SUMMARY
Although in vitro studies of embryonic stem cells have identified polycomb repressor complexes (PRCs) as key regulators of differentiation, it remains unclear as to how PRC-mediated mechanisms control fates of multipotent progenitors in developing tissues. Here, we show that an essential PRC component, Ezh2, is expressed in epidermal progenitors but diminishes concomitant with embryonic differentiation and with postnatal decline in proliferative activity. We show that Ezh2 controls proliferative potential of basal progenitors by repressing the Ink4A-Ink4B locus and tempers the developmental rate of differentiation by preventing premature recruitment of AP1 transcriptional activator to the structural genes that are required for epidermal differentiation. Together, our studies reveal that PRCs control epigenetic modifications temporally and spatially in tissue-restricted stem cells. They maintain their proliferative potential and globally repressing undesirable differentiation programs while selectively establishing a specific terminal differentiation program in a stepwise fashion.
INTRODUCTION
One of the fundamental goals of modern biology is to understand the molecular mechanisms by which multipotent progenitor cells control tissue development and maintenance. Increasing evidence has pointed to a possible role for polycomb group (PcG) proteins in this process. PcG proteins form chromatin-remodeling complexes referred to as polycomb repressor complexes (PRCs) (Ringrose and Paro, 2004). Comprised of Ezh2, Eed, and Suz12, PRC2 is recruited to chromatin, where methyltransferase Ezh2 catalyzes H3 trimethylation on lysine 27 (triMeK27-H3) (Cao et al., 2002). This histone mark then provides a platform to recruit PRC1 (Cao et al., 2002; Min et al., 2003), which aids in PcG-mediated repression either by chromatin compaction or by interfering with the transcription machinery (Francis et al., 2004; Ringrose and Paro, 2004; Sarma et al., 2008). Without Ezh2 activity, PRC1 cannot be recruited to chromatin, and PcG-mediated repression is not established (Cao et al., 2002; Rastelli et al., 1993).
In vitro studies of pluripotent mouse and human embryonic stem cells (ESCs) have shown that PRC2 proteins and their triMeK27-H3 marks reside at and transcriptionally repress many regulatory genes that control specific developmental lineages (Boyer et al., 2006; Lee et al., 2006; Pietersen and van Lohuizen, 2008). Establishing functional significance, Eed null ESCs have elevated expression of PcG-repressed differentiation genes (Boyer et al., 2006; Chamberlain et al., 2008).
Intriguingly, the genes in ESCs that are repressed by triMeK27-H3 frequently contain the additional H3 modification, lysine 4 trimethylation (triMeK4-H3), often associated with active chromatin (Bernstein et al., 2006). This has led to speculation that, through these bivalent marks, differentiation genes controlled by PRC2 may be poised for activation upon removal of their repressive epigenetic marks (Bernstein et al., 2006; Boyer et al., 2006). That said, the role of PRC-mediated chromatin repression in regulating ESC differentiation is complex. Thus, cultured Suz12 null ESCs treated with retinoic acid do not execute normal neuronal differentiation but, rather, fail to suppress pluripotent genes and only partially activate neuronal genes (Pasini et al., 2007). This has led to speculation that PRCs are required for both suppression and activation of differentiation programs in ESCs.
It remains an important challenge to determine whether these epigenetic mechanisms unveiled in vitro operate in vivo to govern fates of the more restricted progenitors that develop and maintain tissues (Spivakov and Fisher, 2007). Assessing the roles of PcG components in tissue organogenesis has been hampered by the early embryonic lethality caused by loss-of-function mutants of core PRC2 components. Conversely and further complicating interpretation is that conditional ablation of Ezh2 in adult bone marrow cells does not seem to affect either hematopoietic SC survival or lineage determination, suggesting either functional redundancy and/or compensation by paralogous PRC genes in at least some tissues (Su et al., 2003, 2005). This also seems to be the case for PRC1 genes such as Bmi1, in which mutations do not appear to affect embryogenesis (Sauvageau and Sauvageau, 2008; Valk-Lingbeek et al., 2004). Ironically, Bmi1 mutants malfunction in maintaining hematopoietic and neuronal adult SC renewal, in part due to misregulation of the Ink4A/Arf locus (Bruggeman et al., 2005; Molofsky et al., 2003, 2005; Park et al., 2003). That said, triMeK27-H3 epigenetic marks are still apparent in Bmi1 null cells (Cao et al., 2005), suggesting that the phenotype does not reflect complete abrogation of PcG-repressive functions. These findings underscore the importance of analyzing PcG functions in other in vivo biological systems in order to understand their physiological relevance in tissue development and maintenance.
Mammalian epidermis is an excellent model to address this problem. Epidermal lineages originate from a single layer of multi-potent progenitors, basal cells, that adhere to an underlying basement membrane separating epidermis from dermis (Fuchs, 2007). In mice, epidermal stratification and fate specification initiate at approximately embryonic day 14 (E14) and complete shortly before birth, when the skin surface barrier is required to keep harmful microbes out and prevent dehydration (Fuchs, 2007).
Basal cells continually fuel the production of ~10 suprabasal layers. Once cells exit the basal layer, they downregulate proliferation-associated genes and execute a terminal differentiation program that is marked by a stepwise transcriptional transition from early differentiation spinous layers to late differentiation granular layers. In the last step, all metabolic activity ceases as dead squames of the protective stratum corneum are formed and subsequently sloughed from the skin surface (Watt et al., 2006).
In this report, we use epidermis as a model to explore the functional significance and physiological relevance of PcG-mediated chromatin repression during embryonic tissue development and homeostasis. We unveil Ezh2 as a critical mediator of chromatin repression in embryonic basal cells and show that its expression diminishes with age and with commitment to terminally differentiate in vivo and in vitro. Using chromatin immunoprecipitation (ChIP) performed on purified embryonic epidermal progenitors, we show that its triMeK27-H3 mark is present both on silent epidermal genes activated late in terminal differentiation and on nonepidermal genes, e.g., neuronal and muscle transcriptional regulators. Interestingly, however, despite this broad association of triMeK27-H3 with silenced genes of different tissue lineages, loss of PcG-mediated repression does not alter the fate of epidermal cells. Rather, it reduces their proliferation through upregulation of the Ink4A-Ink4B locus and tempers differentiation rates and barrier function acquisition by upregulating late-stage epidermal differentiation genes, many of which are controlled by AP1 transcriptional activators. We show that the triMeK27-H3 mark prevents AP1 proteins from binding to these target genes in basal cells and that, during terminal differentiation, these late differentiation genes lose their histone mark, associate with AP1 components, and become activated suprabasally. Our findings provide new mechanistic insights into PcG-mediated repression of tissue-specific differentiation genes and suggest that the spatial balance between expression of PcG epigenetic repressors in undifferentiated cells and transcriptional activators like AP1 in differentiated cells controls proper establishment of the stepwise mechanism of epidermal differentiation during skin development.
RESULTS
Ezh2 Marks the Progenitor Cell Nuclei in Developing Epidermis
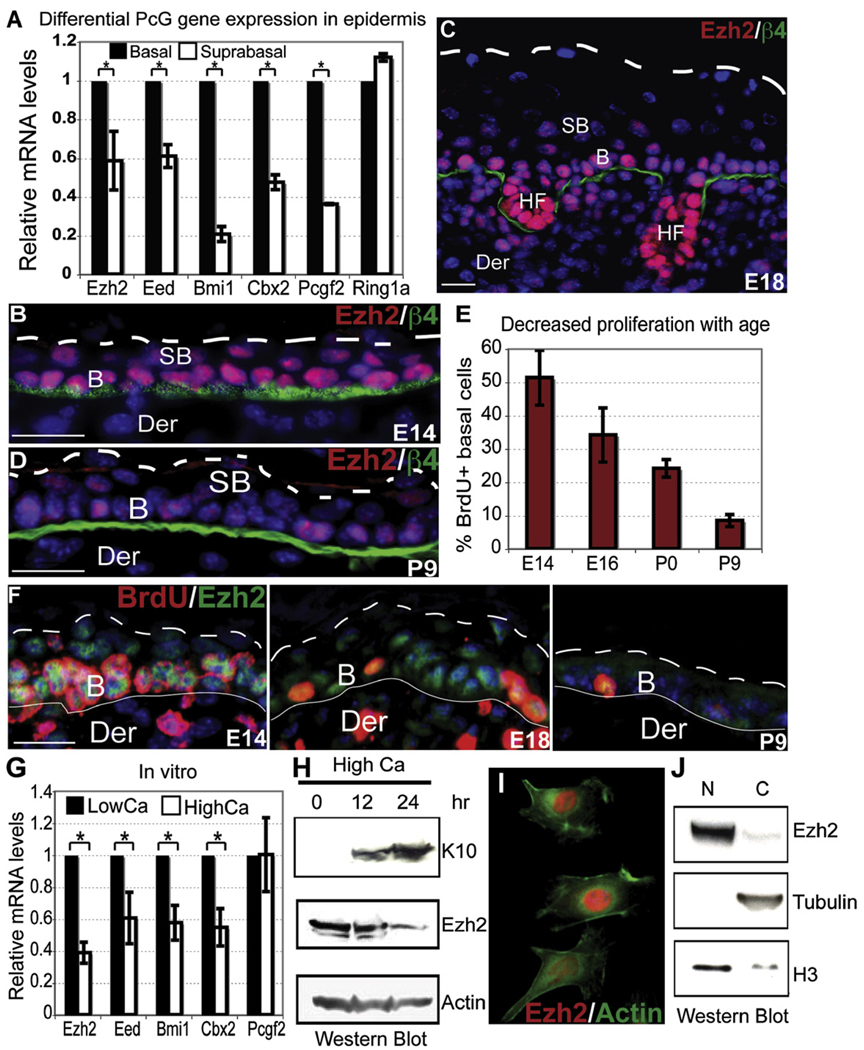
At E14, epidermis consists of a basal layer of progenitors and a layer of suprabasal cells that have initiated a program of terminal differentiation but are still proliferative (Figures S1A and Figure S1B available online) (Lechler and Fuchs, 2005; Watt et al., 2006). Purification of these two populations by fluorescence-activated cell sorting (FACS) and transcriptional profiling revealed that PcG components of PRC2 (Ezh2 and Eed) and PRC1 (Bmi1, Cbx2, and Pcgf2) are expressed in basal cells and downregulated suprabasally. Semiquantitative PCR (RT-PCR) and anti-Ezh2 immunofluorescence confirmed these patterns (Figures 1A and 1B and data not shown).
Figure 1. Ezh2 Marks Basal Cell Progenitors in Developing Epidermis.
(A) Differential expression of PcG components in E14 epidermis. FACS-purified basal and suprabasal cells were analyzed by semiquantitative RT-PCR of their isolated mRNAs. Data are mean ± SD. n = 3. *p < 0.05.
(B–F) Age-related decline in basal Ezh2 correlates with reduced proliferative potential. BrdU was administered 4 hr prior to sacrificing mice. Frozen skin sections were subjected to immunofluorescence microscopy. Primary Abs are color coded according to their secondary Abs. Nuclei are stained with Dapi (blue).
(G) Semiquantitative RT-PCR reveals decreased expression of PcG components with epidermal terminal differentiation in vitro. Primers against HPRT were used for normalization. Data are mean ± SD. n = 2. *p < 0.05.
(H) Ezh2 protein diminishes upon calcium (Ca)-induced MK differentiation in vitro. Controls are actin (unchanged) and K10 (spinous marker). (I–J) Immunofluorescence and western blot analyses show Ezh2 nuclear localization and partitioning. Note that the cytosolic fraction (C) contains minor residual nuclear material as judged by H3, whereas the nuclear fraction (N) is clean as judged by the absence of β-tubulin.
B, basal. SB, suprabasal. Der, dermis. HF, hair follicle. β4, β4 integrin. Scale bars, 30 µm.
By E16–E18, proliferation is restricted to the basal layer, and the epidermis consists of multiple layers of differentiating suprabasal cells (Figures S1C and S1D). At these stages, Ezh2 was strong in basal but weak in spinous, granular, and stratum corneum layers (Figures 1C). After birth, Ezh2 waned, and by postnatal day 9 (P9), Ezh2 was barely detected even in basal cells (Figure 1D). The 5-Bromo-2′-deoxyuridine (BrdU) pulses administered at different developmental ages revealed that the%BrdU(+) basal cells also decreased steadily with age (Figure 1E) and that the few remaining Ezh2(+) cells were also BrdU(+) (Figure 1F).
In vitro, cultured basal epidermal cells exposed to elevated calcium stop proliferating and initiate terminal differentiation (Hennings et al., 1980). Similar to their behavior in vivo, basal mouse keratinocytes (MK) expressed high levels of PcG mRNAs and protein, which progressively declined upon calcium-induced differentiation (Figures 1G and 1H). Both in vivo and in vitro, Ezh2 in proliferating, undifferentiated MK was largely, if not solely, nuclear (Figures 1I and 1J). Therefore, we pursued the notion that, in epidermal MK, Ezh2 functions in the nucleus.
PcG-Dependent triMeK27-H3 Globally Marks Genes of Nonepidermal Lineages and Selectively Marks Genes of Epidermal Lineages
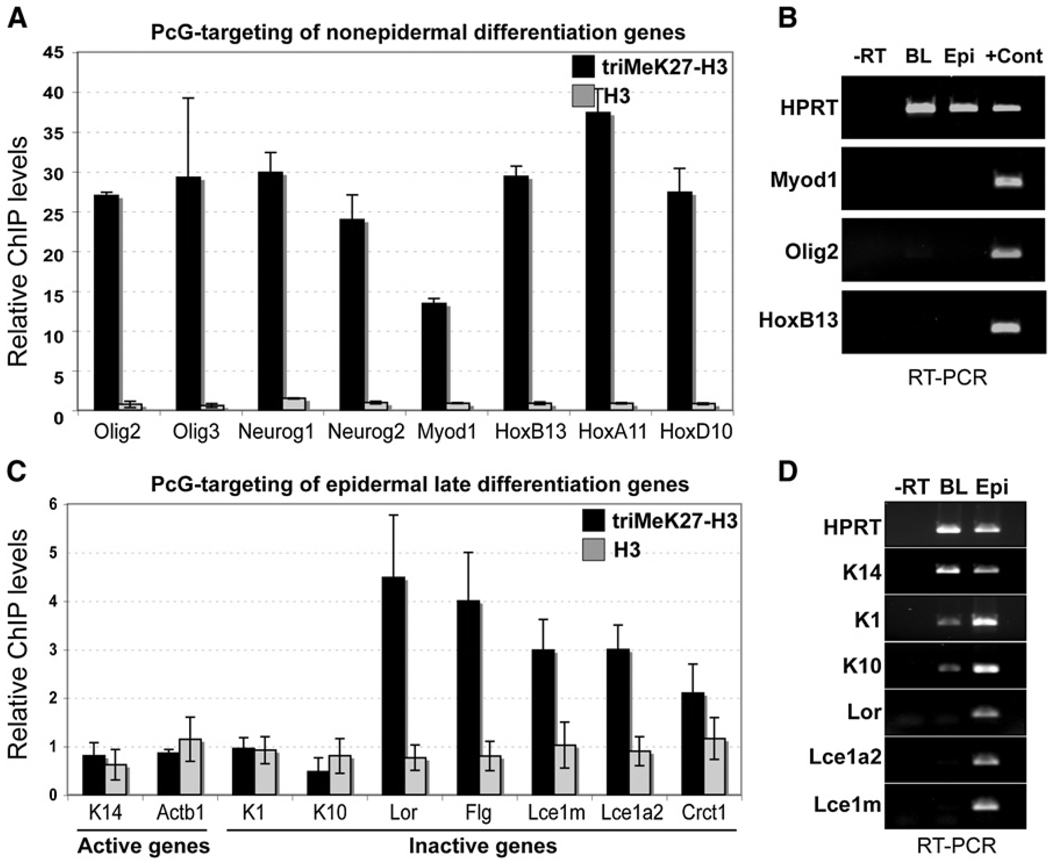
In mouse ESCs, PRC2 marks and globally represses genes involved in the specification of lineages, including those of the epidermal lineage (Boyer et al., 2006). To evaluate the extent to which the repressed PRC2 targets of mouse ESCs are maintained in Ezh2(+) embryonic epidermis, we performed ChIP assays with FACS-purified embryonic epidermal basal cells and Abs against triMeK27-H3 and unmodified histone H3 (Figures S2A and S2B). We first focused on nonepidermal “master” transcriptional regulators of the Hox, neuronal (Neurog1, Neurog2, Olig2, and Olig3), and muscle (Myod1) families that are marked for PcG-mediated repression in ESCs (Boyer et al., 2006). Many of these genes were also transcriptionally repressed by triMeK27-H3 in basal epidermal cells (Figures 2A and 2B).
Figure 2. In Embryonic Basal Progenitors, triMeK27-H3 Marks Silenced Differentiation-Specific Genes of Both Epidermal and Nonepidermal Lineages.
(A and C) ChIP analysis showing association of tri-MeK27-H3 and bulk H3 with promoters of nonepidermal and epidermal genes in FACS-purified E16 basal progenitors. Semiquantitative PCR of the basal K5 gene promoter region was used for normalization. Data are mean ± SD. n = 2.
(B and D) RT-PCR of nonepidermal and epidermal mRNAs in E16 basal cells (BL) versus total epidermis from P0 embryos. Negative control is PCR with total RNA (−RT) as a template. RT-PCR with RNAs isolated from skeletal muscle and brain served as positive controls (+Cont) for Myod1 (muscle) and Olig2 (brain). Control is HPRT (unchanged).
We next focused on identifying epidermal lineage genes targeted by PcG repression in Ezh2(+) embryonic basal cells. For these studies, we analyzed promoters of basal genes, which are transcriptionally active, as well as spinous and granular layer genes, which are not expressed at this stage of differentiation. Our tests included genes previously found in ESCs to be enriched for PcG-mediated repression, as well as other epidermal genes.
Interestingly, genes enriched for triMeK27-H3 in embryonic basal cells showed a good inverse correlation between Ezh2 and terminal differentiation (Figures 2C and 2D) (Fuchs, 2007). Genes coexpressed with Ezh2 in either embryonic basal or early (spinous) layers (actinb1, K14, K1, and K10) were not enriched for triMeK27-H3 in basal progenitors, whereas genes expressed in granular cells (loricrin, filaggrin, Lce1, Lce1a2, and Crct1) showed significant triMeK27-H3 enrichment in embryonic basal cells.
Intriguingly, the triMeK27-H3 status of genes in the embryonic basal epidermal layer in vivo differed from that in cultured ESCs, in which both early (K1) and late epidermal differentiation genes (loricrin and Lce1a2) scored as targets of PcG-mediated repression (Boyer et al., 2006). Thus, whereas some PcG targets established in pluripotent cells appear to be maintained in the epidermal lineage, others may lose their repressive mark during tissue development.
Ezh2 Controls Proliferation of Basal Cells through Repression of Ink4A/Ink4B
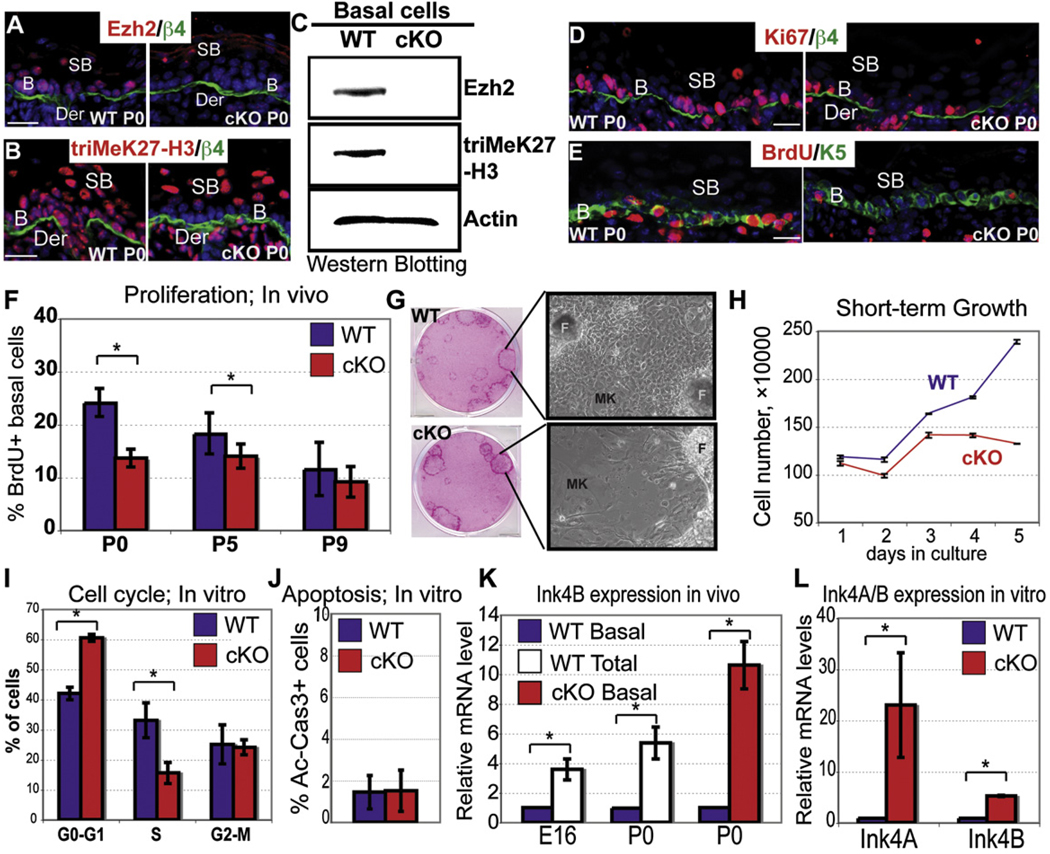
To address the physiological significance of Ezh2’s interesting pattern of gene expression, we conducted conditional loss-of-function studies. Mice harboring the floxed Ezh2 allele (Su et al., 2003) were bred to mice expressing Cre recombinase under the keratin 14 (K14) promoter in embryonic epidermal progenitors (Vasioukhin et al., 1999). Ezh2 conditional knockout (Ezh2 cKO) mice were born alive and in the expected Mendelian ratios. Immunofluorescence and western blot analyses revealed a loss of Ezh2 and triMeK27-H3 marks in basal cells (Figure 3A–3C and Figure S3A), underscoring their reliance upon Ezh2 for establishment and maintenance of this mark. TriMeK27-H3 was still detected in suprabasal cells, most likely because the Ezh2 paralog, Ezh1, is expressed throughout embryonic epidermis and in basal cells of postnatal skin (Figure S8A). We return to this issue later.
Figure 3. A Developmental Role for Ezh2 in the Proliferative Potential of Epidermal Progenitors.
(A–C) Immunofluorescence and western blot reveal the absence of Ezh2 and triMeK27-H3 in P0 Ezh2 cKO basal cells.
(D and E) Immunofluorescence shows fewer Ki67 and BrdU(+) basal cells in P0 Ezh2 cKO epidermis.
(F) Quantification of BrdU(+) basal cells from mice receiving a 4 hr BrdU pulse at indicated times. Data are mean ± SD. WT, cKO n = 3–6. *p < 0.05.
(G–J) Differences in morphologies, growth rates, and cell cycles, but not apoptosis (active caspase 3, Ac-Cas3), of WT versus Ezh2 cKO epidermal cells in vitro. Cell-cycle data are mean ± SD. WT, cKO n = 3. *p < 0.05.
(K–L) Semiquantitative RT-PCR to measure Ink4A and/or Ink4B expression in FACS-purified basal cells in vivo (K) or cultured 1° MK in vitro (L). Data are mean ± SD. n = 2 for in vivo and n = 3 for in vitro analysis. *p < 0.05. For comparative purposes, WT expression in differentiating cells was determined by analyzing total epidermal mRNA. The HPRT gene was used for all normalizations.
MK, mouse keratinocytes. F, feeders. Scale bar, 30 µm.
In the absence of Ezh2, basal epidermal cells were less proliferative, exhibiting reductions in Ki67(+) cells and BrdU labeling (Figures 3D–3F). During early postnatal development when Ezh2 was still expressed in WT basal cells, the % BrdU(+) basal cells in Ezh2 cKO skin was more than 2-fold diminished. As Ezh2 waned in postnatal WT basal cells, the % BrdU(+) basal cells within cKO and WT skins became comparable (Figure 3F).
When cultured, P0 Ezh2 cKO and WT basal cells both formed similar-sized colonies with comparable efficiencies (Figures S3B and S3C). However, whereas WT colonies were composed of tightly packed small cells, cells within cKO colonies were often larger, flat, and loosely packed (Figure 3G). After several days in culture, the growth of Ezh2 cKO cultures began to slow (Figure 3H). This reduction did not appear to be attributable to increased apoptosis (Figure 3J) nor to altered growth factor-stimulated actin dynamics (Su et al., 2005) (Figure 1). Instead, cell-cycle analysis revealed an increase in G1/G0 and decrease in S phase cKO cells compared to WT (Figure 3I). Consistent with a reduction in S phase entry, the phosphorylation of retinoblastoma protein was reduced in cKO cells (Figure S3D).
The proliferation defect in Ezh2 cKO skin appeared to be attributed, at least in part, to derepression of the Ink4B-Arf-Ink4A tumor suppressor locus that encodes potent cell-cycle inhibitors of the G1-to-S transition in a variety of cell types, including epidermis (Bracken et al., 2007; Bruggeman et al., 2007; Kim and Sharpless, 2006). Consistent with prior studies that established Ink4B-Arf-Ink4A as a PcG target, we observed an ~10-fold upregulation of Ink4B in FACS-purified Ezh2 cKO cells in vivo (Figure 3K) and a similar marked upregulation of both Ink4A and Ink4B genes in Ezh2 null MK in vitro (Figure 3L). Finally, in WT skin, Ink4B expression inversely correlated with Ezh2 (Figure 3K, basal versus total).
Ezh2 Tempers Epidermal Development and Barrier Acquisition
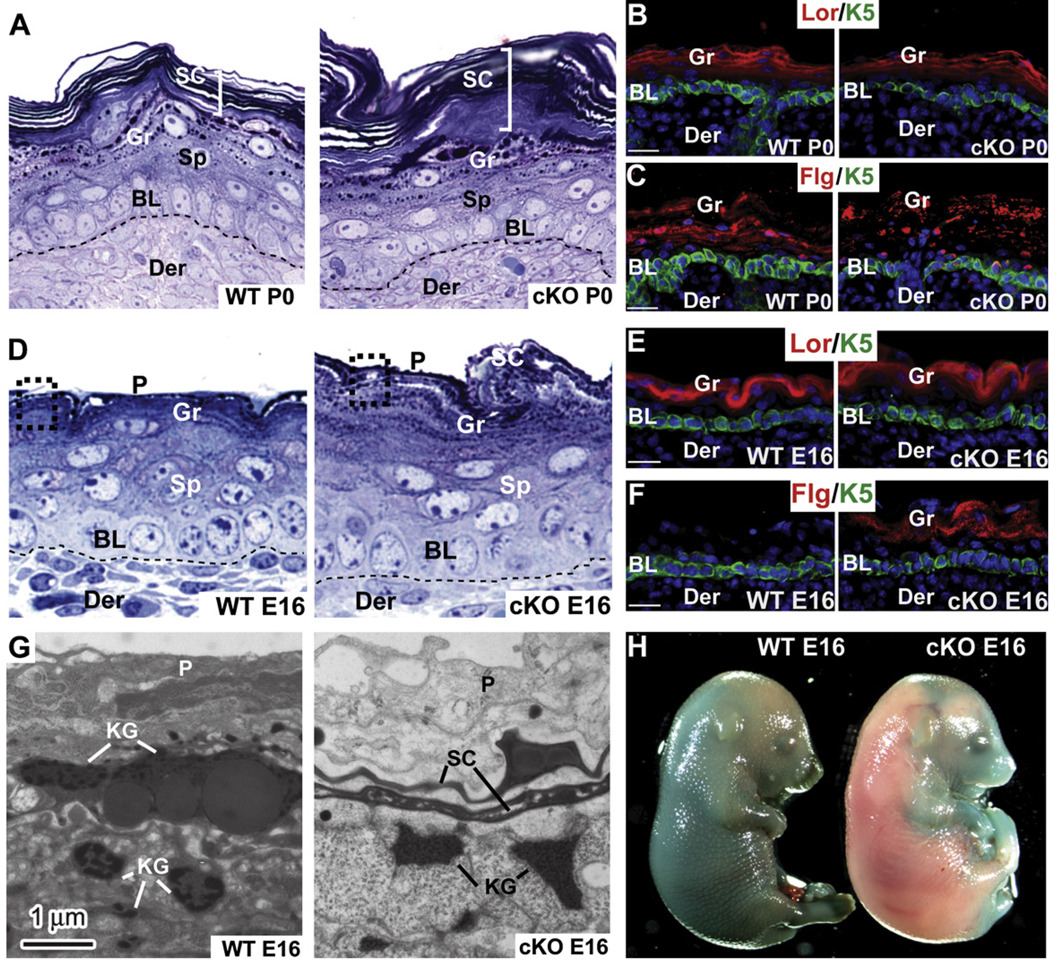
P0 Ezh2 cKO epidermis displayed a hyperthickened stratum corneum and pronounced granular layer accompanied by the expansion of the granular markers loricrin and filaggrin (Figures 4A–4C). These defects were also apparent in E16 cKO skin concomitant with loss of Ezh2 and basal triMeK27-H3 (Figure 4D–4F, Figure S4A, and S4B). Ultrastructurally, the immature large keratohyalin granules (KG) of WT E16 skin were already dispersed in cKO skin (Figure 4G and Figure S4C) (Fuchs, 2007; Steven et al., 1990). A well-formed stratum corneum was also evident, and with proper preservation, highly ordered lipid lamellae interconnected these cells only within the E16 Ezh2 cKO (Figure S4D). Correspondingly, the nonepidermal periderm (P) layer that normally provides a temporary barrier during epidermal maturation was already degenerated (Figure 4D, 4G, and Figure S4C). These various signs of accelerated development (Jaubert et al., 2003; Patel et al., 2006) were manifested functionally in the precocious acquisition of the epidermal barrier capable of excluding dye (Figure 4H).
Figure 4. A Role for Ezh2 in Regulating Temporal Differentiation and Epidermal Barrier Acquisition during Skin Development.
(A–F) Histological and immunofluorescence microscopy reveals signs of accelerated epidermal differentiation in P0 and E16 Ezh2 cKO epidermis. Semithin sections in (A) and (D) are toluidine blue stained. Frozen sections are labeled with Abs as indicated (color coding according to secondary Abs).
(G) Ultrastructural analyses. Note the presence of mature keratohyalin granules (KG), a thin stratum corneum layer (SC), and a partial loss of periderm (P) in Ezh2 cKO E16 epidermis, all lacking in the WT counterpart.
(H) Blue dye exclusion assay to measure skin barrier.
Lor, Loricrin. Flg, filaggrin. BL, basal layer. Sp, spinous layer. Gr, granular layer. Der, dermis. Scale bar, 30 µm.
Ezh2 Is Required to Repress Precocious Epidermal Late Differentiation Genes in Basal Cells
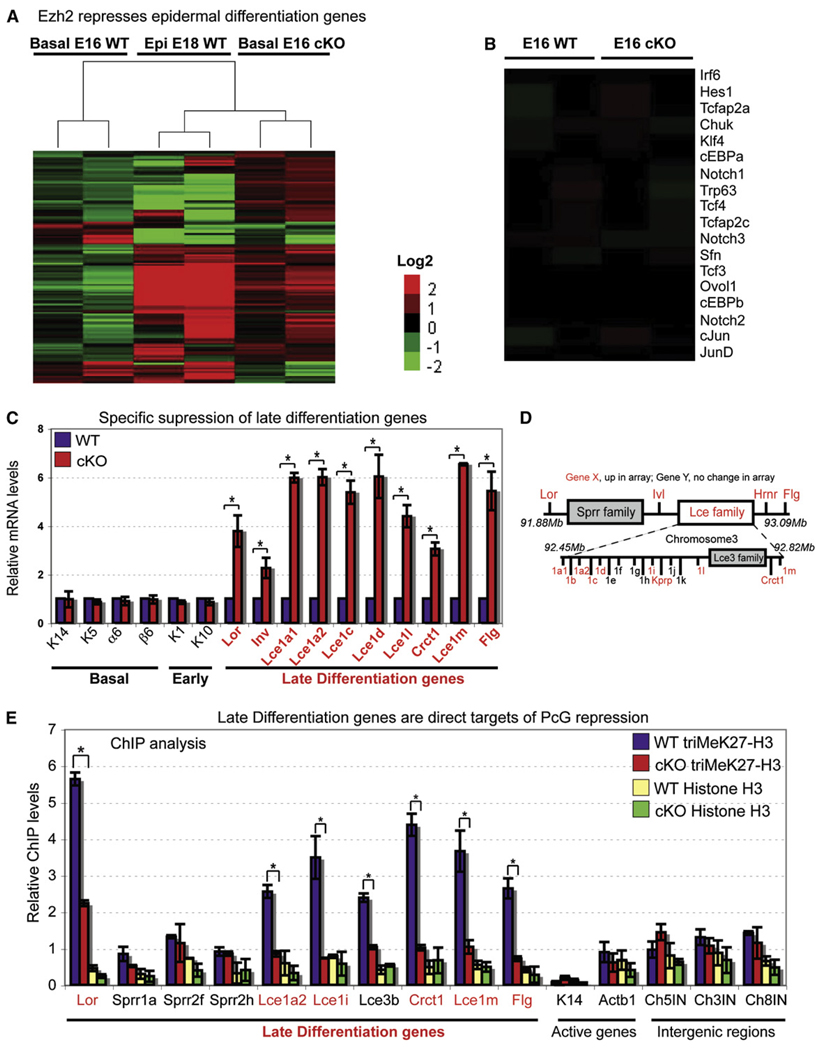
To understand how loss of Ezh2 might result in accelerated epidermal differentiation, we conducted transcriptional profiling of FACS-purified basal cells from E16 Ezh2 cKO and WT skins to determine the global consequences of Ezh2 loss to epidermal gene expression. The data are compiled in Figure 5A, with upregulated genes in red and downregulated genes in green.
Figure 5. Transcriptional Profiling and Chromatin Analyses Reveal Late Differentiation Genes as Direct Functional Targets of Ezh2 Repression in Embryonic Basal Cells.
(A and B) Heatmap derived from cluster analysis of microarray data of E16 basal progenitors shows that genes whose expression is normally associated with late-stage terminal differentiation are elevated in the absence of Ezh2. E18 WT epidermis is used as a comparative source of genes active in late-stage terminal differentiation. Microarray hybridizations for each sample were done in duplicate and are shown as separate columns. Probesets were selected that demonstrate a 2 log change (in either direction) and present (“P”) in both instances of a condition (P in both KO or P in both WT). Red, overexpressed genes. Green, under-expressed genes. No obvious changes were revealed in the basal expression of key epidermal transcriptional regulators when Ezh2 was absent (B).
(C) Semiquantitative RT-PCR of mRNAs from FACS-purified E16 basal progenitors confirms the precocious induction of late differentiation genes when Ezh2 is absent. Semiquantitative RT-PCR with primers against the HPRT gene was used for normalization. Data are mean ± SD. n = 2. *p < 0.05.
(D) Schematic representation of the epidermal differentiation cluster (EDC) on mouse chromosome 3. Late-stage differentiation EDC genes that were upregulated in Ezh2-deficient basal cells are marked in red.
(E) ChIP analyses reveal the presence of triMeK27-H3 at promoters of many genes within the EDC in WT and reduction in Ezh2 cKO cells. Semiquantitative PCR with primers against the promoter of the active K5 gene was used for normalization. Data are mean ± SD. n = 2. *p < 0.05.
As expected, loss of Ezh2 resulted mostly in gene upregulation in basal cells. Surprisingly, however, only a small fraction of the genes analyzed showed a more than 2-fold change compared to WT. Of the 94 genes (0.2%) upregulated, 75% were epidermal genes, and among the others, there did not appear to be a significant bias toward any specific developmental lineage (Table S1). Thus, despite the global association of triMeK27-H3 marks with epidermal and nonepidermal genes (Figures 2A and 2C), the loss of Ezh2 in basal cells led primarily to selective upregulation of epidermal genes. In this regard, PRC-mediated repression in basal epidermal cells appeared to differ markedly from ESCs, in which PRC target genes encompassing multiple lineages were upregulated when PcG function was compromised (Boyer et al., 2006).
As judged by array and RT-PCR, genes associated with basal and/or early (spinous) differentiation were unaffected by loss of Ezh2 (Figures 5B and 5C), and this was consistent with the lack of triMeK27-H3 marks at their promoters (Figure 2C). Notably, this included transcription factors, e.g., Trp63, Tcf3, Notch, Hes1, AP2, cEBP, and Klf4, that have been implicated in either maintenance of the undifferentiated state or induction of terminal differentiation (Fuchs, 2007; Watt et al., 2006). This finding also contrasted with ESCs, in which many targets for global PcG repression of lineages are transcriptional regulators of fate specification and differentiation (Boyer et al., 2006).
Interestingly, 77% of genes upregulated in E16 Ezh2 cKO basal cells were expressed more than 2-fold higher in mature WT epidermis compared to WT basal cells (Figure 5A). More-over, genes upregulated in Ezh2 null basal cells were primarily those expressed during the normal course of terminal differentiation. The largest differences were among genes encoding major granular proteins, e.g., filaggrin (Flg), loricrin (Lor), involucrin (Ivl), and late cornified envelope (Lce) proteins (Figures 5A and 5C and Table S1) (Marshall et al., 2001; Watt et al., 2006). Notably, these genes are within a 2Mb locus encompassing an epidermal differentiation complex (EDC) on mouse chromosome 3 (Figure 5D). These data suggest a role for Ezh2 in ensuring that granular genes within this cluster are repressed until the appropriate stage in development and differentiation.
Studies with cultured Ezh2 null MK provided further evidence to support this notion. Thus, under low calcium conditions that favor the basal fate, filaggrin and loricrin levels were comparable to those of WT cells exposed to high calcium, a trigger of terminal differentiation (Figure S5A). In Ca2+-stimulated Ezh2 null MK, filaggrin and loricrin were even more highly expressed than normal.
To determine the extent to which precocious induction of late-stage differentiation-specific genes in Ezh2 null basal cells is a direct cell autonomous consequence of loss of Ezh2-mediated repression, we conducted ChIP assays with Ezh2, triMeK27-H3, and pan-H3 Abs and primers encompassing loricrin, filaggrin, and other EDC gene promoters. In WT MK, both Ezh2 and triMeK27-H3 were enriched near transcriptional initiation sites (Figure 5E and Figure S5B). The signals for both Ezh2 and triMeK27-H3 appeared to be specific, as judged by comparisons with Ezh2 null cells or control ChIPs in which Abs were omitted (Figure 5E, Figure S5B–S5D).
Basal cell PcG complexes were detected over many, but not all, terminal differentiation-specific genes across the EDC locus (Figure 5E). The distal region harboring the Lce genes was particularly rich in triMeK27H3, whereas the small proline-rich (Sprr) family of genes exhibited only background levels of this modification (Figure 5E). Correspondingly, expression of Sprr genes was unchanged (Figure 5A), suggesting that Ezh2 may differentially regulate EDC genes. Promoters of basally expressed K14 and actinb1 genes and nonexpressed intergenic regions did not display the Ezh2-mediated triMeK27-H3 mark (Figure 5E). These findings underscored the specificity of the ChIP signals and PcGs for late-stage differentiation genes and established them as direct targets for Ezh2-histone methyltransferase.
More to Gene Repression Than triMeK27-H3: Analysis of Nonepidermal Genes
If expression of genes that receive triMeK27-H3 marks is governed solely by PRC-mediated chromatin modifications, then relief of these marks should result in their aberrant activation. We tested this by first examining the expression status of none-pidermal genes harboring this histone mark in WT epidermal cells and in ESCs (Figure 2A). Consistent with the global reduction of triMeK27-H3 when Ezh2 is absent, most of the nonepidermal genes that were analyzed showed a significant reduction in triMeK27-H3 in Ezh2 null MK (Figure S5E). Despite the loss of PcG repression, however, these genes were not activated (Figure S5F).
More to Gene Expression Than triMeK27-H3: Analysis of Epidermal Genes
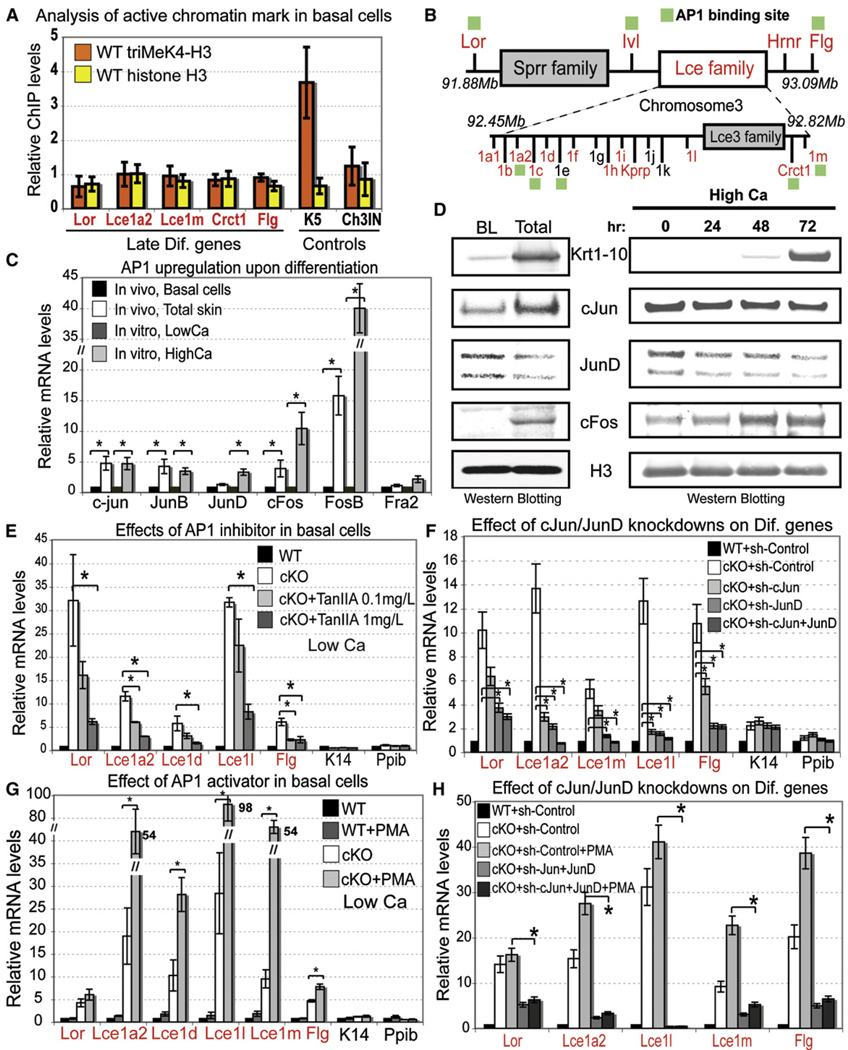
To understand why relief of PcG repression leads to selective activation of epidermal, but not nonepidermal, lineage genes in our Ezh2 cKO mice, we began by investigating whether EDC genes in basal cells selectively contain the active epigenetic mark triMeK4-H3, known to function antagonistically in PcG-regulated genes in ESCs (Bernstein et al., 2006). Although triMeK4-H3 was readily detected at the promoters of naturally expressed basal genes, we did not detect it in the EDC promoters that we analyzed in embryonic basal cells (Figure 6A).
Figure 6. Transcriptional Activator AP1, Rather Than Activating Histone Modifications, Governs Upregulation of EDC Genes in Ezh2 cKO Basal Cells.
(A) ChIP analyses show no association of the active triMeK4-H3 mark with EDC genes in WT embryonic basal cells, in which the EDC is silent. Semiquantitative PCR with primers against the intergenic region on chromosome 5 was used for normalization. The basal K5 gene was used as a positive control. Data are mean ± SD. n = 2.
(B) AP1 consensus sites are found at promoters of EDC genes.
(C and D) Semiquantitative RT-PCR and western blotting detects Jun and Fos AP1 proteins in epidermis and cultured MK. Expression of some members is enhanced in terminally differentiating cells. Positive control is HPRT (unchanged) and Flg (differentiated layers). Data are mean ± SD. n = 2. *p < 0.05.
(E and F) Semiquantitative RT-PCR shows that the precocious activation of EDC genes in Ezh2 cKO basal progenitors is largely abrogated by either the AP1 inhibitor, TanIIA, or RNAi-mediated attenuation of cJun/JunD expression. Data are mean ± SD. n = 2. *p < 0.05.
(G and H) By contrast, PMA, which promotes AP1 activity, enhances precocious expression of EDC genes in Ezh2 cKO, but not WT basal cells (G). EDC upregulation in PMA-treated Ezh2 null cells is significantly lower when Jun and JunD are knocked down than in sh control (H). Data are mean ± SD. n = 3. *p < 0.05. Active K14 and Ppib genes served as controls.
BL, basal layer cells. LowCa, in vitro conditions that prevent differentiation of basal progenitors. HighCa, conditions that promote terminal differentiation.
We next turned to the possibility that the counteractive mark in epidermal basal cells might be a transcriptional activator involved in regulating EDC gene expression. In searching for key EDC gene regulator(s) that might be expressed basally in WT cells, we began to focus on AP1 family members, which are known to regulate loricrin and filaggrin promoter activities (DiSepio et al., 1995; Jang et al., 1996). Genome scans revealed AP1 consensus binding sites throughout the EDC cluster (Figure 6B). Importantly, Jun and Fos AP1 proteins were detected both in basal cells and total epidermis (Figure 6C, 6D, and Figure S6A), and terminal differentiation appeared to elicit an upregulation of some, but not all, AP1 proteins in vivo and in vitro (Figures 6C and 6D).
To test whether AP1 factors might account for upregulation of EDC genes upon loss of Ezh2, we used the chemical inhibitor Tanshinone IIA (TanIIA), which is thought to abrogate all AP1 binding to DNA. In the absence of TanIIA and in low calcium, only Ezh2 null and not WT basal MK expressed the EDC genes (Figure 6E). The addition of TanIIA revealed a dose-dependent reduction in expression of EDC genes that harbored AP1 consensus sites, but not in K14 and Ppib genes that lacked them (Figure 6E). The inhibitory response to TanIIA was observed even for some EDC genes that lacked obvious AP1 sequence motifs. Although consensus sequences are not always necessary for DNA binding, an alternative possibility is that close proximity of genes within this cluster contributes to their coordinate regulation through shared regulatory elements. Although pursuing this further is outside the scope of the present study, such a mechanism has been shown for other gene complexes (Dillon and Grosveld, 1993; Lomvardas et al., 2006).
To verify that EDC gene expression in Ezh2 null basal cells is AP1 dependent, we used RNAi to attenuate expression of Jun proteins, which can homodimerize or form heterodimers with Fos. cJun and JunD shRNAs each led to specific downregulation of EDC genes without affecting K14 or Ppib expression (Figure 6F and Figure S6B). Combination of both shRNAs was even more dramatic, yielding comparable EDC gene expression levels between WT and cKO basal cells (Figure 6F).
Conversely PMA, which facilitates AP1 activity (Eckert et al., 1997), had little or no effect on EDC gene expression in WT basal cells, but in Ezh2 null cells, PMA further enhanced activation of EDC genes in low-calcium conditions (Figure 6G). By contrast, expression of PcG-repressed nonepidermal genes Myod1, Olig2, and HoxB13 were unaffected by PMA (Figure S6C). Finally, PMA resulted only in a very modest upregulation of EDC genes when Ezh2 cKO basal cells were exposed to Jun shRNAs (Figure 6H). Together, these loss- and gain-of-function studies demonstrate that, in basal epidermal cells, EDC genes are repressed by Ezh2 even though AP1 transactivating factors are present.
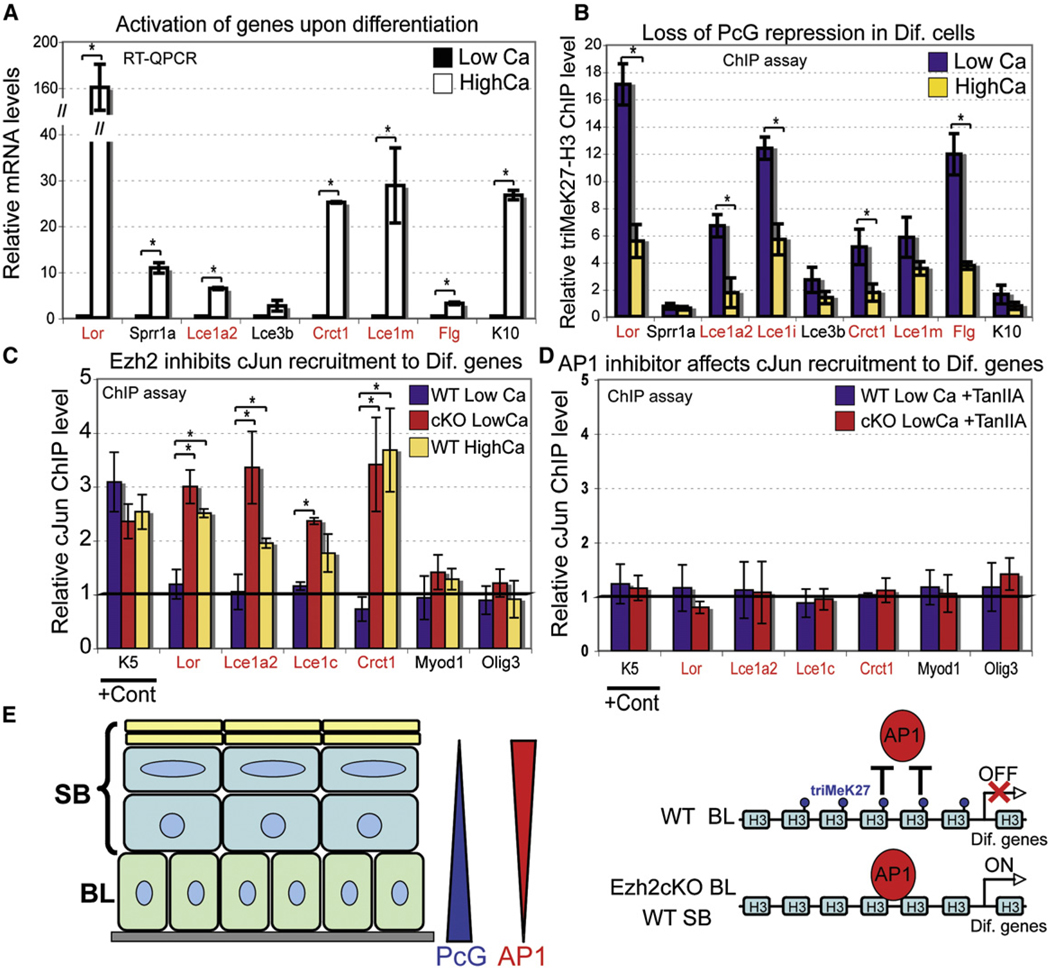
Consistent with our array data (Figure 5B), Ezh2 did not markedly affect AP1 mRNA or protein levels either in vivo or in vitro (Figure S7). Thus, we speculated that triMeK27-H3 marks on EDC genes in WT basal cells may interfere with AP1 recruitment, as it has been shown for Myod1 in muscle cells in vitro (Caretti et al., 2004). To test this hypothesis, we first analyzed whether coordinated relief of triMeK27-H3 and recruitment of AP1 factors control terminal differentiation and EDC gene expression under normal physiological conditions. Of the EDC genes marked by Ezh2 and triMeK27-H3, all but Lce3b were significantly upregulated when basal MK were induced to differentiate (Figure 7A) (Jackson et al., 2005). ChIP analysis of triMeK27-H3 at the EDC cluster detected a significant decrease in this repressive mark concomitant with EDC gene activation (Figure 7B).
Figure 7. Opposing Roles for PcG Repressors and AP1 Transcriptional Activators in Epidermal Development and Differentiation.
(A and B) Semiquantitative RT-PCR shows that EDC genes are induced when WT basal cells commit to terminally differentiate (A), and ChIP analysis (B) shows that triMeK27-H3 marks associated with EDC genes in basal cells decline upon calcium-induced differentiation. Semiquantitative PCR with primers against promoter of the active K5 gene was used for normalization (B).
(C and D) ChIP analysis shows cJun recruitment to promoters of late differentiation genes upon removal of Ezh2-dependent triMeK27-H3 mark in cKO basal cells or WT cells upon differentiation (C), but this recruitment is inhibited upon treatment of cells with AP1 inhibitor TanIIA (D). Presence of cJun at the K5 promoter served as a positive control. No recruitment of cJun to Myod1 and Olig3 promoters that lack obvious AP1 sites was detected. Semiquantitative PCR with primers against intergenic region on chromosome 5 was used for normalization. Data are mean ± SD. n = 2. *p < 0.05.
(E) Differential expression of PcG repressor and Ap1 activator ensures spatial and temporal program of epidermal differentiation. In WT basal cells, the presence of triM3K27-H3 mark prevents AP1 from binding and activation of late differentiation genes. In Ezh2cKO or in differentiated WT cells, loss of triMeK27-H3 mark allows Ap1 to bind and activate transcription of late differentiation genes.
Finally, ChIP analysis detected cJun at several EDC gene promoters only in cKO and not WT basal cells (Figure 7C). By contrast, cJun was not found at Myod1 and Olig3 promoters (Figure 7C). Moreover, upon induction of differentiation, cJun was now detected at promoters of these EDC genes in WT cells (Figure 7C). Importantly, inhibition of AP1 binding by TanIIA reduced the cJun ChIP signal at EDC genes underlying its specificity (Figure 7D).
Ezh1 Increases in the Epidermis as Ezh2 Wanes: Partially Overlapping Activities of Ezh2 and Ezh1
Whereas Ezh2 is often expressed during embryogenesis, Ezh1 is found in adult tissues (Laible et al., 1997; Margueron et al., 2008; Su et al., 2003). This also holds true for epidermis in which postnatally, as basal Ezh2 declined, Ezh1 rose (Figure 1 and Figure S8A). Additionally, although triMeK27-H3 was barely detected in primary P0 Ezh2 null MK, it increased with passage (Figure S8B).
Recently, Ezh1 was shown to function similarly to Ezh2 (Shen et al., 2008), which could explain the residual triMeK27-H3 modification in Ezh2 cKO epidermis. Although an effective Ezh1 shRNA did not appreciably affect the overall high levels of triMeK27-H3 in WT MK, it did show a marked effect on further diminishing triMeK27-H3 marks in passaged Ezh2 null MK (Figure S8C). Together, these findings suggest that the bulk of triMeK27-H3 in embryonic and neonatal basal cells is due to Ezh2 but that Ezh1 can establish this histone mark in the absence of Ezh2.
Analysis of gene expression further suggested that certain functions of Ezh1 and Ezh2 in MK might be similar (Figure S8D). Thus, terminal differentiation genes were upregulated in both Ezh2 null and Ezh1 shRNA cells, and this effect was even stronger in passaged Ezh2 null/Ezh1 shRNA MK. Ezh1 was not upregulated in Ezh2 cKO cells either in vivo or in vitro (Figure S8E), suggesting that both Ezh2 and Ezh1 contribute to repression of terminal differentiation genes. This could explain why no obvious morphological defects were noted in the mature skin of our Ezh2 cKO mice in which Ezh1 is expressed (Figure 3B and Figure S8A). Supportive evidence for this notion stems from engraftments of primary WT and Ezh2 cKO MK infected with control or Ezh1 shRNAs. Cells infected selectively with Ezh1, but not control, shRNAs failed to reconstitute skin in adult mice (Figure S8F). Generation of Ezh1 cKO and Ezh1/Ezh2 cKO mice will be necessary to further dissect roles for these developmentally regulated histone methylases in skin and in other tissues.
DISCUSSION
Molecular mechanisms involving PcG proteins have attracted significant attention in the stem cell field due to their critical role in controlling cultured ESCs (Boyer et al., 2006; Lee et al., 2006; Pasini et al., 2007). By studying the role of Ezh2 in epidermal development and differentiation, we have uncovered new insights into the in vivo relevance of PRCs in the context of lineage determination and differentiation within a tissue. We have shown that Ezh2-dependent histone modifications act in embryonic epidermal progenitors to control both proliferative potential and differentiation. Our results are consistent with a role for PRCs in mediating repression of Ink4A/Ink4B in epidermal stem cells and further unveil a role for PRCs in regulating terminal differentiation of these cells by repressing genes governing epidermal barrier formation.
Our findings are intriguing in light of a recent study reporting enhanced differentiation of organotypic human epidermal cultures by overexpressing or RNAi knockdown of the histone demethylase Jmjd3 (Agger et al., 2007; Sen et al., 2008). In contrast to Ezh2, Jmjd3 levels do not appear to change in basal versus suprabasal cells of mouse epidermis. It is tempting to speculate that suprabasal Jmjd3 might ensure proper differentiation through efficient removal of the Ezh-mediated triMeK27-H3 mark, and in the future, it will be interesting to explore the possible interplay between histone methylases and demethylases in this process. However, from our physiological and molecular evidence, we favor the view that the primary mechanism for controlling epidermal differentiation in mouse development is by temporal expression of histone methylase Ezh2, which in basal cells, interferes with the recruitment of transcriptional activators to their differentiation-specific targets.
Our study also uncovered several notable differences in the mechanisms by which PcGs control differentiation in ESCs versus embryonic epidermal progenitors. In ESCs, triMeK27-H3 is present at and globally represses key transcriptional regulators that promote diverse cell lineages (Boyer et al., 2006; Lee et al., 2006). Such widespread repression is critical since ESCs are pluripotent. That said, ESCs must also retain the ability to respond to external cues and activate any one of the differentiation lineages repressed by PcG complexes. Once they make a commitment to differentiate, they must repress pluripotent regulators, e.g., Oct4, Nanog, and Sox2, and this process also appears to be governed either indirectly or directly by PcG-mediated events (Pasini et al., 2007). By contrast, embryonic epidermal stem cells maintain permanent repression of pluripotent regulators and selectively activate only the terminal differentiation program of the skin barrier.
Our results suggest that, like ESCs, embryonic basal progenitors use PcG-mediated repression to maintain repression of both nonepidermal and epidermal differentiation programs. However, in ESCs, bivalent active and repressive marks exist over many of the lineage-regulatory genes that are governed by PRCs, such that once repressive marks are relieved, the genes are poised to become activated (Pasini et al., 2007). By contrast, although Ezh2 loss in embryonic basal cells reduced triMeK27-H3 on many of the same genes as in ESCs, this was not sufficient for either maintaining or relieving their transcriptional repression. Rather than use a bivalent chromatin mark, embryonic basal epidermal progenitors appear to employ PRCs to prevent recruitment of AP1 and perhaps other key transcriptional activators required for terminal differentiation (Figure 7E). This additional layer of regulation ensures that, as development proceeds, most lineages remain permanently silenced while permitting the epidermal lineage to be selectively activated.
By invoking additional mechanisms to restrict activation of differentiation programs, developing tissues are poised to take advantage of relieving PRC modifications as a means to fine-tune the transcriptional program of the desired lineage. In this regard, it was notable that Ezh2 was highest in proliferative, undifferentiated basal cells. Upon induction of the basal-to-spinous switch, Ezh2 was reduced, and by the time the spinous-to-granular transcriptional switch was executed, little or no Ezh2 was detected. By employing gene-specific transcriptional activators that can operate when Ezh2 and triMeK27-H3 wane, the correct differentiation stage genes can be induced, and the undesirable nonepidermal lineages can remain silenced.
Controlling the balance between proliferation and differentiation requires different challenges in embryonic versus adult skin. In the rapidly growing embryo, basal proliferation must be high, and the program of epidermal differentiation and barrier acquisition must be orchestrated from scratch. By contrast, during homeostasis of the postnatal epidermis, proliferation rates are considerably reduced, and the program of terminal differentiation merely requires maintenance and not establishment. In this regard, it may be relevant that postnatal human basal epidermal cells exhibit low immunoreactivity to the active chromatin mark acetylated-H4 and appreciable reactivity to the repressive chromatin mark triMeK9-H3 (Frye et al., 2007). Additionally, our studies showing that Ezh2 expression wanes after birth suggest that either the Ezh2 paralog Ezh1 or alternative mechanisms come into play to control homeostasis in the mature epidermis. Although distinguishing between these possible models in vivo is predicated upon Ezh1 and Ezh1/Ezh2 double cKO mice, our in vitro and engraftment studies suggest that Ezh2 may provide the bulk of triMeK27-H3 modification in basal epidermal cells, and Ezh1 may function in fine-tuning histone modifications at later stages of terminal differentiation during embryogenesis and in homeostasis of postnatal epidermis.
In closing, it is worth considering the potential clinical impact of our findings. The epidermal barrier does not form until shortly before birth. Prematurely born infants lack this essential shield, and hence, accelerating barrier acquisition becomes a critical necessity in reducing the medical risks of these infants. Our discovery that conditional loss of Ezh2 accelerates epidermal barrier formation in the embryo but does not impair postnatal development offers a hitherto unanticipated target for the development of therapies that might be useful to infant survival. More-over, Ezh2’s exquisite ability to function in rapidly proliferating, tissue-restricted progenitor cells provides an intriguing explanation for why Ezh2 is overexpressed in many tumors wherein hyperproliferation and reduced differentiation are often likened to an embryonic state.
EXPERIMENTAL PROCEDURES
Mice, BrdU Injections, and Barrier Assays
Mice were housed in the ALAAC accredited CBC animal facility at The Rockefeller University in accordance with university and NIH guidelines. Mice conditionally ablated for Ezh2 in skin were generated by mating Ezh2fl/fl mice (Su et al., 2003) with K14-Cre mice (Vasioukhin et al., 1999). Genotyping was confirmed by PCR of tail skin DNAs. BrdU (Sigma-Aldrich) was administered to pregnant mice or to neonatal pups by peritoneal injection (50 µg/g BrdU). Dye exclusion assays were performed as described (Hardman et al., 1998).
Fluorescence-Activated Cell Sorting
FACS purification of epidermal cells from K14-H2B-GFP embryos was performed on a FACSVantage SE system equipped with FACS DiVa software (BD Biosciences). Cells were gated for single events and viability and then sorted according to α6 integrin expression and GFP. Cell-cycle/BrdU incorporation was analyzed using the BrdU-FITC Flow Kit (PharMingen) (see Blanpain et al., 2004), and cell-cycle status was analyzed using FlowJo program.
Microarray Analysis
RNAs from FACS-purified WT and Ezh2 cKO basal cells (see Supplemental Experimental Procedures) were provided to the Genomics Core Facility, MSKCC for quality control, quantification, reverse transcription, labeling, and hybridization to MOE430A 2.0 microarray chips (Affymetrix). Arrays were scanned per the manufacturer’s specifications for the Affymetrix MOE430v2 chip. Images were background subtracted, and probeset expression estimates were generated using the MAS5 algorithm implemented in Bio-conductor. A detection p value of p = 0.05 was used to threshold probesets into present (p ≤ 0.05) or absent (p > 0.05). Probesets were identified as differentially expressed when the absolute fold change was > 2, and the probeset is present in both samples of the relatively overexpressed group. Probesets selected for visualization were log2 transformed and normalized to the replicate mean (estimated from basal cells). Intensity bias between replicates necessitated this within replicate normalization, which leaves the data for each gene centered around zero in the basal cells. These normalized data were analyzed with hierarchical clustering (Pearson correlation, average linkage) and visualized with heatmaps to assist in interpretation.
Statistics
For all graphs, mean value ± one standard deviation was presented with the number of replicates indicated in figure legends. To determine significance between two groups indicated in figures by a bracket, comparisons were made using Student’s t test. For all statistical tests, the 0.05 level of confidence was accepted for statistical significance.
Cell Culture
Enzymatic separation of epidermis from skins and primary P0 MK culturing were as described (Blanpain et al., 2004). To visualize colonies, we fixed cells and stained them with 1% Rhodamine B (Sigma). To induce differentiation in culture, calcium was raised from 0.05 to 1.5 mM. To assess cell growth, we plated equal numbers of live cells on collagen-coated 6-well plates. Cell numbers were determined by hemocytometer.
To assess AP1 function, MK were treated either with Tanshinone IIA (Biomol) for 48 hr with indicated concentrations or with PMA (Sigma) 10 ng/ml for 5–10 hr. DMSO or Ethanol were used as vehicle controls, correspondingly. To attenuate cJun and JunD, cells were infected with lentiviruses that carry shRNA constructs (Sigma). Selection with puromyocin (2 mg/ml) was done 2 days upon infection, and MK were analyzed a week after infection.
Chromatin Immunoprecipitation Assays
Assays were performed with the EZ ChIP kit (Millipore). Briefly, 2 × 106 cells were crosslinked with 1% formaldehyde for 10 min at room temperature (for ChIPs with Ezh2, H3, and triMeK27-H3 Abs) or for 15 min at 37°C (for cJun Abs). Cells were collected, lysed, and sonicated for a total of 200 s using Branson Sonifier 450 equipped with microtip (setting 3; 50% input). Chromatin lysates were used for immunoprecipitations with either Ezh2 (Lake Placid Biologicals), triMeK27-He (Upstate), H3 (Abcam), triMeK4-H3 (Abcam), cJun (Santa Cruz), or no Ab control. Immunoprecipitates were collected with Protein A/G agarose beads (Millipore), protein/DNA crosslinks were reversed by incubating at 65°C, and DNA was purified using QIAgen PCR purification kit (QIAgen). Treatment with TanIIa drug for ChIP was done for 2 hr with 100 µM of TanIIa or DMSO (vehicle control).
ChIP DNA was analyzed by performing quantitative PCR as described above. Primers against K5 and/or intergenic region on chromosome 5 genes were used for normalization. Sequences of primers are available upon request.
Supplementary Material
The Supplemental Data include Supplemental Experimental Procedures, eight figures, and two tables and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(09)00006-3.
ACKNOWLEDGMENTS
We thank Markus Schober, Scott Williams, Greg Wang (Allis’s lab), Maria Nikolova, June Racelis, Svetlana Mazel, Christopher Bare, Xiaoxuan Fan, and Agnes Viale for technical assistance and our colleagues in the Fuchs laboratory, especially Jonathan Nowak, Rui Yi, and Valerie Horsley, for constructive discussion and criticisms. E.E. is a NY Stem Cell Foundation postdoctoral fellow of the Life Sciences Research Foundation. E.F. is an HHMI Investigator.
Footnotes
ACCESSION NUMBERS
Microarray data of genes expressed in basal WT and Ezh2cKO cells at E16 have been deposited to the GEO repository under GSE14045 Series Accession Number.
REFERENCES
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman SW, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, van Tellingen O, van Lohuizen M. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–341. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N, Grosveld F. Transcriptional regulation of multigene loci: Multilevel control. Trends Genet. 1993;9:134–137. doi: 10.1016/0168-9525(93)90208-y. [DOI] [PubMed] [Google Scholar]
- DiSepio D, Jones A, Longley MA, Bundman D, Rothnagel JA, Roop DR. The proximal promoter of the mouse loricrin gene contains a functional AP-1 element and directs keratinocyte-specific but not differentiation-specific expression. J. Biol. Chem. 1995;270:10792–10799. doi: 10.1074/jbc.270.18.10792. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Crish JF, Banks EB, Welter JF. The epidermis: Genes on-genes off. J. Invest. Dermatol. 1997;109:501–509. doi: 10.1111/1523-1747.ep12336477. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Frye M, Fisher AG, Watt FM. Epidermal stem cells are defined by global histone modifications that are altered by Myc-induced differentiation. PLoS ONE. 2007;2:e763. doi: 10.1371/journal.pone.0000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, Yuspa SH. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 1980;19:245–254. doi: 10.1016/0092-8674(80)90406-7. [DOI] [PubMed] [Google Scholar]
- Jackson B, Tilli CM, Hardman MJ, Avilion AA, MacLeod MC, Ashcroft GS, Byrne C. Late cornified envelope family in differentiating epithelia–Response to calcium and ultraviolet irradiation. J. Invest. Dermatol. 2005;124:1062–1070. doi: 10.1111/j.0022-202X.2005.23699.x. [DOI] [PubMed] [Google Scholar]
- Jang SI, Steinert PM, Markova NG. Activator protein 1 activity is involved in the regulation of the cell type-specific expression from the proximal promoter of the human profilaggrin gene. J. Biol. Chem. 1996;271:24105–24114. doi: 10.1074/jbc.271.39.24105. [DOI] [PubMed] [Google Scholar]
- Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–2777. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall D, Hardman MJ, Nield KM, Byrne C. Differentially expressed late constituents of the epidermal cornified envelope. Proc. Natl. Acad. Sci. USA. 2001;98:13031–13036. doi: 10.1073/pnas.231489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Xi ZF, Seo EY, McGaughey D, Segre JA. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc. Natl. Acad. Sci. USA. 2006;103:18668–18673. doi: 10.1073/pnas.0608658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by poly-comb repressors: Postponing commitment. Curr. Opin. Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Rastelli L, Chan CS, Pirrotta V. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 1993;12:1513–1522. doi: 10.1002/j.1460-2075.1993.tb05795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol. Cell. Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group genes: Keeping stem cell activity in balance. PLoS Biol. 2008;6:e113. doi: 10.1371/journal.pbio.0060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Steven AC, Bisher ME, Roop DR, Steinert PM. Biosynthetic pathways of filaggrin and loricrin-Two major proteins expressed by terminally differentiated epidermal keratinocytes. J. Struct. Biol. 1990;104:150–162. doi: 10.1016/1047-8477(90)90071-j. [DOI] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wulfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Lo Celso C, Silva-Vargas V. Epidermal stem cells: an update. Curr. Opin. Genet. Dev. 2006;16:518–524. doi: 10.1016/j.gde.2006.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplemental Data include Supplemental Experimental Procedures, eight figures, and two tables and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(09)00006-3.