excitation laser on the electro-active microwell. The 488 nm line of an Ar+ ion laser was used as optical excitation source and the scattered signal was collected by a Peltier-cooled CCD detector. A 50× (NA = 0.55) objective lens was used to focus the laser beam spot onto the sample surface with diameter of about 2 µm. Wave-numbers ranging from 1100 to 1800 cm−1 were examined here.
Results and Discussion
Description of SERS-Enhanced LDR Reaction
As mentioned in the introduction, we report here the development of a new reporter system for LDR-based SNP detection reaction based on the use of SERS. The SERS effect is related to the phenomenon of plasmon resonance, wherein metal nanostructures exhibit a pronounced optical resonance due to the collective excitation of conduction electrons in the metal, in response to incident electromagnetic radiation.46 The plasmons result in a significant localized enhancement in the magnitude of the electromagnetic field surrounding the particle.31,33,47 SERS-active molecules located in the near-field region of the optical nanostructures are therefore exposed to a larger electromagnetic intensity than that of the excitation light and thus enhancing the strength of its Raman scattered light. Most current SERS-based detection schemes involve the immobilization of the fluorophore-labeled nucleic acids on a solid surface.46–48 The SERS spectra are then generated on the basis of the proximity of the DNA and its fluorophore to the surface. Challenges with surface-tethered systems include steric hindrance (and therefore a limitation in the accessibility of the target DNA) and the longer reaction time required for heterogeneous reactions. As is described in detail in Huh et al.44 the advantage of the electrokinetically active microwell used here is that it enables active mixing to enhance the rate of binding between the SERS enhancers and the biomolecular targets, as well as rapid concentration of the product for surface-phase optical interrogation and enhanced sensitivity. It is important to mention that this is not the only method by which solution-phase sensitivity could be increased, as a number of researchers have demonstrated the use of unique nanoparticle shapes49 and assemblies,40,50 resonance effects51 and multiple wavelength interogation52 to enhance sensitivity and specificity. We note, however, that many of these effects could be used in conjunction with our technique for further enhancement.
In our approach, a nanoparticle Raman enhancer is incorporated directly into one of the LDR probes allowing us to perform the reaction homogenously. Figure 1 illustrates an overview of the SERS-enhanced SNP-LDR reaction developed here. For simplicity a single set of PCR products is presented with two LDR probes. As is shown, one LDR probe is internally modified to contain a fluorophore reporter and the other is internally modified with an exposed amine group. Following enzymatic ligation the exposed amine group reacts with a single silver nanoparticle as shown. If the two fragments match exactly the template sequence (Figure 1a), the ligase with ligate them and the fluorophore and Raman enhancer are held in close proximity. Since the Raman enhancement is dependent upon the distance between the fluorophore and the nanoparticle, a strong SERS signal can be detected in the case where positive ligation occurs. In the case where a base pair mismatch exists (Figure 1b) the probes are not ligated and the fluorophore’s Raman spectrum cannot be detected.
LDR-Based Detection of Point Mutations in the K-ras Oncogene
The efficacy of the LDR primers against their respective SNP targets were initially tested by using traditional gel electrophoresis (Figure 2). Each LDR reaction contained the amplified template DNA, the wild type or mutant LDR primer with a fluorescein-modified deoxythymidine tag, and the common LDR primer. For the upstream primer and downstream primer, a band of the predicted size (20 bp) was observed at lanes 2 and 3. As shown in Figure 2b, lane 2 was brighter than lane 3 with the same molarity because the mutant LDR primer (lane 2) contained the fluorophore. To verify the presence of the expected LDR product, two LDR reaction products reacted using the mutant template (MT, lane 4) and wild type template (WT, lane 5). For the positive sample, the band of LDR reaction size (40 bp) was observed (lane 4) because the two fragments match exactly to the template sequence, resulting in the generation of a longer oligonucleotide, compared to the starting primers. For negative control (lane 5), however, a band of the expected LDR reaction size (40 bp) was not detected since the mutant primer and WT were mismatched at the discriminating base of the upstream primer. After the LDR reaction, in order to obtain the higher SERS detecting sensitivity, the reacted samples were purified by size exclusion column.
Electroactive Microwells for Enhanced SERS Signal Detection
The current chip (Figure 3) comprises a glass substrate with lithographically patterned electrodes and is a modification of that presented in Huh et al.43 The substrate and electrodes are covered with an electrically insulating polyimide layer into which 10 µm diameter wells are etched (Figure 3). To deliver nanoparticles to the wells, a microfluidic structure is then defined in PI using standard lithography techniques. After completion we align and bond the PDMS fluidics to the bottom substrate such that the wells align with the spaces in the upper electrodes, as shown in Figure 3b and c. By applying electric potential between the upper and lower electrodes, we can concentrate the solution-phase targets into the wells as they flow over them. Once concentrated, the well can be interrogated optically through the upper PDMS, as shown in Figure 3a. When the SERS-active LDR products were introduced into the chip via two inlet ports into the chamber, concentration was performed by applying potential of 1.5 V. This concentration step was found to increase the reproducibility and intensity of the SERS signal to the point where the spectrum could be recorded in as little as 15 s.
On-Chip LDR-SERS Detection of Point Mutations in the KRAS Oncogene
In this study, SERS detection was carried out using LDR reaction of low-abundant DNA point mutations in KRAS oncogenes with the allelic composition evaluated at one locus. As mentioned in the Introduction, this oncogene has been associated with a variety of cancers including lung, colorectal, and pancreatic malignancies. To characterize the reaction experiments were conducted using the K-RAS mutation described in Table 1 which has been shown to possess a high diagnostic value for colorectal cancers.53 In our first series of experiments, a downstream primer containing the amine was reacted with the NHS ester of thioctic acid (see Figure 1). The resulting chemical reaction served as a linker between the silver nanoparticles and the DNA primer. The resulting downstream primer was then introduced into an LDR reaction containing the upstream primer and template. In this case the LDR reaction failed. After subsequent testing, it was found that the ligation reaction would not proceed with the nanoparticle attached to the primer so close to the ligation site. Through these initial results, we confirmed that preligation chemistry and postreaction purification were both important to successful detection.
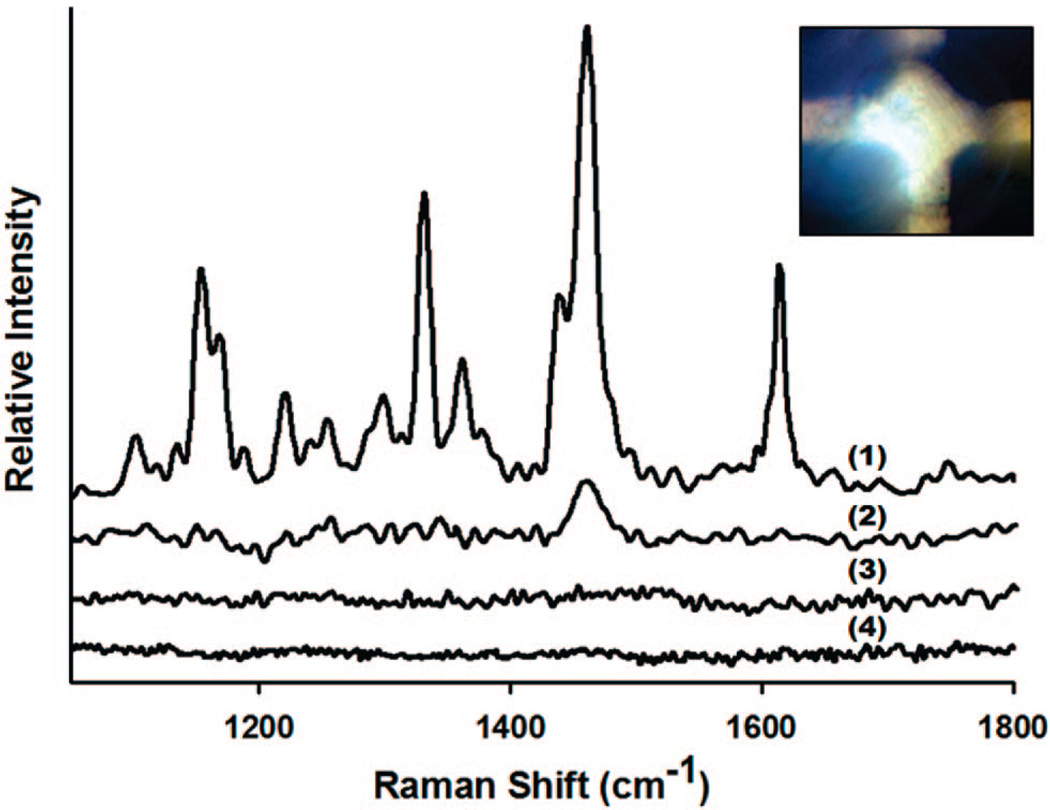
For the on-chip assays, the LDR products described above were introduced into the chip via their respective inlet ports into the central chamber (Figure 3), where the concentration was performed, at a flow rate of 5 µL s−1. After filling the SERS-active LDR products were attracted into the wells for 15 s at an applied potential of 1.5 V. To obtain the SERS signal, the excitation laser was focused at the microwell and the spectrum recorded integration time set to 15 s. In general, chip regeneration could be accomplished by reversing the polarity and rejecting the contents back into the chamber where they can be washed out (in Huh et al.44 we demonstrate as many as 80 attraction repulsion cycles). Here, however, a new device was used for each of the different experiments in order to avoid the potential for cross contamination. Figure 4 shows the SERS spectra collected on-chip for (a) positive sample containing fluorescein-modified deoxythymidine-labeled LDR-SERS products by the mutant template (denoted as FMdT-labeled MT), (b) negative sample reacted by WT, (c) control sample containing only silver particles and the DNA, and (d) background control sample containing silver particles and linker. MT depicts the mutant, functionalized LDR reaction which was expected to show peaks indicative of the dye. As can be seen in Figure 4, our results show that almost no detectable Raman signal was observed from the control sample for random adsorption of the DNA to the particles, nor from background control sample. As expected, Figure 4(1) shows the correct spectroscopic finger-prints corresponding to FMdT-labeled dye suggesting positive detection. In the negative sample the LDR-SERS diagnostic peaks were much weaker suggesting the reaction was successful. On the basis of these results, we used the 1610 cm−1 peak as diagnostic of a successful ligation reaction since it is prominent in the mutated spectrum and completely lost in the wild type spectrum the 1610 cm−1. The remaining peak at 1460 cm−1 in the wild type sample is likely due to fluorescence from nonspecific binding of the unligated primers. The large difference in the melting temperature of the primers as compared to the ligated LDR product meant that even at room temperature a significant difference was observed in the SERS spectra of the two samples.
Figure 4.
SERS spectra collected on-chip for (a) positive sample containing FMdT-labeled LDR-SERS products by the mutant template (denoted as FMdT-labeled MT), (b) negative sample reacted by WT, (c) control sample containing silver particles and DNA, and (d) background control sample containing silver particles and linker. The concentration of each SNP is 100 pM.
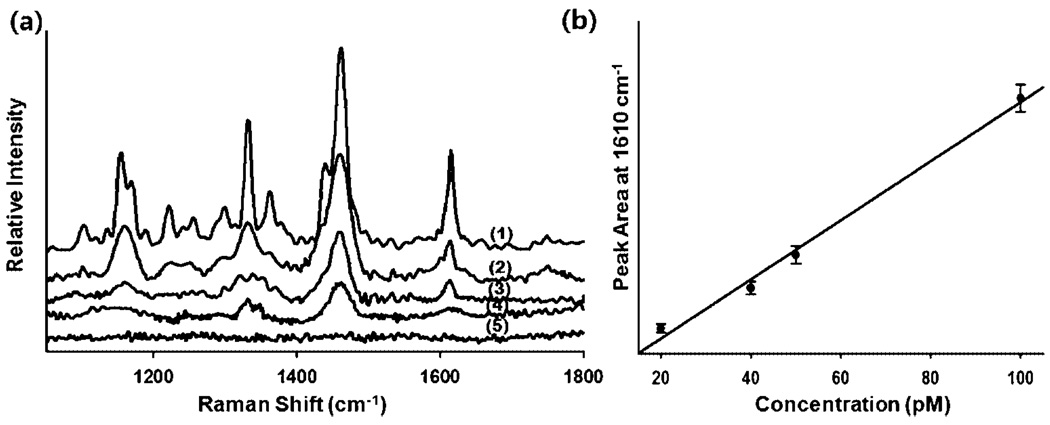
To examine the detection threshold of our reaction and to verify the ability for quantitative analysis, we next conducted a series of experiments at different template concentrations. Figure 5 shows the SERS spectra of FMdT-labeled MT in a microwell for various concentrations of reaction products of (1) 100, (2) 50, (3) 40, (4) 20, and (5) 10 pM. As expected, the intensity of the Raman peak decreases concomitantly with decreasing the concentration of LDR-SERS products (Figure 5a). Consistent with the technique used by Lee et al.,44,54 the concentration response was quantified by observing the change in the area of the SERS peak at 1610 cm−1. The results are plotted in Figure 5b and fitted to a linear curve (correlation coefficient: 0.993). As can be seen, below 20 pM the main diagnostic peak could not be detected and thus we omitted the 10 pM result from the corresponding calibration curve. On the basis of this result we report a limit of detection of 20 pM with this approach.
Figure 5.
(a) SERS spectra of FMdT-labeled MT in a microwell with the different concentrations of LDR-SERS products. (1) 100, (2) 50, (3) 40, (4) 20, and (5) 10 pM LDR-SERS samples. (b) Plot of peak area at 1610 cm−1 as a function of concentration (R = 0.993). Note that the 10 pM result is omitted from (b) since the concentration was below the limit of detection.
Conclusions
In this paper we have reported the development of a novel SERS LDR system for the detection SNPs associated with mutations KRAS oncogene. Using SERS-active LDR products related to KRAS cancer mutation, we successfully detected SERS signals with a limit of detection on the order of 20 pM and demonstrated the ability to quantify the solution concentration on the basis of the intensity of the SERS emission. By relying on detection of the spectrally unique Raman fingerprint, rather than florescence emission this technique could increase the multiplexibility of current homogeneous detection schemes by avoiding the problem of spectral overlap.
Supplementary Material
Supporting Information Available: Complete refs 1, 3, and 10. This material is available free of charge via the Internet at http://pubs.acs.org
Acknowledgment
The authors of this article are thankful for the financial support of the National Institutes of Health—National Institute of Biomedical Imaging and Bioengineering under Grant No. R21EB007031. Funding from the Ludwig Institute for Cancer Research is also acknowledged. The authors thank Francis Barany for his thoughtful insight into LDR.
References
- 1.Wang DG. Science. 1998;280:1077. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 2.Aouacheria A, Navratil V, Wen W, Jiang M, Mouchiroud D, Gautier C, Gouy M, Zhang M. Oncogene. 2005;24:6133. doi: 10.1038/sj.onc.1208745. [DOI] [PubMed] [Google Scholar]
- 3.Sachidanandam R. Nature. 2001;409:928. [Google Scholar]
- 4.Strausberg RL, Simpson AJ, Wooster R. Nat. Rev. Genet. 2003;4:409. doi: 10.1038/nrg1085. [DOI] [PubMed] [Google Scholar]
- 5.Imyanitov EN, Togo AV, Hanson KP. Cancer Lett. 2004;204:3. doi: 10.1016/j.canlet.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 6.Qiu P, Wang L, Kostich M, Ding W, Simon JS, Greene JR. BMC Cancer. 2004;4:4. doi: 10.1186/1471-2407-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakitandwe J, Trognitz F, Trognitz B. Plant Methods. 2007;3:2. doi: 10.1186/1746-4811-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soucek P, Borovanova T, Pohlreich P, Kleibl Z, Novotny J. Breast Cancer Res. Treat. 2007;103:219. doi: 10.1007/s10549-006-9367-9. [DOI] [PubMed] [Google Scholar]
- 9.Sidransky D. Nat. Rev. Cancer. 2002;2:210. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 10.Zheng SL. N. Engl. J. Med. 2008;358:910. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 11.Oh JJ, Koegel AK, Phan DT, Razfar A, Slamon DJ. Lung Cancer. 2007;58:7. doi: 10.1016/j.lungcan.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaffney R, Chakerian A, O’Connell JX, Mathers J, Garner K, Joste N, Viswanatha DS. J. Mol. Diagn. 2003;5:127. doi: 10.1016/S1525-1578(10)60462-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshiya G, Takahata T, Hanada N, Suzuki K, Ishiguro A, Saito M, Sasaki M, Fukuda S. J. Gastroenterol. Hepatol. 2008;23:948. doi: 10.1111/j.1440-1746.2008.05307.x. [DOI] [PubMed] [Google Scholar]
- 14.Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. Nature. 1987;327:298. doi: 10.1038/327298a0. [DOI] [PubMed] [Google Scholar]
- 15.Soussi T, Wiman KG. Cancer Cell. 2007;12:303. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Bond GL, Hu W, Levine A. Cancer Res. 2005;65:5481. doi: 10.1158/0008-5472.CAN-05-0825. [DOI] [PubMed] [Google Scholar]
- 17.Onay VU, Briollais L, Knight JA, Shi E, Wang Y, Wells S, Li H, Rajendram I, Andrulis IL, Ozcelik H. BMC Cancer. 2006;6:114. doi: 10.1186/1471-2407-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ, Jordanova ES. BMC Genomics. 2007;8:53. doi: 10.1186/1471-2164-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun T, Gao Y, Tan W, Ma S, Zhang X, Wang Y, Zhang Q, Guo Y, Zhao D, Zeng C, Lin D. Clin. Cancer Res. 2006;12:7009. doi: 10.1158/1078-0432.CCR-06-0464. [DOI] [PubMed] [Google Scholar]
- 20.Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Cancer Res. 2007;67:9609. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- 21.Risch N, Merikangas K. Science. 1996;273:1516. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 22.Kwok PY. Annu. Rev. Genomics Human Genetics. 2001;2:235. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- 23.Sobrino B, Brion M, Carracedo A. Forensic Sci. Int. 2005;154:181. doi: 10.1016/j.forsciint.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Perkel J. Nat. Methods. 2008;5:447. [Google Scholar]
- 25.Mhlanga MM, Malmberg L. Methods. 2001;25:463. doi: 10.1006/meth.2001.1269. [DOI] [PubMed] [Google Scholar]
- 26.Socher E, Jarikote DV, Knoll A, Roglin L, Burmeister J, Seitz O. Anal. Biochem. 2008;375:318. doi: 10.1016/j.ab.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Satterfield BC, Kulesh DA, Norwood DA, Wasteloski LP, Caplan MR, West JAA. Clinical Chem. 2007;53:2042. doi: 10.1373/clinchem.2007.091488. [DOI] [PubMed] [Google Scholar]
- 28.Kristensen VN, Kelefiotis D, Kristensen T, Borresen-Dale AL. Biotechniques. 2001;30:318. doi: 10.2144/01302tt01. [DOI] [PubMed] [Google Scholar]
- 29.Khanna M, Park P, Zirvi M, Cao WG, Picon A, Day J, Paty P, Barany F. Oncogene. 1999;18:27. doi: 10.1038/sj.onc.1202291. [DOI] [PubMed] [Google Scholar]
- 30.Favis R, Barany F. Circulating Nucleic Acids Plasma Serum. 2000;906:39–43. [Google Scholar]
- 31.Fang C, Agarwal A, Buddharaju KD, Khalid NM, Salim SM, Widjaja E, Garland MV, Balasubramanian N, Kwong D-L. Biosens. Bioelectron. 2008;24:216. doi: 10.1016/j.bios.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 32.Prigent C, Satoh MS, Daly G, Barnes DE, Lindahl T. Mol. Cell. Biol. 1994;14:310. doi: 10.1128/mcb.14.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian X, Zhou X, Nie S. J. Am. Chem. Soc. 2008;130:14934. doi: 10.1021/ja8062502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNamara DT, Kasehagen LJ, Grimberg BT, Cole-Tobian J, Collins WE, Zimmerman PA. Am. J. Trop. Med. Hyg. 2006;74:413. [PMC free article] [PubMed] [Google Scholar]
- 35.Cao YC, Jin R, Mirkin CA. Science. 2002;297:1536. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 36.Fabris L, Dante M, Braun G, Lee SJ, Reich NO, Moskovits M, Nguyen T-Q, Bazan GC. J. Am. Chem. Soc. 2007;129:6086. doi: 10.1021/ja0705184. [DOI] [PubMed] [Google Scholar]
- 37.Vo-Dinh T. IEEE J. Sel. Top Quant. 2008;14:198. doi: 10.1109/JSTQE.2007.914738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wabuyele MB, Vo-Dinh T. Anal. Chem. 2005;77:7810. doi: 10.1021/ac0514671. [DOI] [PubMed] [Google Scholar]
- 39.Jung J, Chen L, Lee S, Kim S, Seong GH, Choo J, Lee EK, Oh CH, Lee S. Anal. Bioanal. Chem. 2007;387:2609. doi: 10.1007/s00216-007-1158-6. [DOI] [PubMed] [Google Scholar]
- 40.Graham D, Thompson DG, Smith WE, Faulds K. Nat. Nano. 2008;3:548. doi: 10.1038/nnano.2008.189. [DOI] [PubMed] [Google Scholar]
- 41.Doering WE, Nie S. J. Phys. Chem. B. 2002;106:311. [Google Scholar]
- 42.Barany F. Proc. Natl. Acad. Sci. U.S.A. 1991;88:189. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huh YS, Chung AJ, Cordovez B, Erickson D. Manuscript in preparation. [Google Scholar]
- 44.Dougan JA, Karlsson C, Smith WE, Graham D. Nucleic Acids Res. 2007;35:3668. doi: 10.1093/nar/gkm237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.K J, Lee K, K A, Fosser RGN. Adv. Funct. Mater. 2005;15:557. [Google Scholar]
- 46.Tuan V-D. IEEE J. Select. Topics Quantum Electron. 2008;14:198. doi: 10.1109/JSTQE.2007.914738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hering K, Cialla D, Ackermann K, Dorfer T, Moller R, Schneidewind H, Mattheis R, Fritzsche W, Rosch P, Popp J. Anal. Bioanal. Chem. 2008;390:113. doi: 10.1007/s00216-007-1667-3. [DOI] [PubMed] [Google Scholar]
- 48.Jung HY, Park YK, Park S, Kim SK. Anal. Chim. Acta. 2007;602:236. doi: 10.1016/j.aca.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 49.Jackson JB, Westcott SL, Hirsch LR, West JL, Halas NJ. Appl. Phys. Lett. 2003;82:257. [Google Scholar]
- 50.Chaney SB, Shanmukh S, Dluhy RA, Zhao YP. Appl. Phys. Lett. 2005;87 [Google Scholar]
- 51.Mahajan S, Baumberg JJ, Russell AE, Bartlett PN. Phys. Chem. Chem. Phys. 2007;9:6016. doi: 10.1039/b712144j. [DOI] [PubMed] [Google Scholar]
- 52.Faulds K, McKenzie F, Smith WE, Graham D. Angew. Chem. 2007;119:1861. doi: 10.1002/anie.200604265. [DOI] [PubMed] [Google Scholar]
- 53.Hashimoto M, Barany F, Soper SA. Biosens. Bioelectron. 2006;21:1915. doi: 10.1016/j.bios.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Lee D, Lee S, Seong GH, Choo J, Lee EK, Gweon DG, Lee S. Appl. Spectrosc. 2006;60:373. doi: 10.1366/000370206776593762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available: Complete refs 1, 3, and 10. This material is available free of charge via the Internet at http://pubs.acs.org