Abstract
Skin epithelia must rejuvenate constantly during normal homeostasis and repair damage after wounding. Fulfilling these roles necessitates reservoirs of stem cells that persist for life. This review focuses on the elusive stem cell niche of the epidermis, long thought to reside within the basal layer that is sandwiched between the basement membrane and the suprabasal interface.
The Epidermal Basal Layer
The skin epidermis serves as the barrier that protects us against the physical, chemical, and thermal assaults of our environment. To achieve these feats, the epidermis generates an elaborate array of supportive appendages, including hair follicles (HFs), sebaceous glands, sweat glands, and nails.
The existence of stem cells within mammalian epidermis is illustrated by the ability to maintain and propagate newborn human (foreskin) epidermal cells in vitro for many generations (reviewed in Green, 2008). Moreover, after culture, these cells are still sufficiently resilient to provide long-term regenerative potential to patients whose skin has been badly burned.
The stem cells of adult epidermis have long been thought to reside within the innermost (basal) layer of this stratified epithelium, which rests upon a basement membrane (BM) rich in extra-cellular matrix (ECM) proteins and growth factors (Figure 1). Common to many stem cell niches, the BM forms a boundary between epidermis and dermis. Basal cells rely upon mesenchymal and BM stimuli to remain proliferative. As they exit this niche and move to suprabasal locations, basal cells terminally differentiate via a spinous cell intermediate, culminating in dead enucleated cells that provide the epidermal barrier until they are shed and replaced by inner layer cells moving outward (reviewed in Blanpain and Fuchs, 2009). The direction of movement is largely columnar, as predicted by Potten many years ago, and supported by mosaic keratin-promoter-driven transgenic mice. Much of the action takes place at the basal-to-spinous cell interface, where basal cells receive microenvironmental cues that influence when to proliferate, differentiate, or function in wound repair. The extent to which individual basal cells experience environmental heterogeneity and whether such variety in external cues impacts epidermal stem cell number remain unclear.
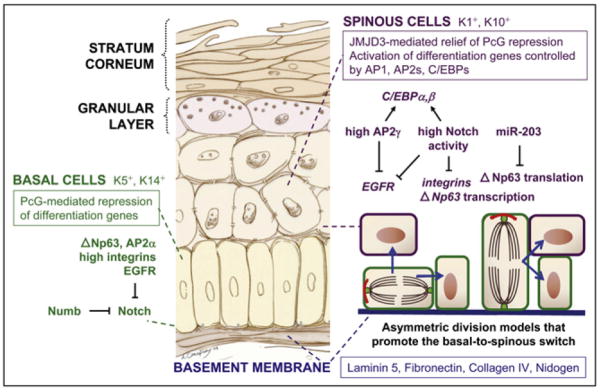
Figure 1. The Architecture of the Basal-to-Spinous Switch.
The proliferative basal cells of the epidermis adhere to an underlying basement membrane, separating the dermis from the epidermis, and differentiate into spinous cells in the suprabasal layer. Integral to this switch are epigenetic changes in chromatin and Notch signaling, which are activated in spinous cells. Notch is thought to be repressed in basal cells by several mechanisms, including the Notch inhibitor Numb, which is frequently asymmetrically inherited after basal cell division. In turn, Notch signaling downregulates integrin and p63 gene expression and, in conjunction with AP2s, activates C/EBP expression. Models of how asymmetric divisions could promote the basal-to-spinous switch are depicted. Epidermal drawing by Ann Canapary.
Regulation of the Basal-to-Suprabasal Switch
Recognized decades ago, the switch between expression of cytoskeletal keratins K5/K14 and K1/K10 is a reliable indicator that an epidermal cell has left its basal niche and committed to terminally differentiate (Blanpain and Fuchs, 2009). One of the main signals orchestrating the basal-to-suprabasal switch is Notch: ligands for Notch reside in the basal layer, and receptors for Notch are expressed suprabasally by spinous cells. Too much Notch activity promotes the fate of K1/K10-expressing spinous cells, while too little results in a diminution of this differentiated state (Blanpain and Fuchs, 2009).
One of Notch’s classical target genes encodes the transcriptional repressor Hes1, shown recently to maintain spinous cell fate in the developing embryo (Moriyama et al., 2008). In addition, Notch signaling induces C/EBPs, which transcriptionally regulate many differentiation-specific genes, including K1 and K10. To fully activate this program, AP2 transcription family factors collaborate with Notch (Wang et al., 2008; Figure 1). Epidermis expresses AP2α (basal preferred) and AP2γ (suprabasal preferred), and most of its genes possess AP2 transcription-factor-binding sites in their 5′ regulatory regions (Blanpain and Fuchs, 2009). When singly targeted, epidermal development and homeostasis are only mildly perturbed, but double targeting of AP2α/γ abrogates C/EBP expression, crippling differentiation (Wang et al., 2008). AP2α promotes spinous cell commitment by tempering epidermal growth factor receptor (EGFR) signaling, a molecular advocate of growth and a repressor of Notch signaling (Kolev et al., 2008).
Further insights into the switch mechanism emanate from studies on p63. In the late 1990s, two groups studying the p53 family of proto-oncogenes independently discovered that mice mutant for p63 are severely compromised in skin development (Koster et al., 2004 and references therein). Uncommitted ectodermal cells covered most of the body surface of p63 mutants, but a few clumps of differentiated epidermal cells remained. This phenotype may reflect an absence of lineage commitment and an early block in epidermal differentiation, or a secondary defect in epithelial stem-cell renewal and/or survival. Discerning between these options is challenging because the p63 gene encodes TAp63 and ΔNp63 isoforms, which arise from differential promoter usage, as well as α, β, and γ subtypes.
ΔNp63α is the most highly expressed and conserved p63 isoform in stratified epithelia, where it is seen mainly in the basal layer (Koster et al., 2004). ΔNp63α lacks the transactivation domain present in p53 but shares its DNA and tetramerization domains. Using an in vitro model of human epidermal regeneration, Truong and Khavari tested whether the isoform functions as a dominant-negative p53 antagonist (Truong and Khavari, 2007). They observed that reducing p63 mRNA levels reduced cell proliferation, and when p53 and p63 small interfering RNAs (siRNAs) were added together, normal cell proliferation was restored. While this study accentuates the opposing effects of ΔNp63α and p53 on cell growth, loss of p63 can also increase apoptosis (Senoo et al., 2007). Thus, it is still unclear whether the proliferation restored by p53 shRNA in p63 shRNA organotypic cultures is achieved byrelieving p63’s dominant-negative action on p53 or via disruption of stress-induced, p53-dependent apoptosis.
Interestingly, p53 siRNA did not rescue terminal differentiation in vitro (Truong and Khavari, 2007), suggesting that p63 governs human epidermal differentiation independent of its role in cell proliferation/survival. These results further highlight the potential significance of p63 isoforms such as ΔNp63α that possess a protein-protein interacting domain not present in p53. It is tempting to speculate that, during homeostasis, ΔNp63α represses key genes and maintains basal cells in their undifferentiated progenitor state. On the other hand, ΔNp63α might endow basal progenitors with the ability to differentiate (Koster et al., 2004). Elucidating ΔNp63α’s interaction partners should shed important new insights into its function(s).
In addition, the 3′UTR of mouse ΔNp63α mRNA possesses seed sequences for miR-203, a microRNA expressed in all transcriptionally active, terminally differentiating skin epithelial cells, but not in proliferative compartments (Yi et al., 2008; Figure 1). Consistent with microRNAs as translational repressors, ΔNp63α and miR-203 display mirror image expression patterns. Importantly, gain- and loss-of-function studies in mice support the view that miR-203 acts at least in part by negatively regulating suprabasal expression of basal genes, thereby refining the basal/spinous boundary and the switch between proliferation and differentiation (Yi et al., 2008). In this regard, Notch signaling not only activates spinous fate, but also represses basal fate through reducing p63 and integrin levels. These effects on basal fate are complex and not always mediated through canonical mechanisms (Blanpain and Fuchs, 2009; Moriyama et al., 2008). An intriguing question is whether Notch signaling activates miR-203 gene expression, further repressing basal fate and sealing commitment in newly suprabasal cells.
The basal-to-spinous switch is also fine-tuned by epigenetic modifications. For example, Frye and Watt observed that β1 integrin-enriched cells are associated with reduced levels of histone H4 acetylation (Frye et al., 2007). This pattern also holds for mouse interfollicular epidermis (IFE) and for HF, suggesting antagonistic roles for H4 acetylation and basal cell behavior. Histone acetylation is associated with active gene expression in general and with c-Myc in particular. In earlier studies, Watt and colleagues suggested a role for c-Myc in driving stem cells to a transient amplifying state en route to differentiation and found that elevated c-Myc expression in basal cells led to widespread enhancement of H4 acetylation as well as additional H4 modifications at lysine 20. These data have led to the view that activated c-Myc may promote a chromatin state permissive for epidermal differentiation (Frye et al., 2007).
In addition, Sen and Khavari globally mapped chromatin from cultured human epidermal keratinocytes for enrichment of the negative histone mark H3K27me3 (Sen et al., 2008). Under growth-promoting conditions, ~10% of the gene promoters assayed were enriched for H3K27me3. Conversely, in differentiation conditions, the promoters of nearly 10% of genes upregulated in keratinocytes had reduced H3K27me3-marked chromatin. The possible functional significance of this opposing correlation in histone modification was underscored by siRNA targeting of JMJD3, the demethylase thought to be responsible for removing the H3K27me3 mark. In human organotypic cultures, reductions in the demethylase correlated with repression of epidermal differentiation, while overexpression of JMJD3 enhanced differentiation (Sen et al., 2008).
These studies are particularly interesting in light of our own recent finding that Ezh2, the H3K27me3 methylase at the core of polycomb complex group (PcG)-mediated chromatin repression, functions in the basal layer of mouse embryonic epidermis in vivo to repress a bank of genes involved in epidermal terminal differentiation (Ezhkova et al., 2009). While the full molecular mechanisms remain uncharacterized, many of these promoters possess binding sites for AP1 transcription factors known to be involved in epidermal differentiation (Zenz and Wagner, 2006). Notably, some AP1 members are expressed basally, albeit at reduced levels compared to suprabasally, and the negative H3K27me3 histone blocked accessibility of AP1 factors to the marked genes in basal cells (Ezhkova et al., 2009).
This regulatory pattern is acutely relevant, in that many genes involved in nonepidermal differentiation programs are also governed by PcG repression in developing epidermis, and they too lose their H3K27me3 mark when Ezh2 is lost (Ezhkova et al., 2009). In contrast to PcG-regulated epidermal genes, however, muscle and neuronal genes remain repressed and don’t bind AP1 factors in Ezh2-depleted basal epidermal cells. Presumably, nonepidermal gene expression depends upon other tissue-specific transcription factors not present in the basal cell population.
Interestingly, loss of Ezh2 (Ezhkova et al., 2009) and JMJD3 (Sen et al., 2008) were both accompanied by precocious epidermal maturation/accelerated differentiation suprabasally, rather than premature differentiation of basal cells. It is tempting to speculate that full-throttle differentiation is dependent upon induction of additional transcription factors, including C/EBPs, which are induced in response to regionally dependent environmental cues, e.g., Notch signaling. Alternatively, it may be that the phenotypes observed to date have not revealed the impact of PcG governance: JMJD3 SiRNA knockdown was only ~80%, and Ezh1 could partially compensate for Ezh2. Other chromatin modifications, including histone deacetylation, may also be involved (Frye et al., 2007).
It is also interesting that epidermal differentiation genes are repressed by PcG in embryonic stem cells (ESCs), which are poised to activate more tissue-specific differentiation programs than their epidermal counterparts (Chi and Bernstein, 2009). ESCs appear to maintain this pliability by marking critical differentiation genes not only with PcG repressive marks but also with an active histone 3 mark, lysine 4 (Chi and Bernstein, 2009). This double marking could explain why loss of PcG repression in ESCs results in activation of many differentiation pathways (Chi and Bernstein, 2009). In contrast, PcG-marked epidermal differentiation genes do not display this active mark in embryonic basal cells (Ezhkova et al., 2009). Rather, epidermal stem cells appear to couple PcG repression with a requirement for tissue-specific transcriptional activators, thereby leading to selective lineage induction only when environmentally cued.
Lift-Off from the BM Launchpad
In addition to signaling cues that are delivered across the basal-spinous interface, the basal layer also receives regulatory input from “below.” Although mechanophysical properties alone are likely to influence the proliferative properties of basal cells, the BM also provides a diverse repertoire of proliferative stimuli for basal cells. Among them is laminin 5, which promotes anchorage, signaling, and migration by acting as ligand for α6β4 at the core of hemidesmosomes and α3β1 integral to focal adhesions (FAs) (Blanpain and Fuchs, 2009; Owens and Watt, 2003). Cultured human epidermal cells that exhibit higher levels of β1 integrin have greater proliferative potential in vitro than other cells within the population (Owens and Watt, 2003). Upon α3β1 integrin activation, a kinase cascade including focal adhesion tyrosine kinase (FAK) and Src tyrosine kinase is unleashed, not only stimulating the Ras-MAPK pathway, but also inducing focal adhesion turnover and epidermal migration (Guasch et al., 2007).
Balancing attachment, detachment, and migration is crucial for the basal epidermal cell. Notably, transgenic suprabasal expression of integrins promotes tumorigenesis in mice (Owens and Watt, 2003). Conversely, conditional loss of FAK leads to an increased resistance to chemically induced skin tumorigenesis, and in vitro, FAK-deficient keratinocytes exhibit defects in cell migration and focal adhesion turnover (Guasch et al., 2007 and references therein).
The BM is also rich in proteoglycans and other proteins, which cast a molecular net for growth-regulatory factors. TGFβs restrict epidermal proliferation, and TGFα, EGFs, and insulin growth factors enhance proliferation (Zenz and Wagner, 2006). When TGFβ receptor signaling is compromised, epidermal homeostasis is maintained, but the apparent normalcy is deceptive (Guasch et al., 2007). In this setting, wounds heal faster, basal cells display increased proliferation counterbalanced by increased apoptosis, and with just one additional oncogenic mutation, the tissue transforms quickly to squamous cell carcinoma of the skin.
The opposing effects of FAK/integrin and TGFβ signaling on basal cell behavior are intertwined. Elevating integrin signaling suppresses TGFβ signaling (Owens and Watt, 2003), and conversely when TGFβ signaling in epidermis is compromised, FAK/integrins are activated and migration is enhanced (Guasch et al., 2007). EGF receptor (EGFR) signaling aligns with activated FAK/integrins in eliciting these responses (Zenz and Wagner, 2006). In addition, the underlying dermis is known to undergo significant crosstalk with the epidermal basal layer in orchestrating its proliferative and migratory behaviors (Blanpain and Fuchs, 2009; Zenz and Wagner, 2006).
How do these signaling pathways participate to preserve stemness? While the underlying mechanisms are still elusive, epidermal basal cells rely heavily upon these regulatory circuits. Mitogen-inducible gene 6 is a suppressor of EGFR signaling, and mice deficient in mitogen-inducible gene 6 display epidermal hyperproliferation and increased tumor susceptibility (Ferby et al., 2006). Another inhibitor of EGFR signaling, Lrig1, is expressed throughout the basal layer of human epidermis but seems to be enriched within less proliferative, β1 integrin-enriched cells (Jensen et al., 2009 and references therein). Mice lacking Lrig1 display a hyperproliferative epidermis, and human keratinocytes faced with an Lrig1 short hairpin RNA produce larger colonies than normal. These observations led Jensen and Watt to posit that Lrig1 might regulate slow cycling features of basal stem cells. When taken together with the AP2α-EGFR-Notch circuitry discussed above, these studies underscore a key role for the EGFR signaling pathway in controlling basal fate and for Notch in regulating suprabasal fate.
As the regulatory roles for additional BM constituents and their associates continue to unfold, the extrinsic signals received by the microenvironment and translated through transmembrane receptors are expected to couple with the intrinsic properties of the basal epidermal cells to define their ability to self-renew and undergo homeostasis and wound repair.
Superimposed on these regulatory pathways is the contribution of the BM and integrins to establish the polarity that enables basal epidermal cells to orient their spindle and divide properly. During embryonic development, divisions occur in a plane parallel to the BM in the single-layered epidermis. Upon stratification, a majority of divisions become asymmetrically oriented, relative to the BM (Lechler and Fuchs, 2005). While this orientation is maintained during development, in adult, asymmetric divisions that leave both daughter cells attached to the BM can occur (Clayton et al., 2007). Specific orientation appears to rely upon asymmetric polarity cues that lead one spindle pole and its astral microtubules to associate with the cortical actin cytoskeleton (Figure 1). Notably, without β1 integrin or α-catenin, spindle orientation becomes randomized (Lechler and Fuchs, 2005).
Several models can explain how the alignment of asymmetric divisions might produce one basal progenitor and one committed cell (Figure 1). If asymmetric to the BM, a division would automatically place one daughter in the suprabasal layer. If parallel, one daughter might inherit factors to reduce the level of integrins, thereby leading to an early departure from the BM niche. In this regard, it is interesting that asymmetric divisions frequently involve asymmetric inheritance of a stronger Notch signal, and in the epidermis, elevated Notch signaling basally results in decreased integrin gene expression (reviewed by Blanpain and Fuchs, 2009).
Is the Basal Layer a Stem Cell Niche?
One of the last frontiers in epidermal biology is the location of its stem cells, and if this is within the basal layer, to what extent the heterogeneity exhibited by basal epidermal cells reflects a difference in their ability to self-renew long-term and generate epidermis. Humans have a thick epidermis with frequent epidermal turnover in vivo and a propensity to yield long-term epidermal cultures in vitro. By contrast, while the mouse is genetically tractable and therefore perfect for addressing the issue, it displays a thinner, less active epidermis, whose cells are difficult to culture long-term.
Recently, a population of murine HF stem cells was identified that is enriched for Lrig1 and Blimp1, and that gives rise to IFE and SGs when stimulated by retinoic acid in vivo (Jensen et al., 2009 and references therein). It has been suggested that these cells might be the elusive IFE stem cells. However, three different lineage tracing studies with Cre recombinase driven by either Shh, Sox9, or cytochrome P450 promoters each document that mouse IFE harbors its own resident progenitors, which can sustain epidermal homeostasis long-term (Levy et al., 2007; Jones et al., 2007; Nowak et al., 2008). Moreover, even though efficient wound repair in mice draws heavily from follicle cells, epidermal cells do contribute, as exemplified by the Edaradd mutant mouse, whose tail skin lacks HFs but is nevertheless able to slowly repair its wounds (Langton et al., 2008).
Upon severe injury, IFE can even regenerate HFs, leading to the view that epidermal basal cells not only self-renew long-term, but are also multipotent (Ito et al., 2007). These findings also raise the possibility that some properties might be shared between epidermal and HF stem cells; however, identifying commonalities has been challenging. A few adult basal cells do show preferential label retention, but they lack defined spatial distribution and aren’t easily traced to embryonic skin (Nowak et al., 2008). Moreover, only a few molecular parallels between HF bulge stem cells and IFE basal cells have been defined, and most, such as K5, K14, and p63, mark all skin epithelial cells with proliferative capacity, whether stem cells or not. The paucity of specific stem cell markers, coupled with their relatively weak display of self-renewal, suggests that either the number or long-term potential of mouse epidermal stem cells within the basal layer is small.
Lineage tracing of single basal cells in tail skin, where mitoses are more frequent than in back skin, suggests that, while the majority of labeled cells are lost within 3 months, some survive and clonally expand in size over time (Clayton et al., 2007). This behavior seems to argue against long-standing models of a discrete epidermal proliferating unit composed of one stem cell surrounded by a steady-state pool of ~10 transit-amplifying progeny that subsequently exit the niche and terminally differentiate (Clayton et al., 2007). Rather, mathematical modeling of the long-term fate of marked basal cells suggests that they could all be equivalent (Clayton et al., 2007). That said, interpretation is confounded on the one hand by environmental assaults that could elicit localized epidermal wound repair, and on the other, by the possibility that longer-term stem cells might escape being marked.
In the future, it will be important to determine whether there is a hierarchy in the relative abilities of individual cells within the basal epidermal layer versus the HF to self-renew and generate epidermal tissue long-term. For now, resolution as to whether basal cells are equivalent and how basal progenitors relate to their better-characterized cousins in the HF awaits identification of the genes that mediate the long-term, self-renewing capacity of the epidermis. Therein lie the clues to the identity of the IFE stem cell and its niche.
Acknowledgments
I thank Cedric Blanpain and Elena Ezhkova for their helpful comments. I also wish to apologize to my colleagues for failing to be inclusive in citations, due to formatting constraints. That said, I thank them for their superb papers that have expanded our understanding of the basal-to-spinous interface.
References
- Blanpain C, Fuchs E. Nat Rev Mol Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AS, Bernstein BE. Science. 2009;323:220–221. doi: 10.1126/science.1166261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K, Sommergruber W, Kraut N, Ullrich A, Fassler R, Klein R. Nat Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- Frye M, Fisher AG, Watt FM. PLoS ONE. 2007;2:e763. doi: 10.1371/journal.pone.0000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H. Bioessays. 2008;30:897–903. doi: 10.1002/bies.20797. [DOI] [PubMed] [Google Scholar]
- Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, Cotsarelis G. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM. Cell Stem Cell. 2009;4:427–439. doi: 10.1016/j.stem.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PH, Simons BD, Watt FM. Cell Stem Cell. 2007;1:371–381. doi: 10.1016/j.stem.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Kolev V, Mandinova A, Guinea-Viniegra J, Hu B, Lefort K, Lambertini C, Neel V, Dummer R, Wagner EF, Dotto GP. Nat Cell Biol. 2008;10:902–911. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton AK, Herrick SE, Headon DJ. J Invest Dermatol. 2008;128:1311–1318. doi: 10.1038/sj.jid.5701178. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy V, Lindon C, Zheng Y, Harfe BD, Morgan BA. FASEB J. 2007;21:1–9. doi: 10.1096/fj.06-6926com. [DOI] [PubMed] [Google Scholar]
- Moriyama M, Durham AD, Moriyama H, Hasegawa K, Nishikawa S, Radtke F, Osawa M. Dev Cell. 2008;14:594–604. doi: 10.1016/j.devcel.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Nowak JA, Polak L, Pasolli HA, Fuchs E. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DM, Watt FM. Nat Rev Cancer. 2003;3:444–451. doi: 10.1038/nrc1096. [DOI] [PubMed] [Google Scholar]
- Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Truong AB, Khavari PA. Cell Cycle. 2007;6:295–299. doi: 10.4161/cc.6.3.3753. [DOI] [PubMed] [Google Scholar]
- Wang X, Pasolli HA, Williams T, Fuchs E. J Cell Biol. 2008;183:37–48. doi: 10.1083/jcb.200804030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz R, Wagner EF. Int J Biochem Cell Biol. 2006;38:1043–1049. doi: 10.1016/j.biocel.2005.11.011. [DOI] [PubMed] [Google Scholar]