Summary
PCR-based surveys of microbial communities commonly use regions of the small subunit ribosomal RNA (SSU rRNA) gene to determine taxonomic membership and estimate total diversity. Here we show that the length of the target amplicon has a significant effect on assessments of microbial richness and community membership. Using OTU- and taxonomy-based tools, we compared the V6 hypervariable region of the bacterial SSU rRNA gene of three amplicon libraries of ca. 100 base pair (bp), 400bp, and 1000bp from each of two hydrothermal vent fluid samples. We found that the smallest amplicon libraries contained more unique sequences, higher diversity estimates, and a different community structure than the other two libraries from each sample. We hypothesize that a combination of polymerase dissociation, cloning bias, and mis-priming due to secondary structure accounts for the differences. While this relationship is not linear, it is clear that the smallest amplicon libraries contained more different types of sequences, and accordingly, more diverse members of the community. Because divergent and lower abundant taxa can be more readily detected with smaller amplicons, they may provide better assessments of total community diversity and taxonomic membership than longer amplicons in molecular studies of microbial communities.
Introduction
Microbial ecologists routinely use PCR-based surveys of microbial communities to catalog the diversity and abundance of microorganisms without requiring cultivation in the laboratory. However, there are many issues that complicate these surveys, including template secondary structure and G+C differences; biased primer annealing and competition within degenerate primer pools; chimera and heteroduplex formation; polymerase error; and differences between reactions in annealing temperature, cycle number, or template concentration (Reysenbach et al., 1992; Farrelly et al., 1995; Suzuki and Giovannoni, 1996; Wintzingerode et al., 1997; Kleter et al., 1998; Polz and Cavanaugh, 1998; Suzuki et al., 1998; Becker et al., 2000; Ishii and Fukui, 2001; Thompson et al., 2002; Hongoh et al., 2003; Acinas et al., 2005; Osborne et al., 2005; Sipos et al., 2007). While shotgun-based metagenomic methods allow one to avoid amplification reactions, investigators still routinely use PCR of SSU rRNA genes to determine the microbial diversity and community structure in environmental samples, regardless of sequencing or screening method used (denaturing gradient gel electrophoresis (Muyzer et al., 1993), terminal restriction fragment length polymorphism (Liu et al., 1997), length heterogeneity-PCR (Suzuki et al., 1998), cloning and sequencing (Giovannoni et al., 1990), and amplicon pyrosequencing (Sogin et al., 2006)). While the design of amplification primers and the use of different cycling conditions can create bias in the composition of amplification libraries, there are no systematic surveys of the effect of PCR amplicon size on assessments of microbial diversity and community structure. In a recent study of microbial diversity in the deep-sea water column of the North Atlantic and hydrothermal vents of the Pacific (Sogin et al., 2006; Huber et al., 2007), massively parallel pyrosequencing of hundreds of thousands of PCR amplicons of the SSU rRNA V6 hypervariable regions revealed extremely high levels of microbial diversity. Deeper sampling of molecular sequences from the microbial population afforded by the massively parallel pyrosequencing approach accounts for some of this increased diversity, but biases associated with longer amplicons might also contribute to differences in diversity estimates based upon full-length versus short PCR amplicon sequences. Here we investigate the effect of PCR amplicon size on assessments of microbial diversity and community structure.
We extracted DNA from two hydrothermal vent fluid samples and constructed amplicon libraries of ca. 100 base pair (bp), 400bp, and 1000bp from each. Four primers (two forward and two reverse) were used in 3 combinations to obtain amplicons of three size classes; all three contained the V6 hypervariable region of the SSU rRNA gene (Table 1). This permitted direct comparisons between the different size libraries by examining only the V6 region, regardless of which primer set was used. We used OTU- and taxonomy-based tools to evaluate results and to elucidate differences in microbial richness and population structure between the three libraries.
Table 1.
Summary of sequencing data for each clone library constructed from samples FS312 and FS396.
| FS312_100bp | FS312_400bp | FS312_1000bp | FS396_100bp | FS396_400bp | FS396_1000bp | |
|---|---|---|---|---|---|---|
| Forward Primer | 967F | 967F | 337F | 967F | 967F | 337F |
| Reverse Primer | 1046R | 1391R | 1391R | 1046R | 1391R | 1391R |
| High Quality Bacterial Sequences | 761 | 860 | 381 | 685 | 866 | 663 |
| Chimeric Sequencesa | ND | 9 | 4 | ND | 3 | 9 |
| High Quality Archaeal Sequences | 0 | 0 | 6 | 0 | 0 | 6 |
| Mis-Primed 16S rRNA Sequences | 0 | 0 | 117 | 0 | 0 | 10 |
| Mis-Primed Non-16S rRNA Sequences | 0 | 0 | 372 | 0 | 0 | 278 |
| % Exact Matchesb | 65.2% | 99.2% | 99.7% | 72.8% | 99.8% | 99.7% |
| % Unique Sequencesc | 37.5% | 14.3% | 17.1% | 25.0% | 10.7% | 11.3% |
Not Determined
Percent of high quality bacterial V6 sequences within library that are exact matches to an existing sequence in the reference database
Perecent of high quality bacterial V6 sequences within library not detected by the other two libraries from the same sample at 100% sequence identity.
Results
Clone Library Construction and Sequence Processing
Clone libraries were constructed from samples FS312 and FS396 for each primer set, resulting in three libraries with inserts of approximately 100 bp, 400 bp, and 1000 bp in size from each site (Table 1). In total, 4,230 high quality, non-chimeric, bacterial ribosomal sequences were obtained from the six libraries. Chimeric sequences were detected in both the 400 and 1000bp libraries, and archaeal sequences were found in the 1000bp libraries (Table 1). We did not search for chimeras in the 100bp libraries due to the small amplicon size. In processing the sequences, we found that both 1000bp libraries contained large numbers of sequences that included the PCR primer 1391R at the 5’ end in sense orientation in addition to its expected position at the 3’ end in the antisense orientation. Of the 880 and 966 1000bp sequences in FS312 and FS396, respectively, 489 and 288 of these mis-primed sequences were identified. In some cases, these sequences contained valid ribosomal RNA sequences, in other cases they were clearly non-ribosomal (Table 1). Because no proper forward PCR primer could be located, the mis-primed sequences were not included in the high quality bacterial dataset. Instead, two datasets were constructed: one that contained the chimeric, archaeal, and mis-primed 16S rRNA sequences (referred to as “With Artifacts”) and one without any of these artifacts (referred to as “High Quality (HQ) Bacteria”). No mis-priming was identified in the 100 and 400bp libraries. The dataset deposited in GenBank includes only the non-chimeric high quality bacterial sequences. For all analyses, the sequences from the 400bp and 1000bp libraries were trimmed to include only the V6 hypervariable region between the 967F and 1046R primer sites of the 100bp library.
Clone Library Comparisons: Diversity
The percent difference between each unique high quality bacterial tag sequence and the closest match in a reference database of V6 sequences (Sogin et al., 2006) was calculated to compare the sequences in each clone library to known microbial sequences. For each sampling site, the 400bp and 1000bp libraries consisted almost entirely of sequences that were exact matches to reference sequences, and both are characterized as containing primarily previously known sequences or sequences similar to known sequences. In contrast, the 100bp library contained a lower percentage of sequences that were exact or close matches to reference sequences, and contained a higher percentage of sequences that were more distant from reference sequences (Table 1, Fig. 1, Supp. Fig. 1).
Figure 1.
Distance between clone sequences and their best match in the reference database and the percent of the clone library each distance represents for each library within samples (a) FS312 and (b) FS396. The y-axis is reduced to show detail below 15% of the clone library.
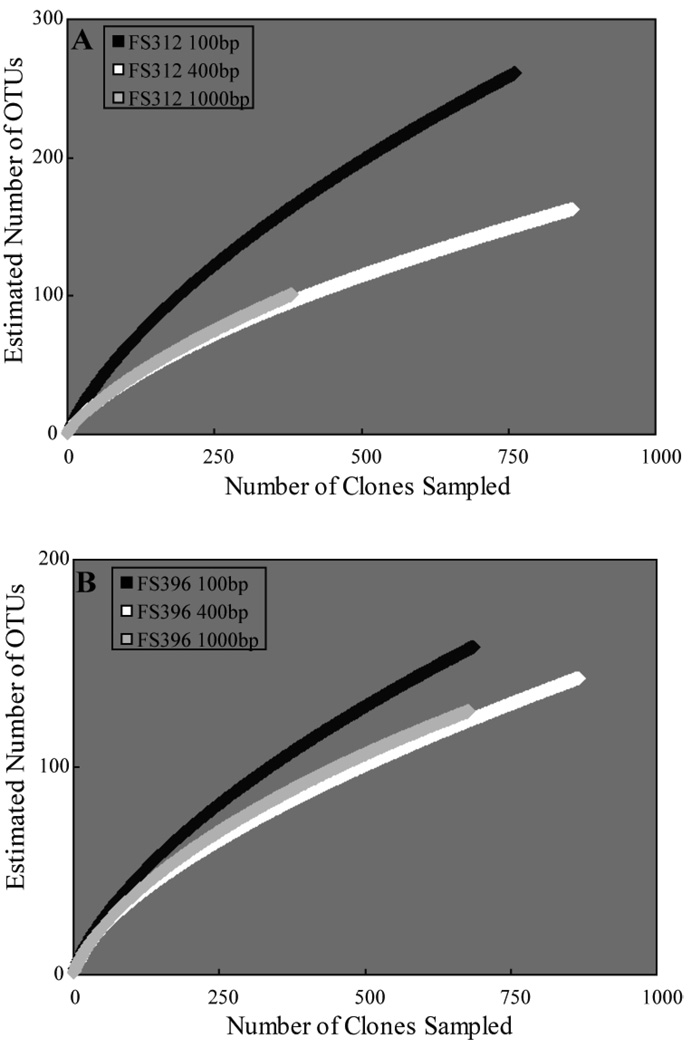
Sequences were assigned to Operational Taxonomic Units (OTUs) based on farthest-neighbor distances between sequences using the program DOTUR (Schloss and Handelsman, 2005). For both samples and at all distances between 0% and 10%, the most OTUs were found in the 100bp library, followed by the 400bp and the 1000bp library (Fig. 2). Similarly, the nonparametric estimators of richness ACE and Chao1 were highest in the 100bp libraries, followed by the 400bp and 1000bp libraries, although there is considerable overlap at the 95% confidence interval within samples (Fig. 2). Rarefaction analysis at all distances from 0 to 10% also predicted that the 100bp libraries contained more diversity than the other two libraries from each sample (Fig. 3). None of the rarefaction curves were near asymptotic, indicating that more sequencing would be necessary for all libraries to capture the full bacterial diversity, as confirmed by deeper surveys of these same samples using pyrosequencing (Huber et al., 2007).
Figure 2.
Non-parametric statistical estimators Chao1 and ACE and the number of OTUs at the 3% difference level for each library within samples (a) FS312 and (b) FS396. Error bars show 95% confidence intervals.
Figure 3.
Rarefaction curves at the 3% difference level for each library within samples (a) FS312 and (b) FS396.
To eliminate the possibility that differences found between libraries were due to the removal of chimeric sequences from the 1000 and 400bp libraries and not the 100bp libraries, DOTUR analyses were also run on the dataset including the chimeric sequences. Inclusion of these sequences did not significantly change the rarefaction (Supp. Fig. 3), ACE, and Chao1 results (Supp. Fig. 2). In addition, because the libraries were unequally sampled and it is known that sampling effort affects non-parametric diversity estimators, we randomly sampled the largest 2 libraries from each site to create pseudolibraries with the same number of clones as were in the smallest library and carried out DOTUR analyses. The results of DOTUR analyses on 10 such pseudolibraries from each library are summarized in Supplementary Figure 2. Even after this correction for potential sampling bias, the diversity estimates for the 100bp library are higher than the other two libraries.
Clone Library Comparisons: Community Membership and Structure
We pooled all of the high quality V6 sequences from each sample and used the program SONS (Schloss and Handelsman, 2006) to compare OTUs between libraries and calculate a variety of measures and estimators of community similarity, overlap, and structure. For both samples, a higher percentage of unique sequences was found in the 100bp libraries in comparison to the other two libraries (Table 1). For sample FS312, the 100bp library contained 285 unique V6 sequences not found in the other two libraries, the 400bp library contained 122 unique sequences, and the 1000bp library contained 65 unique sequences. The same trend was seen for sample FS396, where the 100bp library contained 171 sequences not found in the other two libraries, the 400bp library contained 93 unique sequences, and the 1000bp library contained 81 unique sequences.
We constructed Venn diagrams of the overlap of OTUs for high quality sequences at 3% difference between libraries within each sample to illustrate that there was little overlap of OTUs between the 100 and 1000bp libraries– only 39 of 366 for sample FS312 and 33 of 288 for sample FS396 (Fig 4). Figure 4 also illustrates the small number of core OTUs found in all three libraries. The Yue-Clayton nonparametric maximum likelihood estimator of similarity, which accounts for relative abundances among the OTUs shared between two communities (Schloss and Handelsman, 2006), was calculated to compare community structures between the libraries. As shown in Figure 5, the 400 and 1000bp libraries for each sample have similar community structures that differ from the community structure of the 100bp library.
Figure 4.
Venn diagrams comparing the pooled OTU memberships at the 3% difference level for each library within samples (a) FS312 and (b) FS396.
Figure 5.
Unweighted pair group method with arithmetic mean dendrogram comparing the pairwise Yue-Clayton theta values between the three clone libraries from each sample (a) FS312 and (b) FS396. The length of the reference bar represents a distance of 0.10.
Clone Library Comparisons: Taxonomy and Primer Assessment
The results of taxonomic assignment of V6 sequences at the class level are shown in Figure 6. We used only the V6 region for the taxonomic assignments because our previous work found that the V6 and full-length sequences provide similar taxonomy (Huse et al., 2008). In addition, using the shared section of the sequence across all three amplicon lengths removes any potential for introducing a length-based analytical bias for both sequence-based OTU and taxonomic analyses. For both FS312 and FS396, the major groups detected within all three of the libraries are comparable; however, the relative abundance of some of these groups in the 100bp library is in contrast to the other two libraries from the same sample. For sample FS312, the gamma- and epsilon-proteobacteria dominate in the 1000 and 400bp libraries, while the 100bp library shows a more even distribution of the epsilon-, gamma-, and alpha-proteobacteria. In sample FS396, the epsilon-proteobacteria dominate the 1000 and 400bp libraries, while the epsilon- and delta- proteobacteria appear equally important in the 100bp library (Fig. 6). This result is in agreement with the community similarity index, showing that the 400 and 1000bp libraries are similar to one another in community structure, while the 100bp is different (Fig. 5). We examined our primer sets to look at taxonomic specificity and found that the 337F and 1391R primers were the most “universal,” capturing 93–94% of bacteria in the RDPII (Cole et al., 2007) without a single mismatch in the primer (Table 2). Conversely, 967F and 1046R had many fewer matches to the database, with 1046R only predicted to recover 52% of bacteria represented in the database if not mismatches were allowed.
Figure 6.
Taxonomic breakdown and relative abundance at the bacterial class level of each library within samples (a) FS312 and (b) FS396.
Table 2.
Analysis of primer taxonomic specificity. The percent of each phylum that matched the primer with 0, 1, and 2 errors is shown.
| Phlyum (Total Searched) | 337F | 967F | 1046R | 1391R | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 0 | 1 | 2 | 0 | 1 | 2 | 0 | 1 | 2 | |
| Acidobacteria (2001) | 92% | 94% | 95% | 83% | 91% | 96% | 92% | 100% | 100% | 85% | 90% | 92% |
| Actinobacteria (14748) | 95% | 99% | 99% | 93% | 98% | 99% | 63% | 97% | 100% | 95% | 97% | 98% |
| Aquificae (357) | 95% | 96% | 97% | 0% | 8% | 13% | 1% | 27% | 98% | 95% | 99% | 99% |
| Bacteroidetes (14894) | 96% | 98% | 98% | 1% | 2% | 2% | 13% | 98% | 100% | 94% | 98% | 99% |
| BRC1 (23) | 83% | 87% | 91% | 87% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Chlamydiae (176) | 61% | 63% | 98% | 65% | 97% | 99% | 96% | 99% | 100% | 95% | 97% | 97% |
| Chlorobi (119) | 82% | 88% | 99% | 91% | 98% | 99% | 93% | 99% | 100% | 82% | 89% | 90% |
| Chloroflexi (1175) | 89% | 93% | 98% | 11% | 35% | 88% | 89% | 97% | 98% | 87% | 93% | 94% |
| Chrysiogenetes (4) | 75% | 100% | 100% | 100% | 100% | 100% | 75% | 100% | 100% | 75% | 100% | 100% |
| Cyanobacteria (3253) | 91% | 97% | 99% | 86% | 98% | 99% | 98% | 100% | 100% | 97% | 99% | 99% |
| Deferribacteres (207) | 96% | 99% | 100% | 15% | 17% | 95% | 96% | 99% | 99% | 94% | 99% | 100% |
| Dehalococcoides (87) | 94% | 95% | 95% | 0% | 5% | 99% | 97% | 99% | 100% | 94% | 94% | 95% |
| Deinococcus-Thermus (421) | 97% | 99% | 99% | 81% | 99% | 100% | 46% | 86% | 100% | 96% | 99% | 99% |
| Dictyoglomi (7) | 86% | 86% | 86% | 0% | 0% | 57% | 100% | 100% | 100% | 100% | 100% | 100% |
| Fibrobacteres (89) | 93% | 98% | 99% | 0% | 89% | 98% | 98% | 100% | 100% | 83% | 84% | 84% |
| Firmicutes (44810) | 92% | 99% | 100% | 92% | 98% | 100% | 4% | 96% | 99% | 94% | 98% | 99% |
| Fusobacteria (462) | 96% | 100% | 100% | 0% | 39% | 99% | 97% | 100% | 100% | 98% | 99% | 99% |
| Gemmatimonadetes (233) | 99% | 99% | 99% | 89% | 98% | 99% | 94% | 99% | 99% | 79% | 86% | 90% |
| Lentisphaerae (51) | 92% | 100% | 100% | 98% | 100% | 100% | 84% | 100% | 100% | 92% | 94% | 94% |
| Nitrospira (492) | 95% | 98% | 98% | 56% | 95% | 98% | 96% | 100% | 100% | 83% | 86% | 86% |
| OD1 (22) | 100% | 100% | 100% | 0% | 0% | 23% | 5% | 55% | 95% | 73% | 77% | 77% |
| OP10 (78) | 91% | 95% | 96% | 0% | 0% | 68% | 62% | 76% | 100% | 88% | 95% | 95% |
| OP11 (30) | 77% | 80% | 97% | 0% | 0% | 53% | 13% | 93% | 100% | 47% | 97% | 100% |
| Planctomycetes (1095) | 81% | 84% | 87% | 8% | 48% | 89% | 95% | 99% | 99% | 87% | 92% | 94% |
| Proteobacteria (56663) | 96% | 98% | 99% | 52% | 92% | 97% | 92% | 99% | 100% | 93% | 97% | 98% |
| Spirochaetes (1639) | 95% | 99% | 99% | 0% | 0% | 1% | 72% | 99% | 100% | 96% | 98% | 99% |
| Tenericutes (1349) | 90% | 92% | 99% | 5% | 11% | 15% | 4% | 72% | 100% | 96% | 98% | 99% |
| Thermodesulfobacteria (28) | 100% | 100% | 100% | 4% | 75% | 96% | 96% | 100% | 100% | 100% | 100% | 100% |
| Thermomicrobia (9) | 100% | 100% | 100% | 22% | 89% | 100% | 78% | 100% | 100% | 100% | 100% | 100% |
| Thermotogae (114) | 96% | 99% | 99% | 0% | 0% | 21% | 0% | 0% | 54% | 96% | 99% | 99% |
| TM7 (175) | 94% | 97% | 97% | 0% | 1% | 35% | 1% | 82% | 95% | 90% | 94% | 95% |
| Verrucomicrobia (1587) | 96% | 97% | 98% | 94% | 98% | 99% | 40% | 97% | 100% | 90% | 95% | 96% |
| WS3 (35) | 89% | 100% | 100% | 89% | 94% | 100% | 94% | 100% | 100% | 80% | 83% | 83% |
| Unclassified_Bacteria (3972) | 86% | 90% | 94% | 51% | 66% | 79% | 63% | 94% | 98% | 88% | 94% | 95% |
| All Bacteria (150405) | 94% | 98% | 99% | 63% | 82% | 86% | 52% | 97% | 100% | 93% | 97% | 98% |
Discussion
To examine how the size of PCR amplicons affects estimates of microbial diversity and taxonomic assignments in microbial ecology studies, we constructed three clone libraries each from two hydrothermal vent fluid samples with amplicons of approximately 100bp, 400bp, and 1000bp; all containing the V6 hypervariable region of the SSU rRNA gene. This allowed for direct comparisons between the different size libraries by examining only the V6 region using OTU- and taxonomy-based tools. All results support the conclusion that the 100bp amplicon libraries contained more different types of sequences than the other two libraries and that more of those sequences are different from known sequences in the reference database. In addition, both the taxonomic assessments and community similarity index showed that the 100bp libraries are different in their community structure compared to the other two libraries for each sample, and that those other two libraries are very similar.
There are many possible reasons for the differences seen between the three clone libraries. One obvious difference between the three libraries is that different primer combinations were used to generate each size class. Three primer combinations were used that included the V6 region, and each library from each sample shared one primer in common with another library from the same sample. PCR and cloning conditions were nearly identical between all reactions (amount of template, concentration of primers, dNTPs, Pfu, etc.) with the exception of the annealing temperature and extension time. For the 1000bp primer set, the annealing temperature was 55 °C and the extension time 2 min. For the 100 and 400bp sets, the annealing temperature was 57 °C and the extension time 1 min. For all samples, three individual reactions were pooled for cloning. One possibility for the similarities in community structure between the 400 and 1000bp libraries is that the same reverse primer was used (1391R). However, the same forward primer was used for the 100 and 400bp library (967F), and those libraries are different. More importantly, the 1391R primer appeared to anneal less faithfully in the 1000bp reaction, as suggested by the high number of mis-primed sequences detected in these libraries. Some of the mis-primed and chimeric sequences identified were exact matches to high quality V6 regions, indicating that these artifacts were generated from valid ribosomal RNA sequences. Others were non-ribosomal genes that apparently were amplified by the 1391R primer at both the 5’ and 3’ ends. Even when we included all of the artifactual sequences in our analyses, it is clear that the 100bp dataset contained more diversity than the 1000bp dataset.
All primers used are located in regions of secondary structure, which may affect primer annealing. Polz and Cavanaugh found overamplification of specific templates and determined that the higher the GC content of the priming region, the higher the resulting amplification efficiency (1998). We examined the GC content of the priming region for each primer in E. coli and found that the 967F primer had the lowest GC content (53%), compared to 62–67% for the other three primers, suggesting GC content of priming regions is not a major contributing factor in our study. Polz and Cavanaugh also suggested that degeneracy in primers should be avoided, as it is known that primer degeneracy can reduce specificity and result in particular primers running out as the reaction progresses (1998). However, acknowledging that not one primer fits all, they recommend pooling replicates to decrease variation in PCR reactions. While degenerate primers were used in our experiments, ranging from zero to 64-fold, we also pooled replicates to decrease variation. In addition, even though the 1000bp primer set had a combined 512-fold degeneracy, similar results were found with the 400bp primer set, which only had a combined 8-fold degeneracy. Primer specificity with respect to taxonomy also does not appear to explain our findings, with the least “universal” primer set resulting in the highest diversity estimates (Table 2). We do not believe that primer specificity explains our results.
Another possible explanation for the differences seen between the libraries is cloning bias. There is little published data regarding cloning bias with 16S rRNA genes, but it is plausible that the longer fragments with more secondary structure may interfere with E. coli ribosome assembly or growth. Rainey et al. (1994) found that different taxa were obtained in clone libraries made with the same primer set but different cloning systems. In addition, as previously noted, it is unlikely that mixed communities of amplicons will clone with uniform efficiency, and it is most likely the low abundance genes that will account for this variation (Wintzingerode et al., 1997). Cloning bias remains a possible explanation for our results, particularly with respect to the low abundance members of the community.
An additional source of error in our experiment is related to the kinetics of PCR. It has previously been noted that the PCR kinetics favor smaller amplicons (Kleter et al., 1998). Suzuki and Giovannoni (1996) tested two different primer pairs targeting two different sized amplicons, using 3 cloned ribosomal genes as standards. When the smaller amplicon primer set was used, regardless of starting template concentrations, a bias towards 1:1 product ratio was observed and was dependent on the number of PCR cycles. They attributed this difference to kinetic bias, where the smaller primer set amplified at higher efficiency, resulting in the reaction reaching saturation conditions (Suzuki and Giovannoni, 1996). Saturated templates can then reanneal and inhibit further amplification, while undersaturated targets will continue to amplify, resulting in the skewed product ratio. The other larger amplicon primer set amplified at lower efficiency, but only showed minimal bias in amplification product ratios. However, they note that in highly diverse environmental DNA samples, it is unlikely that any particular gene will reach saturation, and thus the reannealing kinetic bias effect is unlikely. As a follow-up to this work, Suzuki et al. (1998) further examined this kinetic bias in natural populations and found that the template reannealing bias could result in the over-representation of rare members of the microbial community and an under-representation of dominant members. However, others have not observed the same results. Sipos et al. (2007) did not find that reannealing was important in diverse template environmental samples, but instead found that the annealing temperature was key to reducing preferential amplification. This is similar to the findings of Leuders and Friedrich (2003) and Acinas et al. (2005), neither of whom found bias caused by cycle number or the reannealing effect. While the data in Figure 1 and Figure 6 may suggest a kinetic bias, we believe the skew in distribution of the library is due to undersampling of the smallest library, not kinetic bias.
The formation of PCR artifacts, such as heteroduplexes and chimeras, is another known problem in mixed community amplifications (Qiu et al., 2001). Many recommendations for how to minimize these artifacts have been published. For example, Qiu et al. (2001) suggested using fewer PCR cycles, longer extension times, AmpliTaq (over other types of Taq polymerases), and pooling reactions. They also noted that the artifacts increase as the diversity of the mixed community increases. Thompson et al. (2002) demonstrated that heteroduplexes increased with primer limitation, the number of different sequence variants in the original PCR, and the number of variable nucleotides in the target, and they recommended a ‘reconditioning’ step to reduce the possibility of heteroduplex formation (Thompson et al., 2002). No reconditioning to eliminate heteroduplexes was carried out on any of the samples, and all samples were treated identically (with the necessary exceptions of annealing temperature and extension time). One might predict more PCR artifacts in the largest library due to the 512-fold degeneracy of the 337F/1391R primer combination, the large number of nucleotide variants, and the greater chance of the polymerase falling off due to encountering secondary structure. Indeed, we found more artifacts in the largest libraries, as indicated by the high number of sequences flanked by primer 1391 at both the 5’ and 3’ ends of the amplicon. As noted, some of these sequences did contain valid ribosomal RNA sequences. In contrast, the smallest amplicon library containing the V6 region is not a very likely site of recombination due to its high variability. However, because we were unable to screen for artifacts in the 100bp library, we also ran all analyses using artifact sequences and found that the 100bp library contained more diverse, unique sequences than the other 2 libraries.
Finally, the polymerase is a potential source of error in our experiments. All of the amplifications were carried out with the high fidelity, proof-reading PfuTurbo polymerase. It has previously been noted that some polymerases have lower efficiencies when amplifying large fragments (>900bp) or regions of high GC content. However, PfuTurbo does not appear to be as sensitive to amplicon size as other polymerases (Arezi et al., 2003). The inability of polymerases to amplify long fragments as efficiently as short fragments has been noted previously (Suzuki and Giovannoni, 1996; Wintzingerode et al., 1997; Kleter et al., 1998; Becker et al., 2000). This is especially important for the SSU rRNA gene, where encountering problematic secondary structure is quite likely, potentially causing the polymerase to dissociate from the template (Chou, 1992; Suzuki and Giovannoni, 1996; Wintzingerode et al., 1997; Polz and Cavanaugh, 1998; Qiu et al., 2001). We believe this may be an important source for the differences in the diversity estimates and community composition of the libraries. As the polymerase encounters secondary structure in the SSU rRNA gene, it dissociates, and the frequency of dissociation is thus correlated with amplicon length. This relationship is not necessarily linear, as we saw more similarity between the 400 and 1000bp library, suggesting that the secondary structure in the 1046–1391 region of the SSU rRNA may have caused problems for both primer sets. The extremely short length of the 100bp amplicon likely serves as an easier template for PCR to proceed.
The results of this study have important implications for molecular studies of microbial communities. While sequencing large portions of the SSU rRNA gene is essential for detailed phylogenetic analysis, long amplicons may not be the most appropriate tool for measuring total community diversity or taxonomic membership. Regardless of sequencing technology used, the primer set and amplicon size must be considered when designing appropriate molecular microbial ecology experiments. Obviously, if full phylogenetic reconstruction of environmental sequences is desired, larger amplicons are necessary. All three libraries captured the dominant bacterial groups, but the 400 and 1000bp libraries missed the more divergent and possibly low abundance groups, including members of the rare biosphere (Sogin et al., 2006). Therefore, if capturing the most abundant members of a microbial population is the goal, any size amplicon should suffice. However, if a more complete picture of the microbial community structure, membership, and diversity is desired, a smaller amplicon is likely better because it represents a broader sampling of the population, there is little or no systematic loss of specific groups, the PCR proceeds more efficiently, and the opportunity for artifact formation is less. At some point, there is a trade off when the increased diversity detectable by the larger number of informative positions in the longer amplicon is overwhelmed by the number of distinct successful amplicons generated for the smaller length target. Smaller amplicons, however, do require additional sequencing effort because the library contains many more different types of sequences than larger libraries, therefore necessitating deeper sequencing to fully capture the diversity of the library and the microbial community structure. Less sequencing effort is required of larger libraries because there are fewer different sequences present and more modest sequencing efforts should capture the dominant players. All of these parameters need to be taken into consideration when carrying out PCR-based molecular surveys of microbial communities.
Experimental Procedures
Sample Collection and DNA Extraction
Samples were collected from Axial Seamount and DNA extracted as described in Sogin et al. 2006 (2006).
PCR, Clone Library Construction, and Sequencing of Environmental Samples
PCR primers were designed using ARB software (Ludwig et al., 2004) to target the V6 region of the bacterial small subunit ribosomal RNA. These primers were 967F and 1046R modified with the 5’ addition of 454 Life Sciences’ A and B adapters, respectively: 967F- 5’ GCC TCC CTC GCG CCA TCA GCA ACG CGA AGA ACC TTA CC and 1046R- 5’ GCC TTG CCA GCC CGC TCA GCG ACA GCC ATG CAN CAC CT (454 adapter sequence is underlined). Additional primer sets were designed and used to generate PCR amplicons of ∼1000 bp (337 F- 5’ ACN CCT ACG GGN GGC NGC and 1391R- 5’ GAC GGG CGG TGW GTN CA) and ∼400 bp (967FA and 1391R); all included the V6 region.
Amplification reactions were carried out in 30 µl volumes containing 1.5 units Pfu Turbo polymerase (Stratagene, La Jolla CA), 1X Pfu reaction buffer, 200 µM dNTPs (Pierce Nucleic Acid Technologies), 0.2 µM each primer, and 1–3 ng DNA or water as a negative control. Three separate reactions for each sample were carried out to control for stochastic variation in early amplification; amplification reactions containing DNA from Marinobacter aquaeolei and the archaeon Methanococcus jannaschii served as positive and negative controls. For PCR using the 100 and 400 bp primer sets, an initial denaturation step of 3 min at 94°C was followed by 30 cycles of 94 °C for 30 s, 57 °C for 45 s, and 72 °C for 1 min. The final extension step was 72 °C for 2 min. For PCR using the 1000 bp primer set, the annealing temperature was 55 °C and the extension time 2 min. Following PCR, the three reactions for each sample were combined, purified, and concentrated using the MinElute PCR Purification Kit (Qiagen) according to the manufacturer’s instructions. Product quality was assessed on 0.8% agarose gels stained with ethidium bromide. Bands were excised and gel extracted using the MinElute Gel Extraction Kit (Qiagen), followed by the addition of 3’ A-overhangs in 50 µl reactions containing 1X PCR Buffer (Promega), 0.2 mM deoxynucleoside triphosphates (Promega), 1 unit Taq Polymerase (Promega), and 9 µl DNA incubated for 10 minutes at 72 °C. The DNA was immediately cleaned with phenol:chloroform:isoamyl alcohol (25:24:1), followed by an ethanol precipitation and resuspended in 4 µl deionized water. This purified product was ligated to pCR4-TOPO vector for 20 minutes at room temperature and transformed into electrocompetent cells according to manufacturer’s instructions (Invitrogen). For each library, 960 clones were randomly selected and grown in SuperBroth with 50 mg/ml kanamycin in 96 deep-well blocks overnight at 37 °C with vigorous shaking. Cells were collected by centifugation and plasmid DNA was isolated using a standard alkaline-lysis procedure. Plasmids containing the 1000 bp amplicons were sequenced bidirectionally using primers T3 (5’- ATT AAC CCT CAC TAA AGG GA) and T7 (5’- TAA TAC GAC TCA CTA TAG GG); 400 bp products were sequenced in one direction with M13F (5’- GTA AAA CGA CGG CCA G); and 100 bp products were sequenced in one direction with M13R (5’- CAG GAA ACA GCT ATG AC) using AB BigDye3.1 (Applied Biosystems) chemistry and analyzed with an AB 3730xl Genetic Analyzer.
Data Processing
Sequences were processed with an in-house Unix script that incorporates PHRED, cross_match, and PHRAP (Ewing and Green, 1998; Ewing et al., 1998) to translate chromatograms into basecalls and associated quality scores, remove vector sequences, and for the 1000bp amplicons, to assemble forward and reverse reads into full length sequences for each of the cloned PCR amplicons. Sequences were aligned with the program MUSCLE (Edgar, 2004), and PCR primers trimmed from the alignment by hand using BioEdit. The 400 and 1000 base pair clones were examined for chimeras with Pintail (Ashelford et al., 2005) and Mallard (Ashelford et al., 2006). Sequences were then trimmed to contain only the V6 region (here defined as the region in the alignment bounded by the 967F and 1046R primer sequences) and all subsequent analyses were done on this dataset of V6 sequences, including DOTUR (Schloss and Handelsman, 2005), SONS (Schloss and Handelsman, 2006), and taxonomic analyses (Sogin et al., 2006; Huse et al., 2008). V6 sequences were aligned and distance matrices calculated according to Sogin et al. 2006 (2006). Venn diagrams were constructed using the DrawVenn Application courtesy of Stirling Chow (http://apollo.cs.uvic.ca/euler/DrawVenn/index.html). The UPGMA dendrograms were created by converting the pairwise θYC values to distances and constructing a distance matrix that was used as input to the NEIGHBOR program in PHYLIP with the UPGMA clustering algorithm (Version 3.65 package obtained from J. Felsenstein, University of Washington, Seattle). We used the ProbeMatch function in RDPII (Cole et al., 2007) to examine the taxonomic specificity of our primers. We limited our search to only those good quality sequences between E. coli region 330 and 1400 and examined specificity at 0, 1, and 2 errors for each primer.
Sequences are deposited in GenBank under the following Accession Numbers: DQ909090-DQ910173, DQ920704-DQ922483, DQ919167-DQ920703.
Supplementary Material
Acknowledgements
We thank the NOAA Pacific Marine Environmental Laboratory Vents Program, the ROV ROPOS, S. Bolton and D. Butterfield for field support and sample collection, and P. Schloss for assistance in data analysis. This work was supported by a National Research Council Research Associateship Award and L'Oréal USA Fellowship (J.A.H.), NASA Astrobiology Institute Cooperative Agreement NNA04CC04A (M.L.S.), the Alfred P. Sloan Foundation's ICoMM field project, and a subcontract from the Woods Hole Center for Oceans and Human Health from the National Institutes of Health and the National Science Foundation (NIH/NIEHS 1 P50 ES012742-01 and NSF/OCE 0430724; J.Stegeman, PI to HGM and MLS).
References
- Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arezi B, Xing W, Sorge JA, Hogrefe HH. Amplification efficiency of thermostable DNA polymerases. Analytical Biochemistry. 2003;321:226–235. doi: 10.1016/s0003-2697(03)00465-2. [DOI] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Applied and Environmental Microbiology. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S, Boger P, Oehlmann R, Ernst A. PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Applied and Environmental Microbiology. 2000;66:4945–4953. doi: 10.1128/aem.66.11.4945-4953.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Q. Minimizing deletion mutagenesis artifact during Taq DNA polymerase PCR by E.coli SSB. Nucleic Acids Research. 1992;20:4371. doi: 10.1093/nar/20.16.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Research. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using prhed. I. Accuracy assessment. Genome Research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Farrelly V, Rainey FA, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Applied and Environmental Microbiology. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Hongoh Y, Yuzawa H, Ohkuma M, Kudo T. Evaluation of primers and PCR conditions for the analysis of 16S rRNA genes from a natural environment. FEMS Microbiology Letters. 2003;221:299–304. doi: 10.1016/S0378-1097(03)00218-0. [DOI] [PubMed] [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring Microbial Diversity and Taxonomy Using SSU rRNA Hypervariable Tag Sequencing. PLoS Genetics. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Fukui M. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microbiol. 2001;67:3753–3755. doi: 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleter B, van Doorn L-J, ter Schegget J, Schrauwen L, van Krimpen K, Burger M, et al. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of Anogenital Human Papillomaviruses. Am J Pathol. 1998;153:1731–1739. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W-T, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Applied and Environmental Microbiology. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Research. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders T, Friedrich MW. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Applied and Environmental Microbiology. 2003;69:320–326. doi: 10.1128/AEM.69.1.320-326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CA, Galic M, Sangwan P, Janssen PH. PCR-generated artefact from 16S rRNA gene-specific primers. FEMS Microbiology Letters. 2005;248:183–187. doi: 10.1016/j.femsle.2005.05.043. [DOI] [PubMed] [Google Scholar]
- Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Applied and Environmental Microbiology. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Wu L, Huang H, McDonel PE, Palumbo AV, Tiedje JM, Zhou J. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 2001;67:880–887. doi: 10.1128/AEM.67.2.880-887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey FA, Ward N, Sly LI, Stackebrandt E. Dependence on the taxon composition of clone libraries for PCR amplified, naturally occurring 16S rDNA, on the primer pair and the cloning system used. Experientia. 1994;50:796–797. [Google Scholar]
- Reysenbach A-L, Giver LJ, Wicham GS, Pace NR. Differential amplification of rRNA genes by polymerase chain reaction. Applied and Environmental Microbiology. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and Environmental Microbiology. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing SONS, a tool for Operational Taxonomic Unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 2006;72:6773–6779. doi: 10.1128/AEM.00474-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos R, Szekely AJ, Palatinszky M, Revesz S, Marialigeti K, Nikolausz M. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targeting bacterial community analysis. FEMS Microbiology Ecology. 2007;60:341–350. doi: 10.1111/j.1574-6941.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored "rare biosphere". Proceedings of the National Academy of Sciences. 2006;103 doi: 10.1073/pnas.0605127103. 12,115-112,120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Rappe MS, Giovannoni SJ. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of Small-Subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Applied and Environmental Microbiology. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by 'reconditioning PCR'. Nucleic Acids Research. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintzingerode FV, Göbel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiology Reviews. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.